Summary
The chaperone activity of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein (NC) facilitates multiple nucleic acid rearrangements that are critical for reverse transcription of the single-stranded RNA genome into double-stranded DNA. Annealing of the trans-activation response element (TAR) RNA hairpin to a complementary TAR DNA hairpin is an essential step in the minus-strand transfer step of reverse transcription. Previously, we used truncated 27-nucleotide (nt) mini-TAR RNA and DNA constructs to investigate this annealing reaction pathway in the presence and absence of HIV-1 NC. In this work, full-length 59-nt TAR RNA and TAR DNA constructs were used to systematically study TAR hairpin annealing kinetics. In the absence of NC, full-length TAR hairpin annealing is ∼10-fold slower than mini-TAR annealing. Similar to mini-TAR annealing, the reaction pathway for TAR in the absence of NC involves the fast formation of an unstable “kissing” loop intermediate, followed by a slower conversion to an extended duplex. NC facilitates the annealing of TAR by ∼105-fold by stabilizing the bimolecular intermediate (∼104-fold) and promoting the subsequent exchange reaction (∼10-fold). In contrast to the mini-TAR annealing pathway, wherein NC-mediated annealing can initiate through both loop-loop kissing and a distinct “zipper” pathway involving nucleation at the 3′/5′ terminal ends, full-length TAR hairpin annealing switches predominantly to the zipper pathway in the presence of saturated NC.
Keywords: nucleic acid chaperone activity, HIV-1 nucleocapsid protein, TAR RNA/DNA annealing, minus-strand transfer, nucleic acid aggregation, zinc fingers, kissing interactions, zipper pathway
Introduction
Human immunodeficiency virus type-1 (HIV-1) is a retrovirus that contains its genomic information in the form of two identical single-stranded (ss) RNA molecules. Conversion of the RNA genome into double-stranded viral DNA is an essential step of the retroviral life cycle. Reverse transcription involves multiple steps, including several nucleic acid rearrangements catalyzed by the HIV-1 nucleocapsid protein (NC). NC has been shown to facilitate annealing of the tRNA primer onto the primer binding site 1-4, annealing of complementary RNA/DNA repeat regions in minus-strand transfer 5-11, and annealing of complementary DNA sequences in plus-strand transfer 7; 12-14. HIV-1 NC is a short, basic, nucleic acid binding protein, containing two zinc finger domains, each of which has an invariant CCHC metal-ion binding motif 15-18. The mature protein is produced by proteolytic cleavage of the Gag precursor and is found in the interior of the virus particle, where it is tightly associated with genomic RNA 19.
NC has multiple functions during the viral replication cycle. In addition to playing essential roles in reverse transcription, NC or the NC domain of Gag have been shown to facilitate genomic RNA packaging, virus assembly, and integration 20-24. Many of these functions rely on the nucleic acid chaperone activity of NC, which promotes nucleic acid rearrangements leading to thermodynamically more stable structures 22-26. The chaperone function of NC relies on two main activities, nucleic acid (NA) aggregation 27-30 and duplex destabilization 31-38, associated with the cationic N-terminal domain and zinc fingers, respectively.
It has recently been shown that synthesis of long products during reverse transcription in vitro is facilitated under conditions of NA aggregation 39. In addition, DNA synthesis at low RT concentrations became more processive under aggregating conditions, and strand displacement synthesis of the last 99 nucleotides (nt) resulting in production of the DNA flap was shown to be more efficient. Sedimentation analysis and gel-shift annealing studies performed under the same solution conditions as a function of NC concentration, showed that NA aggregation and annealing activities closely parallel each other 30.
Annealing of the trans-activation response element (TAR) DNA hairpin to a complementary TAR RNA hairpin, an essential step in minus-strand transfer, was shown to be accelerated by NC as much as 3000-fold by NC 11. Previous studies used model 27-nt mini-TAR RNA and DNA hairpins derived from the top hairpin loop region of TAR to elucidate the mechanism of NC-mediated TAR RNA/DNA annealing 30. This study showed that the complementary mini-TAR RNA and DNA hairpins anneal via a two-step pathway:
| (1) |
Here, the first bimolecular step leads to the formation of the annealing intermediate RD*, with the forward association rate constant k1, and the intermediate dissociation rate k-1. The annealing intermediate RD* is subsequently converted into the fully annealed RNA/DNA duplex RD via a second monomolecular step, which is characterized by the strand exchange rate k2 and reverse rate k-2. DNA mutagenesis data showed that in the absence of NC, mini-TAR RNA/DNA annealing is nucleated via an extended loop-loop “kissing” interaction 30. The annealing intermediate in this case involves 17 intermolecular base pairs (bp), including nt from the hairpin loop and from the adjacent stem and 3-nt bulge (see Figure 1). In the presence of saturating amounts of NC, an additional annealing pathway was observed, involving nucleation near the 3′/5′ ends 30. The later pathway has also been termed the “zipper” mechanism (Figure 1)40. Multiple annealing pathways have also been proposed for annealing of full-length TAR DNA to complementary DNA oligonucleotides based on single molecule studies 40. However, a kinetic analysis using full-length TAR RNA/DNA hairpins derived from the HIV-1 MAL isolate and NC(12-55), concluded that NC facilitated this annealing reaction primarily via the zipper pathway 41. A recent single molecule spectroscopy study using full-length TAR DNA/cDNA hairpins and full-length HIV-1 NC under non-aggregating conditions is consistent with this conclusion 42.
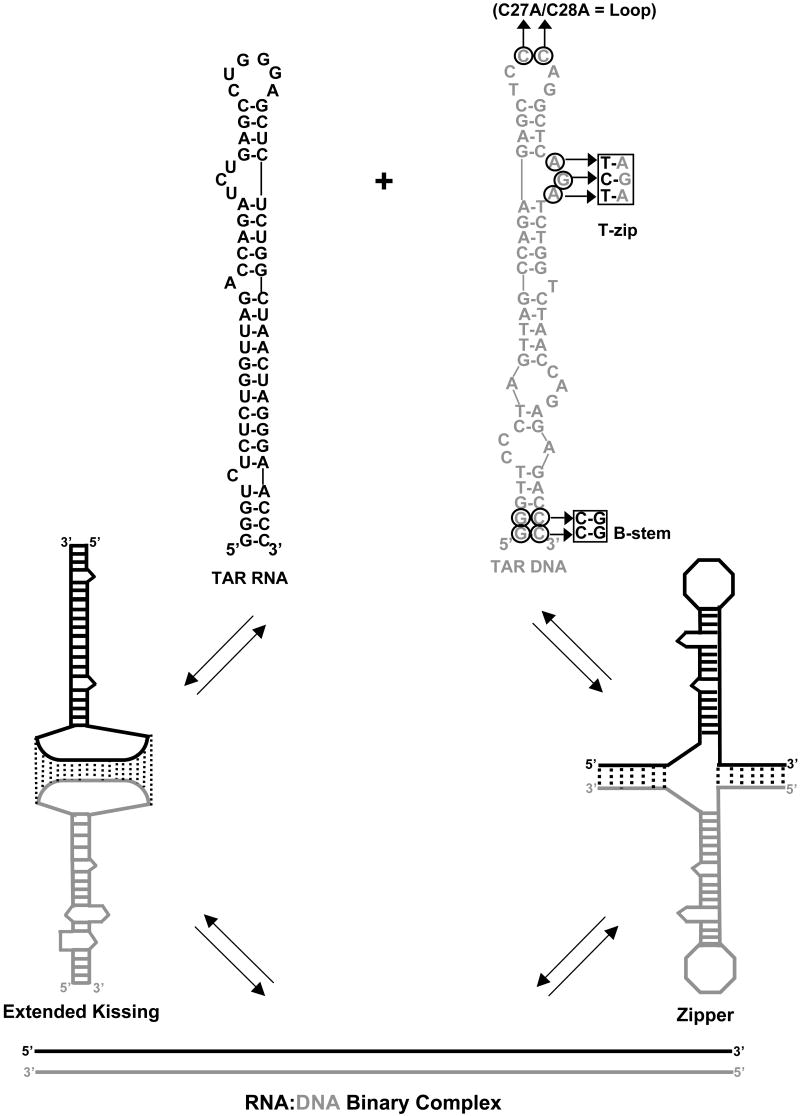
Figure 1.
