Abstract
Biotin influences transcription in organisms from bacteria to humans. The enzyme, biotin protein ligase, which catalyzes post-transcriptional biotin addition to biotin-dependent carboxylases, plays a central roll in transmitting the demand for biotin to gene expression. The molecular mechanism of this communication in bacteria is well understood and involves competing protein:protein interactions. Biochemical measurements indicate that this competition is kinetically controlled. In humans, the biochemistry of biotin sensing at the transcriptional level is not well characterized. However, the biotin holoenzyme ligase (holocarboxylase synthetase) is proposed to both catalyze biotin addition to carboxylases and to histones in its metabolic and transcriptional roles, respectively. Control of human holocarboxylase synthetase function is, however, considerably more complex than the simple competitive protein protein interactions observed in bacterial systems.
Introduction
Communication of nutrient status to transcription provides organisms with mechanisms to respond to metabolic needs at the level of gene expression. Several nutrients impact transcription, including vitamin D (1) in eukaryotic systems and many sugars (2) and metals (3) in prokaryotes. In general, these small molecules bind to transcription factors to influence their binding to target regulatory sites at the genes subject to control by these proteins. Modulation of the binding alters occupancy of the regulatory sites, thereby altering transcription initiation. A new paradigm for linking transcription and metabolism that features a direct role for metabolic enzymes in transcription has recently been discovered (4–6). Systems that have evolved to link biotin status to gene expression in a broad range of organisms provide examples of use of a metabolic enzyme in transcriptional regulation. The molecular mechanism of this communication of biotin demand to transcription is now well understood in bacteria. Although many of the details of the mechanism in humans remain to be elucidated, some parallels can be drawn between the strategies used by the 2 organisms.
In all organisms in which a role for biotin in transcription regulation has been demonstrated, the metabolic enzyme, biotin protein ligase, plays a central role (7). In Escherichia coli, a single protein functions both as the enzyme that funnels biotin into metabolism and as the site-specific DNA binding protein that regulates biotin biosynthesis at the transcription initiation level (8,9). This mechanism has recently been shown to exist in both archae and eubacteria, and is therefore ancient (10). In yeast, low biotin levels induce transcription of several genes (11). Although the role of the ligase in the process is not known, mutations in the gene that codes for the enzyme alter the transcriptional response. In mammals, biotin levels influence levels of transcription of genes that code for biotin-dependent carboxylases, biotin transporters, and the biotin protein ligase itself (12,13). Mutations in the gene that codes for the human biotin holoenzyme ligase result in perturbation of the transcriptional response (12). Thus, in all of the systems in which a transcriptional response to biotin has been observed, a role for the biotin protein ligase in the process is evident. The extent to which this parallel phenomenology extends to molecular mechanism is not yet known.
Bacterial biotin sensing
Molecular mechanisms of bacterial biotin sensing.
Two distinct strategies for biotin sensing have evolved in microorganisms. In the first, a single bifunctional protein both funnels biotin into metabolism and regulates transcription in a manner that reflects biotin demand (8,9). Microorganisms ranging from eubacteria to archaebacteria use this strategy (10). The alternative mechanism of sensing biotin demand is carried out by a 2-polypeptide system (14). In this system, 1 protein functions solely in transferring biotin to biotin-dependent carboxylases, and a second regulates transcription of biotin-linked genes. Whereas the biochemical details of the bifunctional ligase strategy are well understood, the dual protein strategy has only been identified at a sequence level.
Biotin sensing in E. coli: kinetic control by mutually exclusive protein:protein interactions.
