Abstract
Recruitment of substrates to the 26S proteasome usually requires covalent attachment of the Lys48-linked polyubiquitin chain. In contrast, modifications with the Lys63-linked polyubiquitin chain and/or monomeric ubiquitin are generally thought to function in proteasome-independent cellular processes. Nevertheless, the ubiquitin chain-type specificity for the proteasomal targeting is still poorly understood, especially in vivo. Using mass spectrometry, we found that Rsp5, a ubiquitin-ligase in budding yeast, catalyzes the formation of Lys63-linked ubiquitin chains in vitro. Interestingly, the 26S proteasome degraded well the Lys63-linked ubiquitinated substrate in vitro. To examine whether Lys63-linked ubiquitination serves in degradation in vivo, we investigated the ubiquitination of Mga2-p120, a substrate of Rsp5. The polyubiquitinated p120 contained relatively high levels of Lys63-linkages, and the Lys63-linked chains were sufficient for the proteasome-binding and subsequent p120-processing. In addition, Lys63-linked chains as well as Lys48-linked chains were detected in the 26S proteasome-bound polyubiquitinated proteins. These results raise the possibility that Lys63-linked ubiquitin chain also serves as a targeting signal for the 26S proteaseome in vivo.
Keywords: mass spectrometry, proteasome, proteolysis, ubiquitin, yeast
Introduction
Ubiquitin (Ub) is an evolutionarily conserved protein responsible for numerous biologically important processes through its covalent conjugation to client proteins in all eukaryotes (Hershko and Ciechanover, 1998). Protein ubiquitination is regulated by the E1 (Ub-activating enzyme)-E2 (Ub-conjugating enzyme)-E3 (Ub-ligase) cascade reaction. Ub conjugation to lysine residues in a substrate-attached Ub leads to the formation of polymeric Ub chains. In yeast, all seven lysine residues (K6, K11, K27, K29, K33, K48 and K63) of Ub can be used for chain formation, resulting in Ub chains of different topologies (Peng et al, 2003). Of these chains, the best understood type is the polyubiquitin chain linked through K48 of Ubs. K48-linked Ub chains with a length of four or more Ubs serve as the predominant proteasome-targeting signal (Thrower et al, 2000; Pickart and Fushman, 2004). K11- and K29-linked chains are also involved in proteasome-dependent protein degradation (Baboshina and Haas, 1996; Koegl et al, 1999; Jin et al, 2008). In contrast, K63-linked Ub chains and mono-ubiquitination are generally thought to function in proteasome-independent processes such as DNA repair, signal transduction and receptor endocytosis in vivo (Hicke, 2001; Pickart and Fushman, 2004), whereas emerging in vitro studies imply K63-linked chains support the proteasomal degradation (Hofmann and Pickart, 2001; Kim et al, 2007). Much less is known about the functions of chains with other topologies.
Rsp5, an essential HECT-type E3 in Saccharomyces cerevisiae, is involved in various biological processes through both proteasome-dependent and proteasome-independent pathways (Horak, 2003). In the former case, Rsp5 ubiquitinates the largest subunit Rpb1 of RNA polymerase II upon DNA damage and the mRNA export factor Hpr1, leading to their proteasomal degradation (Beaudenon et al, 1999; Gwizdek et al, 2005). Rsp5 also ubiquitinates the membrane-anchored transcriptional factors Spt23 and Mga2 leading to proteasomal processing (Hoppe et al, 2000). In the latter cases, Rsp5 ubiquitinates several plasma membrane proteins for their Ub-dependent endocytosis (Hein et al, 1995; Galan and Haguenauer-Tsapis, 1997; Dunn and Hicke, 2001).
Although Rsp5 participates in multiple cellular processes, the most important function is in the OLE pathway (Hoppe et al, 2000; Jentsch and Rumpf, 2007). In this pathway, the expression of Ole1, a Δ9 fatty acid desaturase, is tightly regulated by two related transcription factors, Spt23 and Mga2. Both proteins are synthesized as inactive precursors (p120s), which are anchored to the endoplasmic reticulum (ER) membrane by their single C-terminal transmembrane domains. Upon unsaturated fatty acid restriction, p120 is ubiquitinated by Rsp5 and subsequently processed by the 26S proteasome to remove the transmembrane domain. The processed N-terminal 90 kDa protein (called p90) is segregated by the Cdc48-Ufd1-Npl4 complex. This allows p90 to enter the nucleus and to activate transcription of target genes (Rape et al, 2001). Importantly, the lethality of Δrsp5 mutations can be suppressed by the addition of oleic acid to the growth media or by overproduction of Spt23-p90 or Mga2-p90, suggesting that the essential role of Rsp5 is ubiquitination of p120 for proteasomal processing (Hoppe et al, 2000).
A major unsolved question in Rsp5 biology is the topology of the Ub chains on its substrate in vivo. Earlier and recent studies suggest that Rsp5 assembles the K63-linked chains on its substrates, which function in proteasome-independent pathways (Galan and Haguenauer-Tsapis, 1997; Kee et al, 2006). As the function of Rsp5 lies on both the proteasome-dependent and -independent pathways, it is possible that Rsp5 attaches the different types of Ub chains to different substrates. In this view, Rsp5 may attach the K48-linked Ub chains to a subset of the substrate to be targeted by the 26S proteasome such as Rpb1, Hpr1, Spt23-p120 and Mga2-p120. Alternatively, it is possible that there are little or no specific functions of each type of Ub chains. Recent studies showed that the ubiquitinated cyclin B with heterogeneous short chains is degraded by the 26S proteasome in vitro (Hanna et al, 2006; Kirkpatrick et al, 2006). Considering this view, it is plausible that Rsp5 may attach exclusively K63-linked Ub chains to the substrate for proteasomal degradation.
In this study, we analysed the Ub chain topologies of several Rsp5 substrates by mass spectrometry (MS) and found that Rsp5 attaches entirely K63-linked Ub chains to its substrates in vitro. Unexpectedly, the 26S proteasome was able to bind and degrade efficiently the substrates with K63-linked chains in vitro. To investigate whether K63-linked ubiquitination is involved in the proteasomal targeting in vivo, we next dissected the ubiquitination of Mga2-p120 by quantitative MS. We found that the ubiquitinated p120 contains relatively high levels of K63-linked chains and Lys48-linkages to a lesser extent, and the K63-linked chains are sufficient for the proteasome-binding and p120-processing. Furthermore, we detected K63-linked chains within the proteasome-bound polyubiquitinated proteins. These results suggest that K63-linked polyubiquitin chains can serve as the proteasomal targeting as well as K48-linked chains.