Two pathways of full-length TAR RNA/DNA annealing. Top: predicted secondary structures of full-length TAR RNA (black) and TAR DNA (gray) hairpins. Sequences are derived from the HIV-1 NL4-3 isolate. The TAR RNA and DNA secondary structures were predicted by m-fold analysis51. Residues mutated in different DNA constructs are circled. Bottom: (left) loop-loop “kissing” pathway, which involves initial formation of an extended “kissing” complex followed by subsequent strand exchange to form the fully annealed duplex; (right) “zipper” pathway involving nucleation through the 3′/5′ termini resulting in formation of a zipper intermediate followed by conversion to the fully annealed duplex.
In this work, we use gel-shift assays to systematically study the annealing kinetics of full-length 59-nt TAR RNA and DNA hairpins in the presence and absence of wild-type HIV-1 NC. The substrates and conditions used here were designed to closely mimic the in vivo process. In particular, the use of full-length NC and gel-based rather than fluorescence-based assays allows reactions to be performed under conditions that promote NA aggregation. In addition, the role of RNA/DNA sequence complementarity and the NC concentration-dependence of the annealing reaction are examined. The data obtained in the absence of NC support an overall slow two-step structure-based annealing mechanism, involving nucleation via a loop-loop kissing interaction followed by conversion to an extended annealed duplex. In contrast, the presence of saturating NC switches the annealing mechanism to the zipper pathway involving intermediate complex formation between the 3′/5′ ends of the hairpin stems.
Results and Discussion
As observed previously for mini-TAR RNA/DNA annealing30, under all conditions examined here the annealing kinetics appears to be bi-exponential, and the percent of the molecules annealed as a function of time, P(t), can be described by the fast and the slow rates, kf, and ks, and the fraction of the fast component f according to:
| (2) |
Previous studies 30 showed that kf and ks correspond to the rate of formation of the intermediate and of the fully annealed duplex, respectively. The fraction of the fast component f is the probability of intermediate formation and P∞ is the final equilibrium percent of RNA annealed. The following equations describe the relationship between these kinetic parameters and the elementary rates of the two-step process 43:
| (3) |
where
| (4) |
Here, and Kd are the equilibrium dissociation constants of the intermediate RD* and the final annealed product RD, respectively. Experimentally, both rates are observed when they are sufficiently different from each other, i.e., kf » ks, and when the intermediate complex is reasonably stable, i.e., 0.1 < f < 1. The annealing of the TAR molecules shown in Figure 1 is strongly driven by formation of 14 new bp, such that the TAR hairpins anneal irreversibly both in the presence or absence of NC, i.e., P∞=100%, and k-2 is negligible. In addition, under all conditions examined here, the slow rate ks is proportional to f, in accord with the second relationship in (3).
According to eqs. (3) and (4), the fitted parameters kf, ks, and f can be used to estimate the elementary rates of the two-step annealing process using the following relationships:
| (5) |
Kinetics of full-length TAR RNA/DNA annealing in the absence of NC
Dependence of full-length TAR RNA/DNA annealing kinetics on the concentration of TAR DNA
Figure 1 shows the full-length 59-nt TAR RNA and TAR DNA substrates used in this work. Gel-shift assays with [32P]-labeled TAR RNA and unlabeled TAR DNA were used to quantitatively monitor complex formation as a function of time. Typical annealing time courses are shown in Figure 2(a). The annealing reactions were performed using several TAR DNA concentrations, with DNA in large excess over TAR RNA. In the absence of NC, the annealing reaction is extremely slow and irreversible. Under these pseudo-first order conditions, the reaction rate increases with increasing DNA concentration, which is consistent with a bimolecular association. To quantitatively analyze the annealing kinetics, the annealing time courses in Figure 2(b) were fit to the general two-exponential expression (2) to obtain ks, kf, and f. P∞ was set to 100% since the reaction is irreversible.
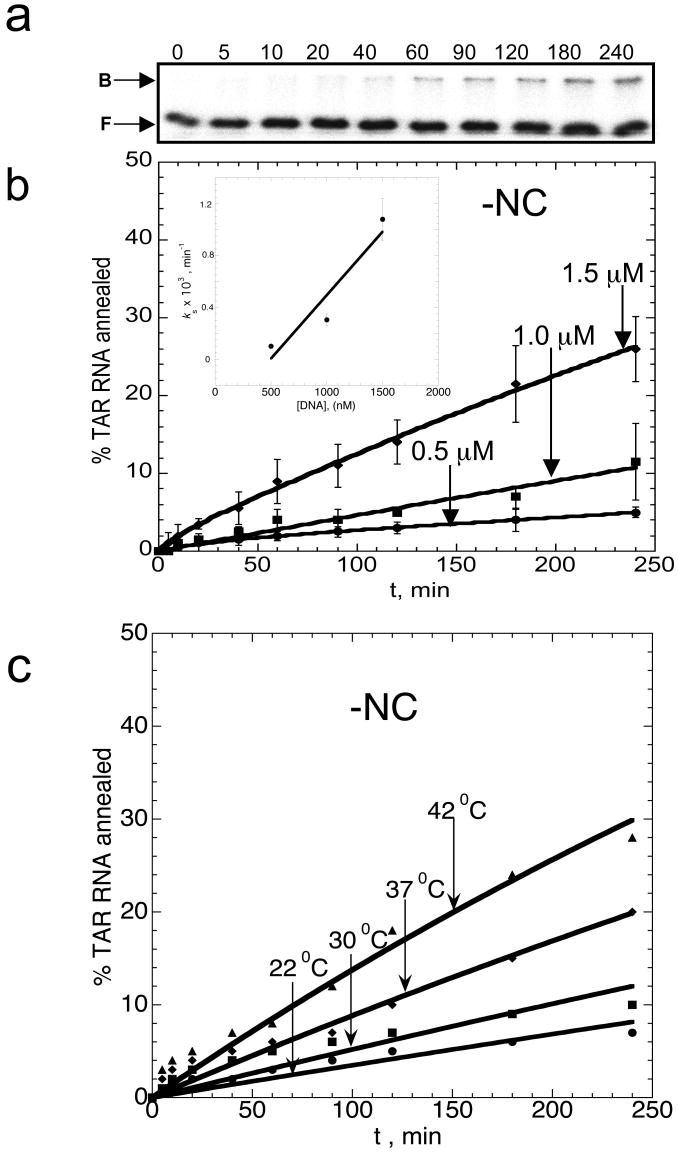
Figure 2.
Time course of TAR RNA/DNA hairpin annealing in the absence of NC. (a) Typical gel-shift analysis performed with 50 nM TAR RNA and 1 μM TAR DNA as a function of time. The numbers at the top of each lane correspond to minutes. Bound (B) and free (F) RNA bands are labeled on the left. (b) Time course showing the percent of TAR RNA (15 nM) annealed to variable concentrations of TAR DNA (as indicated on each curve) at 37 °C. Lines represent double-exponential fits of the data to equation (2), with the equilibrium percent annealed fixed at 100%. The insert shows the dominant slow annealing rate, ks obtained from the fits of the data shown in (b), as a function of TAR DNA concentration, D. The line is a linear fit of the data with a slope corresponding to keff=10±5 M-1s-1. (c) Temperature dependence of TAR RNA/DNA annealing. Time courses for TAR RNA (15 nM) annealing to TAR DNA (1.5 μM) were measured in the absence of NC in 20 mM Na+ at the indicated temperatures.
