Abstract
Current evidence suggests that the length of poly(A) tails of bacterial mRNAs result from a competition between poly(A) polymerase and exoribonucleases that attack the 3′ ends of RNAs. Here, we show that host factor Hfq is also involved in poly(A) tail metabolism. Inactivation of the hfq gene reduces the length of poly(A) tails synthesized at the 3′ end of the rpsO mRNA by poly(A) polymerase I in vivo. In vitro, Hfq stimulates synthesis of long tails by poly(A) polymerase I. The strong binding of Hfq to oligoadenylated RNA probably explains why it stimulates elongation of primers that already harbor tails of 20–35 A. Polyadenylation becomes processive in the presence of Hfq. The similar properties of Hfq and the PABPII poly(A) binding protein, which stimulates poly(A) tail elongation in mammals, indicates that similar mechanisms control poly(A) tail synthesis in prokaryotes and eukaryotes.
Polyadenylation of mRNA plays a role in gene expression in both prokaryotes and eukaryotes (1–4). Long poly(A) tails of eukaryotic RNAs participate in the control of mRNA stability (4) and translation initiation (3). In Escherichia coli, poly(A) tails reduce the stability of regulatory plasmid RNAs (5–7) and of mRNAs (8–11). It has been suggested that poly(A) polymerase I (PAP I) synthesizes short oligo(A) tails that can be either removed by RNase II or used as a binding site by polynucleotide phosphorylase (PNPase) to start degradation of messengers and RNA fragments (12–14). The physical interaction of PAP I with RNase E, a key enzyme of mRNA decay, which is able to cleave poly(A) tails in vitro (15), may also reflect the relationship between polyadenylation and mRNA degradation in bacteria (16). It has also been proposed that ribosomal protein S1 associated with poly(A) tracts affects translation of polyadenylated mRNAs (17).
By analogy with the situation in eukaryotic cells, where the activity of poly(A) polymerase depends on a complex machinery including a poly(A) binding protein (18), we postulated that the abundant Hfq RNA-binding protein of bacteria (also referred to as HF-1), which interacts with A-rich regions of several RNAs (19, 20), may be involved in the metabolism of poly(A) tails. This polypeptide, originally discovered as a host factor involved in phage Qβ RNA replication, is a subunit of the Qβ replicase (21). It is required for the synthesis of the minus strand used as template to generate the original single-stranded RNA genome. Its function in the E. coli cell is still not completely clear. The pleiotropic phenotype of a mutant in which the hfq gene has been interrupted indicates that Hfq is implicated in several metabolic pathways of bacteria (22). The recent discovery that Hfq is required for translation of the σs subunit of RNA polymerase, specific for stationary phase and osmotic upshift, implies that it probably influences the expression of the >40 genes depending on this σ factor (23, 24). Control of phage Qβ RNA replication and rpoS translation both involve the interaction of Hfq with A-rich regions of RNAs (19, 20). Hfq may also affect the stability of the mutS, miaA, and hfq mRNAs (19, 20, 25) and facilitate the degradation of the ompA messenger (26).
Here, we demonstrate that Hfq is an effector of poly(A) metabolism that strongly stimulates the elongation of poly(A) tails catalyzed by PAP I in vitro and affects lengths of oligo(A) tails in vivo.
Experimental Procedures
Bacteria.
The hfq1∷Ω and hfq2∷Ω alleles from strains TX2808 and TX2758 (22) were P1-transduced in strain SK5704 (pnp7 rnb500 rne1) (27) giving rise to strain IBPC922 and IBPC930, respectively. The hfq− genotype of transductants was verified by PCR amplification of chromosomal DNA.
RNA Preparations and Northern Blots.
Bacteria were grown at 30°C and shifted to 44°C (time 0) to inactivate RNase E and RNase II (9). Rifampicin (500 μg⋅ml−1) was added at the time of the shift, and 5 μg of total RNA prepared from aliquots withdrawn at different times was analyzed on Northern blots (9).
Polyadenylation in Vitro.
