Abstract
Background
Alcohol dependence (AD) is costly to societies worldwide, moderately heritable, and genetically complex. Risk loci in several populations have been identified using genetic linkage analysis. To date, there has been no published linkage study of AD focused on African-Americans (AAs).
Methods
We completed a genomewide linkage scan using ~6000 SNP markers to map loci increasing risk for DSM-IV AD in a set of 238 small nuclear families ascertained based on multiple individuals affected with cocaine or opioid dependence. Model free linkage analysis was completed using MERLIN software. A modified marker set was used to avoid bias due to markers in strong linkage disequilibrium.
Results
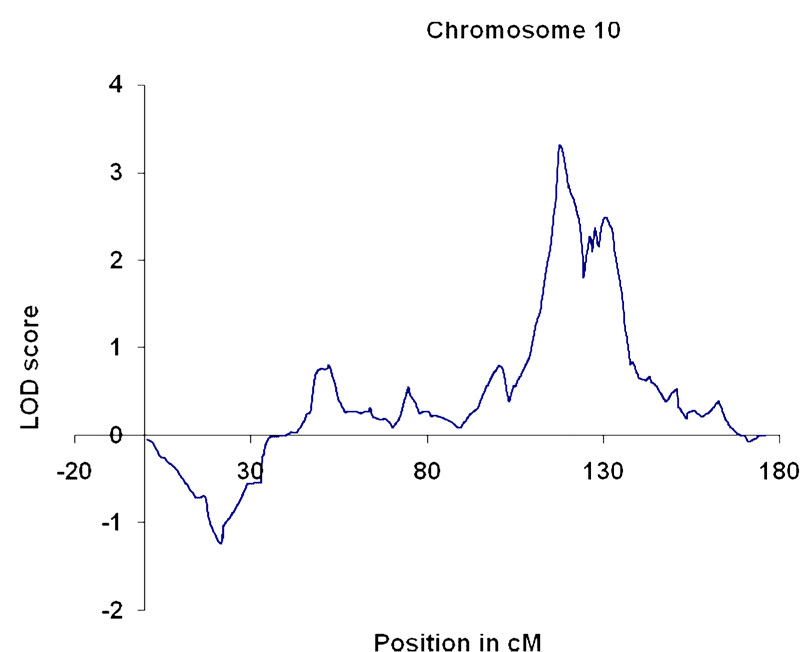
We identified a genomewide-significant linkage to markers near 117.2 cM on chromosome 10q23.3–24.1 (lod score 3.32; p = 5.0E-05; empirical genome wide p=0.033).
Conclusions
These data add to the growing evidence for locations for AD risk loci, and provide the first linkage evidence for such a locus in the AA population.
Keywords: Alcohol dependence, African American population, genomewide linkage, substance dependence genetics
Introduction
Alcohol dependence (AD) is extremely costly to individuals and to society in the US and throughout the world, in terms of morbidity, mortality, and financial burden. Genetic factors are important determinants of the development of AD, as established by twin, family, and adoption studies, with most studies settling on heritability in a range of 0.50–0.60 e.g. (1–3). Numerous other traits related to AD, such as alcohol intake, are themselves heritable (4). However, all studies establishing the basic parameters of heritability for AD have focused on subjects of non-African ancestry, most on subjects of European ancestry. Epidemiologic data show that rates of alcohol use, abuse, and dependence are all somewhat lower in African-Americans (AAs) than in European-Americans (EAs); for AD, the lifetime rates are 13.8% for EAs and 8.4% for AAs (5) Substantial progress has been made in mapping and identifying genes in EA, and to some extent in Native American, populations. It cannot be assumed that the same factors are important in AAs.
Genome scan linkage mapping projects have identified promising map positions for AD susceptibility loci, some of which have already led to the discovery of disease-influencing genes. The first linkage studies of AD (6–8) provided nonsignificant support for several chromosomal locations that could potentially harbor loci influencing risk. Wilhelmsen et al. (9) reported a genomewide linkage study for an alcohol-related trait, low-level response to alcohol; although this study considered a small sample (139 sibling pairs), several “suggestive” linkages were identified. Ehlers et al. (10) conducted linkage analysis in a sample of Mission Indians; several lod scores >2 were identified for the phenotypes of alcohol severity (chromosomes 4 and 12) and alcohol withdrawal (chromosomes 6, 15, and 16). The most recent large study to be reported focused on a set of 474 small families recruited in Ireland (11); this study also reported strongest results (up to a genomewide-significant multipoint lod score of 4.59) on chromosome 4. We are not aware of any previous linkage study that focused on AD in an AA population.