The fraction of the fast component, f, is very small (<0.1), but increases with increasing DNA concentration, D. The D dependence of the dominant slow annealing rate, ks, is presented in the insert in Figure 2(b). As expected, ks increases proportionally to D, and the slope of ks versus D yields an apparent bimolecular association rate constant keff of 10(±5) M-1 s-1. The small probability of intermediate formation (i.e., f«1) and the slow strand exchange rate both contribute to the extremely slow overall rate of annealing observed in the absence of NC. As shown in Figure 2(c), increasing temperature leads to a moderate rate enhancement. Vant Hoff analysis of these data yields an enthalpy for the rate-limiting step of ΔH=27±10 kcal/mol (data not shown). This value is comparable to the ΔH value determined for mini-TAR annealing (ΔH=15±5 kcal/mol) 30. This result is consistent with the hypothesis that mini-TAR and TAR anneal via the same pathway in the absence of NC. Interestingly, TAR annealing is ∼10-fold slower than mini-TAR annealing in the absence of protein chaperone 30. We hypothesize that this is due to the larger entropy loss upon intermediate complex formation in the case of the longer hairpins 44. Alternatively, slower annealing of longer hairpins may be due to a greater electrostatic repulsion. However, an analysis of the salt-dependence of the TAR annealing kinetics does not support the latter explanation 45.
DNA mutational analysis
To determine the pathway of full-length TAR RNA/DNA annealing, kinetic studies were performed using various mutant TAR DNA hairpins. To allow for more accurate quantification of extremely slow annealing kinetics observed in the absence of NC, assays were performed in the presence of 100 mM Mg2+, which is known to facilitate the annealing reaction without altering the annealing pathway 45. The importance of loop-loop complementarity was examined by studying the annealing of TAR RNA to a double (C27A/C28A) loop mutant DNA hairpin (Figure 1, right). Indeed, the C27A/C28A loop mutant displayed dramatically slower annealing compared to WT DNA (Figure 3). In contrast, when the two G:C base pairs at the bottom of the stem are inverted (B-stem mutant, see Figure 1), the rate of annealing is unaffected (Figure 3). These results suggest that loop-loop complementarity is critical for full-length TAR RNA/DNA annealing, while stem end complementarity is not. An additional TAR DNA variant was constructed containing a 3-nt insertion that results in closure of the 3-nt single-stranded bulge located near the top of the hairpin (T-zip, see Figure 1). This change creates 12 continuous bp adjacent to the hairpin loop and results in a dramatic reduction in the annealing rate (Figure 3).
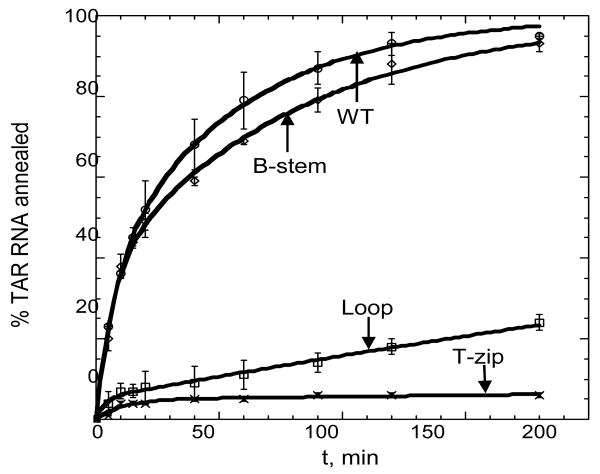
Figure 3.
Mutational analysis of TAR RNA/DNA annealing in the absence of NC. Percent TAR RNA (15 nM) annealed to various TAR DNA molecules (500 nM) as a function of time. The TAR DNA variants used are indicated by each curve (WT, Loop, T-zip or B-stem, see Figure 1). Annealing was performed at 37 °C in the presence of 100 mM Mg2+. Lines represent double-exponential fits of the data to equation (2), with the equilibrium percent annealed fixed at 100%.
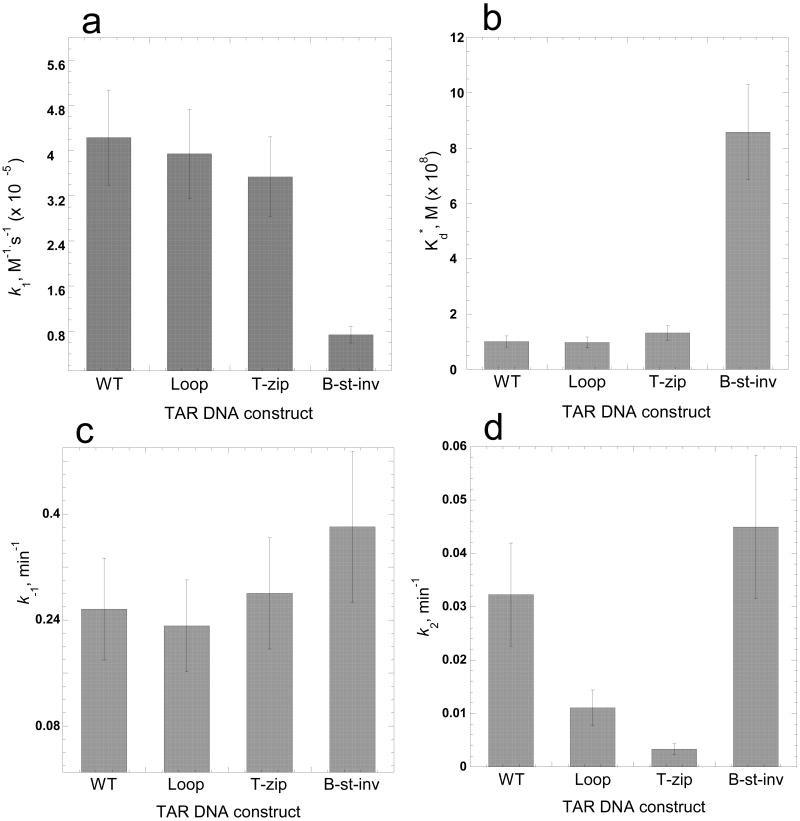
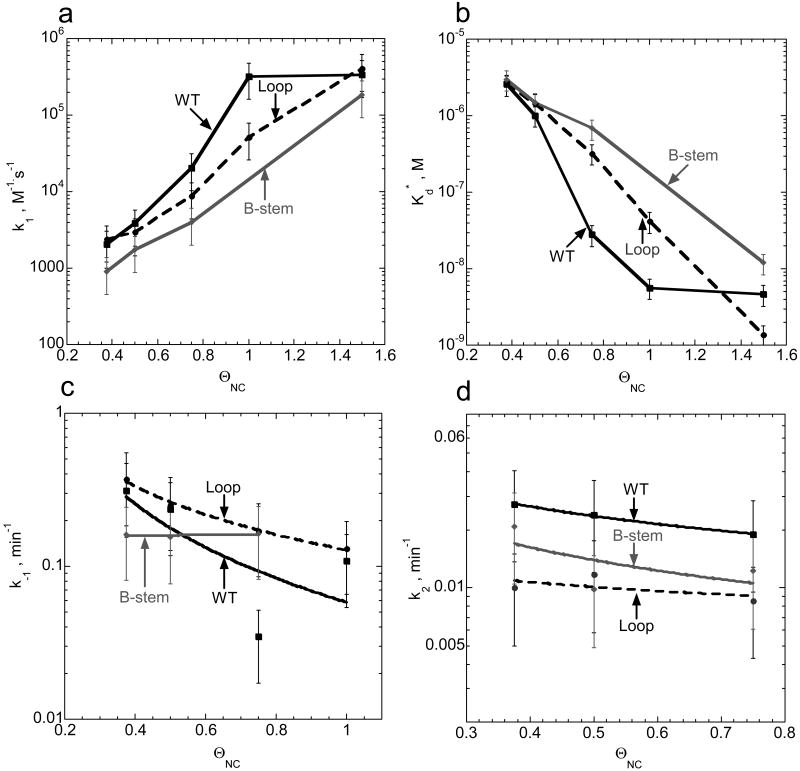
For all DNA mutants, the annealing kinetics measured in the presence of 100 mM Mg2+ was clearly biphasic. The kf, ks and f parameters were determined by fitting the experimental annealing time courses shown in Figure 3 to eq. (2). The elementary reaction rates determined from these fits using eqs. (5) are plotted in Figure 4. As shown in Figure 4(a), the intermediate association rate, k1, decreased by ∼7-fold when mutations were present in the top portion of the hairpin (i.e., Loop or T-zip). In contrast, the B-stem mutation had no effect on this rate. These results are consistent with a mechanism of TAR RNA/DNA annealing involving an extended loop-loop kissing intermediate (see Figure 1, lower left). As expected based on this annealing pathway, the B-stem mutation does not affect k1.
