Abstract
Purpose
Ocular infections caused by fungal organisms can cause significant ocular morbidity, particularly when diagnosis and treatment are delayed. Rapid and accurate identification of Fusarium species at the subgenus level using current diagnostic standards is timely and insensitive. The purpose of this study is to examine the usefulness of polymerase chain reaction (PCR) analysis of the internal transcribed spacer (ITS) regions (ITS1, 5.8S, and ITS2) in detecting and differentiating Fusarium species from isolates of ocular infections, and to assess the correlation between the genotypic and morphologic classification.
Methods
Fifty-eight isolates from 52 patients diagnosed with Fusarium ocular infections were retrieved from storage at the Bascom Palmer Eye Institute’s ocular microbiology laboratory. Morphologic classification was determined at both a general and a reference microbiology laboratory. DNA was extracted and purified, and the ITS region was amplified and sequenced. Following DNA sequences, alignment and phylogenetic analysis were done. Susceptibility to antifungal drugs was measured according to the Clinical and Laboratory Standards Institute reference method.
Results
Sequence analysis demonstrated 15 unique sequences among the 58 isolates. The grouping showed that the 58 isolates were distributed among 4 main species complexes. At the species level, morphologic classification correlated with genotypic classification in 25% and 97% of the isolates in a general microbiology and a reference mycology laboratory, respectively.
Conclusions
The sequence variation within the ITS provides a sufficient quantitative basis for the development of a molecular diagnostic approach to the Fusarium pathogens isolated from ocular infections. Morphology based on microscopic and macroscopic observations yields inconsistent results, particularly at nonreference laboratories, emphasizing the need for a more reproducible test with less user-dependent variability. Fusarium solani tends to be more resistant to certain antifungals (azoles).
INTRODUCTION
Infections of the eye caused by fungi are usually associated with significant loss of visual function. Keratomycosis, or fungal keratitis, can have a deleterious effect on the eye. Despite enormous advances in the field of infectious diseases, the identification of fungi as the cause of an infection is difficult to establish in cases of presumed microbial keratitis because of the nonspecific clinical signs and symptoms and the difficulties encountered in the isolation of these microorganisms in the microbiology laboratory. Delays in identification of fungal pathogens often lead to advanced disease and the timely use of targeted antifungal therapy. To further compound the dilemma, treatment of keratomycosis can be difficult given the paucity of commercial antifungal ophthalmic agents and the not-well-established criteria for antifungal sensitivity testing.
INCIDENCE AND RISK FACTORS
Although still rare compared to bacterial keratitis, keratomycosis seems to be diagnosed more often now than in the past, and its incidence may be on the rise.1 Recent literature indicates that soft contact lens wearers, previously thought to be protected from fungal disease, may be particularly at risk for developing fungal keratitis.2 Other risk factors, such as increased use of corticosteroids and broad-spectrum antibiotics, are also likely to play a role. New classes and newer generations of antifungal therapies are being developed and may be important in combatting this sight-threatening infectious disease.3,4
Fungi are known to be ubiquitous organisms. They make up a portion of the normal flora and are found in 3% to 28% of healthy human conjunctivas.5 Fungal ophthalmic pathogens are usually classified into groups on the basis of laboratory morphologic criteria. Candida species are yeasts and are one of the predominant species found in the northern United States. Septated, nonpigmented filamentous hyphal fungi include Aspergillus, the most common organism isolated worldwide, and Fusarium, the most common organism isolated in the southern United States.1 Pigmented filamentous fungi include Curvularia and Alternaria; whereas Mucor and Rhizopus are nonseptated filamentous fungi.