Polyadenylation was carried out in Mg2+ buffer (28). Template for synthesis of 32P-labeled RNA primer (97RNA) was obtained by PCR amplification of a region of the genome corresponding to the last 97 nucleotides of the rpsO mRNA with the 5′-TAATACGACTCACTATAGGGAGACGTAGCACGTTACACC oligonucleotide containing the sequence of the T7 RNA polymerase promoter and the 5′-GAAAAAAGGGGCCACTCAGG downstream oligonucleotide. The same downstream primer, extended by 18 T at its 5′ end, was used to generate a DNA template encoding polyadenylated RNAs. RNAs transcribed from this latter template have heterogeneous poly(A) tails ranging from 18 to 80 nucleotides presumably resulting from the sliding of the RNA polymerase (29). RNA primers were also polyadenylated by PAP I. Poly(A) RNA fragments of defined sizes were purified from sequencing gels. Purified PAP I overproduced from plasmid pPAP (28) was a gift of S. Cusak and A. J. Carpousis. The stock of PAP I [10 mg⋅ml−1 in 0.3 M ammonium acetate/10 mM Tris⋅HCl (pH 8.5.)] was diluted in 10 mM Tris⋅HCl, pH 8/0.5 mg⋅ml−1 BSA/0.5% Triton X-100/10% glycerol/1 mM DTT/800 mM NaCl. Hfq was purified as described (30).
Gel Shift Assays.
Three fmols of the labeled RNA fragments were incubated with Hfq and, when specified, with a competitor RNA, for 10 min at 37°C in 10 μl of 100 mM NaCl/10 mM Tris⋅HCl (pH 8.1)/0.25% glycerol/0.5 mM DTT/1 mM EDTA/0.06% Triton X-100, and complexes were separated on polyacrylamide gels (20).
Results
Hfq Stimulates Poly(A) Elongation and Destabilizes mRNA in Vivo.
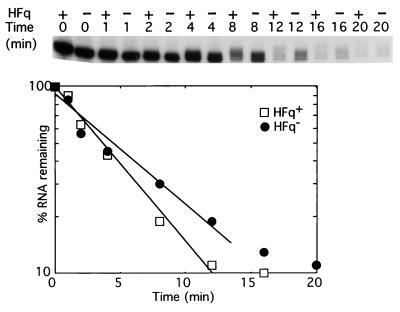
We first investigated whether inactivation of the hfq gene encoding Hfq could affect poly(A) tails synthesized by PAP I in vivo. In bacteria, the length of poly(A) tails is thought to reflect the balance between the polymerization and degradation reactions catalyzed by PAP I and exoribonucleases, respectively (14, 31). In a strain deficient for 3′ to 5′ exonucleases (PNPase and RNase II) and RNase E, where transcription has been inhibited by rifampicin, PAP I progressively synthesizes elongated rpsO transcripts that can be differentiated from nonpolyadenylated mRNAs by Northern blot analysis (9). Introduction of the inactive hfq1∷Ω gene interrupted by a Ω(Kan) cassette that carries transcriptional terminators (22) to the multi-ribonuclease mutant significantly reduces the elongation of the rpsO transcripts (Fig. 1). In the hfq1∷Ω mutant, Northern blot analysis showed that messengers RNAs are extended by about 20 nucleotides, whereas they are extended by 30 nucleotides or more, and become more heterogeneous in length, when hfq is active. Because insertion of the Ω(Kan) cassette is polar, and may affect expression of the downstream genes, we checked that a second polar insertion mutant in hfq: hfq2∷Ω, which does not exhibit the Hfq deficient phenotype (22), had no effect on the elongation of the rpsO mRNA (data not shown). These time course experiments indicate that Hfq is involved in the metabolism of poly(A) tails. Moreover, the half lives of the rpsO mRNA, which were 3.7 and 6.1 min in the hfq+ and the hfq1∷Ω strains, respectively, show that shortening of poly(A) tails in the hfq mutant is correlated with stabilization of the mRNA.
Figure 1.
Hfq affects polyadenylation in vivo. The autoradiograph compares the kinetics of elongation of the poly(A) tail of rpsO mRNA in strains containing (+) or lacking (−) Hfq protein. Times after inhibition of transcription by rifampicin are indicated at the top. Relative amounts of rpsO mRNA were quantified with a PhosphorImager (Molecular Dynamics) and plotted as a function of time to estimate its stability.