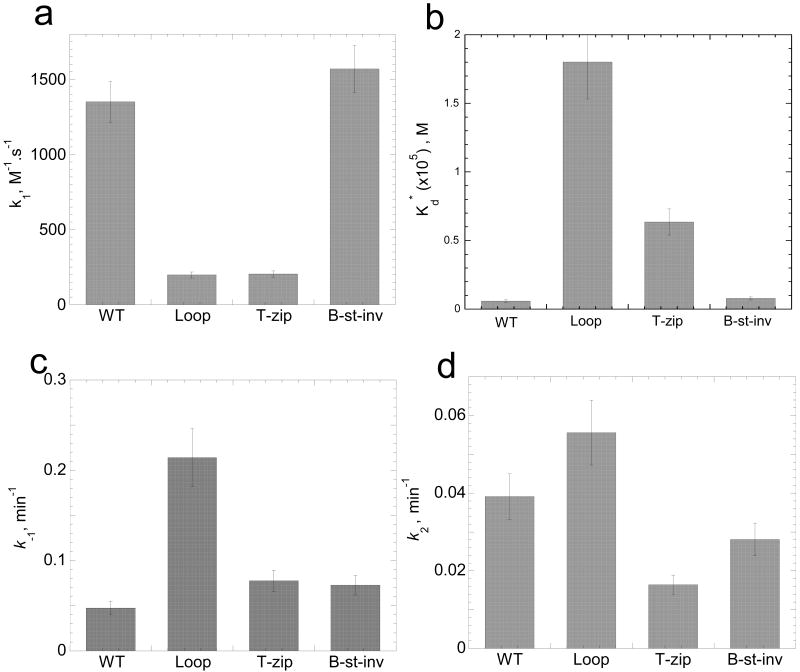
Figure 4.
Elementary reaction rates and intermediate dissociation constant for annealing of TAR RNA to various TAR DNA constructs in the absence of NC. The elementary rates (k1, panel a; k-1, panel c; k2, panel d) and intermediate dissociation constant (panel b) for the two-step annealing process were obtained by analysis of annealing time courses in Figure 3 using equations (2-5).
In agreement with these results, the intermediate dissociation rate, k-1, was not significantly affected by T-zip or B-stem mutations, but increased ∼5-fold in the presence of the Loop mutation (Figure 4(c)). As a result of these changes in the on and off rates for the formation of the reaction intermediate, the intermediate dissociation constant, Kd*, increased ∼30-fold and ∼10-fold in the presence of Loop and T-zip mutations, respectively (Figure 4(b)). Interestingly, the conversion rate, k2, is similar (i.e., within 2-fold of WT) for all of the TAR DNA constructs (Figure 4(d)). The small value of this rate, k2,=0.03±0.02 min-1, suggests that strand exchange is rate-limited by the cooperative melting of a large portion of one or both stems, followed by much faster strand exchange. This hypothesis is consistent with the observation that k2 is essentially the same for mini-TAR and TAR hairpins 45, i.e., that the strand exchange rate does not depend on hairpin stem length. Taken together, these data are consistent with an annealing mechanism in the absence of NC that involves nucleation through an extended loop-loop kissing interaction followed by slower conversion to a fully extended annealed duplex.
Kinetics of full-length TAR RNA/DNA annealing in the presence of NC
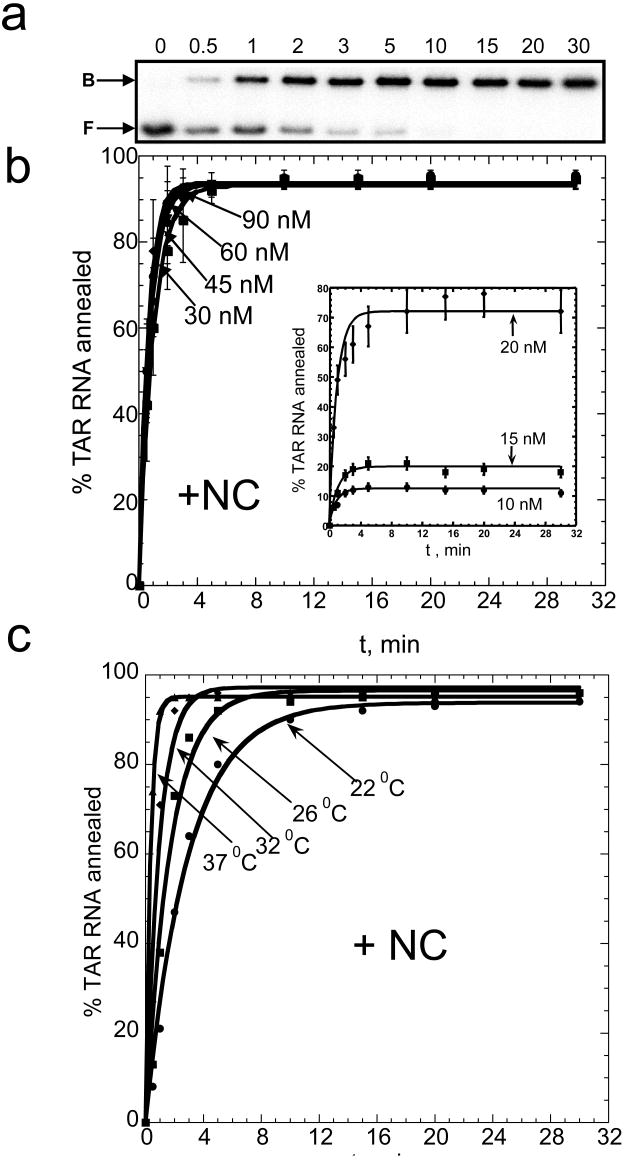
NC greatly facilitates full-length TAR RNA/DNA annealing. The time course of annealing in the presence of saturating amounts of NC (4:1 nt:NC ratio in low salt buffer, i.e., 20 mM Na+ and 0.2 mM Mg2+) is well described by a single-exponential expression (Figure 5). The final yield of annealed product in the reactions shown in Figure 5b (D = 30-90 nM) is close to 100%. Fitting the annealing time courses to a single-exponential expression yields an annealing rate of 4±2 min-1. Although under these conditions, the annealing rate appears to be independent of D (due to the time resolution of the assay), experiments performed at lower D (10-20 nM) show a reduced rate consistent with a bimolecular annealing mechanism (see inset in Figure 5(b)). The equilibrium percent of annealed product also decreases at lower D, suggesting that the annealing reaction is reversible under these conditions, as observed previously30. The annealing rate measured for D = 30-60 nM is ∼10-fold faster than the rate of NC-facilitated mini-TAR RNA/DNA annealing at similar D30. Given that TAR anneals ∼10-fold slower in the absence of NC, the overall NC-facilitated rate enhancement is ∼100-fold greater for full-length TAR annealing compared to mini-TAR.
Figure 5.
Time course of TAR RNA/DNA hairpin annealing in the presence of saturating amounts of NC. (a) Typical gel-shift analysis performed with 15 nM TAR RNA and 45 nM TAR DNA. The numbers at the top of each lane correspond to minutes. The positions of bound (B) and free (F) RNA bands are indicated on the left. (b) Time dependence of the percent of TAR RNA (15 nM) annealed to variable concentration of TAR DNA (as indicated by each curve) at 37 °C and a 4:1 nt:NC ratio. The 10-20 nM DNA curves are shown as an inset. (c) TAR RNA/DNA annealing time courses (15 nM TAR RNA+ 45 nM TAR DNA) obtained at different temperatures, as indicated in the Figure. All lines are single-exponential fits of the data.
