Abstract
Natural killer T (NKT) cells recognize lipid antigens presented by CD1d molecules and play an important role in the regulation of innate and adaptive immune responses. Here we report the identification of a membrane-associated protein, immunoglobulin-like transcript 4 (ILT4), as a novel human CD1d receptor that inhibits CD1d-mediated immune responses. We found that native CD1d tetramer generated by mammalian cells was able to specifically bind human monocytes in the peripheral blood, and this binding was at least partly mediated by monocyte-expressed ILT4. The interaction between ILT4 and CD1d involves the two N-terminal domains of ILT4 and the antigen-binding groove of CD1d (α1/α2 domain). This interaction has been identified on the cell surface as well as in the endosomal and lysosomal compartments. Functional analysis showed that ILT4 could block the loading of lipid antigens such as α-GalCer, and consequently inhibited NKT recognition. The interaction between ILT4 and CD1d may provide new insights into the regulation of NKT-mediated immunity.
Keywords: human, T cells, cell surface molecules, MHC, antigen presentation/processing
Human CD1 family proteins (CD1a–e) are non-polymorphic MHC-I-like molecules and represent a third lineage of antigen presenting molecules capable of intracellular trafficking, processing, and presentation of lipid antigens (1, 2). Natural killer T (NKT)3 cells restricted by CD1d are important in host defense against mycobacterial and viral infections (3–5). Upon TCR engagement, NKT cells rapidly secrete a variety of immunoregulatory cytokines such as IL-4 and IFN-γ, and can exert specific cytotoxicity towards target cells. Consequently, the activation of NKT cells affects the activation of a variety of other cell types, including dendritic cells (DCs), NK cells, B cells and conventional T cells. In vivo, NKT cells are implicated in a broad array of disease conditions ranging from infections to tumors, various forms of autoimmunity, and allergies (4, 6).
In humans, CD1d is mainly expressed on APCs such as DCs and monocytes/macrophages. The mechanism of CD1d antigen presentation is distinct from that of MHC-I and II (1, 2). When the nascent CD1d/β2 microglobulin (β2m) complex is assembled and transported from the endoplasmic reticulum (ER) to the cell surface, it is associated with endogenous lipids such as phosphatidylinositol (PI) or PI-glycan (GPI) (7, 8). Upon reaching the cell surface, CD1d molecules quickly undergo internalization into the endocytic pathway, and CD1d-associated endogenous lipids are replaced with immunogenic lipid antigens in late endosomes and lysosomes. CD1d subsequently recycles to the cell surface to present the newly-bound lipid antigens to NKT cells (2, 9).
A variety of lipid-binding proteins are involved in CD1d trafficking and antigen presentation. Microsomal triglyceride transfer protein (MTP) was shown to play a role in the loading of endogenous lipids in the ER (10) and lipid presentation in lysosomes (11), while saposins reportedly extract membrane-associated lipids and deliver them to CD1d in late endosomes and lysosomes (12, 13). Additionally, CD1e, another member of the CD1 family that is not expressed on the cell surface, might be implicated in lysosomal lipid antigen loading (14).
Lipid-bound CD1d on the surface of APCs is recognized by cognate TCRs expressed on NKT cells. Most NKT cells have a limited TCR repertoire, which mostly consists of Vα24 and Vβ11 chains in humans (6). The molecular interaction between the NKT cell TCR and CD1d has been elucidated in a co-crystal structure with CD1d presenting the synthetic NKT agonist α-Galactosylceramide (αGalCer) (15).
Here we report that, in addition to recognizing cognate TCRs in the presence of lipid antigens, CD1d also interacts with immunoglobulin-like transcript 4 (ILT4), a member of APC-expressed leukocyte receptors. Our results on the binding mechanisms and functions lead us to propose that that this novel interaction may provide a new regulatory mechanism controlling NKT cell stimulation and activation.
Materials and Methods
Tetramers, recombinant proteins and antibodies
The generation of native and αGalCer-CD1d tetramers has been described previously (16). Briefly, human β2m and CD1d fusion protein were produced by a lentivirus-transduced mammalian expression system. Purified protein was biotinylated and tetramerized with PE-conjugated streptavidin to form native CD1d tetramer. αGalCer was loaded at 37°C to generate antigen-loaded tetramer. HLA-A*0201 tetramer was generated in the conventional way, by refolding HLA*0201 heavy chain, β2m and HIV-1 gag-derived peptide SLYNTVATL, and tetramerized with streptavidin-PE (17). ILT4-Fc fusion protein was generated through a lentivirus system similar to that used for β2m-CD1d. ILT4D1-D2 in pET19b vector (Novagen) was expressed in E.Coli, and the inclusion bodies were refolded using the same protocol as was used for HLA-A*0201 production. CD1dX1 mAb was generated from BALB/c mice immunized with CD1d-Fc fusion protein. CD1d42 mAb was purchased from BD Pharmingen, and anti-ILT4 mAb from R&D systems. CD3-FITC (DAKO), CD14-FITC (BD Biosciences) and anti-human IgG-HRP (Sigma-Aldrich) were all purchased commercially. Lamp1-YFP was a kind gift from Dr. Jennifer Lippincott-Schwartz of the Cell Biology and Metabolism Branch, National Institute of Child Health & Human Development (NICHD), National Institutes of Health.
Human PBMCs, monocytes and NKT cells
Fresh human PBMCs were isolated from whole blood of healthy donors by Lymphoprep (Nycomed Pharma AS, Norway) density gradient centrifugation. Monocytes were purified with human CD14 MACS MicroBeads (Miltenyi Biotec) following the manufacturer’s protocol. The NKT cell line was generated by expanding the NKT population from PBMCs following FACS sorting. Briefly, CD1d-tetramers loaded with α-GalCer were used to stain fresh human PBMCs, and tetramer+ CD3+ cells were FACS sorted and expanded by culturing with irradiated mixed human PBMCs from three healthy donors supplemented with 200U/ml IL-2 and 10µg/ml phytohemoagglutinin (PHA) in RPMI1640 with 10% human serum. The expanded cells were tested for αGalCer-CD1d tetramer staining and a further round of FACS sorting and expansion would be performed if less than 95% of the cells were tetramer positive.
Immunoprecipitation
Streptavidin-Dynabeads (Dynal) were coated with biotinylated CD1d or HLA-A*0201 monomer at room temperature for 30 min. The beads were then washed with PBS and added 1% NP-40-lysed HEK293T-ILT2-GFP or HEK293T-ILT4-GFP stable cells to carry out immunoprecipitation at 4°C overnight. The beads were then washed extensively before boiling in loading buffer and subjected to SDS-PAGE. Proteins were transferred to Hybond-C membrane (Amersham) in a Trans-Blot SD Semi-dry Transfer Cell (BIO-RAD) and Western blotting was carried out with HRP labeled anti-hu-IgG-Fc Ab, and blots were visualized by an Enhanced Chemiluminescence (ECL) kit (Amersham).
