Abstract
To analyze the vast number and variety of microorganisms inhabiting the human intestine, emerging metagenomic technologies are extremely powerful. The intestinal microbes are taxonomically complex and constitute an ecologically dynamic community (microbiota) that has long been believed to possess a strong impact on human physiology. Furthermore, they are heavily involved in the maturation and proliferation of human intestinal cells, helping to maintain their homeostasis and can be causative of various diseases, such as inflammatory bowel disease and obesity. A simplified animal model system has provided the mechanistic basis for the molecular interactions that occur at the interface between such microbes and host intestinal epithelia. Through metagenomic analysis, it is now possible to comprehensively explore the genetic nature of the intestinal microbiome, the mutually interacting system comprising the host cells and the residing microbial community. The human microbiome project was recently launched as an international collaborative research effort to further promote this newly developing field and to pave the way to a new frontier of human biology, which will provide new strategies for the maintenance of human health.
Key words: microbiome, microbiota, gut, metagenomics
1. Introduction
An enormous number of microorganisms, the vast majority of which are bacterial species, are known to colonize and form complex communities, or microbiota, at various sites within the human body. It is estimated that the human microbiota is composed of ∼1014 bacterial cells, which is 10 times more than the total number of human cells. The largest and most complex is the one comprised by intestinal bacteria that includes as many as 1012 cells per 1 g of feces in the average human individual.1 Thus, within each human body, intestinal and other microbiota, along with the ‘host’ human cells, form a complex ecosystem that, as a whole, interactively performs various biological processes.2 Therefore, perhaps we should regard ourselves as ‘superorganisms’ together with the indigenous microbes3 and that the composite genome should be referred to as the human ‘metagenome’.
Studies on human intestinal microbiota should include microbial ecology and analysis of the complex metabolism of the microbial community, as well as various host–microbial interactions occurring at the interface between microbes and host intestinal epithelia. Such studies are expected to lead to understanding of the impact of the microbiota on human health and disease.4–6 Along these lines, it should be noted that an international collaborative project, ‘the human microbiome project (HMP)’, was launched7 in 2007 with the aim of collecting and integrating the genomic information from many diverse human microbiomes (the word ‘microbiome’ was first introduced in 2001 to define the collective genomes of microbiota8). This article is intended as an overview of the recent findings in relevant research fields.
2. Microbial diversity of the human gut microbiota
Over the past decade, the bacterial 16S ribosomal RNA gene (16S) sequence (∼1.5 kb) has been a useful landmark for analyzing the microbial diversity of human intestinal microbiota. A large-scale 16S phylotype analysis (grouping only by 16S rRNA sequence similarity) was carried out for three human adult microbiota.9 The analysis of 13 335 bacterial 16S sequences identified 395 phylotypes at the strain level using a threshold of 99% sequence identity (% ID) and a single archaeal phylotype (Methanobrevibacter smithii) within the three samples. Members of the genera Bacteroides, Eubacterium, Clostridium and Ruminococcus were the major species found in the adult microbiota. Of the 395 phylotypes, ∼80% represented sequences from species yet to be cultivated. This analysis also indicated high interindividual variations in microbial composition among the three samples. Another large-scale 16S analysis estimated 4074 phylotypes at the species level (≥97% ID) in 18 348 sequences obtained from 14 subjects including 12 obese adults monitored for over 1 year.10 This and a study using obese mice together revealed the association of the intestinal microbiota with obesity.11 More recently, 16S analysis was performed for 15 172 sequences from 190 samples including subjects with inflammatory bowel disease (IBD) and healthy adults.12 The etiology of IBD is known to largely correlate with the intestinal microbiota or certain microbial members.13–15 These and other studies demonstrated that the intestinal microbiota of IBD patients have reduced microbial diversity compared with those of healthy controls.12,16 The 16S analysis of other disease-afflicted subjects has also been performed in epidemiologic studies involving allergy17–19 and cancer.20,21
The analysis of more than 45 000 bacterial 16S data combined with the three large-scale surveys described above estimated at least 1800 genera (≥90% ID), ∼16 000 phylotypes at the species level (≥97% ID) and ∼36 000 phylotypes at the strain level (≥99% ID) in the human intestinal microbiota, predicting even greater diversity at the species level.12 This analysis also revealed that the majority (98% of all species) belongs to only four bacterial divisions: Firmicutes (64%), Bacteroidetes (23%), Proteobacteria (8%) and Actinobacteria (3%), whereas other minor taxonomic divisions are quite diverse.
Besides these ‘snapshot’ analyses of the intestinal microbiota composition, other long-term surveys have been performed to follow both the overall composition and that of limited members over periods ranging from several months up to 2 years.10,22–24 These longitudinal studies suggested that the composition of intestinal microbiota do not drastically change in adults within the periods examined.