Increasing temperature facilitates TAR annealing further (Figure 5(c)). The Van't Hoff analysis of the temperature dependence of the dominant fast rate (data not shown) yields an enthalpy for the rate-limiting step of 28±8 kcal/mol, consistent with extensive melting occurring prior to complex formation. This enthalpy value is ∼2-fold higher than previously reported for mini-TAR annealing 30. This result implies that in the presence of saturating amounts of NC, annealing of the longer TAR hairpins involves pre-melting of the more stable region than in the case of mini-TAR hairpin annealing.
DNA mutational analysis
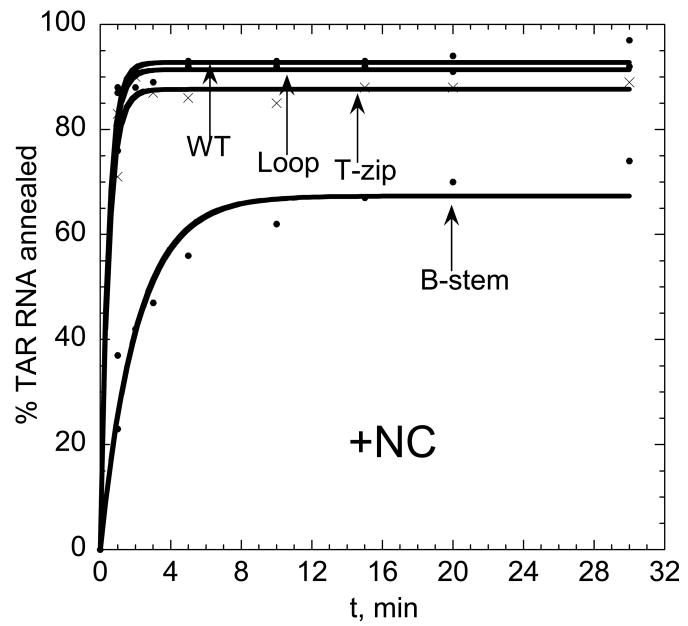
We next used the mutant TAR DNA constructs shown in Figure 1 (right) to investigate the annealing pathway of full-length TAR in the presence of saturating amounts of NC. Under these conditions, annealing in the presence of the Loop or T-zip variants is very fast and similar to that of the WT DNA (Figure 6). In contrast, the annealing rate with the B-stem variant is significantly reduced. The fast component dominates annealing (i.e. f∼1), thus making determination of k-1 and k2 rates less reliable than in the absence of NC. The effects of TAR DNA mutations on the elementary annealing rates are summarized in Figure 7. Relative to the reactions performed in the absence of NC, saturating amounts of NC resulted in 100-fold faster k1 for WT, Loop, as well as T-zip mutant TAR DNA. In contrast, k1 was ∼6-fold slower for the B-stem mutant (Figure 7(a)). The intermediate complex dissociation rate, k-1, was similar for all the TAR DNA constructs, with a slightly faster dissociation measured for the B-stem variant (Figure 7(c)). In part as a result of this, but largely due to the differences in k1, the reaction intermediate was an order of magnitude more stable for the WT, Loop, and T-zip TAR constructs relative to the B-stem variant (Figure 7(b)). These results are consistent with an annealing pathway that involves nucleation via the stem ends. The B-stem mutation introduces non-complementarity between DNA and RNA in this region, thereby making intermediate association slower, and dissociation faster at this nucleation site. The strand exchange rate, k2, is similar for WT, Loop mutant, and B-stem mutant. The ∼10-fold slower exchange rate determined for the T-zip mutant may be due to stabilization of the stem by introduction of 3 additional bp (Figure 7(d)). Taken together, these data are consistent with WT TAR RNA/DNA annealing via the zipper pathway in the presence of saturating amounts of NC.
Figure 6.
Mutational analysis of full-length TAR RNA/DNA annealing in the presence of saturating amounts of NC. Percent TAR RNA (15 nM) annealed to various TAR DNA constructs (45 nM), as indicated on each curve (WT, Loop, T-zip or B-stem, see Figure 1), as a function of time at 37 °C in the presence of a 4:1 nt:NC ratio. Lines represent single-exponential fits of the data.
Figure 7.
Comparison of elementary rates and intermediate dissociation constant of WT and mutant TAR DNA constructs in the presence of saturating concentrations of NC. Elementary rates (k1, panel a; k-1, panel c; k2, panel d) and intermediate dissociation constants (panel b) were obtained by using equations (2-5) to analyze the annealing time courses presented in Figure 6 for TAR RNA (15 nM) annealed to various TAR DNA constructs (45 nM) at 37 °C and a 4:1 nt:NC ratio.
Effect of varying NC concentration on WT and mutant TAR DNA annealing
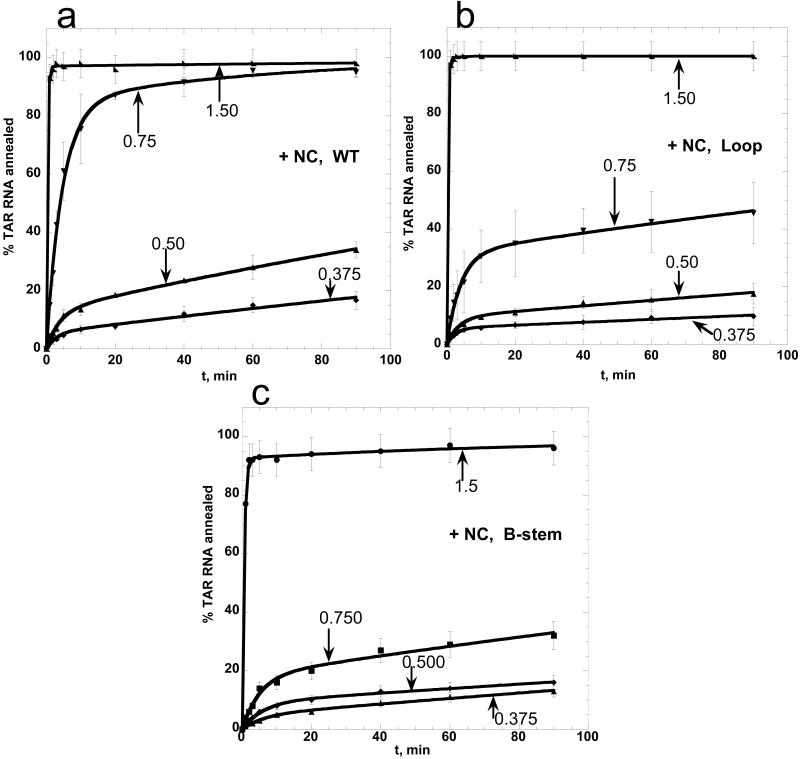
To examine the NC-induced TAR RNA/DNA annealing pathway switch in more detail, the annealing of 15 nM TAR RNA to 150 nM WT or mutant TAR DNA was measured in the presence of varying concentrations of NC (Figure 8). The latter is presented as fractional NA saturation with NC, ΘNC. Under these low salt conditions (20 mM Na+ and 0.2 mM Mg2+) all added NC is bound to NA, and ΘNC can be calculated as
Figure 8.
Annealing of WT and mutant TAR DNA to TAR RNA in the presence of variable amounts of NC. TAR RNA (15 nM) was annealed to 150 nM of WT TAR DNA (a), Loop mutant TAR DNA (b), or B-stem mutant TAR DNA (c) as a function of time in the presence of various concentrations of NC in low salt (20 mM Na+, 0.2 mM Mg2+). Fractional NA saturation with NC, ΘNC, calculated according to eq. (6), is indicated for each curve. Lines represent double-exponential fits of the data.
| (6) |
where NC:nt is the protein to nt concentration ratio and assuming that 1 NC binds to 6±1 nt at saturation 11; 22-24; 36; 46-49. As expected, at saturation (i.e., ΘNC ≥ 1), the annealing in the presence of the Loop mutant DNA (Figure 8(b)) is very fast and similar to that of WT (Figure 8(a)), whereas annealing with the B-stem variant is slightly suppressed (Figure 8(c)). Further decrease in ΘNC (ΘNC = 0.75) leads to a significant reduction of the annealing rate for both B-stem and the Loop mutants compared to WT DNA, with a more pronounced effect on the latter. These results suggest that under slightly sub-saturating concentrations of NC, full-length TAR RNA/DNA annealing occurs with comparable efficiency via both kissing and zipper pathways, with annealing via the stem ends slightly favored. This is consistent with the preferential annealing of WT TAR RNA/DNA via the zipper pathway under saturated NC binding conditions.