Hfq Strongly Stimulates Elongation of Poly(A) Tails by PAP I in Vitro.
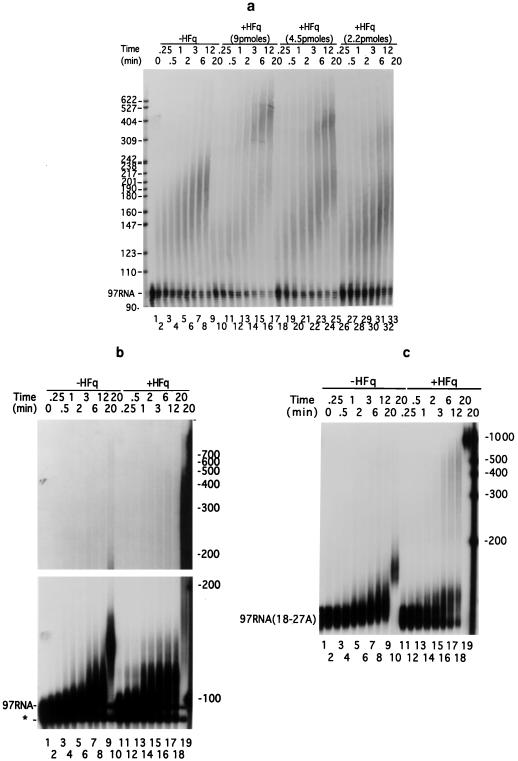
We then verified that the reduction of poly(A) tail elongation observed in vivo after hfq inactivation reflected a direct effect of Hfq on the activity of PAP I. We used purified preparations of Hfq and PAP I to investigate whether Hfq affects polyadenylation of a 97-nucleotide RNA primer (97RNA), corresponding to the 3′ end of the rpsO transcript. The kinetics of elongation of the radioactive primer by PAP I, in the presence of yeast RNA, showed that Hfq favors the appearance of molecules harboring long tails (200–900 A), while it reduces the accumulation of RNA harboring shorter tails (20–150 nucleotides) without significantly affecting the disappearance of the RNA primer (Fig. 2a). The production of RNA harboring long poly(A) tails increased in parallel with the concentration of Hfq. A control experiment showed that Hfq does not catalyze polyadenylation of primers in the absence of PAP I. In another experiment, designed to analyze the initial phase of polyadenylation of primers, we used a low amount of enzyme and omitted yeast RNA. As in the previous experiment, Hfq induced synthesis of elongated molecules ranging from 200 to 900 nucleotides (Fig. 2b, lanes 14–19). Moreover, the progressive elongation of the primer by PAP I (Fig. 2b, lanes 1–10) became biphasic in the presence of Hfq. In the first part of the time course, the radioactive primers were progressively elongated by about 20 A (Fig. 2b, lanes 11–13). Then, Hfq induced the synthesis of much longer polyadenylated molecules with tails ranging from 100 to 600–900 residues (Fig. 2b, lanes 14–18). It is striking that intermediates with tails longer than 20 A are almost undetectable during this second phase of the reaction. Our interpretation is that Hfq begins to stimulate polyadenylation strongly once poly(A) tails reach about 20 A. During the first phase, Hfq slightly affected synthesis of primers extended by 20 A, which appear in 2 min instead of 6 in the absence of Hfq (Fig. 2b, compare lanes 2–7 and 11–14). Then, Hfq caused the very rapid elongation of the tails from 20 to 100–900 A. The rapidity of this second phase of the reaction probably explains why the amount of intermediates decreases abruptly when tails become longer than 20 A. The progressive elongation of RNA primers up to 250 residues in the presence of Hfq, observed in Fig. 2a, may be explained by assuming that the catalytic properties of a fraction of the PAP I molecules remain unaffected when Hfq is at low concentration (the [Hfq]/[PAP I] ratio is 12-fold lower in Fig. 2a than in Fig. 2b). The weak smear of long RNAs appearing in the absence of Hfq (Fig. 2b, lanes 7–10) suggests that PAP I alone can synthesize long poly(A) extensions.
Figure 2.