Flow Cytometry (FACS)
All FACS staining was carried out in 50 µl of FACS washing buffer (PBS with 2% fetal calf serum and 0.02% azide). After the addition of Abs, cell suspensions were incubated on ice for 30 min and washed twice with washing buffer before fixation with 1% paraformaldehyde/PBS. For tetramer staining, cells were incubated with native or α-GalCer-loaded CD1d tetramers at a final concentration of 10 µg/ml at 37°C for 30 min before staining with other fluorescence-labeled mAbs on ice. For blocking experiments, blocking reagents were added at 100 µg/ml to the cell suspension before adding the tetramers. FACS data acquisition and analysis were performed on a FACSCalibur™ machine using CELLQuest™ software (Becton Dickinson).
Domain-swap constructs and transfections
ILT4 domains were replaced with corresponding domains from ILT2 by stitch PCR mutagenesis, and the resulting fusion DNA fragments were then cloned into pEGFP-N1 vector (Clontech) in-frame with the C-terminal EGFP. CD1d domain-swap mutants were generated using similar approaches. DNA constructs were transiently transfected into HEK293T cells by FuGENE6 (Roche) according to the supplier’s manual, and cells were collected at 24h after transfection for FACS analysis.
Confocal microscopy
NKR cells (Normal Rat Kidney Epithelial Cell) were transfected with ILT4-CFP /Lamp1-YFP, CD1D-CFP/Lamp1-YFP, and ILT4-CFP/CD1D-YFP (1 µg) via Amaxa nucleofection™ (T buffer, program X-001) and grown in glass chambers (Lab-TEK). Twenty-four hours after transfection, live cell images were obtained using a Leica SP5 confocal microscope.
NKT responses to plate-bound CD1d
Ninety-six well microtiter plates were coated with 10 µg/ml streptavidin (Invitrogen) at 4°C overnight before binding with 10 µg/ml biotinylated CD1d monomer for 3h. After extensive washing, ILT4-Fc, control Fc fusion protein DR5-Fc or solvent PBS were added to the wells and incubated for 3 more hours before the addition of αGalCer for overnight antigen loading at 37°C. NKT cells were added at 5×104/well and incubated for 24 h in a 37°C incubator with 5% CO2, and the supernatants were harvested and analyzed for cytokine release with an IFNγ ELISA kit (R&D Systems).
Results
Binding of native CD1d tetramers to monocytes
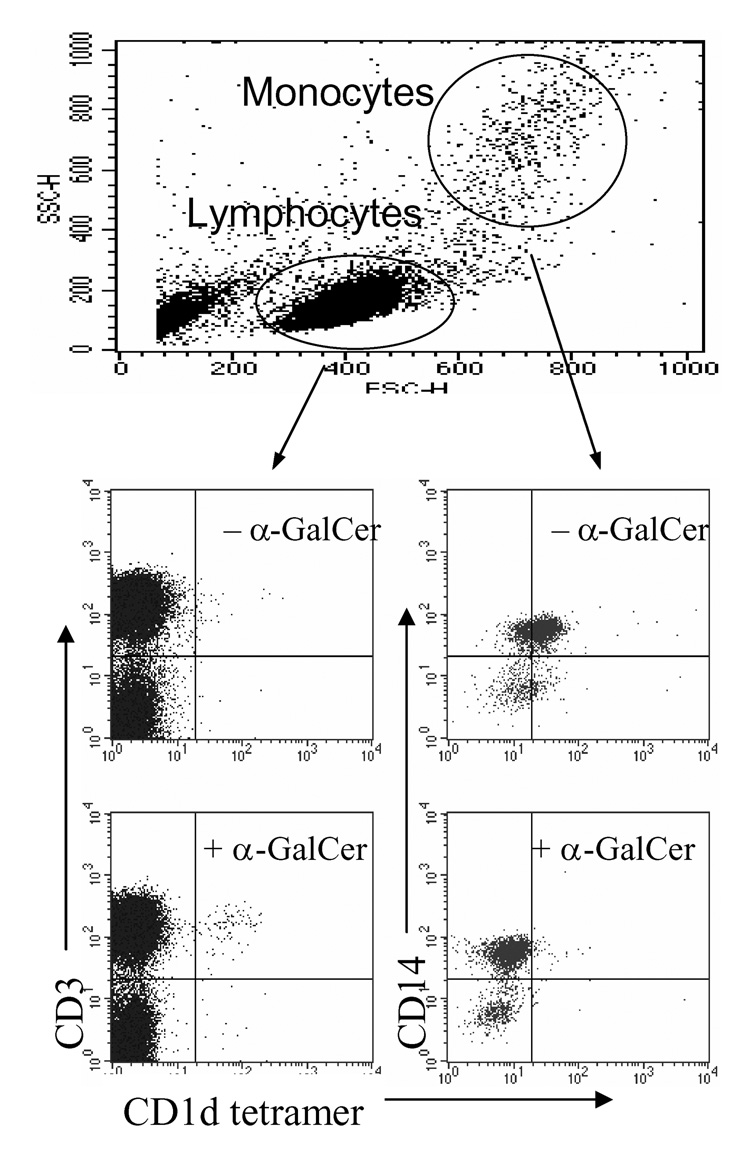
We have previously reported the generation of CD1d tetramers through a lentivirus-transduced mammalian expression system (16). While attempting to detect the NKT subset in human PBMC, we unexpectedly discovered that, although the native CD1d tetramers (tetramers without αGalCer, but potentially with endogenous lipids) did not stain any lymphocytes distinctly, they did show specific binding to monocytes (middle panel, Fig. 1). This binding occurred within both CD14+ and CD14− subsets of monocytes, although the former population had slightly higher staining than the latter. More surprisingly, after loading with αGalCer, tetramers could detect NKT cells in the lymphocyte population as predicted, but they were no longer able to bind monocytes (low panel, Fig. 1). These results prompted us to investigate the molecular mechanism behind this interaction.
Figure 1.
CD1d tetramers differentially bind monocytes. Human PBMCs were stained with PE-conjugated native CD1d-tetramers (upper panel) or α-GalCer-pulsed CD1d-tetramers. Cells were co-stained with anti-CD3 and -CD14 mAbs.