The dissociation constant for the annealing intermediate, as well as elementary rates for the two-step reaction, determined based on the data shown in Figure 8, are presented in Figure 9. The elementary rates could only be determined for ΘNC ≥ 0.375, since at lower ΘNC the annealing is extremely slow and has a negligible fast component. In addition, k2 can not be determined at a ΘNC > 0.75, since the annealing is dominated by the fast component. Figure 9(a) shows that the B-stem mutant has the slowest association rates over the range of 0.375 ≤ ΘNC ≤ 1.5. The Loop mutant has a slightly faster k1 than the B-stem mutant but remains slower than the WT. While k1 is very fast for all the TAR DNA constructs tested at high ΘNC, WT TAR DNA requires less NC to reach its maximum k1. These results are consistent with the existence of multiple annealing pathways, with a greater contribution from the zipper pathway when ΘNC ≥ 0.4.
Figure 9.
Comparison of elementary rates (panels a, c, and d) and intermediate dissociation constants (panel b) for annealing of WT and mutant TAR DNA hairpins to TAR RNA in the presence of variable amounts of NC. Fractional NA saturation with NC, ΘNC, was calculated according to eq. (6). Parameters were obtained using equations (2-5) for the analysis of the annealing time courses presented in Figure 8.
There is no significant difference in the intermediate dissociation rate, k-1, between WT TAR DNA and the two mutant constructs (Figure 9(c)), suggesting that dissociation of the intermediate formed with either the Loop or B-stem variants is comparable in the range of ΘNC studied here. The same conclusion holds for k2, which also appears to be independent of ΘNC (Figure 9(d)). Based on the known duplex destabilizing ability of NC, one would expect k2 to increase with increasing NC concentration. However, we hypothesize that this dependence is most likely obscured by the NC-induced annealing pathway switch from the extended loop-loop kissing pathway to the zipper pathway at ΘNC ∼0.4. This switch is not abrupt; both pathways contribute to annealing, with the zipper contribution taking over completely upon NC saturation.
Role of NC-induced nucleic acid aggregation
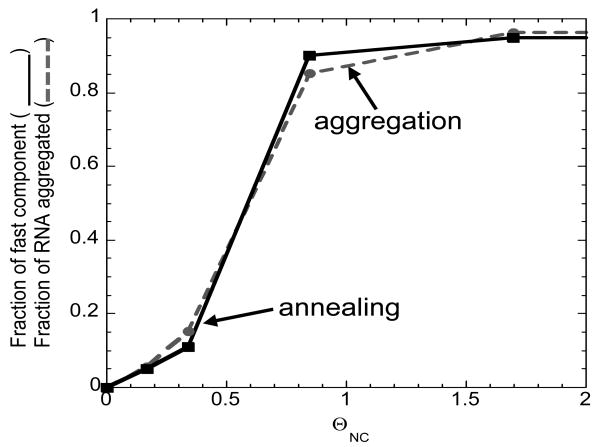
The major effect of increasing NC concentration is to enhance the bimolecular association step of annealing (see Figure 9(a)). This, in turn, results in strong stabilization of the annealing intermediate (Figure 9(b)). To examine the role of NC-induced NA aggregation in bimolecular intermediate stabilization, a sedimentation assay was performed to measure the fraction of aggregated full-length TAR RNA molecules, fa, as a function of fractional NC binding under conditions similar to those of the annealing assays. Figure 10 shows that the amount of NC required to stabilize the reaction intermediate (continuous line, f(ΘNC)) is virtually equivalent to the amount needed to induce NA aggregation (dotted line, fa(ΘNC)). These data are consistent with the results obtained for mini-TAR30, and provide further support for the notion that NC-induced NA aggregation constitutes a major component of NC's chaperone function.
Figure 10.
Fraction of fast annealing component (f) and aggregated fraction of TAR RNA (fa) as a function of fractional NA saturation with NC. f was obtained from the analysis of the data presented in Figure 8 using eqs. (2-5). fa was obtained by sedimentation assays as described in the Materials and Methods carried out under the same solution conditions as the annealing experiments.
Summary and Conclusions
In this work, the mechanism of 59-nt TAR RNA/DNA hairpin annealing was examined as a function of NA saturation with HIV-1 NC. Under all solution conditions tested, TAR hairpins anneal via the two-step pathway described by eq. (1), involving formation of a bimolecular annealing intermediate in pre-equilibrium to the subsequent strand exchange step resulting in the fully annealed RNA/DNA duplex. Interestingly, the nature of the annealing intermediate, i.e., the annealing pathway, changes as more NC is added. DNA mutational analysis showed that in the absence of NC or at low NC binding levels, the TAR RNA/DNA annealing proceeds via an extended loop-loop kissing intermediate (see Figure 1, lower left). In contrast, when levels of NC binding approach saturation, TAR RNA/DNA anneal through the stem ends (see Figure 1, lower right). Both pathways contribute to annealing at intermediate levels of NC binding (see Figure 9 (a) and (b)).
We hypothesize that the existence of two competing pathways for annealing of the TAR RNA/DNA hairpins is due to the comparable stabilities of the two 4-bp helical segments in TAR RNA (see Figure 1, top left). In particular, based on theoretical predictions, the 4-bp helix adjacent to the hairpin loop is ∼1 kcal/mol more stable than the 4-bp helix proximal to the 5′/3′ termini 50; 51. While melting of the more stable helix is less probable, it allows for formation of a 17-bp extended loop-loop intermediate (Figure 1, lower left). In contrast, melting of the terminal 4-bp helix leads to formation of an RNA/DNA intermediate containing only 9 bp. Whereas the former pathway is plausible in the absence of NC, in the presence of saturated NC, which stabilizes both intermediates, the annealing switches to the pathway that requires the least structure pre-melting, i.e. the zipper pathway.
Previous studies of the annealing of mini-TAR RNA/DNA were also consistent with two annealing pathways, with the loop pathway dominating in the absence of NC 30. However, in case of the shorter hairpin, which only contains two helical regions of similar stability, the stem pathway does not dominate even at saturated NC 30.
A recent study examined the annealing of small complementary DNA hairpins, PBS(+) and PBS(-), representing the rate-limiting step of plus-strand transfer during reverse transcription 52. A truncated version of NC, NC(12-55), which lacks the N-terminal basic domain and is therefore a poor aggregating agent, was also able to induce a pathway switch when compared to the reaction performed in the absence of NC 52. Another study from the same group examined the effect of NC(12-55) on the annealing of TAR DNA to its DNA complement cTAR 41. As in the PBS annealing study52, NC(12-55) stimulated the TAR annealing reaction by ∼100-fold. This is lower than the ∼103–fold and ∼105–fold stimulation observed for mini-TAR 30 and TAR annealing (this work), respectively, using full-length NC(1-55). Indeed, we have shown that the duplex destabilizing and minor aggregation activities of NC(12-55) each contribute ∼10-fold to the overall rate enhancement (M.-N. Vo, M. Mitra, I. Rouzina, and K. Musier-Forsyth, manuscript in preparation). Taken together, these results support a major role for NC-induced nucleic acid aggregation in its chaperone activity.
NC modulates the annealing pathway of structured complementary NA via both of its major activities, NA aggregation and destabilization 4; 26; 30. In vivo these diverse capabilities of NC may ensure efficient obligatory and random strand transfer events during reverse transcription 53; 54. The major effect of NC on the efficiency of strand transfers may also explain the increased production of full-length RNA transcripts during in vitro reverse transcription 39, as well as improved strand displacement synthesis of the central DNA flap 55 and greater reverse transcriptase processivity 55, which are both observed under conditions that also promote NC-induced NA aggregation 39; 55.