Until recently the only genes established to affect risk for AD were those encoding several alcohol metabolizing enzymes, but several other genes can now be regarded as confirmed risk loci (GABRA2; (refs. 12–15)), or strong candidates based on published data (e.g., CHRM2; (16, 17)). Further, the data supporting a relationship of ADH and ALDH loci to AD risk are now much richer, and effects at these loci are recognized in many more populations than previously (17–22). Although the mechanism of action of the effects of alcohol-metabolizing enzymes on AD risk is thought to be well understood, we are still in the early stages of understanding the core physiology of other risk loci. It is clear that only a small number of the many genes that influence risk for AD have been identified; and that the effects of many of the identified loci vary by population.
We collected a set of small nuclear pedigrees suitable for linkage analyses of cocaine dependence and opioid dependence (23, 24). Families were selected on the basis of containing at least two siblings each affected with either cocaine or opioid dependence (or both); recruitment was not conditioned on AD. We have also published results from this sample for nicotine dependence (25). We evaluated these subjects with the SSADDA (Semi-Structured Assessment for Drug Dependence and Alcoholism (23, 26, 27), a polydiagnostic instrument that assesses a range of psychiatric diagnoses, including DSM-IV AD. We describe here the results of a genomewide linkage analysis for AD in the AA families in this clinical sample.
Methods
Subjects for this study were recruited at four sites: Yale University School of Medicine (APT Foundation; New Haven, CT), University of Connecticut Health Center (UConn; Farmington, CT), McLean Hospital (Harvard Medical School; Belmont, MA), and Medical University of South Carolina (MUSC; Charleston, SC). Subjects were originally ascertained for affected sibling pair linkage studies of the genetics of cocaine or opioid dependence (23, 24). Families were recruited based on screening suggesting that two siblings would meet diagnostic criteria for opioid dependence (at the UConn and Yale sites) or cocaine dependence (all sites). Alcohol use played no role in proband selection or pedigree extension. Subjects were classified as African- American (AA) based on a Bayesian model-based clustering method using genetic marker information, with STRUCTURE (28). We used 100,000 iterations for the burn-in, with a run length of 100,000. Included in the STRUCTURE analyses were 1408 SNPs from the genomewide scan. The program implements a model-based clustering method for inferring population structure using genotype data. Individuals in the sample are assigned (probabilistically) to populations, or jointly to two or more populations if their genotypes indicate that they are admixed. We allowed for up to five populations, but found the best fit at two: one mostly AA and one mostly EA. We found that only 1% of the subjects self-reporting to be of AA descent clustered in the EA group, and of the subjects self-reported to be EA, only 1.5% clustered in the AA group. Of subjects identifying themselves as White Hispanic, 45% clustered in the AA group versus 55% of subjects who identified themselves as Black Hispanic. Structure reports the posterior probability that individual i is from population k with a value between 0 and 1. A large majority of subjects had a posterior probability of about 0.9 and only a small minority, mostly mixed-ethnicity families, clustered near the middle. The threshold we used in those cases was 0.5. Individuals with a clinical diagnosis of a major psychotic illness (schizophrenia or schizoaffective disorder) were excluded as probands. When an affected sibling pair was recruited, we also recruited additional siblings and parents, whenever possible, regardless of affection status. Out of a total of 339 AA families, 238 families had at least one subject affected with AD. The sample included 840 genotyped subjects (and a total of 1642 for linkage analysis, including, e.g., ungenotyped parents). 50.4% were male, and age range was 18–79, with a mean of 40.8±7.3 (SD). Family size ranged from 3–13 subjects, with a mean of 4.84. Subjects gave informed consent as approved by the institutional review board at each clinical site, and a certificate of confidentiality for the work was obtained from NIH (NIDA). Family characteristics are shown in Tables 1.
Table 1.