Monocyte-expressed ILT4 is a receptor for CD1d
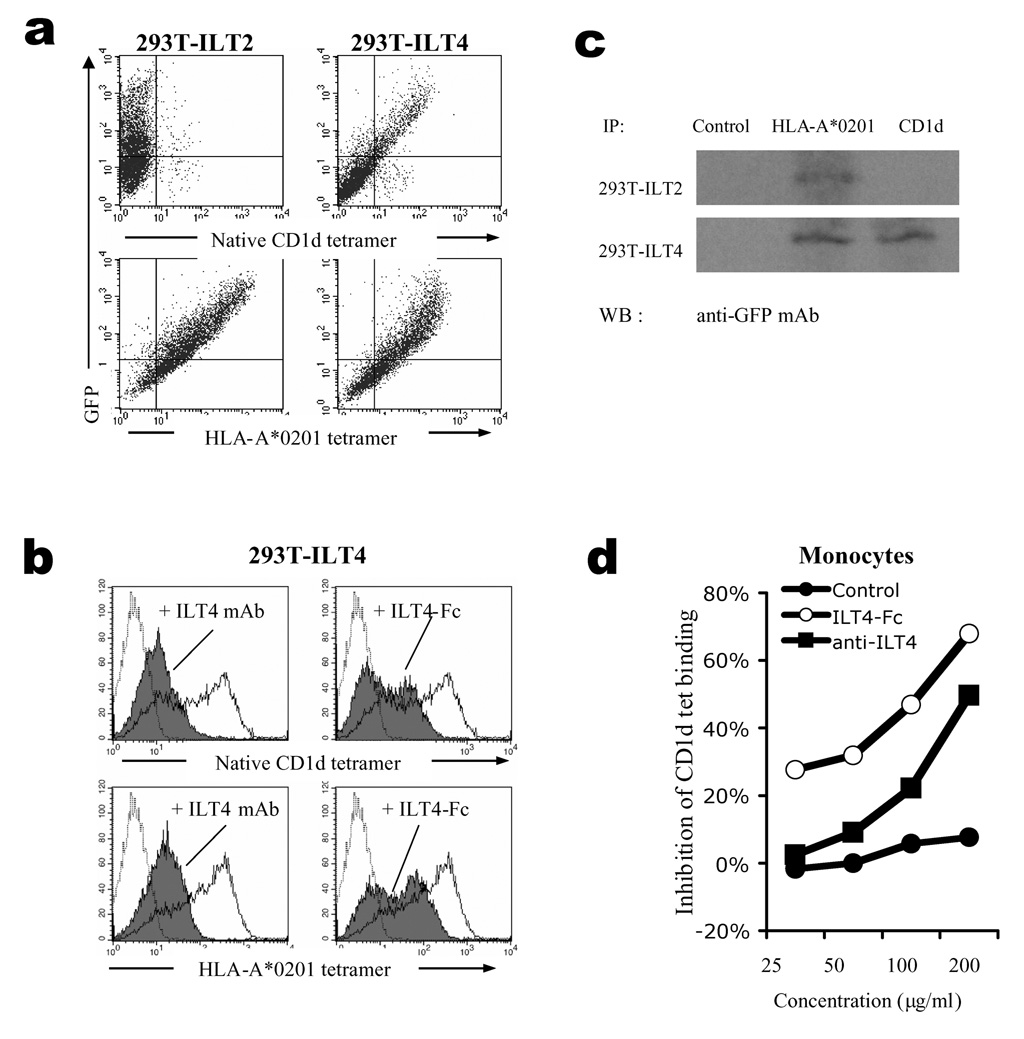
Previous studies have shown that a number of monocyte/macrophage-expressed cell receptors can recognize classical and non-classical MHC-I molecules. Examples include members of the ILT family, such as ILT2 and ILT4 which interact with HLA-G, HLA-B27 and HLA-A2 molecules (18–21). To search for the putative CD1d receptor(s) expressed by monocytes, we tested whether CD1d tetramers were able to interact with transfectants of individual receptors from the ILT family. The results showed that CD1d tetramers only recognized ILT4-transfected HEK293T cells, whereas tetramers of a classical MHC-I molecule, HLA-A*0201, bound to both ILT2 and ILT4-transfected cells (Fig. 2a). The specificities of these interactions were further demonstrated by blocking experiments, in which an ILT4-specific mAb or a soluble ILT4-Fc fusion protein inhibited binding of HLA-A*0201 as well as CD1d tetramers to ILT4 (Fig. 2b). Co-immunoprecipitation experiments showed that ILT4-Fc protein and CD1d interacted with each other in vitro (Fig. 2c). Taken together, these results demonstrated that ILT4 is a receptor for CD1d.
Figure 2.
Monocyte-expressed immunoglobulin-like transcript 4 (ILT4) is partially responsible for CD1d binding to monocytes. (a) Binding of CD1d to ILT4 transfectants. HEK293T cells transfected with ILT2- or ILT4-GFP fusion constructs were stained with PE-conjugated native CD1d-tetramer (upper panel) or HLA-A*0201-tetramer (lower panel). (b) Blocking of CD1d binding by ILT4-specific mAb and ILT4-Fc fusion protein. HEK293T cells transfected with ILT4-GFP were stained with CD1d tetramer (upper panel) or HLA-A*0201 tetramer (lower panel) in the presence (shaded histograms) or absence (solid lines) of anti-ILT4 mAb (left panel) or ILT4-Fc fusion protein (right panel). Streptavidin staining was used as negative control (dotted lines). (c) Immunoprecipitation of ILT4 with CD1d. Dynabeads coated with HLA-A*0201 or CD1d tetramers were used to immunoprecipitate ILT4 or ILT2 protein from HEK293T stable transfectants cell lysate, and the precipitations were subsequently detected by anti-GFP mAb and HRP labelled anti-mouse-IgG secondary Ab. Uncoated Dynabeads were used as control. (d) Blocking of CD1d binding to monocytes. Human monocytes were stained with CD1d tetramers in the presence of various concentrations of anti-ILT4 mAb or ILT4-Fc fusion protein. The inhibition was calculated based on mean fluorescence intensity. Representing results from one out of three repeating experiments were shown.
Next, we tested whether ILT4 is involved in CD1d’s binding to monocytes. We found that ILT4-specific mAb could partially inhibit the binding in a concentration-dependent manner, whereas ILT4-Fc showed even higher blocking efficiency, suggesting that ILT4 is at least partially responsible for the interaction of CD1d with monocytes, although other factor(s) may also be involved (Fig. 2d).