Results
Rsp5 assembles K63-linked ubiquitin chains in vitro
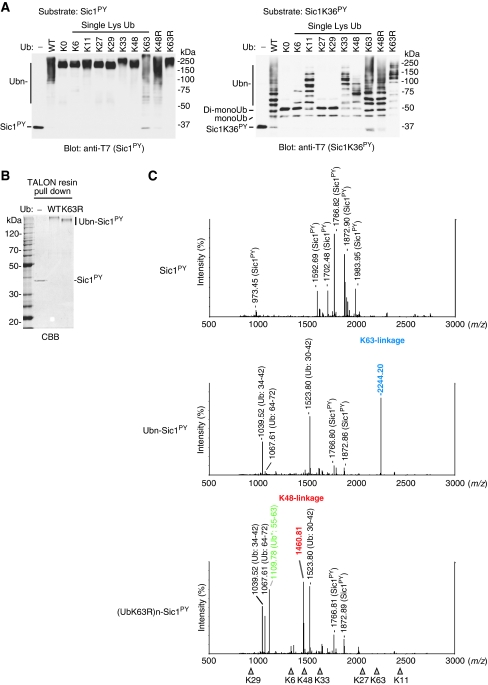
We reported earliera convenient method for preparing polyubiquitinated substrates for the 26S proteasome using Rsp5 in vitro (Saeki et al, 2005). For this device, the PY motif, a Rsp5-recognition site, was introduced to a natural proteasome substrate Sic1, a CDK inhibitor, termed ‘Sic1PY'. The Sic1PY was efficiently ubiquitinated by Rsp5 and degraded rapidly by the purified 26S proteasome in vitro (Saeki et al, 2005). Considering the present scenario, it is critically important to know the topology of the Ub chains that are formed by Rsp5. To this end, we first performed the Sic1PY ubiquitination assay using a series of Ub mutants. A Sic1PY mutant in which all the lysine residues except for K36 were replaced to arginine (Sic1K36PY) was also tested to monitor the number of attached Ub (Figure 1A). Unexpectedly, both Sic1PY and Sic1K36PY were attached with K11-, K33-, K48- and K63-linked Ub chains as judged by their gel mobilities compared with lysine-less Ub (UbK0). Two UbK0 molecules were attached to Sic1K36PY, suggesting that the N terminus, in addition to the K36 residue, is utilized for ubiquitination. On the basis of this consideration, the average length of each Ub chain on Sic1PY was estimated as follows K11- (3 to 4 Ubs), K33- (3 to 4 Ubs), K48- (2 or 3 Ubs) and K63-linked Ub chain (3 or more Ubs).
Figure 1.
Rsp5 assembles K63-linked ubiquitin chains on Sic1PY. (A) Multiple types of Ub chains can be formed on Sic1PY when using Ub mutants. T7-Sic1PY-His6 (Sic1PY, left) or its single lysine construct (Sic1K36PY, right) was ubiquitinated by E1, E2 and Rsp5, with the indicated Ub mutants and analysed by western blotting with anti-T7 antibody. (B) Purification of the ubiquitinated-Sic1PY. After ubiquitination as in (A), the samples were denatured with 6 M urea and the Sic1PY Ub conjugates were pulled down with TALON resin. As a control, Ub-omitted reaction was conducted. Gels were stained with Coomassie brilliant blue (CBB). (C) MS analysis of the purified ubiquitinated-Sic1PY. Gel regions of the ubiquitinated-Sic1PY (Ubn-Sic1PY in A) were excised and subjected to in gel-digestion with trypsin. The resulting peptides were analysed by MALDI-TOF mass spectrometry (MS). The major and specific peaks are labeled. The peak corresponding to K63- and K48-linkages are indicated in blue and red, respectively. The Ub fragments derived from the mutants are indicated by Ub* in green. The ideal masses (m/z) of all seven specific ubiquitin linkages are indicated by the triangles.
To determine more accurately the Ub chain-type specificity of Rsp5, we analysed the ubiquitinated Sic1PY by MS. The method is based on the detection of specific linkages in the tryptic digests of Ub chains (Peng et al, 2003). We found that the peptide mass fingerprinting by MALDI-TOF-MS can be applied to determine the relative abundance of Ub-linkages (Supplementary Figures S1–S3). To analyse the ubiquitinated Sic1PY by MS, the ubiquitinated Sic1PY was isolated from the reaction mixtures by using the hexahistidine-tag of Sic1PY in denatured condition (Figure 1B). The gel portion containing the ubiquitinated Sic1PY was exercised and subjected to in gel digestion with trypsin. The following MALDI-MS analysis revealed that the ubiquitinated Sic1PY with wild-type Ub contains only K63-linkage. In contrast, only K48-linkage was detected in the preparation using UbK63R (Figure 1C). These results indicate that Rsp5 preferentially assembles the K63-linked Ub chains on Sic1PY under normal conditions without forming other types of linkages. It is worth noting that when K63 of Ub is mutated, Rsp5 catalyzes K48-linked chain synthesis, indicating that Rsp5 uses other lysine residues for the chain formation when the preferred site is missing. In this study, we used Ubc4 as E2 because Ubc1, Ubc4 and Ubc5 were found to work equivalently with Rsp5 (Supplementary Figures S4–S6). We further determined the topology of Ub chains of the self-ubiquitinated Rsp5 and two native Rsp5 substrates, Mga2 and Rpb1, in vitro. MS analysis revealed that these ubiquitinated substrates contained only K63-linkages (Supplementary Figures S1, S6 and S7). These results suggest that Rsp5 ubiquitinates its substrates exclusively with K63-linked chains in vitro.
26S proteasome efficiently degrades the ubiquitinated Sic1PY with K63-linked chains
We have showed previously that the ubiquitinated Sic1PY is degraded by the yeast 26S proteasome, and this system was utilized for evaluating the activity of mutant proteasomes (Sone et al, 2004; Isono et al, 2005; Saeki et al, 2005). In this study, we found that this ubiquitinated Sic1PY contains only K63-linked chains (Figure 1). To confirm this unexpected result, we employed the following systematic degradation assays. The wild-type 26S proteasome was affinity purified from the RPN11-3xFLAG cells, in which the RPN11 gene was tagged with three tandem Flag epitopes (Saeki et al, 2005). As a control for degradation assay, we also prepared the 26S proteasome lacking Rpn10, an intrinsic Ub receptor, from the RPN11-3xFLAG Δrpn10 mutant cells (Figure 2A).
Figure 2.
The 26S proteasome degrades K63-linked polyubiquitinated-Sic1PY. (A) The purified 26S proteasomes from RPN11-3xFLAG strain (WT) or Δrpn10 RPN11-3xFLAG strain (Δrpn10) and the mock purified materials from the parent wild-type strain (no-tag) were subjected to SDS–PAGE followed by staining with CBB (left) or analysed by western blot with the indicated antibodies (right). (B) Degradation of the ubiquitinated-Sic1PY by the wild-type 26S proteasome. Sic1PY was ubiquitinated by Rsp5 with wild-type Ub (Ubn-Sic1PY, resulting K63-linked polyUb chain), with methylated Ub (mUb, multiple mono-ubiquitination), or with Ub mutants; UbK48R (K63-linked polyUb chains), UbK63R (K48-linked polyUb chains), or UbK48R K63R (short Ub chains, probably contains K33-linkage). The Ub-omitted reaction was also tested (Sic1PY) as a control. Each substrate (200 nM) was incubated with 25 to 100 nM of the wild-type 26S proteasome at 25°C for 10 min. The reaction was terminated by the addition of SDS-loading buffer and was analysed by western blotting with T7-antibody. (C) Kinetic analysis of Sic1PY degradation. Each substrate (200 nM) was incubated with the wild-type 26S proteasome (50 nM) at 25°C. The reaction was terminated at the indicated time points and analysed as in (B). (D) Degradation assay of Sic1PY Ub conjugates with lysine-less (K0), single-lysine only (K48 or K63) and double-lysines (K48+K63) Ub mutants. Each substrate (200 nM) was incubated with the wild-type 26S proteasome (50 nM) at 25°C for 10 min and analysed as in (B).