An intact corneal epithelium serves as a barrier to mycotic penetration into the corneal stroma. Defects of this epithelial barrier will allow fungal organisms to penetrate into the corneal stroma, where the host inflammatory response, fungal proliferation, and production of mycotoxins lead to tissue necrosis. Keratomycosis can spread peripherally into sclera or can penetrate an intact Descemet membrane and gain access to the anterior chamber with subsequent development of endophthalmitis.6 Invasion of the lens by penetration through an intact lens capsule can also occur. Fungi can grow into the trabecular meshwork, causing obstruction of aqueous outflow, leading to glaucoma. In addition, fungal mycotoxins can cause a trabeculitis, leading to intractable glaucoma.7 This disease progression can be difficult to arrest, given the avascularity of the cornea, sclera, and anterior chamber, thus decreasing the access of immune defense mechanisms as well as penetration of systemically administered antifungals.
Fusarium has been associated with invasive disease, although this characteristic may not be unique. Histopathologic specimens obtained at the time of penetrating keratoplasty, early in the course of keratomycosis, tend to show marked fungal hyphal proliferation with a comparatively minimal inflammatory response. Conversely, corneal buttons, obtained later in the course of keratomycosis, reveal fewer hyphae with a more vigorous inflammatory infiltration.8 Clinically, Fusarium exhibits a similar dichotomous tendency for either aggressive, rapidly enlarging infiltration or indolent, slowly progressing disease.2 Further study into these mechanisms and behaviors is needed, in particular, better species classification of the organisms.
CAUSES
The increase in the incidence of keratomycosis is likely due to a number of recent changes in the contributing risk factors. These risk factors for fungal keratitis can be categorized into 2 areas: factors that disrupt the corneal epithelium and factors that impair the immunologic response to mycosis. Trauma remains the most common risk factor for fungal keratitis. The most commonly encountered history is exposure to outdoor organic matter, such as plants and soil, leading to the development of ocular disease.1 Presumably, the traumatic disruption of corneal epithelium exposes vulnerable stroma to materials contaminated with fungal elements. Given the ubiquity of fungi, direct contact with organic material is not a requirement, as metallic and other foreign bodies may also be involved.
Breakdown of the corneal epithelium due to ocular surgery can also lead to fungal keratitis. Patients who undergo penetrating keratoplasty will be exposed to additional risk factors, including immunosuppression due to the use of corticosteroids or cyclosporin A, contact lens wear, persistent corneal epithelial defects, and broad-spectrum antibiotic use.9 Patients who undergo incisional refractive surgery, such as LASIK or keratotomies, are also at risk.10 The access of fungal organisms to the corneal stroma through the surgical defect in Bowman’s layer may play a role.
Recently, there has been an increase in fungal ulcers in patients wearing soft contact lenses. Previously, soft contact lens wear was associated with a lower rate of fungal corneal ulcers compared to no contact lens wear.11 A potential protective effect from the soft contact lens and fewer associated risk factors in soft contact lens wearers were presumed mechanisms for this lower incidence. However, evidence has emerged incriminating soft contact lens wear in an increase in fungal keratitis, particularly from Fusarium.2 Contact lens solutions have been implicated in the rise, and ReNu with MoistureLoc (Bausch & Lomb, Rochester, New York) was withdrawn from the market for this reason. These solutions may play a role in the disease process, although the details of the interaction have not been fully elucidated. Studies suggest a complex interaction of preservatives capable of deactivating antiseptics used in some lens storage solutions.12 The additional microtrauma to the corneal epithelium by the contact lens and the toxicity of the solution, combined with poor lens care,12 likely have a multifactorial effect, predisposing the eyes to fungal infections.
Risk factors that can affect the normal environment of the ocular surface can lead to fungal infections. Conditions such as lagophthalmos and neurotrophic ulceration can cause a weakened ocular surface and are risk factors. Chronic inflammation of the cornea, such as that seen in dry eye, vernal keratoconjunctivitis, and herpetic eye disease, is also associated with fungal keratitis.13
A link has been established between fungal keratitis and patients with systemic diseases associated with immunosuppression, such as diabetes mellitus and human immunodeficiency virus, which may predispose to keratomycosis.14 Localized immunosuppression caused by the use of topical corticosteroids has been linked to the onset and exacerbation of fungal keratitis.15 As corticosteroids act to reduce the production of phagocytes and the ingestion by phagocytes, as well as inhibit chemotaxis and degranulation,16 their immunomodulatory effects can deter adequate fungal elimination. Corticosteroids appear to alter mycotic strains as well, increasing their pathogenicity to human tissues.17 Future studies will continue to look at the association of corticosteroids and fungal keratitis. It is not known which Fusarium species are more prone to be affected by exposure to corticosteroids.