Families with two or more affected subjects
| Families (n) | 97 |
| Subjects (n) | 516 |
| Family size (avg) | 5.32 |
| Subjects with SSADDA assessment (n) | 277 |
| Subjects meeting DSMIV AD dx (n) | 263 |
| AD subjects per family (avg) | 2.71 |
| Average parental age | 62.4 ± 9.9 |
| Average offspring (sibling pair) age | 39.5 ± 6.6 |
Diagnosis
Subjects were interviewed using the SSADDA for psychiatric diagnosis as described previously (23, 26, 27). The diagnosis of AD was based on application to the SSADDA data set of a computer algorithm using DSM-IV diagnostic criteria. 355 subjects were affected with AD, and a further 184 met criteria for alcohol abuse. For the purpose of these analyses, we considered subjects who met the diagnosis of alcohol abuse to be diagnostically unknown; this was intended as a conservative way to treat subjects who could not be definitively characterized as affected or unaffected.
Laboratory Methods
In most cases DNA was obtained from immortalized cell lines, although for some subjects DNA was extracted directly from blood or saliva. Single nucleotide polymorphisms (SNPs) were genotyped at the Center for Inherited Disease Research (CIDR) using the Illumina Linkage IVb Marker Panel with SNP markers with average 0.64 cM spacing and an average marker heterozygosity of 0.38 in African Americans (http://www.cidr.jhmi.edu/human_snp.html). We previously reported association analyses with this dataset for the major substance dependence traits (29).
Statistical Analysis
PREST (30) was used to validate the reported family relationship assignments and PedCheck (310) was used to detect and correct the Mendelian inconsistencies in the data. Based on this analysis eight changes were made; four reported full-sibs were reclassified as half-sibs; three reported full-sibs and one reported half-sib were actually unrelated and excluded from further analysis. Uninformative SNPs (i.e., those with minor allele frequency [MAF] <0.1) and SNPs not in Hardy–Weinberg equilibrium (HWE) (with (p value < 0.001)) were excluded from the analyses (cf. ref 29). Six thousand eight SNP markers were genotyped. Thirty-six SNP markers that displayed excessive replicate or Mendelian errors, had more than 50% missing data, or were monomorphic were excluded. We limited our analysis to the 5,633 remaining autosomal markers. The average rate of missing data among the remaining markers was 0.10%. By subject, 1669 subjects (total dataset including EA subjects who were not included in the linkage analysis) had ≤1% missing data, and 30 subjects had 1–5% missing data. No subject had >5% missing data. By marker, 5573 markers had <1% missing data; 40 had 1–2% missing; 9 had 2–3% missing; 5 had 3–4% missing; 2 had 4–5% missing; 2 had 5–6% missing; and 2 had 6–8% missing. No marker had >8% missing data. Linkage analyses were performed using the package Merlin (32). To address the issue of marker-marker linkage disequilibrium (LD) in the data set that has the potential to spuriously inflate linkage results, SNPs showing LD were grouped into clusters (33). Clusters were created when the pairwise r2 between neighboring SNPs was greater than 0.10. Analyses were repeated with r2 thresholds of 0.05, 0.2, and 0.3 to gauge the effect of LD on the linkage results. Map positions for the clusters were calculated as the midpoint between the positions of the outer-most SNPs in the cluster. In the few instances of an obligate recombination among SNPs in a cluster, genotypes for SNPs in the cluster were recoded as missing for the recombinant individuals. Population haplotype frequencies were calculated using the available genotype data and a maximum-likelihood E-M algorithm. No linkage disequilibrium was assumed between the clusters.
Multipoint nonparametric linkage analysis was then performed using the haplotypes based on the SNPs comprising these clusters as unlinked (composite) markers, thus gaining linkage information while avoiding inaccuracies caused by ignoring the tight intermarker LD between the SNPs.
To estimate the type I error rate in the analysis, 1000 unlinked genome scans were simulated and analyzed in the same way as the original data. This provided us with empirical p-values and genome-wide p-values for the data set.
Results
Simulations
From 1000 genomewide simulations (for model-free multipoint linkage), we observed 1166 lod scores >1.9, 75 lod scores >3, 33 lod scores >3.32, and 2 lod scores >4.