To optimize our degradation assay system, the wild-type 26S proteasome was titrated in the degradation assay with 200 nM of substrates. Strikingly, low concentrations (25 nM) of the 26S proteasome were sufficient to degrade the K63-linked ubiquitinated Sic1PY with wild-type Ub (Figure 2B). In contrast, the Δrpn10 26S proteasome cannot degrade the K63-linked ubiquitinated Sic1PY (Supplementary Figure S8) as observed for the K48-linked ubiquitinated Sic1 (Verma et al, 2004).
To exclude the possibility that the ubiquitinated Sic1PY contains undetectable levels of K48-linkages, which may stimulate proteasomal degradation, we performed the degradation assay using the ubiquitinated Sic1PY with UbK48R, which contains only K63-linkages (Figure 2C). The Sic1PY ubiquitinated with UbK48R was degraded at a comparable dose and rate with wild-type Ub (Figure 2B and C). Thus, the results suggested that K63-linked Ub chains promote the degradation by the 26S proteasome. We believe that the concentration of the 26 proteasome is biologically relevant: The concentration of the 26S proteasome in yeast cells is estimated to be ∼60 nM, if the proteasomes are distributed evenly throughout cells and to be ∼8.6 μM, if all the proteasomes are present in nucleus (Russell et al, 1999; Ghaemmaghami et al, 2003; Jorgensen et al, 2007).
To investigate the types of ubiquitination that undergo degradation, we prepared the ubiquitinated Sic1PY with different topologies by a series of mutant Ubs; K48-linked chains with UbK63R (K63R), short Ub chains possibly containing K33-linkage with UbK48R K63R (K48R K63R), and multiple mono-ubiquitination with methylated Ub (m) and lysine-less Ub (K0). In comparison to the K63-linked ubiquitinated Sic1PY, higher concentrations (100 nM) of the 26S proteasome were required for the degradation of the K48-linked ubiquitinated Sic1PY. On the other hand, the ubiquitinated Sic1PY with short chains were only slightly degraded (Figure 2B), indicating that the short Ub chains are a less favorable signal for proteasomal degradation. As expected, the 26S proteasome failed to degrade the multiply mono-ubiquitinated Sic1PY with methylated Ub (mUb) and UbK0 (Figure 2B and D). Thus, the K63-linked Ub chains are efficient signal for degradation, whereas multiple mono-Ubs and short chains are not. To simplify this, we further prepared the ubiquitinated Sic1PY with single lysine Ub mutants. Although a single- or double-lysine Ub mutants (e.g., K48, K63, K48, K63) were polymerized efficiently on Sic1PY by Rsp5, the Sic1PY Ub conjugates were incompetent to proteasomal degradation (Figure 2D), which is apparently consistent with the lack of their interaction with the 26S proteasome as described below.
As Sic1PY is an artificially devised substrate for Rsp5, we also examined the degradation of the Rpb1, a subunit of the RNA polymerase II complex, as a native Rsp5 substrate (Beaudenon et al, 1999; Somesh et al, 2005). Rpb1 was efficiently polyubiquitinated by Rsp5 and was degraded by the 26S proteasome in vitro (Supplementary Figure S7). MS analysis showed that the ubiquitinated Rpb1 in the reaction contained only K63-linkage (data not shown).
26S proteasome and its ubiquitin receptors bind K63-linked ubiquitin chains
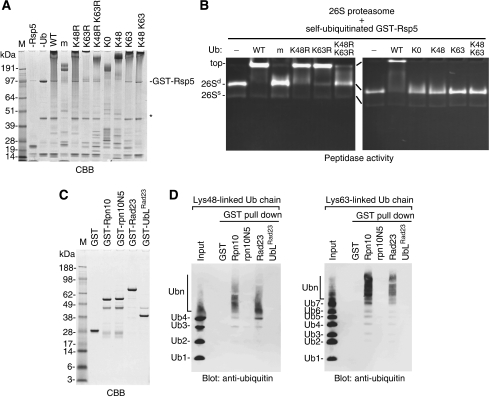
We further investigated whether K63-linked Ub chains are suitable for the targeting signal of the 26S proteasome. We noticed that the self-ubiquitinated Rsp5 is neither degraded nor deubiquitinated by the 26S proteasome (data not shown), like the self-ubiquitinated Cdc34 (Elsasser et al, 2002). By using this property, we analysed the interaction between the purified 26S proteasome and the Ub chains on GST-Rsp5 by native-PAGE (Figure 3A and B). In the presence of the ubiquitinated Rsp5 with wild-type Ub, the migration of the 26S proteasome was retarded due to their binding, whereas Rsp5- or Ub-omitted reactions had no effects (Figure 3B). These data indicate that the 26S proteasome binds directly with the K63-linked Ub chains on GST-Rsp5. Expectedly, the 26S proteasome also binds efficiently the ubiquitinated Rsp5 with UbK63R, which contains K48-linkages. In contrast, the proteasome binds only weakly with multiple mono-ubiquitinated and short polyubiquitinated Rsp5, which were generated by methylated Ub (mUb) or UbK48R K63R (K48R K63R), respectively (Figure 3B). The gel shift by the respective Ub conjugates occurred in a dose-dependent manner (Supplementary Figure S9), suggesting that the 26S proteasome binds with high affinity to both K48- and K63-linked Ub chains almost equivalently, but with low affinity to the multiple mono-Ubs or short Ub chains (Figure 3B, left). Although polyubiquitination occurred with a single- or double-lysine Ub mutants (UbK48, UbK63 or UbK48+UbK63) on Rsp5 (Figure 3A), the assembled chains were incompetent for proteasome-binding irrespective of K48- and/or K63-linked chains (Figure 3B, right). These results clearly indicate that certain lysine residue(s) other than the remaining lysine residue is required for their proper conformations. Consequently, the 26S proteasome was not able to degrade the polyubiquitinated Sic1PY with single-lysine Ub mutants (Figure 2D).
Figure 3.
The 26S proteasome and its ubiquitin receptors bind K63-linked polyubiquitin chains. (A) Self-ubiquitination of GST-Rsp5 with Ub mutants. GST-Rsp5 was self-ubiquitinated with a series of Ub mutants. After the reactions, a portion was analysed by SDS–PAGE. The topologies of ubiquitination on Rsp5 are as follow K63-linked chains with wild-type Ub (WT), with UbK48R (K48R), with UbK63 (K63) or with UbK48+K63 (K48 K63); K48-linked Ub chains with UbK63R (K63R) or with UbK48 (K48); short Ub chains that probably contain K33-linkages with UbK48R K63R (K48R K63R); multiple mono-Ub with methylated-Ub (m) or with lysine-less Ub (K0). The degradation products of GST-Rsp5 are indicated by the asterisk. (B) Association of the 26S proteasome and self-ubiquitinated Rsp5 monitored by gel shift assay. The 26S proteasome was preincubated with the self-ubiquitinated GST-Rsp5 produced as in (A), then, the mixtures were subjected to 3.5% native-PAGE followed by in-gel activity assay using suc-LLVY-amc. As a control, Ub-omitted reaction (−) was also tested. Doubly capped and singly capped species of the 26S proteasomes are indicated by 26Sd and 26Ss, respectively. (C) SDS–PAGE analysis of GST-fusion proteins. GST-fusion proteins (1 μg) were subjected to SDS–PAGE followed by CBB-staining. GST-rpn10N5 is an Ub-interacting motif mutant, in which LAMAL sequence was mutated to NNNNN. GST-UbLRad23 is a deletion mutant of C-terminal Ub-associated domains. (D) GST pull-down assays of free K48- and K63-linked Ub chains with the proteasomal Ub receptor proteins. Free K48-linked (left) or K63-linked (right) Ub chains were preincubated with the indicated GST-fusion proteins, then, GST-fusion proteins were pulled down with glutathione-immobilized agarose. The bound materials were eluted with SDS-loading buffer and analysed by western blot with anti-Ub antibody. GST alone, GST-rpn10N5 and GST-UbLRad23 were used as the control.