Characterization of the CD1d-ILT4 interaction
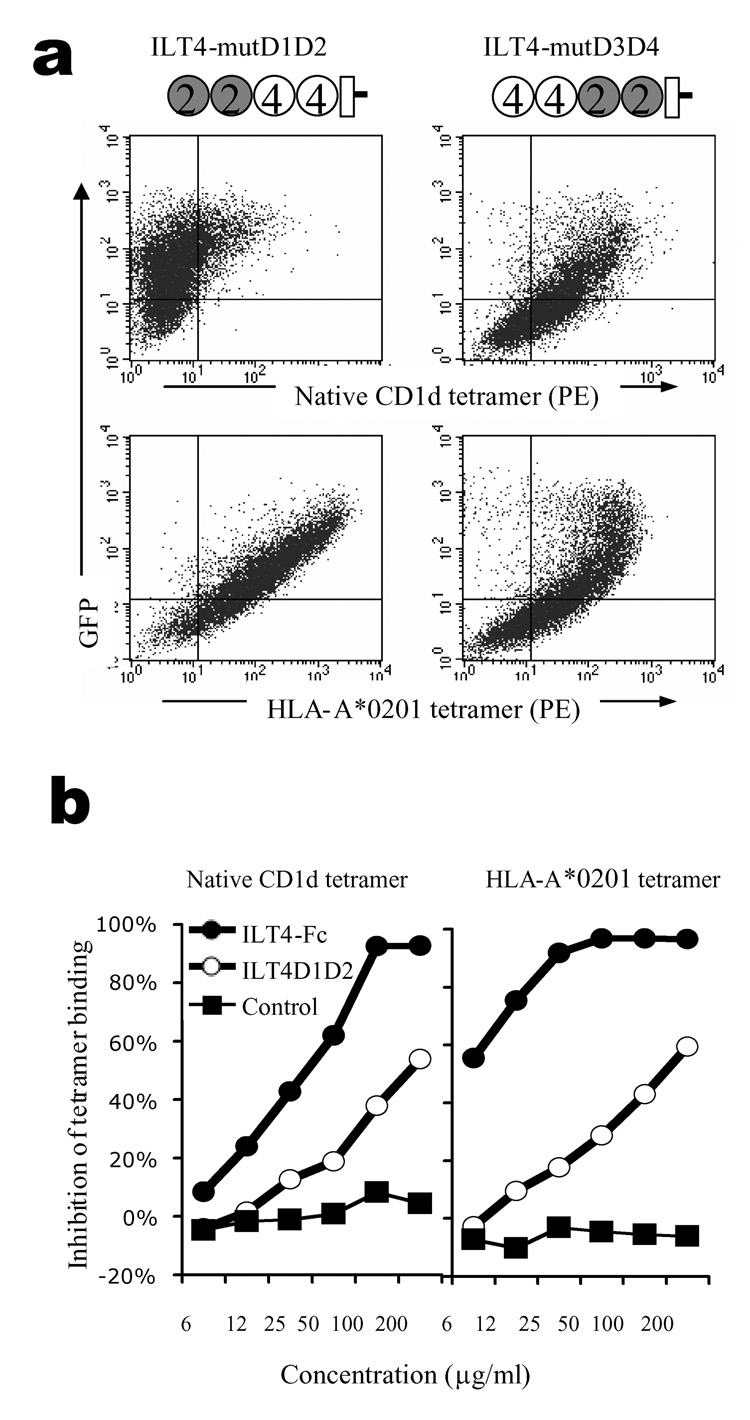
ILT4 is highly homologous to ILT2, both having four Ig domains in their extracellular regions (D1, D2, D3, and D4) that assemble in similar conformations (22, 23). A co-crystal structure of ILT2 with HLA-A2 has been resolved and shows that the contact lies in the D1 and D2 domains of ILT2, and the α3 and β2m domains of MHC-I (24). To examine which part of ILT4 is involved in its interaction with CD1d, we generated domain-swap mutants in which D1–D2 or D3–D4 were exchanged between ILT2 and ILT4. These constructs were transfected into HEK293T cells and tested for their ability to bind CD1d compared with HLA-A*0201 tetramers. Both mutated constructs bound the HLA-A*0201 tetramers, indicating that the mutant proteins were expressed and correctly folded (bottom panel, Fig. 3a). However, only the construct containing D1–D2 from ILT4 was able to bind to CD1d while the other mutant did not show significant binding (top panel, Fig 3a), suggesting D1–D2 of ILT4 plays a dominant role. To further confirm this, we generated a truncated protein which only has D1–D2 domain from ILT4. This protein, here referred as ILT4 D1–D2, was able to block CD1d-ILT4 interaction, although the blocking effect was less potent than that observed with full-length ILT4 Fc-fusion protein (Fig. 3b). The decreased efficiency is presumably due to the facts that the full length protein is better folded/supported than the truncated (D1–D2) form, and that the dimeric Fc-fusion protein has a higher ligand-binding avidity.
Figure 3.
ILT4 D1–D2 are the main binding site for CD1d. (a) Binding of ILT4 mutants by CD1d. HEK293T cells transfected with GFP fusion constructs encoding ILT4 mutants were stained with PE-conjugated CD1d or HLA-A*0201 tetramers. Cartoons illustrate the structure of the constructs. Open circles represent ILT4 domains, and filled circles represent ILT2 domains. (b) Blocking of tetramer binding to ILT4-transfectants. CD1d (left panel) or HLA-A*0201 (right panel) tetramers were used to stain HEK293T-ILT4 cells in the presence of different concentrations of blocking proteins. Results from one out of three repeated experiments are shown.