Linkage analysis
We observed one linkage peak that satisfied empirical genomewide significance criteria (i.e., based on simulations); this corresponded to a maximum lod score of 3.32 on chromosome 10 at 117.4 cM (10q23.3–q24.1) (genomewide empirical p=0.033; point p=0.00005) (Figure 1). The location and the value of this peak did not change when analyses were repeated using different r2 values for grouping SNPs into clusters. The 95% (i.e., one LOD-unit) confidence interval is a 10 cM region between 115.1 cM and 125.3 cM. Lod scores greater than 1 were also observed on chromosome 9 (lod score 1.01 at 66.4 cM); chromosome 14 (a lod score of 1.03 at 32.0 cM); chromosome 16 (lod score 1.2 at 29.9 cM); and chromosome 17 (lod score 1.67 at 48.9 cM). Notably, no lod score >0.33 was observed on chromosome 4, which is the chromosome where positive linkage signals have been observed most consistently in other (i.e., non-AA) populations (6–8, 10, 11).
Figure 1.
Chromosome 10 linkage results. LOD, logarithm of odds.
Linkage results are summarized in Table 2.
Table 2.
Maximum observed lod scores on each chromosome.
| Chromosome | Position | Lod | p-value |
|---|---|---|---|
| 1 | 19.0 | 0.11 | 0.2 |
| 2 | 19.1 | 0.47 | 0.07 |
| 3 | 159.8 | 0.37 | 0.1 |
| 4 | 51.0 | 0.33 | 0.11 |
| 5 | 82.2 | 0.17 | 0.2 |
| 6 | 28.7 | 0.24 | 0.15 |
| 7 | 66.8 | 0.19 | 0.2 |
| 8 | 55.0 | 0.06 | 0.3 |
| 9 | 66.4 | 1.01 | 0.02 |
| 10 | 117.4 | 3.32 | 0.00005 |
| 11 | 58.1 | 0.1 | 0.2 |
| 12 | 50.1 | 0.26 | 0.14 |
| 13 | 108.8 | 0.4 | 0.09 |
| 14 | 32.0 | 1.03 | 0.015 |
| 15 | 112.4 | 0.14 | 0.2 |
| 16 | 29.9 | 1.2 | 0.009 |
| 17 | 48.9 | 1.67 | 0.003 |
| 18 | 0.1 | 0.25 | 0.14 |
| 19 | 5.6 | 0.17 | 0.2 |
| 20 | 79.8 | 0.52 | 0.06 |
| 21 | 4.4 | 0.01 | 0.4 |
| 22 | 35.5 | 0.46 | 0.07 |
Discussion
We completed a dense SNP genomewide linkage scan for the trait of DSM-IV AD in a sample of AA small nuclear families. We observed one genomewide significant linkage result, on chromosome 10, at 117.4 cM. Although we observed other linkage signals that might indicate locations of AD susceptibility genes, most notably a lod score of 1.67 on chromosome 17, the chromosome 10 signal was by far the strongest and the only one meeting genome wide significance. This is the first report of genomewide-significant linkage to the trait of AD in an AA population, and the first report of a significant AD linkage to this chromosomal location, to our knowledge. Li et al. (34) have previously reported on a linkage study for nicotine dependence-related traits in AAs, and they, interestingly, also reported a single genomewide-significant linkage signal, also on chromosome 10. These investigators observed a lod score of 4.17 at 92 cM. Because this location is not within the 1-lod support interval for the locus we observed on the same chromosome, it likely represents a different risk gene. We previously reported linkage results for cocaine and opioid dependence, and cocaine-induced paranoia, in parts of the same AA sample reported upon here (23, 24). As we have reported previously (29), the correlations for diagnoses of alcohol, opioid, and cocaine dependence in this sample are not high; they range between 0.067 (cocaine vs opioid dependence) and 0.295 (cocaine dependence vs AD). Thus, linkage analyses of these traits capture different information. The chromosomal region contains numerous genes potentially relevant to alcohol dependence, of which perhaps the most immediately interesting are the genes encoding synaptic vesicular amine transporter, or VMAT 2 (SLC18A2) (which is however distal to the peak signal); and serotonin 7 receptor (HTR7).