DIAGNOSIS
Because it may be difficult to obtain laboratory verification of the diagnosis, clinical suspicion of fungal keratitis is helpful. Presenting signs and symptoms can be very similar to those of any infectious keratitis, namely, foreign body sensation, pain, redness, and/or decreased vision. At times, signs and symptoms of fungal keratitis present in indolent fashion over several days.
Some of the clinical features of fungal keratitis can suggest the diagnosis. Mycotic infiltrates exhibit irregular, feathery margins with or without an epithelial defect. Satellite lesions, branching patterns, ring infiltrates, endothelial plaques, and anterior chamber reaction with hypopyon are highly suggestive of a fungal etiology. Fungal infiltrates may be deep in the stroma without an overlying epithelial defect. Pigmented hyphae will produce a brownish pigmentation to infiltrates in the cornea, which can aid in diagnosis. As with subjective symptoms, associated inflammatory signs, such as conjunctival injection and lid edema, can be underwhelming compared to the area of corneal infiltration.
Although the presence of risk factors and the associated clinical signs and symptoms may suggest the diagnosis of fungal keratitis, significant overlap exists with microbial keratitis of other etiologies. Fungal keratitis can present without identifying clinical features, and other causes of keratitis can mimic fungal-like clinical features. Basing the diagnosis on clinical findings alone can lead to erroneous treatment.18 The cornerstone of diagnosis remains corneal scraping and culture with the assistance of a microbiology laboratory.
Advantages of corneal scrapings for laboratory investigation include procurement of adequate culture specimen and debridement of the epithelium for better antifungal penetration and decreased fungal load. Corneal scraping with a spatula or surgical blade is recommended to achieve deeper tissue access and appropriate debridement, which may not be achievable through Dacron or calcium alginate swabbings.
In the last 50 years, laboratory diagnosis has been achieved through Gram and Giemsa staining of smears. Reports on sensitivity and specificity of staining methods are varied, but initial recovery of fungal pathogens by corneal smear can be as high as 78%.19 Potassium hydroxide wet-mount preparation can also be performed, often in an office setting for immediate diagnosis.
Material from a corneal scraping should be plated on culture media. Culture plates should include routine media used in infectious keratitis. The initial microbiologic corneal culture should include sheep blood agar, chocolate agar, Sabouraud dextrose agar, and thioglycolate broth. Brain-heart infusion medium may be substituted for Sabouraud agar. Fungal media should be kept at room temperature, and plastic bags may be used to enhance humidity levels, which can aid in fungal growth. Fungi may be identified in culture as soon as 72 hours.1 In the setting of an imbedded foreign body, contact lens wear, or suture abscess, evaluation with cultures of these offending materials can be instrumental in obtaining culture positivity. In contact lens–related keratitis, all paraphernalia, including contact lens, lens case, and lens solution, should be sent to the microbiology laboratory for investigation.
One of the limiting problems of corneal scrapings is the small sample size, making isolation of infectious agents difficult. Newer methods of identifying mycotic pathogens in these small samples include immunofluorescence and polymerase chain reaction (PCR). While sensitivity is high and time to diagnosis is reduced, expense and difficulty of implementation limit their use.20 More commonly, in the setting of repeated negative culture results, the next step in evaluation entails corneal biopsy in order to obtain a larger sample size of the infected tissue.
