EMBO J 28, 383–393 (2009); published online 15 February 2009
The molecular mechanism by which proteins are secreted in gram-negative bacteria is supported by nanomachines called type I to type VI secretion systems (T1SS to T6SS). Previous data suggested that the ClpV ATPase energizes the transport of Hcp and VgrG proteins through the T6SS secretion channel. The identification of a yet unknown tubular structure, which interacts with ClpV, adds another level of complexity and challenges our vision of the T6SS working model.
Gram-negative bacteria evolved multiple strategies to transport protein across membranes. Each strategy involves a distinct and specialized macromolecular complex, frequently called secreton. These nanomachines span the cell envelope and are made with a specific set of proteins. A recent addition to this growing collection of secretion devices is the type VI secretion system (T6SS). The T6SS arises as a fascinating topic, as its impact on the interaction with the host is determinant for a successful infection (Mougous et al., 2006; Pukatzki et al, 2006).
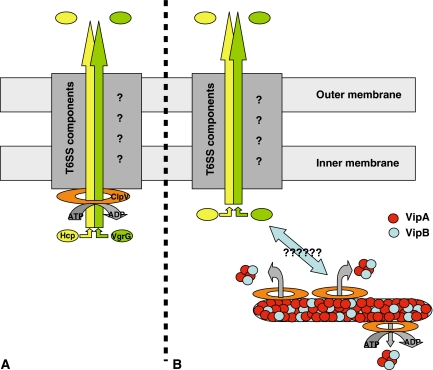
We can point to some usual ‘hot' questions in the field of secretion. What are the substrates of the secretion system and how are they transported across the envelope? How is the secreton assembled? How is the dynamic/kinetic of the substrate movement across the secreton channel supported? There are reviews (Filloux et al, 2008), which describe the distinctive characteristics of the T6SS, but unfortunately, we have no real clue about the role each component may play in the mechanism. However, the movement of proteins across membranes is an energy-demanding process, and most secretion systems use ATP as energy source. The T6SS seems to comply with this rule, as it involves a hexameric ATPase from the AAA+ family, ClpV. Moreover, two T6SS-secreted proteins were previously identified, namely the members of the Hcp and VgrG families. A T6SS working model could be the recognition of the Hcp and VgrG substrates by ClpV, threading through the oligomeric ClpV central channel, unfolding of the substrates while they move across the ClpV pore and possibly towards other channel-forming components of the T6SS (Figure 1A).
Figure 1.
Schematic representation of the T6SS. (A) Model in which Hcp (yellow) and VgrG (green) are secreted through the yet uncharacterized T6SS secreton (dark grey and question marks). The first step in the secretion process is the threading of Hcp and VgrG through the central channel of the hexameric ClpV ATPase (orange). (B) In this panel, the ClpV ATPase is shown involved in the severing of VipA (red)/VipB (blue) tubules, by threading smaller VipA/VipB complexes through the ClpV pore. In this context, it is not understood how this process and the T6SS-dependent secretion of Hcp and VgrG could be linked (bidirectional arrow and question marks).
The work presented by Bönemann and collaborators, in this issue of the EMBO Journal, challenges this concept by describing another intriguing role of the ClpV protein of Vibrio cholerae in the T6SS. The data presented suggest that Hcp or VgrG do not interact with ClpV. Instead, the authors reveal that ClpV interacts with two uncharacterized proteins from the V. cholerae T6SS, VipA and VipB, which are described as cytosolic and not secreted proteins. Strikingly, VipA and VipB are forming microtubules, and the activity of ClpV results in breaking up large VipA/B complexes (2000 kDa) into smaller complexes (100 kDa) (Figure 1B). This disassembly process requires recognition of VipA/VipB by the ClpV N terminus. Very importantly, a ClpV derivative, in which key residue within the central channel have been changed, kept its oligomerization ability and ATPase activity but did not exhibit VipA/B remodelling activity, which suggest that threading through the ClpV pore is also required for severing VipA/VipB tubules.
Up to now, protein secretion mechanisms have been found to share similarities with other biological processes that give access to the bacterial cell surface. It includes type IV pili and flagella assembly or F-pilus-dependent DNA transfer. Biological processes are thus copied and pasted with modifications, which allow adaptation and evolution towards a novel function. T6SS is not an exception, as VgrG components share similarities with gp5/gp27 proteins of the bacteriophage T4 tail spike. The tail spike is used to puncture the bacterial membrane to allow bacteriophage DNA injection. The T6SS may use a bacteriophage-like strategy to puncture membranes and make a route for proteins across the bacterial cell envelope. This is obviously a wild shortcut, but it indicates some direction into which to look at to understand the T6SS molecular mechanism. If VgrG behaves like gp27, it will assemble as a trimer forming a hollow cylinder (Kanamaru et al, 2002). Another T6SS-dependent secreted protein, Hcp, may assemble as nanotubes formed by the stacking of hexameric Hcp rings (Ballister et al, 2008). These tubes may also be considered as part of a channelling structure across the bacterial cell envelope.
The story by Bönemann and colleagues brought another tubular structure in the T6SS landscape, the VipA/VipB oligomeric complex. It is not that often when macromolecular structures, which are part of a bacterial secretion machine, could be visualized under microscopic analysis. One famous example is the needle of the type III secretion system (Kubori et al, 1998). Whereas in this case, it has been easy to spot the function, the T6SS tubules are far more enigmatic. What are the VipA/B tubules needed for and why should they be disassembled by the AAA+ ClpV ATPase to support T6SS function? AAA+ ATPase functions are broad and cover protein degradation, protein disaggregation or protein complex disassembly (Hanson and Whiteheart, 2005). In that last case, an example is the disassembly of SNARE complexes, which are involved in intracellular membrane fusion in eukaryotes. Another example is the ESCRT (endosomal sorting complexes required for transport) machinery, which facilitates sorting of proteins into intraluminal vesicles. The ESCRT-III complex forms a lattice on the surface of the endosomal membrane, and its disassembly by the AAA+ ATPase Vps4 precedes membrane invagination and fission. Even though this is ringing a bell, we are far to speculate that T6SS may involve membrane fusion–fission events that could be relying on AAA+ ATPases and microtubules severing.
In conclusion, the study by Bönemann and colleagues brings a new fact in the T6SS mechanism, but by no means it allows us to better understand how the T6SS is working. Yet, what should be highlighted from this study is that it is the first time that the possibility of a tubular protrusion within the cytoplasm is proposed for a secretion machine. Could this structure be a funnel to feed in the T6SS machine? Could it be a stock of potential T6SS substrates, which have not been characterized as such so far? Is that a structural feature that can be found in T6SSs from other bacteria? Far more easy to raise questions than to get answers, but understanding the role of VipA/B tubules will provide a significant step forward in resolving the T6SS mechanism.
Acknowledgments
Thanks to Abderrahman Hachani for discussion and comments on the manuscript. AF is supported by the Royal Society.
References
- Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD (2008) In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci USA 105: 3733–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A, Hachani A, Bleves S (2008) The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154: 1570–1583 [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW (2005) AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6: 519–529 [DOI] [PubMed] [Google Scholar]
- Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG (2002) Structure of the cell-puncturing device of bacteriophage T4. Nature 415: 553–557 [DOI] [PubMed] [Google Scholar]
- Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán JE, Aizawa SI (1998) Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280: 602–605 [DOI] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312: 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA 103: 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]