16S analyzes of infant intestinal microbiota have also been carried out.25–27 One analysis revealed a dramatic progression in microbial composition until at least 1 year after birth with higher interindividual variations, but less complex than those between adults, converging toward a profile characteristic to the adult type at the end of the first year of life.25 As might be expected, fraternal twins tend to show a significantly high similarity in their temporal microbial composition profiles.25,28 No clear correlation was found in overall microbial composition due to the mode of delivery (Cesarean section or vaginal birth) and feeding with breast milk or formulated milk. Therefore, the source of these early colonizers is not clear, whereas some specific species are known to be transmitted from mother to baby.26,29,30 Infant intestinal microbiota is mostly composed of bacteria such as Staphylococcus, Streptococcus, Bifidobacterium and Enterobacteria. Both adult and infant intestinal microbiota members are restricted to a small subset of species as described above, implying that the intestinal microbiota have evolved to shape overall microbial diversity under strong selective pressures.4,5
3. Sequence-based metagenomics of the human gut microbiome
The 16S analysis disclosed the existence of numbers of unculturable bacteria in the human intestinal microbiota, with only up to 20% of the 16S data able to be assigned to known species that have been successfully cultured in the laboratory during the past four decades.9 Purely culturable bacteria are definitely necessary for comprehensive characterization of their biological and genetic natures. On the other hand, it is obvious that the 16S data do not provide any information on functional features of the microbial community. Thus, both culture-based and 16S-based approaches have crucial limitations for further studies, particularly for functional analysis. Metagenomics, the third and newest approach, has made it possible to comprehensively explore the biological nature of these complex communities.31,32 This culture-independent approach includes shotgun sequencing of microbial DNA prepared from the microbiota containing unculturable species isolated directly from the environments, followed by intensive computational analysis of obtained genomic sequences. Metagenomics is a quite effective and powerful approach for collecting and analyzing the information of genes present in the microbial community, because the proportion of the protein-coding regions in bacterial genomes can be as high as 80%, so that most of the metagenomic sequences obtained contain at least partial gene regions directly related to functions.33 Biased data can be minimized in the metagenomic analysis, but a small fraction of genes may be relatively under-represented in the entire data set mainly due to toxicity of the gene products or sequence regions to the host Escherichia coli during cloning of the microbial DNA.34 In addition, the degree of gene coverage is largely dependent on sequencing depth and complexity of the communities. These problems can be overcome by employing next-generation DNA sequencers based on massively parallel sequencing technologies,35 by which the cloning step is eliminated and sequence quantity is increased by orders-of-magnitude compared with that of conventional Sanger sequencers.
To date, metagenomic data of human and mouse intestinal microbiomes have been published from three separate groups.11,36,37 Gill et al. obtained ∼78 megabases (Mb) unique metagenomic sequence data from the intestinal microbiome of two healthy human adults. Comparison of gene sets annotated in the intestinal microbiomes with human genes identified significant numbers of bacterial genes that are not encoded in the human genome. The function of these gene products contributes largely to the metabolism of glycans, amino acids and xenobiotics, and biosynthesis of vitamins and isoprenoids, which are necessary processes in human biology. These findings indicate the symbiotic relationship with humans and support the concept that we are superorganisms, the union of humans with their microbiota.3 Kurokawa et al.37 analyzed 13 human intestinal microbiomes including adults, children and unweaned infants and obtained 479 Mb unique metagenomic sequence data. Unexpectedly, more than half (up to 90%) of total shotgun reads were assembled to form unique contigs in each sample, which is in sharp contrast to soil microbiota in which <1% of total reads were assembled. These results suggest that the amount of sequence data produced in the two metagenomic studies (around 50 Mb of Sanger sequence data for each sample) could substantially uncover the major species with the highest orders of magnitude in quantity in each microbiota, and that these species may be comprised of a very limited number of strain-level species, perhaps accounting for <20 species in each microbiome. The constitution of major species by strain-level species, not by high species-level diversity, could be inferred from base-inconsistency in the generated contigs, with most of those ≥5 kb displaying high sequence similarity of >99.5% ID between assembled reads. In order to uncover the vast numbers of other species present at lower orders of magnitude, it would be necessary to produce sequence data of several more orders of magnitude.