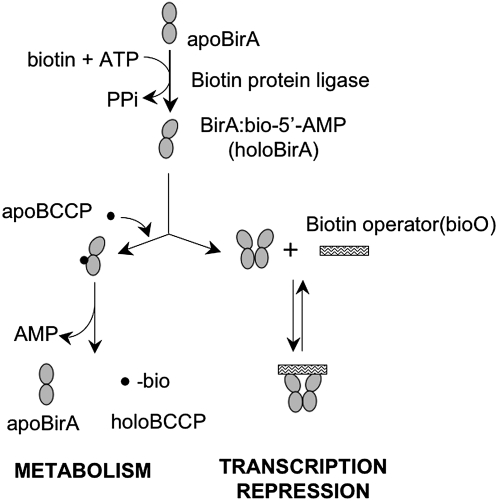
During the past several years, molecular details of biotin sensing in E. coli have been elucidated. In this organism, biotin is either funneled into metabolism via its linkage to the biotin carboxyl carrier protein (BCCP)4 subunit of acetyl CoA carboxylase (ACC), or it is used in transcription repression (Fig. 1). A single protein, E. coli biotin protein ligase (BirA), carries out both of these functions (8,9) by first catalyzing synthesis of activated biotin, biotinoyl-5′-AMP, from substrates biotin and adenosine triphosphate. The resulting enzyme:adenylate complex either interacts with the BCCP subunit of ACC, with consequent biotin transfer, or it can form a homodimer that binds sequence specifically to the biotin operator sequence of the biotin biosynthetic operon. The former interaction funnels biotin into metabolism and the latter causes repression of transcription initiation. This system allows for the transmission of the demand for biotin signaled by the intracellular apoBCCP concentration to biotin production.
FIGURE 1 .
The E. coli biotin regulatory system. BirA binds to substrates biotin and adenosine triphosphate to catalyze synthesis of biotinoyl-5′-AMP. The resulting enzyme-adenylate complex can either interact with the BCCP subunit of ACC or homodimerize to transfer biotin or repress transcription initiation, respectively. Adapted from (23).
In the E. coli biotin regulatory system, the bifunctional ligase switches between its enzymatic and DNA binding functions. Combined in vivo and in vitro measurements reveal that the bifunctional biotin protein ligase has evolved to use a limited amount of sequence information to perform its multiple tasks. Indeed, it is the limited nature of the structural components essential for both biological functions that is the key to functional switching in the system.
Structural data has provided important clues in elucidating the mechanism of functional switching in BirA. The 35.3-kDa polypeptide chain folds into 3 modules, including an amino (N)-terminal DNA binding domain, a central domain, and a carboxyl (C)-terminal domain (Fig. 2) (15). The N-terminal domain is a winged helix-turn-helix module that is characteristic of many DNA binding proteins. The central domain consists of a core β-sheet sandwiched between multiple α-helices and contains the catalytic site, which is also the site for corepressor binding. The C-terminal domain folds into an all β-strand structure that, based on structural information, functions in the homodimerization required for DNA binding by BirA and the heterodimerization with BCCP that precedes biotin transfer (16,17).
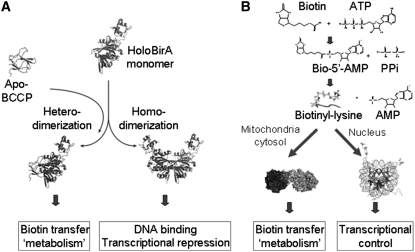
FIGURE 2 .
The alternative protein:protein interactions formed by holoBirA (A). Models were created in MolMol (32) using pdb files 2ewn and a pdb file provided by Zachary Wood (University of Georgia). The alternative functions of HCS in post-translational addition of biotin to carboxylases or histones (B). The carboxylase and histone models were created using Pymol (33) with pdb files 2qf7 and 1kx5, respectively.
In addition to the apo-monomer, structures of 2 dimeric forms of BirA have been determined by X-ray crystallography (17,18). These include the biotin- or substrate-bound and the adenylate-bound species (Fig. 2). Each dimer is formed by side-by-side alignment of the central β-sheets of the contributing monomers, forming an extended intermolecular β-sheet. In addition, 3 of 4 loops that are partially disordered in the aporepressor are located in the monomer-monomer interface. One of the loops, termed the biotin binding loop or BBL, is folded over the biotin molecule. This ligand-induced folding is important for stabilizing the loop for participation in dimerization. In addition to the experimentally determined structures of the homodimers, a structure of the complex of holoBirA bound to the biotin acceptor protein, BCCP, has been modeled (Fig. 2A) (16). Like the homodimer, the heterodimer forms through extension of the central β-sheet of BirA, by formation of a parallel interaction with the acceptor protein. Thus the 2 protein:protein interactions use the same surface of holoBirA, and are, consequently, mutually exclusive. The basic features of this modeled structure have recently been confirmed with the solution of a high-resolution structure of the Pyroccocus horikoshii biotin protein ligase bound to its homologous BCCP (19).
