Abstract
The living organisms may be studied as a whole complex system. The “omics sciences” tend at understanding and describing the global information of genes, mRNA, proteins, and metabolites. The aim of the Bioinformatics should be that of developing methods not only able to study the individual components of a system, but also to represent and simulate the relationships between all these components.
Keywords: Bioinformatics, mRNA, proteins, genes, omics sciences
Description
Until the last century, the approach of biological science was to break down the object of study in its elementary parts and to study all the singular units in order to explain the life processes. This was a typical analytical and reductionist procedure, which allowed the understanding of almost all properties of molecular parts of living organisms, such as genes, proteins, metabolites, and was focused on the study of each single component of the system under consideration but was not able to predict the behavior of the systems as a whole.
A system can be defined as a number of interacting elements existing within a boundary that is surrounded by an environment. Therefore, a complex system is able to create new properties from the interactions between its components, and also to interact and to respond with the external inputs.
When the interactions between the parties are determined by the dynamical processes inducing the emerging properties like adaptability, self-organization and the ability to respond under disturbance, the system becomes complex. In this way these non linear interactions allow a number of possible several states and new emergent behaviors are not predictable from the simple sum of the component parts. These principles were applied to study the living organisms, the stock markets, the ecosystems and the flock of birds.
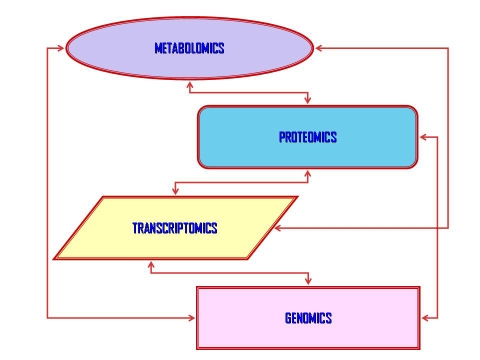
In biology it's necessary to study the living organism as a whole, and the laws of regarding the organizational forces of systems, which yet are not well known, but are essential to solve and to understand the collective phenomena and the framework for the functionality of the systems. The “omics sciences” are a first effort to study the behaviors of the “biology actors” in a environment as whole, that are aimed at understanding and describing the global information of genes (genomics), mRNA (transcriptomics), proteins (proteomics), and metabolites (metabolomics). Nevertheless a method able to describe the relationships between the different “omics” levels was not yet developed (Figure 1).
Figure 1.
Scheme of the relationships between the different omics levels.
Moreover, the growing number of data produced by biological sciences and by the new techniques of cellular and molecular analysis may be analysed concerning not only the single components but also the relationships between them in order to avoid typical analytical and reductionist procedures that were not able to understand natural phenomena, such as protein folding, or to solve some major diseases.
It is now widely accepted that dividing the complex systems into smaller components and simpler units is not sufficient to understanding of organization and of the onset of emerging property. It is necessary to focus on the interactions and on the networks between the components in order to reproduce the characteristics of complexity.
In this context it's necessary to consider that the number of the possible connections between units, from which all possible states of the systems depend, is enormous, and to reproduce a good model of these is not easy. To reproduce the functional networks some mathematic tools, as graphs and neural nodes, are commonly used.
The goal of Bioinformatics and Systems Biology is to create algorithms aimed to highlight and predict the networks between genes, mRNA, proteins and metabolites, and the bases of the functionality of the complex systems. In particular, these algorithms may simulate also the relationships between the different “omic” levels in order to explain the molecular basis of some diseases, to evaluate the modulation effects of some functional mechanisms and to provide useful information in pharmacological studies.
Acknowledgments
This work was partially supported by “RNBIO-Rete Nazionale di Bioinformatica Oncologica”.