Two previous linkage studies reported evidence for linkage of traits related to alcohol dependence and chromosome 10 markers. Agrawal et al. (35) reported suggestive linkages at the 60 and 99 cM on that chromosome to a quantitative trait based on DSM-IV AD symptoms, in a mostly EA sample – quite distant from our observed linkage peak. Schuckit et al. (36) observed suggestive linkage to the trait of “level of response to alcohol” in the region of 120–140 cM on chromosome 10; this linkage signal is far more likely to have its origin at the same genetic locus, or loci, as the one we observed. Kendler et al (37) also found modest evidence for linkage of a conduct disorder and AD-related trait in the same region. Consistent with expectations based on relatively low correlations between different SD diagnoses in our sample, the major linkage peak we observed for AD does not coincide with peaks observed for cocaine (23), nicotine (25), or opioid dependence (24), in our previous studies, with one exception: we observed “suggestive” linkages for Fagerstrom Test for Nicotine Dependence (FTND) scores on both sides of the chromosome 10 peak, i.e., flanking it, under different analytic models, in EA subjects (24).
As we stated above, it cannot be assumed that identical risk factors are important in different populations. There are numerous reported instances that show evidence for different linkage signals in different populations (e.g., genetic linkage studies of cocaine and opioid dependence that we have reported previously, 23, 24). There are also instances where specific risk alleles differ by population. One such instance – where it has been shown that a specific risk allele is present in AAs but not EAs – involves ADH loci and risk for alcohol dependence. ADH2 rs2066702, traditionally called Alcohol Dehydrogenase-2*3, is a functional variant that encodes a high-activity isozyme that is common in AAs but rare in EAs (38). According to the observations of several authors, including ourselves (39), this is a risk variant. (We observed minor allele frequencies for this variant of 0 in EAs, 0.178 in AD AAs, and 0.255 in AA control subjects (39).)
Power is always a concern for genetic linkage studies of complex traits such as AD. Phenotypic ascertainment is time consuming and expensive, collecting families with multiple affected individuals is challenging, and extending sibling pairs to include additional siblings and parents, difficult. Thus, methods that increase the power of linkage studies with such samples provide an efficient means of extracting the largest possible amount of linkage data. Considerably more linkage information may be extracted from a linkage sample using a high-density SNP map, compared to what is now considered a low-density (400 marker) short tandem repeat (STR) map (such as that employed in our previous linkage studies with parts of this sample). The information increment was estimated to be close to 75% in families where parental genotypes were unavailable but <50% where parental information was available (40). Evans and Cardon (41) also emphasized that very dense SNP maps provide the greatest increment in linkage information when parental genotypes are unavailable. Due to the characteristics of our particular sample, which is rich in small nuclear families (in most cases including no more than one parent), the increment in information achieved is of particularly great value, and it is doubtful that we would have identified a statistically-significant linkage in this sample without a highdensity marker set because of the relatively small set of informative families. The fact that AD is less common in AAs than in EAs suggests that a higher genetic loading is required in this population for the trait to be expressed, which may also have worked in our favor to identify a significant linkage. It is difficult to estimate power for a study such as this; power depends on several factors of which only two (sample size and marker density) are known. Other factors include heterogeneity and effect size. Risch and others have published power calculations for affected sibling pair linkage based on sample size and λ. Based on Risch’s power calculations (42) assuming 100 ASPs (approximately the number included here) and close linkage, we had sufficient power to detect a locus with a λs = 3. For a λs = 2, power is only about 40%. Our power was somewhat better than this since we had ~2.7 affected members per family.
In summary, we presented a genomewide linkage scan of AD in an AA population, based on a dense SNP map. We identified one linkage peak empirically established as genomewide-significant, the first such finding in an AA population. This finding, if replicated or otherwise confirmed (by identification of a risk locus that maps within the linkage peak), will provide useful information in the search for genetic loci influencing AD risk specifically in the AA population – either through direct query of the linked region, or by providing a signal that can be used to rank SNPs in a genome wide association analysis, as a way to address the severe multiple testing issues inevitably countered in the course of genome wide association designs (43). Regardless of whether such a risk locus would affect outcome exclusively in AAs or generalizes to other populations, it may be expected to point to mechanisms of action for risk that apply globally.