The analysis by Kurokawa et al. also found 647 novel gene families specific to these intestinal microbiomes when compared with genes present in the metagenomic data of other microbial communities such as sea-surface, deep-sea and soil, and significantly enriched genes in these microbiomes when compared with gene sets in known microbes, except for the gut inhabitants. These gut microbiome-enriched (gut-enriched) genes were assigned to 237 and 136 clusters of orthologous groups (COGs) for the adult/child and the infant microbiomes, respectively, sharing 58 COGs between them for a total of 315 COGs in all. In the 315 enriched COGs, the function related to carbohydrate metabolism was remarkable in both types, but the functional repertories clearly differed between the adult and infant types. The adult type was rich in polysaccharide-degrading enzymes and the infant type was rich in sugar transporters. These data indicate that the functionality of healthy intestinal microbiota relies largely on available nutrients in the diet.38,39
4. Further analysis of metagenomic genes identified in 13 Japanese samples
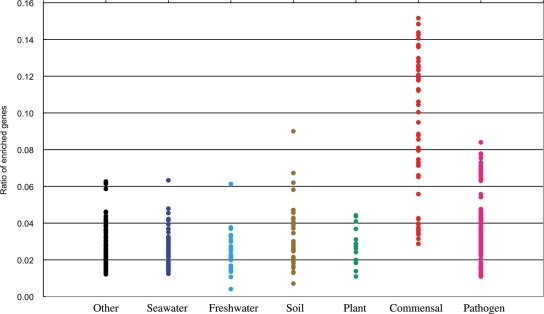
Genome analysis of several Bacteroides strains dominant in adult intestinal microbiota indicated the richness of genes involved in polysaccharide metabolism,40–42 exemplifying the functional adaptation of intestinal microbes to gut habitats rich in polysaccharides, which are metabolized by bacteria to generate short-chain fatty acids such as butyrate, the major energy source for the host.38 It is also valuable to examine and compare the content of genes belonging to the 315 gut-enriched COGs in each genome of bacteria isolated from various environments. Our group recently performed a similarity search analysis of the enriched genes for 371 bacteria whose genomic sequences are already available. The 371 bacteria were classified into seven groups according to their origin of isolation. The ratio of adult gut-enriched genes in each genome is shown in Fig. 1. The average percentage ratios were 3.9% for all 371 microbes, 9.2% for 46 commensal bacteria, 4.0% for 94 pathogens and 2.7% for 231 bacteria from the other five groups. Those of infant gut-enriched genes were 2.8% for all microbes, 6.0% for the commensal bacteria, 3.5% for the pathogens and 1.9% for the other bacteria. These data clearly show that most of the commensal bacteria contain gut-enriched genes at a higher ratio than the other environmental bacteria. Interestingly, many of the commensal bacteria with lower ratios overlap those of pathogens, implying that intestinal microbes with a lower ratio of gut-enriched genes might be primary reservoirs for the corresponding pathogens. Table 1 lists the top 15 species with a higher ratio of adult and infant gut-enriched genes, respectively, which differ considerably from one another, except for the two species, Ruminococcus and Dorea. For the adult gut-enriched genes, members aggregated to three species, Bacteroides, Eubacterium and Ruminococcus, all of which are known dominant species in adult intestinal microbiota. For the infant gut-enriched genes, members are relatively diverse with Clostridium, Bifidobacterium and Lactobacillus species being characteristic. Some of them (e.g. Bifidobacterium and Lactobacillus) are prominent probiotics, living microorganisms having beneficial effects on host health.43 The species with the higher ratios tend to be dominant in intestinal microbiota compared with the species with the lower ratios. For instance, E. coli K12 had a ratio of 3.4% for adult gut-enriched genes, much lower than the 15 species listed, which is consistent with the minority of E. coli in the adult intestinal microbiota. It is also possible that the metabolic mutualism between intestinal members may allow the species with lower ratios, e.g. the human intestinal archaeon (3.6 and 1.2% for the adult and the infant gut-enriched genes, respectively), to stably colonize the gut.44
Figure 1.
Ratio of gut microbiome-enriched genes (adult) in sequenced genomes of bacteria isolated from various environments. The similarity search was performed with a threshold value of ≤1e−8 for 371 bacteria whose genomic sequences were available from public databases. The 371 bacteria were classified into seven groups according to their origin of isolation; commensals, pathogens, plant-related, soil-born, freshwater-born, seawater-born and others, and shown in red, pink, green, brown, light blue, dark blue and black dots, respectively.
Table 1.
Top 15 species with the high ratio of adult and infant gut-enriched genes
| Species having higher ratio of adult gut-enriched genes |
Species having higher ratio of infant gut-enriched genes |
||
|---|---|---|---|
| Ratio | Species | Ratio | Species |
| 0.152 | Bacteroides ovatus | 0.102 | Bifidobacterium longum NCC2705 |
| 0.148 | Bacteroides WH2 | 0.098 | Clostridium ramosum JCM1298 |
| 0.144 | Bacteroides sp. A01 | 0.093 | Bifidobacterium catenulatum JCM1194 |
| 0.143 | Bacteroides vulgatus | 0.092 | Clostridium clostridioforme JCM1291 |
| 0.141 | Bacteroides thetaiotaomicron 3731 | 0.087 | Collinsella aerofaciens |
| 0.137 | Bacteroides thetaiotaomicron VPI-5482 | 0.082 | Lactobacillus johnsonii NCC 533 |
| 0.136 | Bacteroides thetaiotaomicron 7330 | 0.081 | Ruminococcus gnavus |
| 0.130 | Bacteroides uniformis | 0.081 | Enterococcus faecalis V583 |
| 0.128 | Bacteroides caccae | 0.081 | Lactobacillus acidophilus NCFM |
| 0.126 | Eubacterium ventriosum | 0.079 | Dorea longicatena |
| 0.125 | Ruminococcus gnavus | 0.077 | Listeria monocytogenes EGD-e |
| 0.123 | Dorea longicatena | 0.076 | Lactobacillus plantarum WCFS1 |
| 0.121 | Bacteroides sp. A03 | 0.073 | Streptococcus agalactiae A909 |
| 0.121 | Ruminococcus torques | 0.072 | Streptococcus pneumoniae TIGR4 |
| 0.121 | Bacteroides fragilis NCTC 9343 | 0.072 | Streptococcus pneumoniae R6 |
In summary, the abundance of specific gene sets (i.e. gut-enriched genes) in commensal bacteria suggests that their genomes have evolved to accumulate functions advantageous for competitive survival and colonization in the gut habitat, a possible consequence of the functional adaptation to the gut ecosystem.5,42 This distinct feature also suggests the difficulty for these intestinal microbes to survive in other environments where available nutrients and surrounding conditions are different in quantity and quality from those in the gut environment. Therefore, intestinal microbes may have additional properties tolerant to transient, but harsh conditions encountered in the mouth and stomach, through which they must travel to reach the gut. Genes encoding these yet unidentified functions may be included in the 315 gut-enriched COGs that contain many conserved but function-unknown genes, accounting for nearly 30% of all gut-enriched COGs, and in the 647 novel gene families identified.37