The 26S proteasome utilizes multiple Ub receptors such as Rpn10, Rpn13, Rad23 and Dsk2 (Elsasser et al, 2002; Verma et al, 2004; Richly et al, 2005; Husnjak et al, 2008; Schreiner et al, 2008). To investigate the binding properties of the Ub receptors against K63-linked Ub chains, we performed a GST pull-down assay with a mixture of free K63-linked chains. We found that both Rpn10 and Rad23 have markedly high affinity with long chains, more than seven Ubs, but low affinity with short chains. This property was also observed in a comparative analysis using free K48-linked chains (Figure 3D). The results are consistent with a previous study, which showed that the maximum affinity of human Rad23 for K48-linked Ub chains is reached with a chain length of six or more Ubs (Raasi et al, 2004).
Mga2-p120 is modified mainly with K63-linked ubiquitin chains in vivo
Is the K63-linked Ub chain recognized and targeted by the 26S proteasome in vivo? One way to show this is to isolate and dissect Ub conjugates of Rsp5 substrate. Among the known substrates of Rsp5, we selected Mga2 because the essential role of Rsp5 is the ubiquitination of Spt23-p120 and Mga2-p120 and their regulations have been characterized extensively (Hoppe et al, 2000; Rape et al, 2001; Shcherbik et al, 2003; Shcherbik and Haines, 2007).
To purify the Mga2-p120 Ub conjugates in enough amounts for MS analysis, we investigated optimal conditions, in which the ubiquitination levels were enhanced and the ubiquitinated proteins were stabilized in vivo. Because Mga2 is a protein of a very low abundance, ∼300 copies per cell (Ghaemmaghami et al, 2003), we produced N-terminally Flag-tagged Mga2 under the GAL1 promoter. The ubiquitination and the processing of overproduced Mga2 is governed by Rsp5, Ubp2, Cdc48 and the proteasome (Supplementary Figure S10), indicating that the overexpressed Mga2 seems to be functionally equivalent to endogenous one. In addition, the PDR (pleiotropic drug registance) 5 gene was deleted to increase sensitivity to the proteasome inhibitor MG132 (Fleming et al, 2002). To enhance the ubiquitination levels of Mga2, we used a plasmid that constitutively expresses wild-type Ub (Supplementary Figure S12).
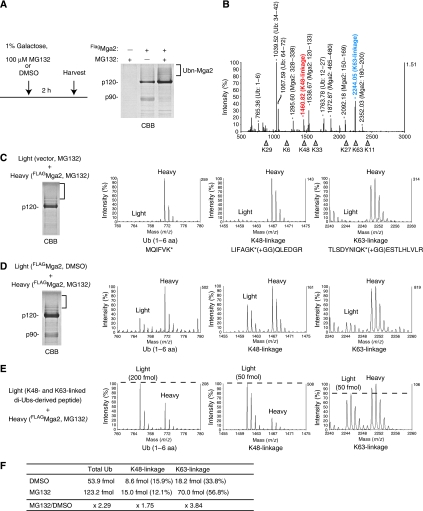
Under these conditions, we successfully isolated the Mga2-p120 Ub conjugates to a detectable level in a CBB stained gel (Figure 4A). The gel portion of the Mga2-p120 Ub conjugates was excised and subjected to MS analysis (Figure 4B). Within the multiple ion peaks corresponding to Mga2 and Ub peptides, a strong peak of K63-linkage and a relatively weak peak of K48-linkage were detected.
Figure 4.
Mga2-p120 is modified with mainly K63-linked ubiquitin chains in vivo. (A) SDS–PAGE analysis of the Mga2 Ub conjugates from MG132-treated cells. Wild-type cells (YYS1246, Δpdr5 Δlys2 background) carrying PGAL1-FLAGMGA2 (FlagMga2) and PTDH3-Ub plasmids were cultured in SRaf medium. Mga2 expression was induced by 1% galactose for 2 h in the presence of 100 μM MG132 or DMSO. Mga2 and its Ub conjugates were affinity-purified by anti-Flag M2 agarose, and subjected to SDS–PAGE analysis followed by CBB staining. (B) MS spectrum of the ubiquitinated-Mga2. The gel portion of the Mga2-p120 Ub conjugates from MG132-treated cells, indicated by a blanket in (A), was excised and subjected to in-gel digestion with trypsin. The resulting peptides were analysed by MALDI-TOF MS. Peaks corresponding to K48- and K63-linkages are indicated in red and blue text, respectively. (C) Detection of the heavy isotope-labeled Ub chains of the Mga2 Ub conjugates using SILAC. The cells (YYS1301) carrying a control plasmid grew in light medium and the cells (YYS1303) carrying the PGAL1-FLAGMGA2 (FlagMga2) grew in heavy medium. After the addition of 1% galactose for 2 h in the presence of 100 μM MG132, the two cultures were mixed and analysed as in (A). MS spectra of the SILAC ion pairs are presented: a linear Ub peptide (1–6 amino acids), Ub K48-, and K63-linkages. (D) Stabilizations of both the K48- and K63-linked Ub chains on the Mga2 by proteasome-inhibition. The cells (YYS1303) were grown in heavy or light medium. Mga2 expression was induced by 1% galactose for 2 h in the presence of 100 μM MG132 for heavy culture or DMSO for light culture. Then, the two cultures were mixed and analysed as in (C). (E, F) Absolute quantitation of the Ub chains on the Mga2. The tryptic digests of light Ub chains (a 1:1 mixture of K48- and K63-linked di-Ubs) were used as internal standards. Note that the mixture of di-Ubs generates a Ub (1–6 amino acid) and each linkages at a 4:1 ratio. The heavy lysine-labeled Mga2 Ub conjugates were prepared as in (C). Then, the peptides were mixed and the relative ion intensities were analysed by MS. The amount of the total Ub, K48-linkage and K63-linkage were calculated. See also, Supplementary Figure S13.
To quantify the Ub linkages by MS, we next used SILAC (stable isotope labeling by amino acids in cell culture): the Δlys2 background strains were grown in SILAC medium supplemented with ‘heavy' lysine (13C6-Lys) (de Godoy et al, 2006). To exclude the possibility that the Ub-linkages detected in MS is due to contaminants, we first grew the cells with a control plasmid in ‘light' medium, whereas the cells carrying the PGAL1-MGA2 plasmid in heavy medium. After incubation with 1% galactose and 100 μM MG132 for 2 h, equal amounts of the cells from the two cultures were mixed together. The Mga2 Ub conjugates were then purified and subjected to MS analysis. As the incorporation of heavy lysine results in a mass shift of 6 Da, K-containing peptides are detected as heavy ions by MS. As shown in Figure 4C, the heavy lysine-labeled Ub linkages were only detected as heavy ions. The absence of light ions indicates that the detected Ub chains were not contaminant.