Acknowledgments
We thank the families who volunteered to participate in this research study. This work was supported by NIH grants R01 DA12690, R01 DA12849, K24 DA15105, K24 DA022288, R01 AA11330, K24 AA13736 and M01 RR06192; and the U.S. Department of Veterans Affairs (the VA Connecticut–Massachusetts Mental Illness Research, Education and Clinical Center [MIRECC]). Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University contract number N01-HG-65403. Ann Marie Lacobelle and Greg Kay provided excellent technical assistance. John Farrell provided excellent database support. We also thank the Rutgers University Cell and DNA Repository, the contractor for the NIDA Center for Genetic Studies, co-directed by Dr. Jay Tischfield and Dr. John Rice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
The authors reported no biomedical financial interests or potential conflicts of interest.
References
- 1.Kendler KS, Heath AC, Neale MC, Kessler RC, Eaves LJ. A population-based twin study of alcoholism in women. JAMA. 1992;268:1877–1882. [PubMed] [Google Scholar]
- 2.Kendler KS, Prescott CA, Neale MC, Pedersen NL. Temperance board registration for alcohol abuse in a national sample of Swedish male twins, born 1902 to 1949. Arch Gen Psychiatry. 1997;54:178–184. doi: 10.1001/archpsyc.1997.01830140090015. [DOI] [PubMed] [Google Scholar]
- 3.Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;56:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- 4.Whitfield JB, Zhu G, Madden PA, Neale MC, Heath AC, Martin NG. The Genetics of Alcohol Intake and of Alcohol Dependence. Alcoholism: Clin Exp Res. 2004;8:1153–1160. doi: 10.1097/01.alc.0000134221.32773.69. [DOI] [PubMed] [Google Scholar]
- 5.Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2007;64:830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- 6.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet (Neuropsychiat Genet) 1998;81:207–215. [PubMed] [Google Scholar]
- 7.Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, et al. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–945. [PubMed] [Google Scholar]
- 8.Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, et al. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J of Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 9.Wilhelmsen KC, Schuckit M, Smith TL, Lee JV, Segall SK, Feiler HS, et al. The search for genes related to a low-level response to alcohol determined by alcohol challenges. Alcohol Clin Exp Res. 2003;27:1041–1047. doi: 10.1097/01.ALC.0000075551.02714.63. [DOI] [PubMed] [Google Scholar]
- 10.Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC. Genomic screen for loci associated with alcohol dependence in Mission Indians. Am J Med Genet (Neuropsychiatr Genet) 2004;129:110–115. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- 11.Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG et al. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol Psychiatry. 2006;11:603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- 12.Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO. Variations in GABRA2, Encoding the α2 Subunit of the GABAA Receptor, Are Associated with Alcohol Dependence and with Brain Oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Covault C, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet. 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- 14.Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, et al. Association between alcoholism and GABRA2 in a Russian population. Alcohol Clin Expl Res. 2005;29:493–498. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- 15.Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, et al. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- 16.Wang JC, Hinrichs AL, Stock H, Budde J, Allen R, Bertelsen S, et al. Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Hum Mol Genet. 2004;13:1903–1911. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- 17.Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence in European Americans: results from family-controlled and population-structured association studies. Pharmacogenet and Genomics. 2005;15:755–768. doi: 10.1097/01.fpc.0000180141.77036.dc. [DOI] [PubMed] [Google Scholar]
- 18.Hasin D, Aharonovich E, Liu X, Maman Z, Matseoane K, Carr L, Li T. Alcohol and ADH2 in Israel: Ashkenazis, Sephardics and recent Russian Immigrants. Am Journal Psychiatry. 2002;159:1432–1434. doi: 10.1176/appi.ajp.159.8.1432. [DOI] [PubMed] [Google Scholar]
- 19.Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology online publication. 2006a;31:1085–1095. doi: 10.1038/sj.npp.1300925. [DOI] [PubMed] [Google Scholar]