The mutually exclusive dimerization reactions, one between 2 holoBirA monomers and the other between holoBirA and apoBCCP, provide a mechanism for communicating biotin demand to transcription through competition between homo- and heterodimerization. Before discussing the biochemical evidence that supports this model, in vivo data on the regulation will be briefly summarized. In E. coli, there is only 1 biotin-dependent carboxylase, ACC, which catalyzes the first committed step of fatty acid synthesis. Transcription of the genes that code for the 4 ACC subunits is transcriptionally regulated in response to growth rate (20). This is physiologically reasonable, because fatty acid biosynthesis feeds into membrane biosynthesis. Therefore, rapid growth rate places a high demand on biotin, because ACC is only functional in its post-translationally biotinylated form. The funneling of the system into metabolism has been investigated in vivo by monitoring the effect of increasing intracellular apoBCCP on transcription repression at the regulatory region of the biotin biosynthetic operon (21). The measurements indicate that increased apoBCCP concentration results in derepression of transcription initiation, consistent with a system in which the switch from metabolism to transcription repression reflects a competition homo- and heterodimerization by holoBirA. The structural data indicate that the molecular basis of this competition lies in the formation of mutually exclusive protein:protein interactions by holoBirA.
In principle, the partitioning between the 2 interactions in the biotin regulatory system could be controlled at the equilibrium or kinetic level. To determine the mechanism, partitioning of holoBirA into its 2 alternative interactions has been directly measured. Homodimerization of holoBirA measured using sedimentation equilibrium measurements is characterized by an equilibrium dissociation constant of ∼10 μM (22). Because the heterodimerization reaction results in chemical transfer of biotin to apoBCCP, it cannot be directly measured using an equilibrium technique. Rather, a competition method in which the effect of apoBCCP on holoBirA dimerization was monitored by sedimentation velocity was used (23). Results of the measurements reveal that the 2 protein:protein interactions occur with similar energetics, which suggests that the system is not controlled by equilibrium. The sedimentation velocity measurements also revealed that homodimerization of holoBirA is characterized by very slow kinetics (23). This has led to the conclusion that the system has evolved kinetically so that, if available, apoBCCP has a window of opportunity to engage holoBirA, with the resulting partitioning of the control system toward metabolism. Alternatively, upon depletion of apoBCCP, holoBirA has sufficient time to dimerize and bind to the operator sequence of the biotin biosynthetic operon with concomitant repression of transcription initiation.
Biotin sensing in humans.
Like bacteria, humans can respond to biotin levels by altering transcription of several genes. The biochemistry of the human system has not yet been elucidated at the detailed level of the bacterial system. However, at a superficial level, some parallels between the 2 systems are evident. Most significantly, the human biotin regulatory system features a role for the biotin protein ligase, or holocarboxylase synthetase (HCS), in both metabolism and transcription. At a biochemical level, this means that, like the bacterial ligase, the human enzyme must be capable of interacting with distinct partners relevant to its metabolic and transcriptional functions. However, in addition to this parallel requirement of the 2 enzymes, complexities in the human system render regulation of HCS function considerably more elaborate than regulation of BirA.