Next, the yeast cells carrying the PGAL1-MGA2 plasmid grown in heavy medium were treated with MG132, whereas the same cells grown in light medium were treated with DMSO. Then, the p120 Ub conjugates were purified and subjected to MS analysis. Comparing the intensities of SILAC ion pairs, the relative amounts of Ub-linkages were determined both the K63- and K48-linkages were increased to 1.75- and 3.84-fold, respectively, by inhibition of the proteasome (Figure 4D). To quantify the absolute amount of the Ub chains on Mga2-p120, we prepared a control Ub peptides that consist of a 1:1 mixture of K48- and K63-linked di-ubiquitins. The tryptic digests of the Mga2 Ub conjugates from the MG132-treated cells (heavy) were mixed with the control Ub peptides (light), then, analysed by MS (Figure 4E and Supplementary Figure S13). The absolute amounts of total Ub and the linkages were determined by respective ion pairs, and the percentages of chains were then calculated (Figure 4F). Strikingly, the K63-linked chains occupied ∼57% of total Ubs and existed in 4.7-fold larger amount than K48-linkaged chains.
K63-linked ubiquitin chains are sufficient for the p120-processing in vivo
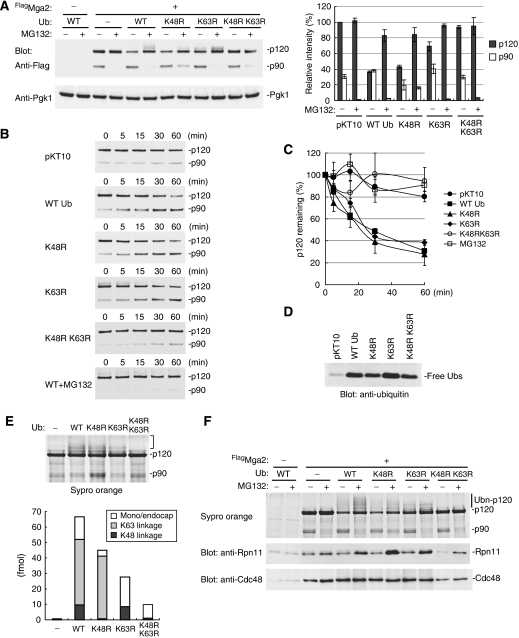
To investigate the functional significance of the K63-linked Ub chains, we first analysed steady-state levels of Mga2 in mutant Ub-expressing cells. In the wild-type Ub-overexpressing cells, surprisingly, the p120 levels were greatly decreased to ∼38% as compared with that in the cells carrying a control plasmid (Figure 5A). The decrease was abrogated by MG132, suggesting that the overproduced Ub stimulates the degradation of Mga2. Importantly, the Mga2 degradation was also observed in both the UbK48R- and the UbK63R-expressing cells, ∼58% decrease and ∼32% decrease, respectively (Figure 5A). In contrast, the p120 protein levels remained unchanged in the UbK48K63R-expressing cells.
Figure 5.
K63-linked Ub chains promote Mga2-p120 processing in vivo. (A) Steady-state levels of Mga2 in the Ub mutant-expressed cells. The cells expressing FlagMga2 and respective Ubs (YYS1301-1306) were cultured in SRaf medium. Mga2 was expressed by 1% galactose for 3 h in the presence or in the absence of 100 μM MG132. The total cell extracts were analysed by western blot with anti-Flag antibody for the detection of both Mga2-p120 and Mga2-p90 (upper) and anti-Pgk1 antibody as a loading control (lower). The protein levels of p120 and p90 are shown with mean±s.d. values of three experiments in the right graph. (B, C) Cycloheximide-chase analysis of Mga2. The respective cells as in (A) were cultured in SRaf medium. After Mga2 expression by 1% galactose for 1 h, translation was inhibited by cycloheximide at a final concentration of 0.4 mg/ml. Aliquots were taken at the indicated time points after cycloheximide addition. The total cell extracts were analysed by western blot with anti-Flag antibody for the detection of both Mga2-p120 and Mga2-p90 (upper) and anti-Pgk1 antibody as a loading control (lower). To inhibit proteasome activity, cells were treated with MG132 (100 μM) upon the Mga2 expression. The protein levels of p120 are shown with mean+SD values of three experiments in the graph. (D) Expression levels of wild type and mutant Ubs. Cell extracts were analysed by western blot with anti-Ub. (E) Absolute quantitation of the Ub chains on the Mga2 from the mutant Ub-expressed cells. The Mga2 Ub conjugates were purified from the MG132-treated cells cultured in SILAC medium. The ubiquitination of Mga2 was confirmed by Sypro Orange-staining and was quantified by MS as in Figure 4E. See also Supplementary Figure S14. (F) The 26S proteasome binds the ubiquitinated Mga2 in vivo. Mga2 and its Ub conjugates were immunoprecipitated under a mild condition and were analysed by western blot analysis with anti-Rpn11 and anti-Cdc48 antibodies. The ubiquitination levels of Mga2 were monitored by Sypro Orange-staining.
Next, we performed a chase experiment to monitor the rate of p120-processing (Figure 5B). In the wild-type Ub-overexpressing cells, the half-life of the p120 was determined to ∼30 min. Concomitant with decrease of the p120 levels, the p90 levels were increased and subsequently degraded after 60 min (data not shown). Similarly, in both the UbK48R- and UbK63R-expressing cells, the p120-processing also occurred as fast as in the wild-type Ub-expressing cells. In contrast, the p120-processing only slightly occurred in the cells expressing the UbK48R K63R mutant or in the MG132-treated cells.
Because each Ub mutants were expressed at levels 25- to 50-fold higher than endogenous Ub (Figure 5D), the mutants should inhibit the formation of chains. To confirm this, we quantified the Ub linkages in the ubiquitinated-Mga2 from the mutant Ub-expressing cells by MS. As expected, only K63-linkage was detected in the ubiquitinated Mga2 from the UbK48R-expressing cells, whereas only K48-linkage was detected from the UbK63R-expressing cells (Figure 5E and Supplementary Figure S14). These results indicate that each Ub chains can be formed on p120 independently and are functionally equivalent in the p120-processing.
The 26S proteasome binds K63-linked ubiquitin chains in vivo
If K63-linked Ub chains serve as a targeting signal for the 26S proteasome, the interaction between the 26S proteasome and the p120 Ub conjugates should be observed in vivo. To test this, we performed the Mga2 immunoprecipitation assay under mild conditions, without high salt wash. As shown in Figure 5F, the 26S proteasome was co-precipitated with K63-linked ubiquitinated Mga2 from the UbK48R-expressed cells in which the proteasome activity was inhibited by MG132. The interaction was detected at background levels from DMSO (mock)-treated cells, suggesting that the 26S proteasome continuously processes the K63-linked ubiquitinated p120. In contrast, Cdc48, the Ub-dependent segregase for p90, was also detected in the Mga2 precipitates, but the binding was apparently not correlated with the ubiquitination levels of p120 and was not affected by MG132. Possibly, Cdc48 may recognize only a subset of the ubiquitinated Mga2 and/or may be resting until the p90 generation. Nonetheless, the result suggests that the K63-linked Ub chains are sufficient to the proteasomal targeting.