Hfq affects PAP I activity in vitro. (a) The labeled 97RNA and 1 μg yeast RNA were incubated with 500 fmol PAP I. Purified Hfq was added as indicated. (b) and (c) 360 fmol of labeled 97RNA (b) or polyadenylated 97RNA(18–27A) (c) RNA primers were incubated with 20 fmol PAP I. A total of 4.5 pmol of purified Hfq was added. RNAs withdrawn at different times were analyzed on sequencing gels. Lanes 1 (a–c) show the nonelongated primers. Addition of Hfq, times of incubation, positions, and the nature of the RNA primers and markers are indicated. The star in b shows a shorter RNA produced by T7 RNA polymerase. The upper part of b was overexposed to show the smears of long molecules (lanes 14–18). Reactions of lanes 10 and 19 of b and c contained 10 times more PAP I than lanes 9 and 18.
Hfq Causes the Processive Elongation of Poly(A) Tails by PAP I.
The partition of the RNA substrate into elongated and nonelongated molecules, occurring in the absence of Hfq (Fig. 2a, lanes 1–9 and Fig. 2b, lanes 1–9), might be due either to the processivity of the reaction, i.e., PAP I remains associated with the molecule that it polyadenylates, or to a preferential adenylation by PAP I of molecules harboring a poly(A) extension (32, 33). To discriminate between these two possibilities, we used an oligoadenylated 97RNA harboring tails ranging from 18 to 27 A, referred to as 97RNA(18–27A), as primer. The fact that the 97RNA(18–27A) primer rapidly disappeared and that all molecules of primer were elongated at approximately the same rate in spite of the very low molar concentration of the enzyme (18 times lower than that of primer), demonstrates that PAP I is a distributive enzyme, i.e., the enzyme dissociates from poly(A) after addition of each (or few) nucleotide(s), and that it preferentially polyadenylates RNA harboring a 3′ poly(A) extension (Fig. 2c, lanes 1–9) (34). It was therefore possible to use the 97RNA(18–27A) primer to investigate whether Hfq could transform PAP I into a rapid processive enzyme as previously demonstrated for the PABP II poly(A) binding protein and the PAP of the mammalian nucleus (33). The fact that addition of Hfq caused partition of the oligoadenylated RNA substrate into rapidly elongated and nonelongated molecules demonstrates that Hfq switches PAP I to a processive mode of polyadenylation (Fig. 2c). In this experiment, the rapid phase of elongation began when RNAs harbored tails of 30–35 A. The early appearance of smears of slightly elongated 97RNA in the presence of Hfq (Fig. 2b, compare lanes 2 and 3 to lanes 11 and 12) suggests that Hfq-stimulated polyadenylation begins to exhibit some processivity once primers have acquired a tail of about 5 A. The processivity of the Hfq-stimulated polyadenylation probably explains why PAP I elongates only a portion of the RNA molecules harboring tails of 20–150 A in the presence of Hfq (Fig. 2a).
Hfq Preferentially Binds RNA Harboring a Poly(A) Extension.