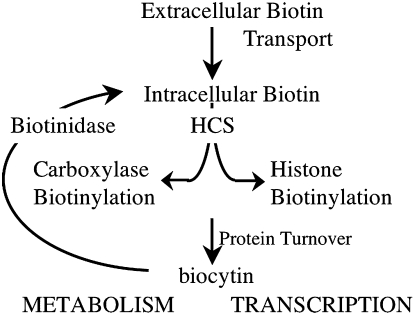
The human biotin cycle (Fig. 3) is the system by which biotin that is consumed by humans is used and recycled (24). Once the biotin is digested, it is transported into the cell via the sodium-dependent multivitamin transporter. In the cell, the vitamin is post-translationally added to the biotin-dependent carboxylases in a reaction catalyzed by HCS. The 5 carboxylases catalyze reactions that are critical to several metabolic pathways, including fatty acid synthesis, amino acid breakdown, and gluconeogenesis. Like other proteins in the cell, the carboxylases have limited lifetimes and are subject to turnover. Proteolysis of proteins with biotin modification yields biocytin (biotinoyl lysine), the hydrolysis of which is catalyzed the enzyme biotinidase, with the resulting biotin reentering the biotin cycle. A biotin molecule can pass several times through the cycle before being excreted.
FIGURE 3 .
The proposed biotin cycle in which biotin is first consumed and transported into cells, where it is linked to either biotin-dependent carboxylases or histone proteins in nucleosomes. Upon turnover of the biotinylated proteins, biocytin is released and hydrolyzed by biotinidase. The resulting free biotin reenters the biotin cycle.
In addition to the role that it plays in modifying metabolic enzymes, a role for HCS in transcription regulation has been demonstrated. The influence of biotin on levels of specific proteins has been recognized since the 1960s (25). Recently, it has been shown that when cells in tissue culture are subjected to biotin starvation and then administered biotin replete media, levels of transcription of several genes first decrease in biotin starvation and then increase back to normal levels when the cells are replenished with biotin (12). Among the genes subject to transcriptional control by biotin are those that encode the ACC I, the α-subunit of propionyl CoA carboxylase, the sodium-dependent multivitamin transporter, and the HCS (12,13). This transcriptional response to changing biotin concentration requires a wild-type holoenzyme carboxylase. Thus, in addition to its function in metabolism, as found in E. coli, the HCS functions in the transcriptional response to biotin.
The biochemical mechanism by which the HCS switches from its metabolic and transcriptional roles is not known. However, to consider how it switches, it is first important to describe how HCS is thought to function in transcription. In contrast to the bifunctional bacterial biotin ligase, the human ligase possesses no sequence that is capable of folding into a DNA binding domain (26,27). Thus, unlike the bifunctional bacterial enzyme, HCS is not a site-specific DNA binding protein. Several years ago it was shown that histones can be biotinylated in a reaction catalyzed by biotinidase (28). Moreover, more recently, HCS was found to be capable of catalyzing linkage of biotin to histones in vitro (29). In addition to the cytosol, HCS, but not biotinidase, has been shown by immunofluorescence to be localized to the nucleus (29). Based on the combined data, the proposed mechanism by which HCS functions in transcription is by translocating to the nucleus and biotinylating histones in response to biotin supply/demand. The biotinylation of histones is proposed to alter the structure/dynamics of the chromatin to change the levels of transcription at specific loci.
In the context of this proposed mechanism, one can speculate on how HCS function in metabolism and transcription might be regulated. Obvious potential points of regulation include localization of the enzyme to the nucleus and relative preference for carboxylase and histone substrates. However, it is also necessary to recognize that at least 2 forms of the enzyme, products of alternatively spliced pre-mRNAs encoded by a single gene, exist in human cells (26,27,30). They encode a full-length 82-kDa polypeptide and N-58, a 76-kDa polypeptide that is initiated at the methionine codon that is 58 triplets downstream of the initiation codon in the full-length message. The C-terminal segments, which are homologous to sequences of other known biotin protein ligases, of the 2 proteins are identical and encode the region that carries out biotin transfer (26,31). The structure at the N-terminus may be significant for regulating HCS function either at the level of nuclear localization or substrate preference in biotin transfer.
The E. coli biotin protein ligase and the human paralog, HCS, function in both metabolism and transcription initiation. In both systems, the demand for biotin influences the enzyme's function. In the bacterial system, a simple molecular mechanism involving formation of alternative protein:protein interactions relevant to the 2 functions operates. In the human system, like the bacterial, the proposed molecular mechanism relies on the ability of the HCS to recognize different protein partners (Fig. 2B). The contributions of complicating factors, including the availability of 2 HCS forms and nuclear localization to regulation of biotin sensing in humans are the subjects of ongoing investigations.