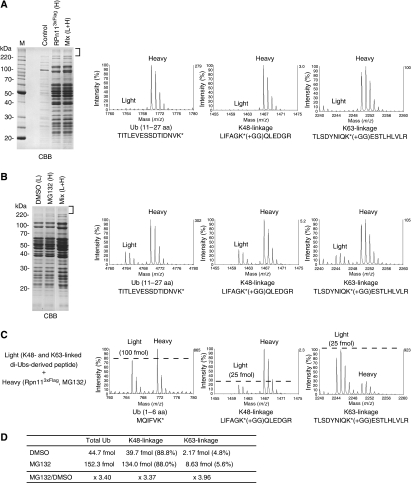
A recent study showed that total levels of K63-linked Ub chains as well as K48- and K11-linked chains were elevated by the treatment of MG132 in mammalian cells (Bennett et al, 2007). Their observation and our results raise the possibility that K63-linked Ub chains are widely utilized for the proteasome-dependent protein degradation. To investigate this, we analysed the proteasome-bound ubiquitinated proteins by MS using SILAC. Under a mild condition, the polyubiquitinated proteins were co-purified with the 26S proteasome from the Ub-overexpressed cells (Supplementary Figure S15). By using MS, we found that K63-linkages in addition to K48-linkages within the ubiquitinated proteins (Figure 6A and Supplementary Figure S15). The other types of Ub linkages including K11-linkage were not detected even when analysed by using a nano LC-coupled MALDI-MS/MS (data not shown). Importantly, the levels of both the K48- and K63-linkages were increased to 3∼4-fold by MG132-treatment (Figure 6B and Supplementary Figure S15). By using the control Ub peptides, the absolute amount of the K63-linkages was calculated as 4∼5% of total Ub, and the K48-linkages were predominant, ∼88%, as expected (Figure 6C, D and Supplementary Figure S16). It is known that Hul5, a proteasome-bound E4, assembles K63-linked chains onto the proteasome-bound ubiquitinated substrate in vitro (Crosas et al, 2006), suggesting the possibility that the presence of the K63-linkages is due to the Hul5 activity. However, K63-linkage was still detected in the proteasome-bound Ub conjugates from Δhul5 cells (Supplementary Figure S17), indicating that the K63-linked chains are formed by an E3(s) other than Hul5. Collectively, these results suggest that K63-linked Ub chains may be widely utilized for the proteasome-dependent degradation.
Figure 6.
Quantitation of the proteasome-bound Ub chains. (A) Identification of the K63-linked Ub chains from the purified 26S proteasome. The RPN11-3xFLAG cells (YYS1320) grew in heavy medium and the control cells (YYS1314) grew in light medium were treated with 100 μM MG132 for 2.5 h. Then, the 26S proteasome was purified under a mild condition and analysed as in Figure 4C. MS spectra of SILAC pair ions are presented: a Ub peptide (11-27 amino acid), Ub K48-, and K63-linkages. (B) Both the K48- and K63-linkages were accumulated by proteasome-inhibition. The RPN11-3xFLAG cells (YYS1320) were grown in heavy or light medium. The heavy culture was treated with 100 μM MG132 for 3 h, whereas the light culture was treated with DMSO for 2.5 h. The proteasomes were purified and relative ion intensities were measured as in Figure 4D. (C, D) Absolute quantitation of the Ub chains of the proteasome-bound ubiquitinated proteins. Tryptic digests of the heavy lysine-labeled Ub conjugates in (A) were mixed with the internal standard Ub peptides and analysed by MS. The amount of the total Ub, K48-linkage, and K63-linkage were calculated. See also, Supplementary Figure S15 and S16.
Discussion
Rsp5-mediated ubiquitination results in K63-linked polyubiquitin chains
We started this study by investigating the chain topology of the ubiquitinated Sic1PY, which was used to evaluate the proteasome activity in vitro (Sone et al, 2004; Isono et al, 2005; Saeki et al, 2005; Kriegenburg et al, 2008). Although four different types of Ub chains can be assembled by Rsp5 using Ub mutants (Figure 1 and Supplementary Figure S5), direct analysis by MS revealed that the ubiquitinated Sic1PY with wild-type Ub contained only K63-linked chains (Figure 1). In addition, we found that Rsp5 itself and two native Rsp5 substrates were also modified with K63-linked Ub chains in vitro (Supplementary Figures S1, S6 and S7). Thus, Rsp5 seems to assemble exclusively the K63-linked Ub chains on substrate, as reported previously (Galan and Haguenauer-Tsapis, 1997; Kee et al, 2006).
In our in vivo analysis, we used Mga2-p120 processing in an experimental setting because this process has been proven to be solely dependent on ubiquitination by Rsp5 followed by cleavage by the 26S proteasome. Relatively low levels of K48-linked Ub chains in addition to K63-linked chains were detected in the ubiquitinated Mga2-p120 (Figure 4). How K48-linked chains were introduced remains unclear. After the processing, the released p90 (active form) is subsequently ubiquitinated by Ufd2, an E4 that can extend K48-linked chains on ubiquitinated substrates (Johnson et al, 1995; Saeki et al, 2004), and is degraded by the 26S proteasome in the nucleus (Richly et al, 2005). As both Mga2 and Ub were overexpressed in our experimental setting, Ufd2 could target the Mga2-p120 under such nonphysiological conditions. However, the Mga2-p120 Ub conjugates from Δufd2 cells still contained the K48-linked chains (Supplementary Figure S11), indicating that Ufd2 is not responsible for the K48-linked chain formation. Another possibility is that Ubc1 could preassemble the K48-linked chains prior to Rsp5-mediated ubiquitination of Mga2-p120 because Ubc1 itself can form K48-linked Ub chains (Hodgins et al, 1996; Rodrigo-Brenni and Morgan, 2007). However, only K63-linkage was detected in the Mga2-p120 Ub conjugates made by Rsp5 even if Ubc1 was used as E2 in vitro (Supplementary Figure S6). Ubp2 is known as a deubiquitinating enzyme that regulates Rsp5-mediated ubiquitination (Kee et al, 2005, 2006). We found that the ubiquitination levels of Mga2-p120 were markedly increased in Δubp2 cells (Supplementary Figure S11). Therefore, there is no doubt that Rsp5 ubiquitinates the overexpressed Mga2 used in this study, but it is plausible that there is an additional E3 for Mga2-p120 in the cells as proposed previously (Shcherbik et al, 2003).
Why are not all Rsp5 substrates degraded by the 26S proteasome in vivo?
Although there are many Rsp5 substrates in the cells, only a subset of substrates are degraded by the 26S proteasome (Horak, 2003). One possibility is that the length of K63-linked Ub chains is also a key determinant for proteasomal degradation. We showed that the proteasomal Ub receptors efficiently bound long K63-linked Ub chains (Figure 3D). Most plasma membrane proteins are attached by Rsp5 with multiple mono-Ubs or a short chain with up to four Ubs (Rotin et al, 2000), in which Ub lengths might simply be enough to escape proteasomal targeting. Ubp2 might maintain short Ub chains on such Rsp5 substrates (Kee et al, 2005, 2006). Another possibility is the structural property of the substrate itself. It has been proposed that a loosely folded region is required for efficient proteasomal degradation (Prakash et al, 2004; Piwko and Jentsch, 2006). Some degradable Rsp5 substrates may contain this ‘engagement' site. Our results support this notion, judging from the findings that physiological substrates (e.g., Sic1 and Rpb1) were readily degraded by the 26S proteasome, whereas nonphysiological substrates (e.g., Rsp5 and GFP) were not (data not shown).