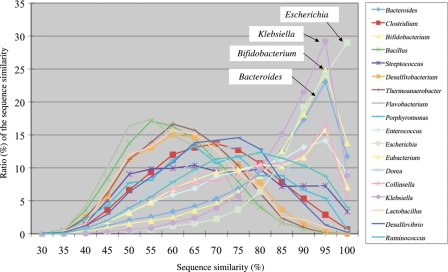
About 75% of the genes annotated in the metagenomic sequences of the 13 human intestinal microbiomes showed sequence similarity ranging from 40 to 100% ID at the amino acid level with known genes.37 When, for each gene, the best blastp-hit was used to tentatively assign the gene to a species, the distribution of sequence similarity for each species assigned could be depicted as shown in Fig. 2. Fig. 2 shows two distinct distribution patterns; those that peaked at high similarity of over 80% ID and those with low similarity between 50 and 80% ID. Typical species with the first pattern are Escherichia, Klebsiella, Bifidobacterium and Bacteroides, all of which were previously isolated from humans and fully sequenced, so it is reasonable that many of the metagenomic genes showed high sequence similarity with those in these microbes. On the other hand, the metagenomic genes showing low similarity with known genes in tentatively assigned species, such as Bacillus, Clostridium and Streptococcus, probably originate from species that are close to the assigned taxa but have not yet been isolated or sequenced. These data suggest that human intestinal microbes constitute distinct phylogenies from those of other environmental bacteria and have evolved with their own unique histories including co-evolution with the human host and its ancestors.2,5
Figure 2.
Distribution of sequence similarity of genes identified in the human intestinal microbiome. Only the data for the species that had sufficient numbers of best blastp-hit to known genes were represented. Species names are indicated in the box. Of these, four typical distribution patterns, which peaked at high sequence similarity (≥80% ID), are indicated by name in the figure.
5. Issues to be considered in 16S and metagenomic studies
In both cases of 16S rRNA phylotyping and metagenomic analysis, establishment of a reliable method for microbial DNA isolation from any given microbiota is a critical issue. This is because the intestinal microbiota comprise Gram-positive and Gram-negative bacteria and a small fraction of archaea, some of which may be hard-to-lyse species. So far, methods based on mechanical disruption using zirconium/silica beads11,22,36,45 and those based on enzymatic lysis9,25,37,46,47 have been developed and employed for the isolation of microbial DNA from various bacterial sources.48–52 The DNA extraction method has been assessed with respect to the quality and quantity of obtained DNA as well as fecal sample preservation.53 Our group recently assessed and compared the published lysis methods using fecal samples. Our preliminary results, including UniFrac54 analysis of enumerated 16S data, shape and yield of obtained DNA, showed that many of the methods examined gave similar results in quality, but varied by more than 10-fold in quantity and in the degree of DNA fragmentation (data not shown).
The 16S analysis also has an intrinsic problem in quantitative evaluation of the microbial composition because of the existence of multiple heterogeneous copies of the 16S rRNA genes within a genome along with uneven PCR-amplification of the 16S region. The range in copy number of the 16S rRNA gene varies from 1 to as many as 15 in prokaryotic species.55 In addition, the PCR primer Bact-8F,56 often used for amplification of the nearly full-length 16S sequence, might not be suitable at least for the quantification of Bifidobacteria, of which the 16S sequence has three base mismatches with the primer, underscoring the composition of this species.25 Recently, an improved primer pool was developed.57 Next-generation DNA sequencers guarantee the rapid collection of genomic data but provide less read-length than that of conventional Sanger sequencers.35 The feasibility of pyrosequencing reads of 200–300 bases for the 16S phylotype analysis58 was evaluated by collecting 141 000 reads from rhesus macaque intestinal microbiota.59 The results showed high reproducibility of the phylogenetic assignments and similarity of the major types and relative numbers of taxa to those obtained from Sanger sequences.
6. Functions of the gut microbiota
Deciphering biological features of a taxonomically complex and ecologically dynamic microbial community is a challenging issue in gut microbiome research. Germ-free and gnotobiotic mice,60 pig61 and zebrafish62 provide simplified model ecosystems that allow detailed evaluation of functions of colonized microbiota or microbes and the corresponding host responses in vivo,63,64 as well as their impact on various host physiologies.11,65–69 Immunity in germ-free mice is premature, but is restored by colonization of commensal bacteria and even by a single microbe.70,71 Protection against pathogenic infection is also a characteristic feature of commensal bacteria.72–74
Investigations have been made regarding the functions of commensal bacterial genes on their colonization in the mouse intestine. Flexible transcriptional regulation for adaptation to changes in available nutrients, including those during weaning, was found in Bacteroides thetaiotaomicron.39,75 Surface glycans expressed by Bacteroides fragilis are essential for its colonization.76 The analysis of certain Lactobacillus strains, which are thought to have health-promoting properties as probiotics, identified genes inducible upon their colonization.77 Expression profiling of both bacterial and host genes in mono-associated mice colonized by either Bifidobacterium longum or B. thetaiotaomicron and di-associated mice colonized by both bacteria were examined.78 The co-existence of B. longum expanded diversification in the carbohydrate substrates accessed by B. thetaiotaomicron in a host-independent manner. On the other hand, the presence of B. longum significantly reduced the expression levels of host genes responsible for antimicrobial activity against Gram-positive bacteria compared with that by B. thetaiotaomicron alone, suggesting the involvement of host responses in competitive colonization between these bacteria.