- 20.Luo X, Kranzler HR, Zuo L, Wang S, Schork NJ, Gelernter J. Diplotype trend regression (DTR) analysis of the ADH gene cluster and ALDH2 gene: Multiple significant associations for alcohol dependence. Am J Hum Genet. 2006b;78:973–987. doi: 10.1086/504113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, et al. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- 22.Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, van den Oord EJ, et al. Association of ADH and ALDH Genes With Alcohol Dependence in the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD) Sample. Alcohol Clin Exp Res. 2008 Mar 4; doi: 10.1111/j.1530-0277.2008.00642.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, et al. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet. 2005;136:45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- 24.Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, Weiss R, Sonne S, Zhao H, Farrer L, Kranzler HR. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006;78:759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gelernter J, Panhuysen C, Weiss R, Brady K, Poling J, Krauthammer M, Farrer L, Kranzler HR. Genomewide linkage scan for nicotine dependence: Identification of a chromosome 5 risk locus. Biol Psychiatry. 2007;61:119–126. doi: 10.1016/j.biopsych.2006.08.023. Published online Nov. 1, 2006. [DOI] [PubMed] [Google Scholar]
- 26.Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, et al. Reliability of DSM-IV diagnostic criteria using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2007;91:85–90. doi: 10.1016/j.drugalcdep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Y, Kranzler HR, Panhuysen C, Weiss RD, Poling J, Farrer LA, Gelernter J. Substance Dependence Low Density Whole Genome Association Study In Two Distinct American Populations. Human Genetics. 2008;123:495–506. doi: 10.1007/s00439-008-0501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McPeek and Sun. Statistical Tests for Detection of Misspecified Relationships by Use of Genome-Screen Data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 33.Abecasis GR, Wigginton JE. Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet. 2005;77:754–767. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li MD, Payne TJ, Ma JZ, Lou X-Y, Zhang D, Dupont RT, et al. A genomewide search finds major susceptibility loci for nicotine dependence on chromosome 10 in African Americans. Am J Hum Genet. 2006;79:745–751. doi: 10.1086/508208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agrawal A, Hinrichs AL, Dunn G, Bertelsen S, Dick DM, Saccone SF, et al. Linkage scan for quantitative traits identifies new regions of interest for substance dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. Drug Alcohol Depend. 2008;93:12–20. doi: 10.1016/j.drugalcdep.2007.08.015. Epub 2007 Oct 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuckit MA, Wilhelmsen K, Smith TL, Feiler HS, Lind P, Lange LA. Autosomal linkage analysis for the level of response to alcohol. Alcohol Clin Exp Res. 2005;29:1976–1982. doi: 10.1097/01.alc.0000187598.82921.27. [DOI] [PubMed] [Google Scholar]
- 37.Kendler KS, Kuo P-H, Webb BT, Kalsi G, Neale MC, Sullivan PF, Walsh D, Patterson DG, Riley BP, Prescott CA. A Joint Genomewide Linkage Analysis of Symptoms of Alcohol Dependence and Conduct Disorder. Alcoholism: Clinical and Experimental Research. 2006;30:1972–1977. doi: 10.1111/j.1530-0277.2006.00243.x. [DOI] [PubMed] [Google Scholar]
- 38.Thomasson M, Beard JD, Li T-K. ADH2 gene polymorphisms are determinants of alcohol pharmacokinetics. Alcohol Clin Exp Res. 1995;19:1494–1499. doi: 10.1111/j.1530-0277.1995.tb01013.x. [DOI] [PubMed] [Google Scholar]
- 39.Luo X, Kranzler HR, Zuo L, Wang S, Schork NJ, Gelernter J. Multiple ADH genes modulate risk for drug dependence in both African- and European-Americans. Human Molecular Genetics. 2007;16:380–390. doi: 10.1093/hmg/ddl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The International Multiple Sclerosis Genetics Consortium: Enhancing linkage analysis of complex disorders: an evaluation of high-density genotyping. Hum Mol Genet. 2004;13:1943–1949. doi: 10.1093/hmg/ddh202. [DOI] [PubMed] [Google Scholar]
- 41.Evans DM, Cardon LR. Guidelines for genotyping in genomewide linkage studies: single-nucleotide-polymorphism maps versus microsatellite maps. Am J Hum Genet. 2004;75:687–692. doi: 10.1086/424696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Risch N. Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet. 1990;46:229–241. [PMC free article] [PubMed] [Google Scholar]
- 43.Roeder K, Bacanu S-A, Wasserman L, Devlin B. Using Linkage Genome Scans to Improve Power of Association in Genome Scans. Am J Hum Genet. 2006;78:243–252. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]