Ubiquitin chain topologies for proteasomal targeting signal
It is widely accepted that K48-linked Ub chains play a central role in the proteasome-dependent proteolysis, whereas K63-linked chains function in proteasome-independent processes (Chau et al, 1989; Finley et al, 1994; Hershko and Ciechanover, 1998; Pickart and Fushman, 2004). However, previous and recent studies have raised possibilities that K11- and/or K63-linked Ub chains also serve as proteolytic signals (Baboshina and Haas, 1996; Hofmann and Pickart, 2001; Kirkpatrick et al, 2006; Kim et al, 2007; Jin et al, 2008). In this study, we showed that homogeneous K63-linked chains with sufficient length served as the proteasomal targeting signal in vitro (Figures 2, 3 and Supplementary Figures S7–S9). Moreover, our experiments, although carried out under a nonphysiological condition in which Ub was overexpressed, suggested that K63-linked Ub chains can be utilized to the proteasome in vivo (Figures 5, 6 and Supplementary Figures S13–S17). Direct analysis of the proteasome-bound ubiquitinated proteins suggests that K63-linked Ub chains, even at a small fraction, are certainly utilized in the proteasome-dependent protein degradation (Figure 6). Curiously, we failed to detect K11-linked chains in this assay, possibly, this type of Ub conjugates may be delivered to Cdc48/p97 via UBX-UBA proteins as recently reported (Alexandru et al, 2008). In contrast to previous reports (Guterman and Glickman, 2004; Kirkpatrick et al, 2006; Boutet et al, 2007), multiple-mono Ubs and short Ub chains are unlikely to be signals for the proteasomal targeting (Figures 2 and 3).
Our work is reminiscent of the earlier study in yeast; overexpression of UbK63R in Δubi4 cells caused hypersensitivity to various stresses including heat, amino acid analogues and UV (Arnason and Ellison, 1994). It is known that the ubiquitin-proteasome pathway degrades damaged cellular proteins by heat stress and amino acid analogues (Ciechanover et al, 1984). It is likely that K63-linked Ub chains are involved in protein quality control in cells. In this context, it should be noted that Rsp5 was shown to be also involved in degradation of stress-induced abnormal proteins (Hoshikawa et al, 2003). Our results are consistent with these observations in the sense that K63-linked Ub chains would serve as the proteasomal degradation signal. Further studies are needed to solve more precisely this longstanding and fundamental question.
Materials and methods
Plasmids, strains, and protein purifications
The plasmids, yeast strains and protein purification methods are described in the Supplementary data.
In vitro degradation assay
The ubiquitination of Sic1PY was performed as described previously (Saeki et al, 2005).
The degradation was initiated by adding the polyubiquitinated substrate to the purified 26S proteasome in buffer A (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 10% glycerol) containing 1 mM DTT, 2 mM ATP and 5 mM MgCl2 at 25°C. The reaction was terminated by adding SDS sample buffer and analysed by western blotting with anti-T7 antibody.
Mass spectrometry
Protein samples were analysed by MALDI-TOF mass spectrometry (Voyager DE-PRO or 4800 MALDI TOF/TOF, Applied Biosystems) as described previously (Saeki et al, 2004; Tanaka et al, 2008) with minor modifications. Briefly, CBB-stained protein bands were excised from the SDS–polyacrylamide gel, destained and in-gel digested with 10 μg/ml of modified trypsin (Trypsin Gold, Mass Spec Grade, Promega). For SILAC analysis, the excised gel was treated with 0.5% RapiGest (Waters) for improvement of tryptic digestion. To prepare the Ub internal standard peptides, a 1:1 mixture of K48-linked and K63-linked di-ubiquitins (Boston Biochem) was subjected to in gel-digestion with trypsin. MS and MS/MS data were obtained according to the instructions provided by the manufacturer and then analysed by ProteinPilot software 2.0 (Applied Biosystems) or manually.
Gel shift assay
The self-ubiquitinated GST-Rsp5 (2 pmol) was incubated with a purified 26S proteasome (2 pmol) in 5 μl total volume of buffer A plus 1 mM DTT, 2 mM ATP and 5 mM MgCl2 for 5 min on ice, mixed with dye and then subjected to 4% native-PAGE (Elsasser et al, 2002). The gel was incubated with 0.1 mM succinyl-Leu-Leu-Val-Tyr-7-amide-4-methyl-coumarin (suc-LLVY-amc) in buffer A plus 2 mM ATP and 5 mM MgCl2 for 10 min at 25°C. The proteasome bands were visualized under UV light (360 nm) and analysed by the gel documentation system (UVP Inc.) equipped with the UV cut-off filter (410 nm cut off, Kenko, Japan).
Purification of the ubiquitinated Mga2-p120 from yeast cells
Yeast cells (YYS1246; Δpdr5 Δlys2) were transformed with PGAL1-FLAGMGA2 (pOKA606), which expresses Flag-tagged Mga2 under the galactose-inducible promoter, and pKT10-Ub plasmid (pOKA601), which constitutively express wild-type Ub. The transformants were cultured to an OD600 between 0.6 and 0.8 in SRaf-Ura-Trp medium (0.67% yeast nitrogen base without amino acids, 0.5% casamino acids, 2% raffinose, 400 mg/l adenine, 10 mM phosphate buffer, pH 7.5). For stable isotope labeling experiments, the cells were grown in SILAC medium (0.67% yeast nitrogen base without amino acids, 2% raffinose, 20 mg/ml 13C-lysine, amino acid mixtures omitting appropriate nutrients, 400 mg/l adenine, 10 mM phosphate buffer, pH 7.5). Then, FlagMga2 was produced by the addition of galactose (1% final) for 2 h in the presence of 100 μM MG132 (20 mM stock in DMSO, Peptide Institute, Japan). The cells (corresponding to 100 OD600) were lysed by glass beads in lysis buffer, 50 mM HEPES-Na, pH 7.5, 100 mM NaCl, 10% glycerol, 10 mM iodoacetamide, 1 mM 1,10-Phenanthroline, 100 μM MG132, and 2 × concentration of protease complete inhibitor cocktail (-EDTA, Roche). After removal of the glass beads, Triton-X100 (1% final) was added and incubated for 30 min at 0°C. The extracts were cleared by centrifugation and incubated with anti-FLAG M2 agarose beads (Sigma) for 1.5 h at 4°C. Beads were washed twice with lysis buffer containing 1% Triton-X100 and twice with lysis buffer containing 1 M NaCl, then, twice with lysis buffer containing 0.2% Triton-X100. The Mga2 and its ubiquitinated species were eluted with 400 μg/ml Flag peptide (Sigma) in the same buffer. For immunoprecipitation experiments, the washing step with high salt buffer was omitted.
Purification of the proteasome-bound ubiquitinated proteins
The RPN11-3xFLAG tagged cells (YYS1338) were cultured an OD600 between 0.6 and 0.8 in SILAC medium (0.67% yeast nitrogen base without amino acids, 2% glucose, 20 mg/ml 13C-lysine, amino acid mixtures omitting appropriate nutrients, 400 mg/l adenine, 10 mM phosphate buffer, pH 7.5). Then, the cells were treated with 100 μM MG132 or DMSO for 2.5 h, and lysed by glass beads in lysis buffer, 50 mM HEPES-Na, pH 7.5, 50 mM NaCl, 10% glycerol, 10 mM iodoacetamide, 1 mM 1,10-Phenanthroline, 100 μM MG132 and 2 × concentration of protease complete inhibitor cocktail (-EDTA, Roche). After removal of the glass beads, the extracts were cleared by centrifugation and incubated with anti-FLAG M2 agarose beads (Sigma) for 1.5 h at 4°C. Beads were washed three-times with same buffer and the proteasomes were eluted with 400 μg/ml 3 × Flag peptide (Sigma) in the same buffer.