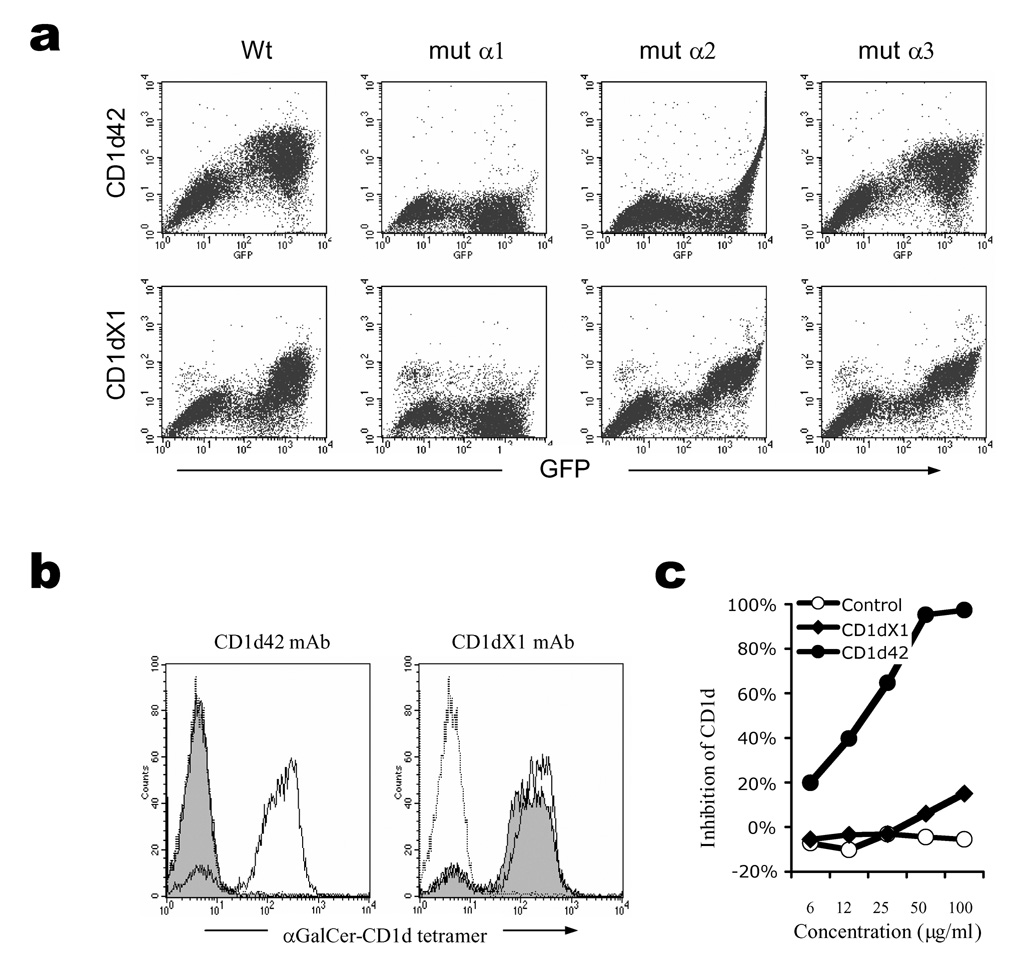
Next, we attempted to map the binding site for ILT4 on CD1d by using CD1d mAbs recognizing different epitopes. We generated a series of CD1d mutants in which every domain in the heavy chain was replaced by the corresponding domain of CD1b. The GFP-fusion constructs of these mutants were transfected into HEK293T cells, and a panel of CD1d mAbs were used to stain the transfectants in order to map their epitopes. Using this approach, the CD1d42 mAb was found to interact with the α1 and α2 domains of CD1d, since mutating either the α1 or α2 domain abolished its binding. On the other hand, mAb CD1dX1 was shown to recognize the α1 domain only, and mutating either α2 or α3 domain showed no effect on its binding (Fig. 4a). The α1 and α2 domains are the antigen-binding domains for CD1d; we observed that the CD1d42 mAb efficiently blocked recognition of NKT cells by αGalCer-loaded CD1d-tetramer, whereas CD1dX1 mAb did not show any effect (Fig. 4b). We anticipated these mAbs would have differential effects on CD1d binding to ILT4. Indeed, when mAb CD1d42 was incubated with HEK293T-ILT4 cells before native CD1d-tetramer staining, it showed dose-dependent inhibition of CD1d-tetramer binding, while mAb CD1dX1 did not show any effect (Fig. 4c). These results demonstrate that the antigen-binding α1 and α2 domains of CD1d are involved in its interaction with ILT4, and therefore, ILT4 might play a role in antigen presentation by CD1d.
Figure 4.
CD1d α1 and α2 domains are involved in ILT4 interaction. (a) Epitope mapping of CD1d mAbs. HEK293T cells were transfected with wild type (wt), or single domain mutated CD1d-GFP fusion constructs. The cells were then stained with CD1d42 or CD1dX1 mAb. (b) Blocking of CD1d binding to NKT cells by CD1d mAbs. αGalCer-CD1d tetramers were used to stain NKT cells in the presence (shaded histograms) or absence (solid lines) of CD1d mAbs. Streptavidin-PE stainings were used as control (dotted line). (c) Blocking of CD1d binding to ILT4 transfectants by CD1d mAbs. HEK293T-ILT4 cells were stained with CD1d tetramers in the presence of different concentrations of CD1d mAbs or control mAb (W6/32). Results from one out of three repeated experiments are shown.
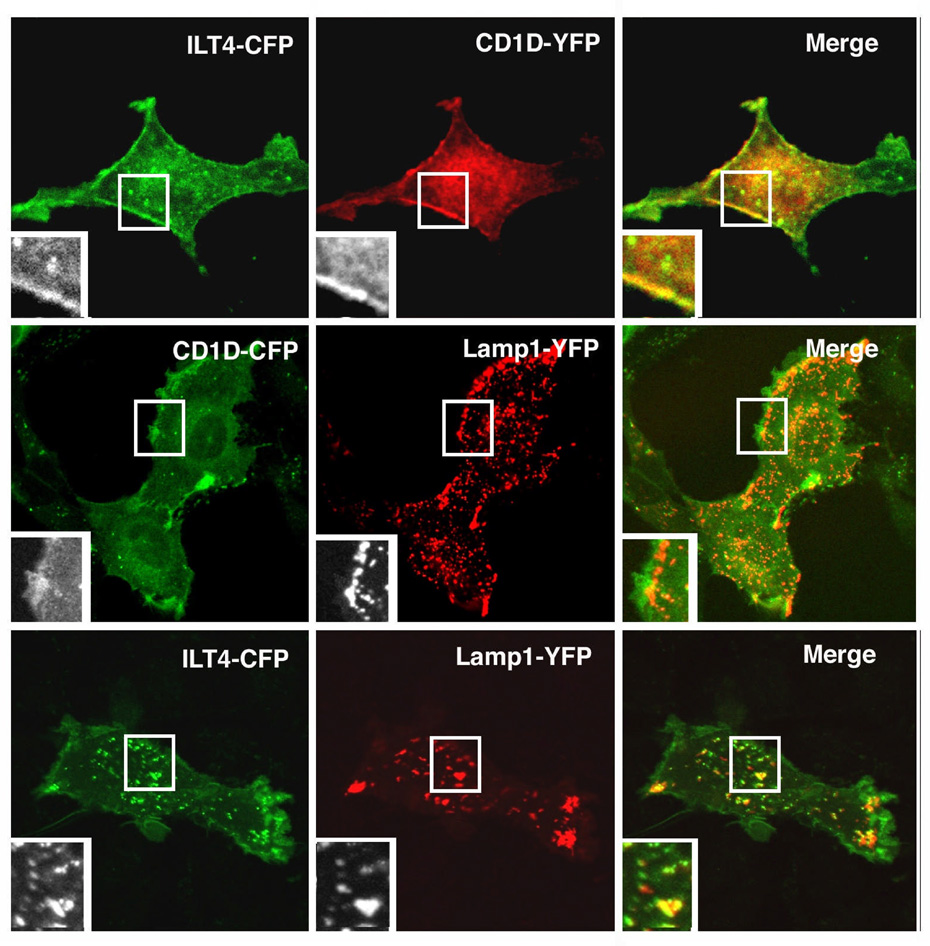
Co-localization of CD1d and ILT4
In general, ILT4 is expressed on antigen presenting cells (APC) such as monocytes/macrophages and DC, which also express CD1d, but not expressed on T cells, including NKT cells (25). It has been established that human CD1d trafficking to the endocytic system is essential for lipid antigen presentation. Therefore, to understand how ILT4 regulates CD1d-mediated antigen presentation, it is necessary to investigate the subcellular localization and/or co-localization of these two molecules. By co-transfection and confocal microscopy we observed that the sub-cellular distribution of CD1d and ILT4 largely overlap with each other both at the cell surface and in the cytoplasm (Fig. 5, upper panel). Furthermore, both molecules could be found in the LAMP-1+ compartments (Fig. 5, middle and lower panels), suggesting ILT4 also co-localizes with CD1d during its intracellular recycling. The co-trafficking of CD1d with ILT4 further suggests that ILT4 might be involved in regulating CD1d antigen presentation.