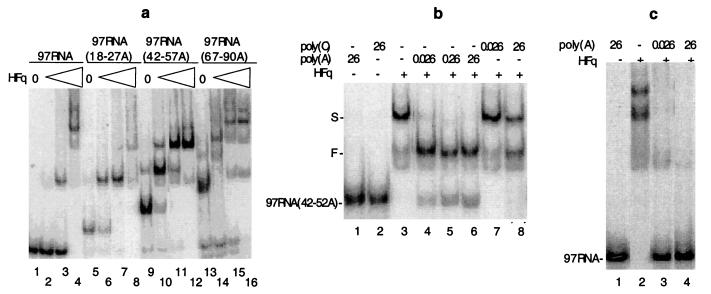
Previous characterization of A-rich regions of several RNAs bound by Hfq (19, 20) suggests that oligo(A) tails are preferential binding sites for this protein. Formation of complexes with poly(A) may be sufficient to explain why Hfq selectively stimulates the elongation of RNA fragments harboring poly(A) extensions. In agreement with this hypothesis, gel shift experiments showed that low concentrations of Hfq slow down the migration of the rpsO 97RNA(18–28A) fragment harboring a 18–28(A) tail, but have little or no effect on the migration of the tail-less RNA (Fig. 3a, compare lanes 2 and 3 to lanes 6 and 7). This demonstrates that Hfq exhibits a higher affinity for oligoadenylated RNAs. The apparent association constant (Ka), measured in the presence of 4 mM Mg2+, is two orders of magnitude higher for the RNA fragment harboring a tail of 18–22 A (≈2 × 108 M−1) than for the tail-less RNA (≈3.5 × 106 M−1) (data not shown). Moreover, competition experiments, in which unlabeled poly(A) was used to prevent the association of Hfq with the labeled RNA fragment, confirms that complexes between Hfq and the tail-less RNA are unstable compared with those involving the polyadenylated 97RNA(42–52A). A concentration of poly(A) 11 times lower than that of the RNA fragment is sufficient to abolish formation of a complex with the tail-less fragment (Fig. 3c, lanes 2–4). In contrast, binding of Hfq to the 97RNA(42–52A) is not inhibited when poly(A) is at a 60 times higher concentration than the RNA (Fig. 3b, lanes 2–6). Concentrations of the 97- to 145-nucleotide RNA fragments and of the 300- to 6,000-nucleotide poly(A) were both expressed in nucleotide molarity for comparison. The low affinity of Hfq for the tail-less RNA fragment and the failure to compete formation of a complex between Hfq and the polyadenylated RNA with a high poly(A) concentration indicates that neither the RNA nor the poly(A) moieties alone can ensure formation of a stable Hfq-poly(A) RNA complex. This implies that Hfq probably binds strongly to a composite structure comprising a stretch of A and an mRNA structural determinant. The fact that a high concentration of poly(C) (Fig. 3b, lanes 3, 7, and 8) and several mRNA fragments (not shown) do not prevent formation of the weak slow migrating 97RNA(42–52A)-Hfq complexes as efficiently as poly(A) confirms that Hfq preferentially binds poly(A) (35). The multiple bands formed at high Hfq concentration could result from the binding of several Hfq monomers or multimers (35) (Fig. 3a). The fact that the number of slowly migrating complexes increases upon elongation of the oligo(A) tails suggests that Hfq coats the poly(A) extensions (Fig. 3a). This may explain why the yield of mRNAs with long poly(A) tails increases in parallel with the amounts of Hfq added to the polyadenylation mixture (Fig. 2a).
Figure 3.
Hfq strongly binds poly(A) RNA. Gel shift experiments were performed with RNA primers with or without a poly(A) extension indicated in a (Top) or b and c (Left). (a) Increasing amounts of Hfq (55 fmol in lanes 2, 6, 10, and 14; 275 fmol in lanes 3, 7, 11, and 15 and 1100 fmol in lanes 4, 8, 12, and 16) were added to the different primers. (b and c) The complexes formed by the RNA primers and 1100 fmol of Hfq were competed by poly(A≈300–6000) or poly(C) (Sigma) at concentrations indicated in pmols of nucleotides (Top). S and F in b show positions of the slow and fast-migrating complexes, respectively.
Discussion
In this report, we provide the evidence that Hfq is an effector of poly(A) metabolism which stimulates the elongation of bacterial poly(A) tails by PAP I. This property probably explains the shortening of poly(A) tails observed in vivo when Hfq is inactivated. However, it is also possible that these shorter tails reflect the fact that Hfq is a poly(A) binding protein that protects poly(A) from attack by ribonucleases. Kinetic polyadenylation studies demonstrate that Hfq converts PAP I into a processive enzyme, which rapidly extends RNAs harboring oligo(A) tails. In the absence of effector, PAP I is a distributive polymerase that exhibits a preference for polyadenylated primers (34). In contrast, in the presence of Hfq, poly(A) elongation becomes processive, and tails are synthesized very rapidly once RNA primers have acquired a tail of 20–35 residues. The preferential binding of Hfq to RNA fragments harboring oligo(A) extensions indicates that PAP I stimulation begins once Hfq can strongly bind the mRNA. Accordingly, RNA with tails of 5–10 A, whose elongation is only slightly stimulated by Hfq (Fig. 2b), do not form stable complexes with the protein (data not shown). These properties of Hfq are reminiscent of those of the poly(A) binding protein PABP II of mammals, which causes the transition from a slow phase of polyadenylation to a rapid processive elongation of poly(A) once the poly(A) tails are long enough to permit binding of the protein (32). Sequence comparisons with programs gapped blast and psi-blast (36) and 434100 sequences of databases did not find significant homologies between Hfq and RNA binding proteins. Moreover, global and local sequence alignments (37, 38) did not show similarities between Hfq and PABPII. Comparison of the affinities of Hfq for poly(A) RNA, tail-less RNA, and poly(A) led us to the conclusion that strong Hfq binding sites consist of a poly(A) track and an mRNA motif. Accordingly, Hfq binding to the rpoS, oxyS, and Qβ RNAs occurs at A-rich regions interrupted by other nucleotides (19, 20). Hfq could bind at the junction between mRNA and oligo(A) or at hybrid sites resulting from an intramolecular interaction between the 3′ oligo(A) and a remote upstream mRNA segment. Simultaneous binding of Hfq to an internal segment and the 3′ extremity of the minus RNA strand of phage Qβ supports this hypothesis (39, 40).