Supplementary Material
Supplementary Information
Acknowledgments
The authors thank Dr RJ Deshaies for kindly providing single Lys Sic1 plasmids and Dr Y Kimura for cdc48 mutant and anti-Cdc48 antibody. We are grateful to members of the Tanaka's lab for useful discussions. This work was supported by grants from the Ministry of Education, Science, Sports, Culture and Technology (MEXT) of Japan (to AT and KT) and the Target Protein Project of MEXT (YS and KT). YS was supported by the Japanese Society for the Promotion of Science.
References
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ (2008) UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell 134: 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnason T, Ellison MJ (1994) Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol 14: 7876–7883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baboshina OV, Haas AL (1996) Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem 271: 2823–2831 [DOI] [PubMed] [Google Scholar]
- Beaudenon SL, Huacani MR, Wang G, McDonnell DP, Huibregtse JM (1999) Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol 19: 6972–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR (2007) Global changes to the ubiquitin system in Huntington's disease. Nature 448: 704–708 [DOI] [PubMed] [Google Scholar]
- Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA (2007) Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell 130: 349–362 [DOI] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Finley D, Varshavsky A (1984) Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell 37: 57–66 [DOI] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127: 1401–1413 [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, de Souza GA, Li G, Mortensen P, Mann M (2006) Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol 7: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R, Hicke L (2001) Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J Biol Chem 276: 25974–25981 [DOI] [PubMed] [Google Scholar]
- Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol 4: 725–730 [DOI] [PubMed] [Google Scholar]
- Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V (1994) Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol 14: 5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JA, Lightcap ES, Sadis S, Thoroddsen V, Bulawa CE, Blackman RK (2002) Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc Natl Acad Sci USA 99: 1461–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JM, Haguenauer-Tsapis R (1997) Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. Embo J 16: 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Guterman A, Glickman MH (2004) Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J Biol Chem 279: 1729–1738 [DOI] [PubMed] [Google Scholar]
- Gwizdek C, Hobeika M, Kus B, Ossareh-Nazari B, Dargemont C, Rodriguez MS (2005) The mRNA nuclear export factor Hpr1 is regulated by Rsp5-mediated ubiquitylation. J Biol Chem 280: 13401–13405 [DOI] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D (2006) Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127: 99–111 [DOI] [PubMed] [Google Scholar]
- Hein C, Springael JY, Volland C, Haguenauer-Tsapis R, Andre B (1995) NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol 18: 77–87 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hicke L (2001) Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol 2: 195–201 [DOI] [PubMed] [Google Scholar]
- Hodgins R, Gwozd C, Arnason T, Cummings M, Ellison MJ (1996) The tail of a ubiquitin-conjugating enzyme redirects multi-ubiquitin chain synthesis from the lysine 48-linked configuration to a novel nonlysine-linked form. J Biol Chem 271: 28766–28771 [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM (2001) In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem 276: 27936–27943 [DOI] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S (2000) Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102: 577–586 [DOI] [PubMed] [Google Scholar]
- Horak J (2003) The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: insights from yeast. Biochim Biophys Acta 1614: 139–155 [DOI] [PubMed] [Google Scholar]
- Hoshikawa C, Shichiri M, Nakamori S, Takagi H (2003) A nonconserved Ala401 in the yeast Rsp5 ubiquitin ligase is involved in degradation of Gap1 permease and stress-induced abnormal proteins. Proc Natl Acad Sci USA 100: 11505–11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453: 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E, Saito N, Kamata N, Saeki Y, Toh EA (2005) Functional analysis of Rpn6p, a lid component of the 26 S proteasome, using temperature-sensitive rpn6 mutants of the yeast Saccharomyces cerevisiae. J Biol Chem 280: 6537–6547 [DOI] [PubMed] [Google Scholar]
- Jentsch S, Rumpf S (2007) Cdc48 (p97): a ‘molecular gearbox' in the ubiquitin pathway? Trends Biochem Sci 32: 6–11 [DOI] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270: 17442–17456 [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B (2007) The size of the nucleus increases as yeast cells grow. Mol Biol Cell 18: 3523–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Lyon N, Huibregtse JM (2005) The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. Embo J 24: 2414–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Munoz W, Lyon N, Huibregtse JM (2006) The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J Biol Chem 281: 36724–36731 [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL (2007) Certain pairs of ubiquitin-conjugating enzymes (E2 s) and ubiquitin-protein ligases (E3 s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem 282: 17375–17386 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP (2006) Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol 8: 700–710 [DOI] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96: 635–644 [DOI] [PubMed] [Google Scholar]
- Kriegenburg F, Seeger M, Saeki Y, Tanaka K, Lauridsen A-MB, Hartmann-Petersen R, Hendil KB (2008) Mammalian 26S proteasomes remain intact during protein degradation. Cell 135: 355–365 [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D (2004) Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 8: 610–616 [DOI] [PubMed] [Google Scholar]
- Piwko W, Jentsch S (2006) Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat Struct Mol Biol 13: 691–697 [DOI] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A (2004) An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol 11: 830–837 [DOI] [PubMed] [Google Scholar]
- Raasi S, Orlov I, Fleming KG, Pickart CM (2004) Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J Mol Biol 341: 1367–1379 [DOI] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107: 667–677 [DOI] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S (2005) A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120: 73–84 [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO (2007) Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell 130: 127–139 [DOI] [PubMed] [Google Scholar]
- Rotin D, Staub O, Haguenauer-Tsapis R (2000) Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J Membr Biol 176: 1–17 [DOI] [PubMed] [Google Scholar]
- Russell SJ, Steger KA, Johnston SA (1999) Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J Biol Chem 274: 21943–21952 [DOI] [PubMed] [Google Scholar]
- Saeki Y, Isono E, Toh-e A (2005) Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol 399: 215–227 [DOI] [PubMed] [Google Scholar]
- Saeki Y, Tayama Y, Toh-e A, Yokosawa H (2004) Definitive evidence for Ufd2-catalyzed elongation of the ubiquitin chain through Lys48 linkage. Biochem Biophys Res Commun 320: 840–845 [DOI] [PubMed] [Google Scholar]
- Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M (2008) Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453: 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik N, Haines DS (2007) Cdc48p(Npl4p/Ufd1p) binds and segregates membrane-anchored/tethered complexes via a polyubiquitin signal present on the anchors. Mol Cell 25: 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik N, Zoladek T, Nickels JT, Haines DS (2003) Rsp5p is required for ER bound Mga2p120 polyubiquitination and release of the processed/tethered transactivator Mga2p90. Curr Biol 13: 1227–1233 [DOI] [PubMed] [Google Scholar]
- Somesh BP, Reid J, Liu WF, Sogaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2005) Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121: 913–923 [DOI] [PubMed] [Google Scholar]
- Sone T, Saeki Y, Toh-e A, Yokosawa H (2004) Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J Biol Chem 279: 28807–28816 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Tanaka N, Saeki Y, Tanaka K, Murakami M, Hirano T, Ishii N, Sugamura K (2008) c-Cbl-dependent monoubiquitination and lysosomal degradation of gp130. Mol Cell Biol 28: 4805–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J 19: 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118: 99–110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information