Commensal bacteria share many indistinguishable features with pathogenic bacteria relating to host immune response.79–81 For example, lipopolysaccharide (LPS), lipoteichoic acid (LTA) and peptidoglycan, major cell wall components of all bacteria, are well known ligands recognized by membrane-bound toll-like receptor 4 (TLR4) and TLR2 (TLR2), that serve as sensors of bacterial infection and lead to the production of pro-inflammatory cytokines such as TNFα and IL-6.82 Commensal bacteria have the ability to activate pro-inflammatory responses leading to harmful effects on the host via TLR signaling in mice lacking IL-10,83 an anti-inflammatory cytokine.84 Commensal bacteria also have the ability to activate anti-inflammatory responses leading to beneficial effects on the host via TLR signaling when the cascade to pro-inflammatory responses is lacking.85 These results indicate that the impact of intestinal microbiota on the host physiology largely depends on the state of host immunity and that host-commensal bacteria interactions are considered to be placed at the exquisitely equilibrated state between pro-inflammatory and anti-inflammatory responses,81,86 where the host preserves intestinal microbes while still being able to sense the bacteria that penetrate across intestinal borders.
Mammalian epithelia including Paneth and dendric cells are major sources of endogenous antimicrobial substances, including: lysozymes, phospholipases and various antimicrobial peptides such as α-defensins,87 angiogenin488 and RegIIIγ.89 Several studies have examined the relationships between these antimicrobial peptides and their effect on various intestinal microbes. RegIIIγ is a C-type lectin, of which the expression is mediated by TLR signaling90 and down-regulated by a Gram-positive Bifidobacterium strain.78 The expression of mouse cryptdins, a counterpart of human α-defensins, requires Nod2 (nucleotide-binding oligomerization domain containing 2), a cytoplasmic pattern recognition receptor expressed in Paneth cells that senses for bacterial peptidoglycans.91,92 Mutations in the Nod2 gene are highly correlated with the etiological risk of a subset of Crohn’s disease (CD).14,93 The expression of α-defensins at the ileal of patients with CD of the ileal was significantly down-regulated, but not at the ileal of patients with CD of the colon.94 Certain Enterococcus strains have been shown to regulate the phosphorylation of peroxisome proliferators-activated receptor γ (PPARγ) to induce the expression of downstream target genes including interleukin-10 (IL-10).95 PPARγ-deficient mice exhibited dysfunction on the maintenance of gut homeostasis.96 Bacteroides thetaiotaomicron induced PPARγ-mediated cytoplasmic re-distribution of the NF-κB subunit RelA in intestinal cells, selectively attenuating the inflammatory response.97 These findings indicate that PPARγ is a nuclear factor associated with anti-inflammatory response. The expression of antimicrobial cathelicidin LL-37 is induced by butyrate, the product of polysaccharide fermentation by intestinal microbes.98 These antimicrobial peptides may suppress microbial overgrowth and excessive contact of bacteria to the epithelia by directly killing them, resulting in minimizing inadequate stimulation of inflammatory responses.80,99
Some antimicrobial peptides were shown to dramatically increase in expression during the post-natal period.88,89,99,100 And, it has been shown that TLR signaling by LPS is activated in vaginally delivered newborn mice immediately after birth but not in newborns delivered by Cesarean section.101,102 These findings suggest that exposure of maternal-derived commensals to the intestinal epithelia in neonates is involved in initiating the development of intestinal homeostasis. Secreted (or mucosal) IgA produced by gut-associated lymphoid tissues is largely involved in shaping the intestinal microbiota composition, whereas lack of IgA expression can lead to adaptive immune response.103–105 Intestinal alkaline phosphatase (IAP) was found to dephospholyrate the phosphate moiety in LPS, resulting in detoxification of LPS and prevention of bacterial penetration across the epithelial barrier, suggesting that IAP plays an important role in maintenance of gut homeostasis.106,107 Recently, a study on gene expression of the TLR4-mediated signaling cascade in LPS-stimulated macrophages identified two classes of genes: those responding only to initial LPS stimulation and those responding to repeated LPS stimulation. The former class of genes includes pro-inflammatory cytokine genes, whereas the latter includes antimicrobial genes such as cathelicidin-related peptides. Histone modifications are involved in this regulation,108 which could also be linked to disease susceptibility from environmental factors.109
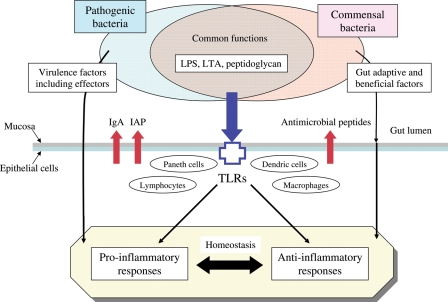
In summary, the host has evolved to establish many processes that sustain unresponsiveness toward the commensal bacteria while at the same time maintaining responsiveness toward pathogens (Fig. 3). These processes include the production of IgA, IAP and various antimicrobial peptides and epigenetic control of pro-inflammatory responses, all of which sever routes leading to excessive inflammatory response. On the other hand, pathogens also have evolved to equip various virulence factors, including effectors, that confer additional abilities for evading the host defense system, eventually inducing pro-inflammatory responses79,110 via change of the microbiota composition in favor of the pathogens.72 In contrast, commensal bacteria may also have evolved not only to acquire specific functions adaptive to the gut habitat, e.g. carbohydrate metabolism, energy production, cell maturation and proliferation toward intestinal homeostasis,111 but also to eliminate undesired appendages that could result in sensing for pro-inflammatory responses, e.g. profound depletion of genes for cell motility function in the metagenomic data of human intestinal microbiomes37 and attenuation of host immune response by loss of flagellar function.112
Figure 3.