Figure 5.
Confocal microscopy analysis of colocalization of ILT4 and CD1d. Rat kidney epithelial cell line NKR was co-transfected with ILT4-CFP and CD1D-YFP (upper panel), CD1D-CFP and Lamp1-YFP (middle panel), or ILT4-CFP and Lamp1-YFP (lower panel) for confocal microscopy analysis.
ILT4 blocks loading of αGalCer onto CD1d and inhibits NKT cell recognition
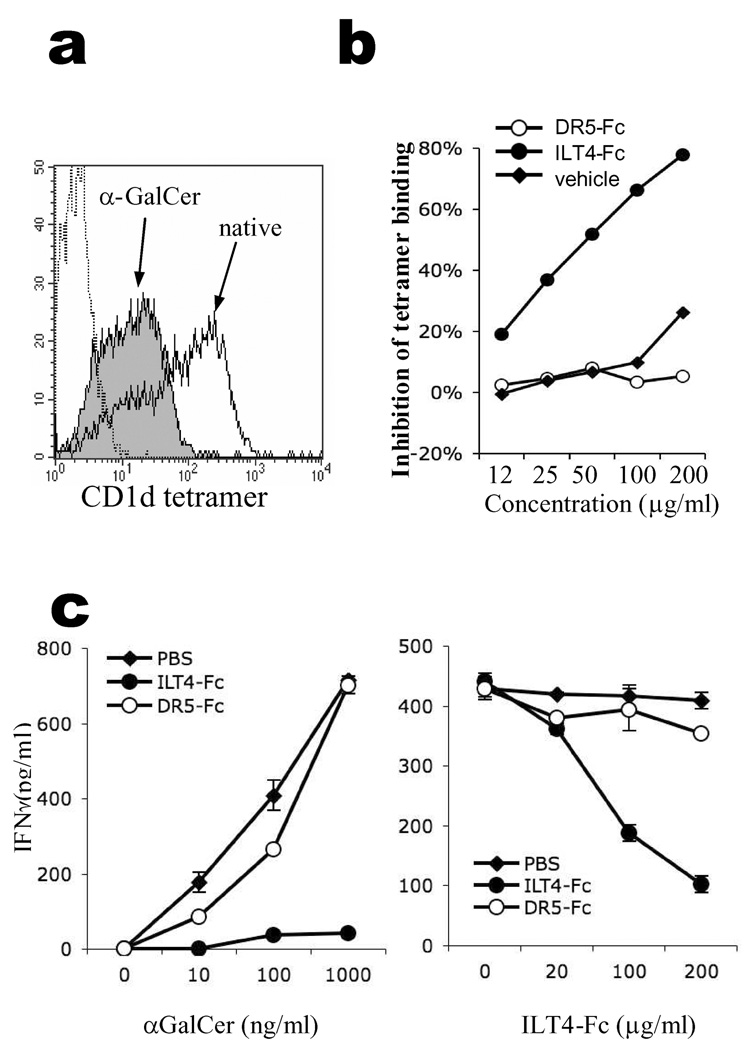
We have shown that native CD1d tetramer binds ILT4 on monocytes in an antigen-dependent manner (Fig. 1). To confirm that αGalCer-loading affects CD1d’s interaction with ILT4, we stained ILT4-transfected HEK293T cells with native or αGalCer-loaded CD1d tetramers, and discovered that αGalCer-loading had a negative effect on CD1d binding to ILT4. As shown in Fig. 6a, binding of αGalCer-CD1d to ILT4 was significantly attenuated compared to native CD1d. Since it is generally accepted that native CD1d contains tissue-derived lipid antigens (7, 8) (our unpublished data), this result suggests that the nature of the lipid antigen associated within CD1d has an influence on ILT4 recognition.
Figure 6.
ILT4 regulates APC-NKT recognition in an antigen-dependent manner. (a) Blocking of CD1d binding to ILT4 by αGalCer. HEK293T-ILT4 cells were stained with native (open histogram) or αGalCer-loaded CD1d tetramers (filled histogram). PE-conjugated Streptavidin was used as negative control staining (dotted line). (b) Blocking of αGalCer loading to CD1d by ILT4. Native CD1d-tetramers were pre-incubated with different concentrations of ILT4-Fc (filled circles), or control protein DR5-Fc (open circles) before being pulsed with αGalCer antigen. The resultant αGalCer-CD1d tetramers were used to stain NKT cells and MFI of the stainings were compared to calculate the inhibition rates. As a control, pre-generated αGalCer-CD1d tetramers were used to stain NKT cells in the presence of various concentrations of ILT4-Fc protein (filled diamonds). Results from one out of three repeated experiments are shown. (c) IFNγ secretion by a NKT cell line (NKN) stimulated by plate-bound CD1d protein. Plate-bound CD1d were incubated with 200µg/ml of ILT4-Fc, control protein DR5-Fc or solvent PBS, before being pulsed with different concentrations of αGalCer (left panel). In the right panel, the plates were incubated with different concentrations of blocking reagents before being pulsed with 100ng/ml of αGalCer.