Materials and Methods
Protein, RNA and DNA preparation
The NC protein used in this work was prepared by solid-phase synthesis as described 40, and its purity was estimated to be > 95% by SDS-polyacrylamide gel electrophoresis. The concentration of NC was determined by measuring its absorbance at 280 nm and using ε280 = 6050 mol-1 cm-1.
All DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Full-length TAR RNA was generated by in vitro transcription using a PCR-amplified template encoding the 59-nt TAR sequence downstream of a T7 RNA polymerase promoter. The T7 promoter was appended during the PCR reaction and the sequences of DNA template and primers for the PCR reaction were: TAR DNA template, 5′-GGGTCTCTCTGGTTAGACCAGATCTGAGCCTGGGAGCTCTCTGGCTAACTAGGGAACCC-3′ and its complementary sequence 5′-GGGTTCCCTAGTTAGCCAGAGAGCTCCCAGGCTCAGATCTGGTCTAACCAGAGAGACCC-3′; T7-TAR primer 1, 5′-CTGTAATACGACTCACTATAGGGTCTCTCTGGTTAGACCAG-3′ and TAR primer 2, 5′-GGGTTCCCTAGTTAGCCAGAGAGC-3′. PCR reactions were performed with PfuTurbo DNA polymerase (Stratagene, La Jolla, CA) according to the enzyme manufacturer's protocol using the provided buffer. Briefly, each reaction included 30 cycles of denaturation, annealing and extension at temperatures of 94 °C for 45 sec, 50 °C for 45 sec and 72 °C for 2 min, respectively, followed by one cycle of extension at 72 °C for 10 min. PCR-generated DNA template was extracted with phenol:chloroform:isoamyl alcohol (25:24:1, by volume) and precipitated with ethanol prior to use. The purified DNA template (5 μg per 100 μL reaction) was in vitro transcribed according to standard procedures, followed by DNase treatment to remove the DNA template. All RNA and DNA oligonucleotides were gel purified on 12% (w/v) denaturing polyacrylamide gels, dissolved in diethyl pyrocarbonate-treated water, and stored at −20 °C Full-length TAR RNA was internally [32P]-labeled via in vitro transcription under similar conditions, except that the GTP concentration was lowered from 4 mM to 1 mM and 17 mCi/ml [α-32P]-GTP was added to the reaction.
The concentrations of the RNA and DNA oligonucleotides were determined by measuring their absorbances at 260 nm and using the following extinction coefficients: TAR RNA (59-mer), 5.34 × 105 M-1 cm-1; TAR DNA (59-mer), 5.65 × 105 M-1 cm-1; C27A/C28A TAR DNA (59-mer), 5.74 × 105 M-1 cm-1; B-stem TAR DNA (59-mer), 5.65 × 105 M-1 cm-1; T-zip TAR DNA (62-mer), 5.80 × 105 M-1 cm-1.
Prior to use, all oligonucleotides were refolded in 25 mM Hepes (pH 7.5) and 100 mM NaCl at a concentration 100× greater than that used in the annealing reactions. This was accomplished by incubation at 80 °C for two min, cooling to 60 °C for two min, followed by addition of MgCl2 to a final concentration of 10 mM and placement on ice.
Annealing assays
For annealing assays, refolded [32P]-labeled full-length TAR RNA was combined with refolded unlabeled complementary WT or mutant TAR DNA constructs in a solution containing 20 mM Hepes (pH 7.5), 20 mM NaCl and 0.2 mM MgCl2 (this MgCl2 concentration comes from the refolding buffer) at 37 °C, except when otherwise indicated.
In the absence of NC, DNA concentration-dependence studies were performed with 50 nM RNA and various concentrations of DNA, as indicated in the figure legends. Reactions were initiated by adding DNA to RNA, followed by incubation in the reaction buffer for the indicated time. Reactions were quenched by placing solutions on ice followed by addition of glycerol to 5% final concentration. Samples were loaded onto 12% SDS polyacrylamide gels (375 mM Tris-HCl, pH 8.8, 0.1% SDS; 19:1 acrylamide/bisacrylamide (w/v)) run at 25 °C in Tris-glycine (25 mM Tris, 250 mM glycine, pH 8.3) running buffer.
In the presence of NC, studies of the DNA concentration-dependence of TAR RNA/DNA annealing were performed by mixing 15 nM RNA with various concentrations of DNA prior to the addition of NC to achieve a 4:1 nt:NC ratio. Thus, final NC concentrations were 0.664 μM, 0.885 μM, 1.11 μM and 1.55 μM in the presence of 30 nM, 45 nM, 60 nM and 90 nM TAR DNA, respectively. Annealing reactions were quenched at the indicated times by incubation with 1% (w/v) SDS and immediate placement on ice for 5 min. Samples were subjected to phenol/chloroform-extraction (2×), followed by addition of glycerol to 5% final concentration and separation on 12% SDS-polyacrylamide gels as described earlier. Gels were visualized using a Bio-Rad Molecular Imager FX and quantified with Bio-Rad Quantity One Software. NC concentration-dependence studies were carried out similarly with 15 nM RNA and 150 nM DNA in the presence of various concentrations of NC, as indicated in the figure legends.
Sedimentation/aggregation assays
Refolded [32P]-labeled full-length TAR RNA (15 nM) was combined with TAR DNA (45 nM) in a solution containing 20 mM Hepes, pH 7.5, 20 mM NaCl and 0.2 mM MgCl2. Upon addition of NC to final concentrations of 0.1, 0.2, 0.5, 1.0, or 2.0 μM, reactions (40 μL) were incubated at 37 °C for 30 min. At the end of the incubation period, solutions were centrifuged at 12,000 rpm in a microcentrifuge for 20 min. Supernatant (5 μL) was collected and analyzed by scintillation counting. The percent radioactivity remaining in the supernatant relative to the RNA-only sample (set to 100%) was plotted as a function of NC concentration.
Acknowledgments
We would like to thank Dr. Robert Gorelick (NCI, Frederick) for NC purification, and Drs. Daniel G. Mullen and Brandie Kovaleski (University of Minnesota) for chemical synthesis of NC. This research was supported by NIH grant GM065056 (to K.M.-F.) and by NIH predoctoral training grant T32 GM008700 (to M.-N.V).