Molecular interactions at the frontline between the host, intestinal commensal bacteria and pathogenic bacteria. Commensal bacteria possess specific functions adaptive to the gut habitat and are beneficial to host cells, including maintenance of intestinal homeostasis, but also include functions such as those of TLRs that signal immune responses to pathogenic infection.81,86 Host cells produce various antimicrobial substances such as IgA, IAP and antimicrobial peptides at the frontline to suppress excessive immune response to commensal bacteria, while maintaining responsiveness to pathogens equipped with various virulence factors to evade the host defense system.
7. HMP and future directions
The HMP aims at a better understanding of the roles of human microbes on human biology including their relationship with health and disease.7 The project includes metagenomic and 16S analysis of the microbiota inhabiting various body sites such as the oral and nasal cavities, the gastrointestinal and urogenital tracts, and skin in several hundreds of healthy and disease-afflicted subjects, as well as genome sequencing of nearly 1000 human commensal microbes. In addition, various metadata concerning the host are needed, including variations in host genotypes113 and metabolic phenotypes114 that largely influence host–microbe interactions. In this regards, one challenging issue is the construction of an integrated metabolic map, of both human and microbiota,115 which will become the new frontier for medical purposes such as the development of biomarkers for prediction of disease predisposition of individuals, extensive drug design targeting the intestinal microbiota and personalized drug therapies.116,117
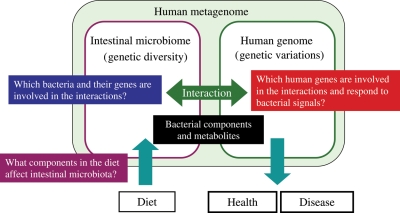
A simplified model system using gnotobiotic animals has provided fundamental knowledge of the molecular mechanisms involved in intestinal host–microbe interactions, in which active bacterial components have been identified.118,119 However, an enormous number and variety of bacterial components and products must participate in these interactions, and most remain unknown. Future studies will include those to explore and identify intestinal bacteria and their gene products (including metabolites) that are involved in host–microbial interactions, to identify human genes that respond to bacterial signals crucial for human physiology and to identify dietary components that influence and shape the intestinal microbiota composition. These scientific challenges will be achieved by using advanced ‘omics’ technologies coupled with the vast quantities of genomic data that are already being accumulated by the HMP. Thus, the human intestinal microbiome will pave the way leading to a new frontier in human biology, in which the human genome and the intestinal microbiome are tightly linked together as an integral part of the ‘human metagenome’ (Fig. 4).
Figure 4.
Future direction of human intestinal microbiota research. Human intestinal microbiota function is responsible for both human health and disease in accordance with its own genetic diversity and in association with human genetic variation. The study of human microbes, especially the vastly abundant intestinal microbes, is a new frontier in human biology. Many questions remain to be answered about host–microbe interactions, including: what factors and dietary components shape microbiota diversity, which bacteria and their components interact with host cells, which human genes respond to and how do they react to bacterial signals affecting human physiology.
Funding
This research was supported in part by the Grant-in-Aid for Scientific Research on Priority Areas ‘Comprehensive Genomics’ (17020007) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Acknowledgements
The authors thank all members of the Human MetaGenome Consortium Japan (HMGJ) for collaborative research on the human intestinal microbiota and microbiome.