What is the role of Hfq in RNA metabolism? The greater heterogeneity of tails synthesized in cells containing Hfq compared with the shorter tails detected in cells lacking Hfq suggests that PAP I exhibits some processivity in vivo. Assuming that the balance between PAP I, which begins to add A distributively, and exonucleolytic activities maintain poly(A) tails shorter than 20 A (9, 11), we propose that modifications of these activities and/or preferential polyadenylation of certain RNAs favor appearance of slightly longer poly(A) extensions that can be used by Hfq to convert PAP I to the processive mode of synthesis of long poly(A) tails. In spite of the fact that most accessible RNA extremities can probably be polyadenylated by PAP I (8), it appears that only 1.3% of E. coli mRNA (41) and as low as 0.011% of the mRNA of the filamentous phage f1 (42) gain tails long enough to be retained on oligo(dT). Maybe some mRNAs harboring strong Hfq binding sites are preferentially elongated by PAP I. The processivity of polyadenylation probably accounts for the synthesis of poly(A) tails longer than 50 nucleotides in the presence of RNase II (11). Elongation of destabilizing poly(A) tails in the presence of Hfq may explain why some mRNAs become more stable upon Hfq inactivation (25, 26). In vitro data showing that length of poly(A) does not affect degradation of structured RNA by degradosome (43) and our previous observation that poly(A) elongation is correlated with mRNAs destabilization in a strain deficient for PNPase and RNase II (9) indicate that the mechanism of degradation activated by long poly(A) tails is probably different from the exonucleolytic pathway of decay catalyzed by degradosome and/or PNPase (13). Long tails may, for example, facilitate access of poly(A)-bound RNases (or the degradosome) to cutting sites internal to the RNA (14). In the case of ompA, Hfq associated with the 5′ RNA leader could stabilize the interaction of poly(A)-bound RNases with the body of the molecule (26). Interestingly, Hfq shares the capability of interfering with translation with the yeast poly(A) binding protein, Pab1p (3, 23). The interaction of Hfq with rpoS mRNA unmasks the ribosome binding site trapped in a stable secondary structure (20, 44). By analogy with eukaryotes, one could also speculate that poly(A) tails, synthesized in the presence of Hfq, may facilitate the interaction of a poly(A)-bound translation activator to its 5′ operator (3).