Other articles in this supplement include references (34–37).
Published as a supplement to The Journal of Nutrition. Presented as part of the symposium “Advances in Understanding of the Biological Role of Biotin at the Clinical, Biochemical, and Molecular Level” given at the 2008 Experimental Biology meeting on April 7, 2008, in San Diego, CA. The symposium was sponsored by the American Society for Nutrition and supported by an educational grant from Mead Johnson Nutritionals. The symposium was chaired by Donald Mock and Hamid M. Said.
Supported by NIH Grant R01-GM46511.
Author disclosure: D. Beckett, no conflicts of interest.
Abbreviations used: ACC, acetyl CoA carboxylase; BCCP, biotin carboxyl carrier protein; BirA, Escherichia coli biotin protein ligase; C, carboxyl; HCS, holocarboxylase synthetase; N, amino.
References
- 1.Norman AW. Minireview: vitamin D receptor: new assignments for an already busy receptor. Endocrinology. 2006;147:5542–8. [DOI] [PubMed] [Google Scholar]
- 2.Weickert MJ, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–74. [PubMed] [Google Scholar]
- 3.Pennella MA, Giedroc DP. Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals. 2005;18:413–28. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y. Metabolic enzymes and coenzymes in transcription–a direct link between metabolism and transcription? Trends Genet. 2004;20:445–52. [DOI] [PubMed] [Google Scholar]
- 5.Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–4. [DOI] [PubMed] [Google Scholar]
- 6.Commichau FM, Stulke J. Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression. Mol Microbiol. 2008;67:692–702. [DOI] [PubMed] [Google Scholar]
- 7.Beckett D. Biotin sensing: universal influence of biotin status on transcription. Annu Rev Genet. 2007;41:443–64. [DOI] [PubMed] [Google Scholar]
- 8.Barker DF, Campbell AM. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J Mol Biol. 1981;146:451–67. [DOI] [PubMed] [Google Scholar]
- 9.Barker DF, Campbell AM. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J Mol Biol. 1981;146:469–92. [DOI] [PubMed] [Google Scholar]
- 10.Rodionov DA, Mironov AA, Gelfand MS. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 2002;12:1507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pirner HM, Stolz J. Biotin sensing in Saccharomyces cerevisiae is mediated by a conserved DNA element and requires the activity of biotin-protein ligase. J Biol Chem. 2006;281:12381–9. [DOI] [PubMed] [Google Scholar]
- 12.Solorzano-Vargas RS, Pacheco-Alvarez D, Leon-Del-Rio A. Holocarboxylase synthetase is an obligate participant in biotin-mediated regulation of its own expression and of biotin-dependent carboxylases mRNA levels in human cells. Proc Natl Acad Sci USA. 2002;99:5325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pacheco-Alvarez D, Solorzano-Vargas RS, Gonzalez-Noriega A, Michalak C, Zempleni J, Leon-Del-Rio A. Biotin availability regulates expression of the sodium-dependent multivitamin transporter and the rate of biotin uptake in HepG2 cells. Mol Genet Metab. 2005;85:301–7. [DOI] [PubMed] [Google Scholar]
- 14.Rodionov DA, Gelfand MS. Computational identification of BioR, a transcriptional regulator of biotin metabolism in Alphaproteobacteria, and of its binding signal. FEMS Microbiol Lett. 2006;255:102–7. [DOI] [PubMed] [Google Scholar]
- 15.Wilson KP, Shewchuk LM, Brennan RG, Otsuka AJ, Matthews BW. Escherichia coli biotin holoenzyme ynthetase/bio repressor crystal structure delineates the biotin- and DNA- binding domains. Proc Natl Acad Sci USA. 1992;89:9257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weaver LH, Kwon K, Beckett D, Matthews BW. Competing protein: protein interactions are proposed to control the biological switch of the E coli biotin repressor. Protein Sci. 2001;10:2618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood ZA, Weaver LH, Brown PH, Beckett D, Matthews BW. Co-repressor induced order and biotin repressor dimerization: a case for divergent followed by convergent evolution. J Mol Biol. 2006;357:509–23. [DOI] [PubMed] [Google Scholar]