Footnotes
Edited by katsumi Isono
References
- 1.Savage D. C. Microbial ecology of the gastrointestinal tract. Ann. Rev. Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 2.Dethlefsen L., et al. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lederberg J. Infectious history. Science. 2000;288:287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 4.Bäckhed F., et al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Ley R. E., et al. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 6.Dethlefsen L., et al. Assembly of the human intestinal microbiota. Trends Ecol. Evol. 2006;21:517–523. doi: 10.1016/j.tree.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh P. J., et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooper L. V., et al. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 9.Eckburg P. B., et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ley R. E., et al. Obesity alters gut microbial ecology. Nature. 2006;444:1009–1010. [Google Scholar]
- 11.Turnbaugh P. J., et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 12.Frank D. N., et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xavier R. J., et al. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 14.Schreiber S., et al. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat. Rev. Genet. 2005;6:376–388. doi: 10.1038/nrg1607. [DOI] [PubMed] [Google Scholar]
- 15.Peterson D. A., et al. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manichanh C., et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penders P., et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56:661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacDonald T. T., et al. Immunity, inflammation, and allergy in the gut. Science. 2005;307:1920–1925. doi: 10.1126/science.1106442. [DOI] [PubMed] [Google Scholar]
- 19.Penders J., et al. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62:1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 20.McGarr S. E. Diet, anaerobic bacterial metabolism, and colon cancer: a review of the literature. J. Clin. Gastroenterol. 2005;39:98–109. [PubMed] [Google Scholar]
- 21.Moore W. E. C., et al. Intestinal floras of populations that have a high risk of colon cancer. Appl. Environ. Microbiol. 1995;61:3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuki T., et al. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 2004;70:7220–7228. doi: 10.1128/AEM.70.12.7220-7228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoetendal E. G., et al. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heilig H. G. H. J., et al. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 2002;68:114–123. doi: 10.1128/AEM.68.1.114-123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer C. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:1556–1573. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Favier C. F., et al. Development of bacterial and bifidobacterial communities in feces of newborn babies. Anaerobe. 2003;9:219–229. doi: 10.1016/j.anaerobe.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Favier C. F., et al. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 2002;68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart J. A., et al. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J. Med. Microbiol. 2005;54:1239–1242. doi: 10.1099/jmm.0.46189-0. [DOI] [PubMed] [Google Scholar]
- 29.Matsumiya Y., et al. Molecular epidemiological study of vertical transmission of vaginal Lactobacillus species from mothers to newborn infants in Japanese, by arbitrarily primed polymerase chain reaction. J. Infect. Chemother. 2002;8:43–49. doi: 10.1007/s101560200005. [DOI] [PubMed] [Google Scholar]
- 30.Martín R., et al. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003;143:754–758. doi: 10.1016/j.jpeds.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Tringe S. G., et al. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 2005;6:805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 32.Weng L., et al. Application of sequence-based methods in human microbial ecology. Genome Res. 2006;16:316–322. doi: 10.1101/gr.3676406. [DOI] [PubMed] [Google Scholar]
- 33.Noguchi H., et al. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorek R., et al. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- 35.Mardis E. R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Gill S. R., et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurokawa K., et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flint H. J., et al. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature Rev. Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 39.Bjursell M. K., et al. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the sucking period. J. Biol. Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- 40.Xu J., et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 41.Xu J., et al. Honor thy symbionts. Proc. Natl. Acad. Sci. USA. 2003;100:10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J., et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:1574–1586. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saier M. H., Jr, et al. Probiotics and prebiotics in human health. J. Mol. Microbiol. Biotechnol. 2005;10:22–25. doi: 10.1159/000090345. [DOI] [PubMed] [Google Scholar]
- 44.Samuel B. S., et al. A humanized gnotobiotic mouse model of host–archaeal–bacterial mutualism. Proc. Natl. Acad. Sci. USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zoetendal E. G., et al. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat. Protoc. 2006;1:870–873. doi: 10.1038/nprot.2006.142. [DOI] [PubMed] [Google Scholar]
- 46.Pei Z., et al. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. USA. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morita H., et al. An improved DNA isolation method for metagenomic analysis of the microbial flora of the human intestine. Microbes Environ. 2007;22:214–222. [Google Scholar]
- 48.Aas J. A., et al. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bik E. M., et al. Molecular analysis of the bacteria microbiota in the human stomach. Proc. Natl. Acad. Sci. USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao Z., et al. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grice E. A., et al. A diversity profile of the human skin microbiota. Genome Res. 2008 doi: 10.1101/gr.075549.107. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin R., et al. Diversity of the Lactobacillus group in breast milk and vaginal of healthy women and potential role in the colonization of the infant gut. J. Appl. Microbiol. 2007;103:2638–2644. doi: 10.1111/j.1365-2672.2007.03497.x. [DOI] [PubMed] [Google Scholar]