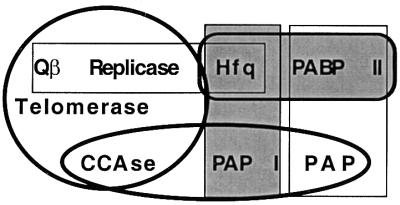
In conclusion, we would like to point out that the association of Hfq with polyadenylation establishes a link between modern polyadenylation machineries and the catalysts of the ancient RNA world (45) (Fig. 4). In addition to the structural homologies between the eukaryotic and prokaryotic PAPs, which all belong to the nucleotidyltransferase family, we have discovered here that the E. coli and mammalian enzymes are affected very similarly by Hfq and PABP II, respectively, i.e., they show increased processivity. Hfq is a common effector of PAP I and of the phage Qβ RNA replicase, which probably originates from a primitive telomerase (35, 45). In addition to the PAPs, the nucleotidyltransferase family includes the tRNA nucleotidyl transferase CCA-adding enzymes, which also are functionally related to this primitive telomerase (46). Like telomerases, PAP I adds A residues downstream of 3′ terminal hairpins, in this case transcription terminators (9), reminiscent of the tRNA-like 3′ tags recognized by telomerase-related catalysts. Loss of the replication function of 3′ tags that are used as transcription terminators and protecting structures in bacteria may explain why the CCA motif has not been conserved. Moreover, that a paradoxical interplay of exonucleases and nucleotidyltransferases destroy and rebuild the tRNA-3′ends (45) as well as prokaryotic mRNA extremities (8, 31) may also be a hint to an evolutionary relationship between CCA addition and polyadenylation.
Figure 4.
Functional and evolutionary relationships between polyadenylation machineries and telomerases. The thick rectangular frame encompasses the Hfq and PABP II polyadenylation stimulatory factors. Hfq and PABP II are linked to enzymes that they modulate by thin rectangular frames. Members of the nucleotidyltransferase family are surrounded by an oval, and telomerase-related enzymes that maintain the 5′ extremities of RNA tagged by a hairpin structure are circled. The functional relationships described here are shaded.
Acknowledgments
We thank B. Beltchev for purification of Hfq, S. Cusack and A. J. Carpousis for the gift of PAP I, A. Ishihama for Hfq antibodies used in Hfq purification, M. E. Winkler for strains TX2808 and TX2758, I. Boni for reminding us that Hfq binds poly(A), M. Springer for suggesting that Hfq might relate PAPs to primitive telomerase, Ph. Derreumeaux for help in sequence comparisons, M. Grunberg-Manago, C. Condon and R. Buckingham for reading the manuscript, and H. Weber for advice. We also acknowledge Ministère de l'Education Nationale de la Recherche et de la Technologie, Centre National de la Recherche Scientifique, and Paris7 University for support.
Abbreviations
- PAP I
poly(A)polymerase I
- PNPase
polynucleotide phosphorylase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040549897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040549897
References
- 1.Coller J M, Gray N K, Wickens M P. Genes Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarkar N. Annu Rev Biochem. 1997;66:173–197. doi: 10.1146/annurev.biochem.66.1.173. [DOI] [PubMed] [Google Scholar]
- 3.Sachs A B, Sarnow P, Hentze M W. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 4.Jackson R J, Standart N. Cell. 1990;62:15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- 5.Xu F, Lin-Chao S, Cohen S N. Proc Natl Acad Sci USA. 1993;90:6756–6760. doi: 10.1073/pnas.90.14.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mikkelsen N D, Gerdes K. Mol Microbiol. 1997;26:311–320. doi: 10.1046/j.1365-2958.1997.5751936.x. [DOI] [PubMed] [Google Scholar]
- 7.Söderbom F, Wagner E G H. Microbiology. 1998;144:1907–1917. doi: 10.1099/00221287-144-7-1907. [DOI] [PubMed] [Google Scholar]
- 8.Haugel-Nielsen J, Hajnsdorf E, Régnier P. EMBO J. 1996;15:3144–3152. [PMC free article] [PubMed] [Google Scholar]
- 9.Hajnsdorf E, Braun F, Haugel-Nielsen J, Régnier P. Proc Natl Acad Sci USA. 1995;92:3973–3977. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hajnsdorf E, Régnier P. J Mol Biol. 1999;286:1033–1043. doi: 10.1006/jmbi.1999.2547. [DOI] [PubMed] [Google Scholar]