Abbreviations
- TAR
trans-activation response element
- NC
nucleocapsid protein
- bp
base pair
- NA
nucleic acid
- nt
nucleotide(s)
- ss
single-stranded
- ds
double-stranded
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chan B, Weidemaier K, Yip WT, Barbara PF, Musier-Forsyth K. Intra-tRNA distance measurements for nucleocapsid proteindependent tRNA unwinding during priming of HIV reverse transcription. Proc Natl Acad Sci U S A. 1999;96:459–64. doi: 10.1073/pnas.96.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski MC, Roques B, Darlix JL. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci U S A. 1992;89:6472–6. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng YX, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol. 1999;73:4251–6. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hargittai MR, Gorelick RJ, Rouzina I, Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J Mol Biol. 2004;337:951–68. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 5.Davis WR, Gabbara S, Hupe D, Peliska JA. Actinomycin D inhibition of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase and nucleocapsid protein. Biochemistry. 1998;37:14213–21. doi: 10.1021/bi9814890. [DOI] [PubMed] [Google Scholar]
- 6.Golinelli MP, Hughes SH. Secondary structure in the nucleic acid affects the rate of HIV-1 nucleocapsid-mediated strand annealing. Biochemistry. 2003;42:8153–62. doi: 10.1021/bi027039w. [DOI] [PubMed] [Google Scholar]
- 7.Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J Virol. 2000;74:8980–8. doi: 10.1128/jvi.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo J, Wu T, Kane BF, Johnson DG, Henderson LE, Gorelick RJ, Levin JG. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 2002;76:4370–8. doi: 10.1128/JVI.76.9.4370-4378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong MK, Harbron EJ, O'Connor DB, Guo J, Barbara PF, Levin JG, Musier-Forsyth K. Nucleic acid conformational changes essential for HIV-1 nucleocapsid protein-mediated inhibition of self-priming in minus-strand transfer. J Mol Biol. 2003;325:1–10. doi: 10.1016/s0022-2836(02)01177-4. [DOI] [PubMed] [Google Scholar]
- 10.Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix JL. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–41. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You JC, McHenry CS. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J Biol Chem. 1994;269:31491–5. [PubMed] [Google Scholar]
- 12.Johnson PE, Turner RB, Wu ZR, Hairston L, Guo J, Levin JG, Summers MF. A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–91. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- 13.Muthuswami R, Chen J, Burnett BP, Thimmig RL, Janjic N, McHenry CS. The HIV plus-strand transfer reaction: determination of replication-competent intermediates and identification of a novel lentiviral element, the primer over-extension sequence. J Mol Biol. 2002;315:311–23. doi: 10.1006/jmbi.2001.5205. [DOI] [PubMed] [Google Scholar]
- 14.Wu T, Guo J, Bess J, Henderson LE, Levin JG. Molecular requirements for human immunodeficiency virus type 1 plus-strand transfer: analysis in reconstituted and endogenous reverse transcription systems. J Virol. 1999;73:4794–805. doi: 10.1128/jvi.73.6.4794-4805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berg JM. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–7. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 16.Covey SN. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986;14:623–33. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green LM, Berg JM. Retroviral nucleocapsid protein-metal ion interactions: folding and sequence variants. Proc Natl Acad Sci U S A. 1990;87:6403–7. doi: 10.1073/pnas.87.16.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson LE, Copeland TD, Sowder RC, Smythers GW, Oroszlan S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J Biol Chem. 1981;256:8400–6. [PubMed] [Google Scholar]
- 19.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 20.Carteau S, Gorelick RJ, Bushman FD. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999;73:6670–9. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog Nucleic Acid Res Mol Biol. 2002;72:223–68. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 22.Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–37. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 23.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 24.Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–70. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bampi C, Jacquenet S, Lener D, Decimo D, Darlix JL. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Curr HIV Res. 2004;2:79–92. doi: 10.2174/1570162043485022. [DOI] [PubMed] [Google Scholar]
- 26.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–86. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 27.Le Cam E, Coulaud D, Delain E, Petitjean P, Roques BP, Gerard D, Stoylova E, Vuilleumier C, Stoylov SP, Mély Y. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers. 1998;45:217–29. doi: 10.1002/(SICI)1097-0282(199803)45:3<217::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 28.Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Justome A, Delain E, Gorelick RJ, Le Cam E. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J Mol Biol. 2006;364:496–511. doi: 10.1016/j.jmb.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 29.Stoylov SP, Vuilleumier C, Stoylova E, De Rocquigny H, Roques BP, Gerard D, Mély Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–12. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 30.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J Mol Biol. 2006;363:244–61. doi: 10.1016/j.jmb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 31.Azoulay J, Clamme JP, Darlix JL, Roques BP, Mély Y. Destabilization of the HIV-1 complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J Mol Biol. 2003;326:691–700. doi: 10.1016/s0022-2836(02)01430-4. [DOI] [PubMed] [Google Scholar]
- 32.Beltz H, Azoulay J, Bernacchi S, Clamme JP, Ficheux D, Roques B, Darlix JL, Mély Y. Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J Mol Biol. 2003;328:95–108. doi: 10.1016/s0022-2836(03)00244-4. [DOI] [PubMed] [Google Scholar]
- 33.Beltz H, Piemont E, Schaub E, Ficheux D, Roques B, Darlix JL, Mély Y. Role of the structure of the top half of HIV-1 cTAR DNA on the nucleic acid destabilizing activity of the nucleocapsid protein NCp7. J Mol Biol. 2004;338:711–23. doi: 10.1016/j.jmb.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 34.Bernacchi S, Stoylov S, Piemont E, Ficheux D, Roques BP, Darlix JL, Mély Y. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J Mol Biol. 2002;317:385–99. doi: 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- 35.Kankia BI, Barany G, Musier-Forsyth K. Unfolding of DNA quadruplexes induced by HIV-1 nucleocapsid protein. Nucleic Acids Res. 2005;33:4395–403. doi: 10.1093/nar/gki741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urbaneja MA, Wu M, Casas-Finet JR, Karpel RL. HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity. J Mol Biol. 2002;318:749–64. doi: 10.1016/S0022-2836(02)00043-8. [DOI] [PubMed] [Google Scholar]
- 37.Williams MC, Gorelick RJ, Musier-Forsyth K. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein's nucleic acid chaperone function. Proc Natl Acad Sci U S A. 2002;99:8614–9. doi: 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc Natl Acad Sci U S A. 2001;98:6121–6. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anthony RM, Destefano JJ. In vitro synthesis of long DNA products in reactions with HIV-RT and nucleocapsid protein. J Mol Biol. 2007;365:310–24. doi: 10.1016/j.jmb.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu HW, Cosa G, Landes CF, Zeng Y, Kovaleski BJ, Mullen DG, Barany G, Musier-Forsyth K, Barbara PF. Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription. Biophys J. 2005;89:3470–9. doi: 10.1529/biophysj.105.065326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Godet J, de Rocquigny H, Raja C, Glasser N, Ficheux D, Darlix JL, Mély Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J Mol Biol. 2006;356:1180–92. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 42.Liu HW, Zeng Y, Landes CF, Kim YJ, Zhu Y, Ma X, Vo MN, Musier-Forsyth K, Barbara PF. Insights on the role of nucleic acid/protein interactions in chaperoned nucleic acid rearrangements of HIV-1 reverse transcription. Proc Natl Acad Sci U S A. 2007;104:5261–7. doi: 10.1073/pnas.0700166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantor CR, Schimmel PR. Biophysical Chemistry. Part III. The Behavior of Biological Macromolecules. W. H. Freeman & Co.; San Francisco: 1980. [Google Scholar]
- 44.Shubsda MF, McPike MP, Goodisman J, Dabrowiak JC. Monomer-dimer equilibrium constants of RNA in the dimer initiation site of human immunodeficiency virus type 1. Biochemistry. 1999;38:10147–57. doi: 10.1021/bi990744t. [DOI] [PubMed] [Google Scholar]
- 45.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Effect of Mg2+ and Na+ on the Nucleic Acid Chaperone Activity of HIV-1 Nucleocapsid protein: Implications for Reverse Transcription. 2008 doi: 10.1016/j.jmb.2008.12.073. Submitted to JMB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dib-Hajj F, Khan R, Giedroc DP. Retroviral nucleocapsid proteins possess potent nucleic acid strand renaturation activity. Protein Sci. 1993;2:231–43. doi: 10.1002/pro.5560020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix JL. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–9. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casas-Finet JR, Medaglia M, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–9. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urbaneja MA, Kane BP, Johnson DG, Gorelick RJ, Henderson LE, Casas-Finet JR. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J Mol Biol. 1999;287:59–75. doi: 10.1006/jmbi.1998.2521. [DOI] [PubMed] [Google Scholar]
- 50.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–81. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramalanjaona N, de Rocquigny H, Millet A, Ficheux D, Darlix JL, Mély Y. Investigating the mechanism of the nucleocapsid protein chaperoning of the second strand transfer during HIV-1 DNA synthesis. J Mol Biol. 2007;379:1041–1053. doi: 10.1016/j.jmb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Galetto R, Negroni M. Mechanistic features of recombination in HIV. AIDS Rev. 2005;7:92–102. [PubMed] [Google Scholar]
- 54.Gao L, Balakrishnan M, Roques BP, Bambara RA. Insights into the multiple roles of pausing in HIV-1 reverse transcriptase-promoted strand transfers. J Biol Chem. 2007;282:6222–31. doi: 10.1074/jbc.M610056200. [DOI] [PubMed] [Google Scholar]
- 55.Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Borde I, Reboud-Ravaux M, Restle T, Gorelick RJ, Le Cam E. HIV-1 protease and reverse transcriptase control the architecture of their nucleocapsid partner. PLoS ONE. 2007;2:e669. doi: 10.1371/journal.pone.0000669. [DOI] [PMC free article] [PubMed] [Google Scholar]