Similarly, ILT4 did not affect αGalCer-CD1d tetramer binding to NKT cells (filled diamonds, Fig. 6b). Nevertheless, during the generation of αGalCer-CD1d tetramers, we observed that incubation of ILT4-Fc with native CD1d tetramer prior to αGalCer pulsing resulted in reduced tetramer binding to NKT cells in a concentration-dependent manner (filled circles, Fig. 6b). On the contrary, a control protein death receptor 5 (DR5)-Fc, which does not bind CD1d, did not show any effect on CD1d tetramer binding (open circles, Fig. 6b). This result suggests that, although ILT4 does not bind to αGalCer-loaded CD1d, its binding to native CD1d does prevent further lipid exchange by CD1d.
Finally, we investigated whether the interaction between ILT4 and CD1d had any functional implication for NKT cells. We used αGalCer-pulsed plate-bound CD1d to stimulate NKT cells, and investigated whether ILT4 could affect NKT activation. As shown in Fig. 6c (left panel), if plate-bound CD1d was pre-incubated with ILT4-Fc fusion protein before pulsing with αGalCer, its ability to stimulate NKT cells was dramatically reduced, even at very high concentrations of αGalCer antigen. This effect showed a dose-response with ILT4-Fc protein (Fig. 6c, right panel). As controls, another Fc fusion protein (DR5-Fc) or protein solvent (PBS) did not cause significant reduction in NKT activation. These results suggest that ILT4 could inhibit NKT activation by directly binding to CD1d and preventing antigen loading.
Discussion
The ILT family of proteins are also referred to as leukocyte immunoglobulin-like receptors (LIRs), monocyte/macrophage inhibitory receptor (MIR), or CD85. They are a set of immunoreceptors expressed on the surface of lymphoid and myeloid cells (25). Two of the best characterized members, ILT2 and ILT4, have been found to bind to MHC-I molecules such as HLA-A2, HLA-G, HLA-F, and their viral homologue, UL18 (20, 26–29). Both ILT2 and ILT4 are APC-expressed inhibitory receptors containing ITIM motifs which can abrogate activating signals resulting in a potent inhibition of APC functions (28, 30). In this study, we discovered that CD1d, a member of the non-classical MHC-I family, is also able to interact specifically with ILT4 but not ILT2, unlike MHC-I proteins, which bind both ILT2 and ILT4. This is fairly unusual because the structure of ILT2 and ILT4 are very similar (24, 31). The inability of CD1d to bind to ILT2 suggests that there is a different sub-molecular mechanism involved in this novel interaction.
The co-crystal structure of ILT2 with HLA-A2 demonstrates that ILT2 uses its first two N-terminal Ig domains to interact with the α3 domain of MHC- I heavy chain and β2m, which are essential for CD8 binding (24). As a result, CD8 was found to compete with ILT molecules for MHC-I binding (18). ILT4 structure displays significant similarity to that of ILT2 (22, 23), and its co-crystal structure with HLA-G confirmed that ILT4 uses its D1 and D2 domain to interact with ligand like HLA-G (31). However, the ILT4 contact site on HLA-G is mainly located on the hydrophobic regions of the α3 domain and is largely independent of β2m (31). In parallel, we proved that although the D1 and D2 domains of ILT4 are the binding sites for CD1d, their interactions with CD1d seem to involve the α1 and α2 domains, which happen to be the location of the antigen binding groove.
This observation raises an interesting question of whether ILT4 can influence NKT cell recognition by contacting directly with the antigen-binding groove and its associated lipid antigen. Although a co-crystal structure of ILT4 and CD1d would be ideal to address this question, our binding data do shed some light on this interaction. We found that ILT4 interacts only with native CD1d, which contains endogenously-derived lipid antigens or chaperone lipids (data not shown), but not with CD1d loaded with αGalCer. Two possibilities may explain these observations. One is that the antigen-binding groove-associated lipids, such as endogenous lipids, are in direct contact with ILT4, and the glycan heads protruding from the α-helix plane decide the specificity of ILT4. On the other hand, αGalCer loading may induce conformational changes of CD1d that affect the ILT4 binding sites. The latter is supported by the crystal structures of lipid-free or αGalCer-bound CD1d molecules, which show substantial conformational differences (32). Whichever the case is, the fact that αGalCer can inhibit the binding of ILT4 to CD1d indicates that there is a competition between these two ligands. It is not yet known whether this phenomenon only applies to αGalCer, which is the strongest NKT agonist ever found, or it is a common mechanism affecting other foreign lipid antigens as well.
The functional implication of the ILT4-CD1d interaction is an intriguing topic. Although we observed that ILT4 affects CD1d antigen presentation, and consequently inhibits NKT recognition, there are quite a lot details needing further investigation. By examining the intracellular localization using confocal microscopy, we have shown that ILT4 accompanies CD1d at the cell surface as well as in the endocytic compartments, suggesting that this interaction is very likely to happen on the same cells, and the regulatory function of ILT4 might be an integral component of the CD1d antigen presentation machinery. However, the possibility that APC-expressed CD1d interacts with ILT4 expressed on other cells can not be ruled out, considering that fact that classical HLA-I molecules bind to ILT4 mainly through trans-interaction(23, 25).
The functional implications of CD1d-ILT4 interaction discussed above are based on monocytes and ILT4-transfected cells. Another interesting point to note is the role of CD1d-ILT4 interaction in the functions of other professional APC such as DCs. We and other have shown the abundant expression of ILT4 on the surface of DC (25)(supplemental figure 1). However, binding by CD1d tetramers on monocyte-derived DC is negative. Moreover, the CD1d binding did not improve during the upregulation of ILT4 accompanying DC maturation (supplemental figure 1). This strongly suggests that the DC-expressed ILT4 is in different forms to those expressed by monocytes or ILT4 transfected cells, probably due to differential post-translational modification or to association with accessory molecules. This is an area where we are actively investigating.
In summary, we have established that ILT4 serves as a novel receptor for CD1d, and its interaction modulates CD1d-mediated antigen presentation. This study may provide new insights into the regulation of NKT cell immunity.
Supplementary Material
Acknowledgments
We thank Professor Alain Townsend, Drs Hal Drakesmith, Simon Kollnberger Paul Bowness, Mariolina Salio, Professors Vincenzo Cerundolo, and Andrew J. McMichael for discussions, and Dr Sarah Bangs and Mr Neil Haig for critical reading of the manuscript.
Footnotes
This work was supported by Medical Research Council UK and Chinese National Natural Science Foundation(30371346).
Abbreviations used in this paper: αGalCer, α-galactosylceramide; β2m, β2 microglobulin; DC, dendritic cells; NKT cells, natural killer T cells; ER, endoplasmic reticulum; ILT4, immunoglobulin-like transcript 4.