- 18.Weaver LH, Kwon K, Beckett D, Matthews BW. Corepressor- induced organization and assembly of the biotin repressor:a model for allosteric activation of a transcriptional regulator. Proc Natl Acad Sci USA. 2001;98:6045–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagautdinov B, Matsuura Y, Bagautdinova S, Kunishima N. Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate. J Biol Chem. 2008;283:14739–50. [DOI] [PubMed] [Google Scholar]
- 20.Li SJ, Cronan JE, Jr. Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J Bacteriol. 1993;175:332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cronan JE, Jr. Expression of the biotin biosynthetic operon of Escherichia coli is regulated by the rate of protein biotination. J Biol Chem. 1988;263:10332–6. [PubMed] [Google Scholar]
- 22.Eisenstein E, Beckett D. Dimerization of the Escherichia coli biotin repressor: corepressor function in protein assembly. Biochemistry. 1999;38:13077–84. [DOI] [PubMed] [Google Scholar]
- 23.Zhao H, Beckett D. Kinetic Partitioning Between Alternative Protein-Protein Interactions Controls a Transcriptional Switch. J Mol Biol. 2008;380:223–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahon RJ. Biotin in metabolism and molecular biology. Annu Rev Nutr. 2002;22:221–39. [DOI] [PubMed] [Google Scholar]
- 25.Dakshinamurti K, Cheah-Tan C. Biotin-mediated synthesis of hepatic glucokinase in the rat. Arch Biochem Biophys. 1968;127:17–21. [DOI] [PubMed] [Google Scholar]
- 26.Leon-Del-Rio A, Leclerc D, Akerman B, Wakamatsu N, Gravel RA. Isolation of a cDNA encoding human holocarboxylase synthetase by functional complementation of a biotin auxotroph of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:4626–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki Y, Aoki Y, Ishida Y, Chiba Y, Iwamatsu A, Kishino T, Niikawa N, Matsubara Y, Narisawa K. Isolation and characterization of mutations in the human holocarboxylase synthetase cDNA. Nat Genet. 1994;8:122–8. [DOI] [PubMed] [Google Scholar]
- 28.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem Mol Med. 1995;56:76–83. [DOI] [PubMed] [Google Scholar]
- 29.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum Mol Genet. 2004;13:15–23. [DOI] [PubMed] [Google Scholar]
- 30.Hiratsuka M, Sakamoto O, Li X, Suzuki Y, Aoki Y, Narisawa K. Identification of holocarboxylase synthetase (HCS) proteins in human placenta. Biochim Biophys Acta. 1998;1385:165–71. [DOI] [PubMed] [Google Scholar]
- 31.Campeau E, Gravel RA. Expression in Escherichia coli of N- and C-terminally deleted human holocarboxylase synthetase. Influence of the N-terminus on biotinylation and identification of a minimum functional protein. J Biol Chem. 2001;276:12310–6. [DOI] [PubMed] [Google Scholar]
- 32.Koradi R., Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55,29–32. [DOI] [PubMed] [Google Scholar]
- 33.DeLano WL. “The PyMOL Molecular Graphics System.” DeLano Scientific LLC, http://www.pymol.org, San Carlos, CA, USA.
- 34.Mock DM, Said HM. Introduction to Advances in Understanding of the Biological Role of Biotin at the Clinical, Biochemical, and Molecular Level. J Nutr. 2008;138:152–3. [DOI] [PubMed] [Google Scholar]
- 35.Mock DM. Marginal biotin deficiency is common in normal human pregnancy and is highly teratogenic in mice. J Nutr. 2008;138:154–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Said HM. Cell and molecular aspects of the human intestinal biotin absorption process. J Nutr. 2008;138:158–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zempleni J, Gralla M, Camporeale G, Hassan YI. Sodium-dependent multivitamin transporter gene is regulated at the chromatin level by histone biotinylation in human Jurkat lymphoblastoma cells. J Nutr. 2008;138:163–6. [DOI] [PMC free article] [PubMed] [Google Scholar]