- 53.Nechvatal J. M., et al. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J. Microbiol. Methods. 2008;72:124–132. doi: 10.1016/j.mimet.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 54.Lozupone C., et al. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Acinas S. G., et al. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 2004;186:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edwards U., et al. Isolation and direct complete nucleotide determination of entire gene: characterization of a gene encoding for a 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frank J. A., et al. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008;74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Z., et al. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acid Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKenna P., et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bry L., et al. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 61.Meurens F., et al. Commensal bacteria and expression of two major intestinal chemokines, TECK/CCL25 and MEC/CCL28, and their receptors. PLoS One. 2007;7:e677. doi: 10.1371/journal.pone.0000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rawls J. F., et al. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hooper L. V., et al. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hooper L. V., et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 65.Bäckhed F., et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bäckhed F., et al. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dumas M. -E., et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. USA. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crawford P. A., et al. Microbial regulation of intestinal radiosensitivity. Proc. Natl. Acad. Sci. USA. 2005;102:13254–13259. doi: 10.1073/pnas.0504830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stappenbeck T. S., et al. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andrew J., et al. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology. 2005;115:153–162. doi: 10.1111/j.1365-2567.2005.02159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mazmanian S. K., et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 72.Stecher B., et al. Salmonella enterica Serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Asahara T., et al. Probiotic Bifidobacteria protect mice from lethal infection with Shiga toxin-producing Escherichia coli O157:H7. Infect Immun. 2004;72:2240–2247. doi: 10.1128/IAI.72.4.2240-2247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hudault S., et al. Escherichia coli strains colonising the gastrointestinal tract protect germfree mice against Salmonella typhimurium infection. Gut. 2001;49:47–55. doi: 10.1136/gut.49.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sonnenburg J. L., et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 76.Liu C. H., et al. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc. Natl. Acad. Sci. USA. 2008;105:3951–3956. doi: 10.1073/pnas.0709266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bron P. A., et al. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 2004;186:5721–5729. doi: 10.1128/JB.186.17.5721-5729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sonnenburg J. L., et al. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raskin D. M., et al. Bacterial genomics and pathogen evolution. Cell. 2006;124:703–714. doi: 10.1016/j.cell.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 80.Pamer E. G. Immune responses to commensal and environmental microbes. Nat. Rev. Immunol. 2007;8:1173–1178. doi: 10.1038/ni1526. [DOI] [PubMed] [Google Scholar]
- 81.Sansonetti P. J. War and peace at mucosal surfaces. Nat. Rev. Immunol. 2004;4:953–964. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 82.Akira S., et al. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 83.Kühn R., et al. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 84.Fiorentino D. F., et al. IL-10 inhibits cytokine production by activated macrophages. J. Immunol. 1991;147:3815–3822. [PubMed] [Google Scholar]
- 85.Rakoff-Nahoum S., et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 86.Sansonetti P. J. The innate signaling of dangers and the dangers of innate signaling. Nat. Immunol. 2006;7:1237–1242. doi: 10.1038/ni1420. [DOI] [PubMed] [Google Scholar]
- 87.Ayabe T., et al. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 88.Hooper L. V., et al. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 89.Cash H. L., et al. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brandl K., et al. MyD88-mediated signals induce the bactericidal lectin RegIIIγ and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kobayashi K. S., et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 92.Ogura Y., et al. Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut. 2003;52:1591–1597. doi: 10.1136/gut.52.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ogura Y., et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 94.Wehkamp J., et al. Reduced Paneth cell α-defensins in ileal Crohn’s disease. Proc. Natl. Acad. Sci. USA. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Are A., et al. Enterococcus faecalis from newborn babies regulate endogenous PPARγ activity and IL-10 levels in colonic epithelial cells. Proc. Natl. Acad. Sci. USA. 2008;105:1943–1948. doi: 10.1073/pnas.0711734105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Adachi M., et al. PPARγ in colonic epithelial cells protects against experimental inflammatory bowel disease. Gut. 2006;55:1104–1113. doi: 10.1136/gut.2005.081745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kelly D., et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat. Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 98.Raqib R., et al. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotics. Proc. Natl. Acad. Sci. USA. 2006;103:9178–9183. doi: 10.1073/pnas.0602888103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peschel A., et al. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 100.Ménard S., et al. Developmental switch of intestinal antimicrobial peptide expression. J. Exp. Med. 2008;205:183–193. doi: 10.1084/jem.20071022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lotz M., et al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 2006;203:973–984. doi: 10.1084/jem.20050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kelly D., et al. Importance of microbial colonization of the gut in early life to the development of immunity. Mutation Res. 2007;622:58–69. doi: 10.1016/j.mrfmmm.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 103.Fagarasan S., et al. Intestinal IgA synthesis: regulation of front-line body defences. Nat. Rev. Immunol. 2003;3:63–72. doi: 10.1038/nri982. [DOI] [PubMed] [Google Scholar]
- 104.Suzuki K., et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. USA. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peterson D. A., et al. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 106.Bates J. M., et al. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Goldberg R. F., et al. Intestinal alkaline phosphatase is a gut mucosal defense factor maintained by enteral nutrition. Proc. Natl. Acad. Sci. USA. 2008;105:3551–3556. doi: 10.1073/pnas.0712140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Foster S. L., et al. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 109.Jirtle R. L., et al. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ogawa M., et al. The versatility of Shigella effectors. Nat. Rev. Microbiol. 2008;6:11–16. doi: 10.1038/nrmicro1814. [DOI] [PubMed] [Google Scholar]
- 111.Rawls J. F., et al. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rawls J. F., et al. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc. Natl. Acad. Sci. USA. 2007;104:7622–7627. doi: 10.1073/pnas.0702386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Linzmeier R. M., et al. Human defensin gene copy number polymorphisms: comprehensive analysis of independent variation in α- and β-defensin regions at 8p22–p23. Genomics. 2005;86:423–430. doi: 10.1016/j.ygeno.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 114.Holmes E., et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nicholson J. K., et al. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 116.Jia W., et al. Gut microbiota: a potential new territory for drug targeting. Nat. Rev. Drug Discov. 2008;7:123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- 117.Clayton T. A., et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 118.Mazmanian S. K., et al. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 119.Mazmanian S. K., et al. The love–hate relationship between bacterial polysaccharides and the host immune system. Nat. Rev. Immunol. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]