References
- 1.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 2.Barral DC, Brenner MB. CD1 antigen presentation: how it works. Nat Rev Immunol. 2007;7:929–941. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 3.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 4.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 5.Li D, Xu XN. NKT cells in HIV-1 infection. Cell Res. 2008;18:817–822. doi: 10.1038/cr.2008.85. [DOI] [PubMed] [Google Scholar]
- 6.Van Kaer L. NKT cells: T lymphocytes with innate effector functions. Curr Opin Immunol. 2007;19:354–364. doi: 10.1016/j.coi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Park JJ, Kang SJ, De Silva AD, Stanic AK, Casorati G, Hachey DL, Cresswell P, Joyce S. Lipid-protein interactions: biosynthetic assembly of CD1 with lipids in the endoplasmic reticulum is evolutionarily conserved. Proc Natl Acad Sci U S A. 2004;101:1022–1026. doi: 10.1073/pnas.0307847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joyce S, Woods AS, Yewdell JW, Bennink JR, De Silva AD, Boesteanu A, Balk SP, Cotter RJ, Brutkiewicz RR. Natural ligand of mouse CD1d1: cellular glycosylphosphatidylinositol. Science. 1998;279:1541–1544. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 9.De Libero G, Mori L. Recognition of lipid antigens by T cells. Nat Rev Immunol. 2005;5:485–496. doi: 10.1038/nri1631. [DOI] [PubMed] [Google Scholar]
- 10.Dougan SK, Salas A, Rava P, Agyemang A, Kaser A, Morrison J, Khurana A, Kronenberg M, Johnson C, Exley M, Hussain MM, Blumberg RS. Microsomal triglyceride transfer protein lipidation and control of CD1d on antigen-presenting cells. J Exp Med. 2005;202:529–539. doi: 10.1084/jem.20050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sagiv Y, Bai L, Wei DG, Agami R, Savage PB, Teyton L, Bendelac A. A distal effect of microsomal triglyceride transfer protein deficiency on the lysosomal recycling of CD1d. J Exp Med. 2007;204:921–928. doi: 10.1084/jem.20061568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou D, Cantu C, 3rd, Sagiv Y, Schrantz N, Kulkarni AB, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, Savage P, Bendelac A, Teyton L. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004;5:175–181. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 14.de la Salle H, Mariotti S, Angenieux C, Gilleron M, Garcia-Alles LF, Malm D, Berg T, Paoletti S, Maitre B, Mourey L, Salamero J, Cazenave JP, Hanau D, Mori L, Puzo G, De Libero G. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 15.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 16.Li D, Chen N, McMichael AJ, Screaton GR, Xu XN. Generation and characterisation of CD1d tetramer produced by a lentiviral expression system. J Immunol Methods. 2008;330:57–63. doi: 10.1016/j.jim.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 18.Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, Allan DS, Makadzange A, Rowland-Jones S, Willcox B, Jones EY, van der Merwe PA, Kumagai I, Maenaka K. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci U S A. 2003;100:8856–8861. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen RL, Raine T, Haude A, Trowsdale J, Wilson MJ. Leukocyte receptor complex-encoded immunomodulatory receptors show differing specificity for alternative HLA-B27 structures. J Immunol. 2001;167:5543–5547. doi: 10.4049/jimmunol.167.10.5543. [DOI] [PubMed] [Google Scholar]
- 20.Lepin EJ, Bastin JM, Allan DS, Roncador G, Braud VM, Mason DY, van der Merwe PA, McMichael AJ, Bell JI, Powis SH, O'Callaghan CA. Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur J Immunol. 2000;30:3552–3561. doi: 10.1002/1521-4141(200012)30:12<3552::AID-IMMU3552>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 21.Allan DS, Colonna M, Lanier LL, Churakova TD, Abrams JS, Ellis SA, McMichael AJ, Braud VM. Tetrameric complexes of human histocompatibility leukocyte antigen (HLA)-G bind to peripheral blood myelomonocytic cells. J Exp Med. 1999;189:1149–1156. doi: 10.1084/jem.189.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman TL, Heikema AP, West AP, Jr, Bjorkman PJ. Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT2) Immunity. 2000;13:727–736. doi: 10.1016/s1074-7613(00)00071-6. [DOI] [PubMed] [Google Scholar]
- 23.Willcox BE, Thomas LM, Chapman TL, Heikema AP, West AP, Jr, Bjorkman PJ. Crystal structure of LIR-2 (ILT4) at 1.8 A: differences from LIR-1 (ILT2) in regions implicated in the binding of the Human Cytomegalovirus class I MHC homolog UL18. BMC Struct Biol. 2002;2:6. doi: 10.1186/1472-6807-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4:913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 25.Colonna M, Nakajima H, Cella M. A family of inhibitory and activating Ig-like receptors that modulate function of lymphoid and myeloid cells. Semin Immunol. 2000;12:121–127. doi: 10.1006/smim.2000.0214. [DOI] [PubMed] [Google Scholar]
- 26.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu ML. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 27.Navarro F, Llano M, Bellon T, Colonna M, Geraghty DE, Lopez-Botet M. The ILT2(LIR1) and CD94/NKG2A NK cell receptors respectively recognize HLA-G1 and HLA-E molecules co-expressed on target cells. Eur J Immunol. 1999;29:277–283. doi: 10.1002/(SICI)1521-4141(199901)29:01<277::AID-IMMU277>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 28.Colonna M, Samaridis J, Cella M, Angman L, Allen RL, O'Callaghan CA, Dunbar R, Ogg GS, Cerundolo V, Rolink A. Human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J Immunol. 1998;160:3096–3100. [PubMed] [Google Scholar]
- 29.Colonna M, Navarro F, Bellon T, Llano M, Garcia P, Samaridis J, Angman L, Cella M, Lopez-Botet M. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vitale M, Castriconi R, Parolini S, Pende D, Hsu ML, Moretta L, Cosman D, Moretta A. The leukocyte Ig-like receptor (LIR)-1 for the cytomegalovirus UL18 protein displays a broad specificity for different HLA class I alleles: analysis of LIR-1 + NK cell clones. Int Immunol. 1999;11:29–35. doi: 10.1093/intimm/11.1.29. [DOI] [PubMed] [Google Scholar]
- 31.Shiroishi M, Kuroki K, Rasubala L, Tsumoto K, Kumagai I, Kurimoto E, Kato K, Kohda D, Maenaka K. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc Natl Acad Sci U S A. 2006;103:16412–16417. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch M, Stronge VS, Shepherd D, Gadola SD, Mathew B, Ritter G, Fersht AR, Besra GS, Schmidt RR, Jones EY, Cerundolo V. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol. 2005;6:819–826. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.