- 11.O'Hara E B, Chekanova J A, Ingle C A, Kushner Z R, Peters E, Kushner S R. Proc Natl Acad Sci USA. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen S N. Cell. 1995;80:829–832. doi: 10.1016/0092-8674(95)90284-8. [DOI] [PubMed] [Google Scholar]
- 13.Coburn A G, Mackie A G. J Mol Biol. 1998;279:1061–1074. doi: 10.1006/jmbi.1998.1842. [DOI] [PubMed] [Google Scholar]
- 14.Hajnsdorf E, Braun F, Haugel-Nielsen J, Le Derout J, Régnier P. Biochimie. 1996;78:416–424. doi: 10.1016/0300-9084(96)84748-1. [DOI] [PubMed] [Google Scholar]
- 15.Huang H, Liao J, Cohen S N. Nature (London) 1998;391:99–102. doi: 10.1038/34219. [DOI] [PubMed] [Google Scholar]
- 16.Raynal L C, Carpousis A G. Mol Microbiol. 1999;32:765–775. doi: 10.1046/j.1365-2958.1999.01394.x. [DOI] [PubMed] [Google Scholar]
- 17.Kalapos M P, Paulus H, Sarkar N. Biochimie. 1997;79:493–502. doi: 10.1016/s0300-9084(97)82741-1. [DOI] [PubMed] [Google Scholar]
- 18.Wahle E, Keller W. Trends Biochem Sci. 1996;21:247–250. [PubMed] [Google Scholar]
- 19.Senear A W, Argetsinger Steitz J. J Biol Chem. 1976;251:1902–1912. [PubMed] [Google Scholar]
- 20.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franze de Fernandez M T, Hayward W S, August J T. J Biol Chem. 1972;247:824–821. [PubMed] [Google Scholar]
- 22.Tsui H C, Leung H C, Winkler M E. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 23.Muffler A, Fischer D, Hengge-Aronis R. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 24.Brown L, Elliott T. J Bacteriol. 1996;178:3763–3770. doi: 10.1128/jb.178.13.3763-3770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsui H C, Feng G, Winkler M E. J Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vytvytska O, Jakobsen J S, Balcunaite G, Andersen J S, Baccarini M, von Gabain A. Proc Natl Acad Sci USA. 1998;95:14118–14123. doi: 10.1073/pnas.95.24.14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arraiano M C, Yancey S D, Kushner S R. J Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raynal L C, Krisch H M, Carpousis A J. Biochimie. 1996;78:399–398. [Google Scholar]
- 29.Macdonald L E, Zhou Y, McAllister W T. J Mol Biol. 1993;232:1030–1047. doi: 10.1006/jmbi.1993.1458. [DOI] [PubMed] [Google Scholar]
- 30.Carmichael G G, Weber K, Niveleau A, Wahba A J. J Biol Chem. 1975;250:3607–3612. [PubMed] [Google Scholar]
- 31.Coburn G A, Mackie G A. Prog Nucleic Acid Res Mol Biol. 1999;62:55–108. doi: 10.1016/s0079-6603(08)60505-x. [DOI] [PubMed] [Google Scholar]
- 32.Wahle E. Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- 33.Bienroth S, Keller W, Wahle E. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sano H, Feix G. Eur J Biochem. 1976;71:577–583. doi: 10.1111/j.1432-1033.1976.tb11148.x. [DOI] [PubMed] [Google Scholar]
- 35.Blumenthal T, Carmichael G G. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 36.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang X, Miller W. Adv Appl Math. 1991;12:373–381. [Google Scholar]
- 38.Needleman S B, Wunsch C D. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 39.Miranda G, Schuppli D, Barrera I, Hausherr C, Sogo J M, Weber H. J Mol Biol. 1997;267:1089–1103. doi: 10.1006/jmbi.1997.0939. [DOI] [PubMed] [Google Scholar]
- 40.Klovins J, Berzins V, van Duin J. RNA. 1998;4:948–957. doi: 10.1017/s1355838298980177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao G-J, Sarkar N. Proc Natl Acad Sci USA. 1992;89:7546–7550. doi: 10.1073/pnas.89.16.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frances Goodrich A, Steege D A. RNA. 1999;5:972–985. doi: 10.1017/s1355838299990398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blum E, Carpousis A J, Higgins C F. J Biol Chem. 1999;274:4009–4016. doi: 10.1074/jbc.274.7.4009. [DOI] [PubMed] [Google Scholar]
- 44.Brown L, Elliott T. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maizels N, Weiner A M. In: The RNA World. Gesteland R F, Atkins J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 577–602. [Google Scholar]
- 46.Yu D, Maizels N, Weiner A M. RNA. 1996;2:895–908. [PMC free article] [PubMed] [Google Scholar]