A larger-sized tissue sample can be obtained by performing a diagnostic superficial keratectomy or corneal biopsy. It can usually be performed at the slit lamp with use of a topical anesthetic. Using a 2- to 3-mm sterile disposable dermatologic trephine, tissue with infiltration, as well as adjacent clear stroma, is trephinated at partial thickness, avoiding the visual axis if possible. The lamellar keratectomy is completed with a surgical blade to undermine the trephinated area. Both microbiologic and histopathologic examination should be undertaken, as direct examination of involved tissue is often superior to culture in the setting of keratomycosis.21
Recently, new imaging modalities have become useful in the diagnosis of fungal keratitis. Given the propensity for aggressive invasion of ocular structures, all cases of fungal keratitis with an inadequate posterior segment view should undergo diagnostic B-scan ultrasound to rule out endophthalmitis. Diagnostic corneal imaging in the form of confocal microscopy continues to gain clinical application, and it may play an adjunctive diagnostic role in keratomycosis.22 Because of the paucity of commercially available antifungals and their less than favorable pharmacologic properties, aggressive diagnostic studies are needed when antifungal therapy is being considered, and therapy should rarely be initiated without laboratory or supporting diagnostic confirmation.
TREATMENT
The available antifungal agents continues to evolve. Two primary classes of drugs—the polyenes and azoles—have had the most use, and recently a third class—the echinocandins—was introduced. Natamycin and amphotericin B are the principal polyenes. These agents disrupt fungal cytoplasmic membranes through binding of ergosterol, a sterol unique to fungi.23 While fungal membrane damage appears dose-related, human cell toxicity limits higher concentration levels.
Topical natamycin 5% (Alcon, Fort Worth, Texas) is the first-line agent in the treatment of fungal keratitis.1 It is the only commercially available ophthalmic antifungal agent. Its poor water solubility requires suspension. However, as formulated, natamycin 5% suspension is well tolerated, nontoxic to the cornea, and adherent to the cornea.24 Natamycin demonstrates in vitro activity against a variety of fungal pathogens, including Aspergillus, Candida, and Fusarium, which accounts for its use as a first-line agent in most cases of fungal keratitis.1
Amphotericin B acts as a fungal membrane disruptor, much like natamycin. Amphotericin B must be extemporaneously compounded and most frequently is formulated topically as 0.1% to 0.25%. Amphotericin B does not penetrate an intact epithelium, and thus debridement is crucial. Yeast infections, such as Candida, respond well to amphotericin B, making it a frequent first-line agent in this setting.25 Intracameral and intravitreal routes have been used, although with extreme caution given the drug’s proclivity for toxicity, including retinal necrosis. Intravenous infusion of amphotericin B should likewise be used with caution because of renal toxicity.24
The azoles have had the most activity in terms of new derivatitives of the older molecules. Newer-generation triazoles have followed the prior generation of imidazoles. All azoles act to inhibit cytochrome P-450 14α-demethylase, thereby increasing fungal cell membrane permeability and leading to fungal cell death. All azoles, excluding fluconazole, may also decrease tissue damage through inhibition of the inflammatory response, although this action may adversely affect their in vivo activity.23
Older azoles include miconazole and ketoconazole. Miconazole can be used topically, and subconjunctival administration appears to be better tolerated than with most other antifungal agents.26 Ketoconazole is readily available for oral administration. Newer azoles include fluconazole, a synthetic bis-triazole. Oral administration of fluconazole achieves high ocular tissue and fluid levels in animal models.27 Topical administration likewise results in high penetration in the absence of epithelium.28
Newer-generation triazoles are voriconazole and posaconazole. Voriconazole exhibits excellent in vitro susceptibility profiles against Candida, Aspergillus, and Fusarium,3 although clinical in vivo experience requires further investigation. The drug is available in oral and intravenous forms and can be compounded for topical ophthalmic use. Posaconazole likewise shows promise in the treatment of fungal infection and has activity against Candida strains less responsive to fluconazole. A case demonstrating resolution of ocular Fusarium following administration of posaconazole has been reported.4
An entirely new class of antifungal agents that inhibit fungal cell wall development, the echinocandins, has also been introduced. Caspofungin is in this new class, is available in intravenous formulation, and shows experimental promise for the treatment of keratitis.29
Given this array of choices, a treatment regimen for newly diagnosed keratomycosis can be approached as follows. Initial monotherapy with natamycin 5% is recommended and is likewise convenient, given its commercial availability. Aggressive initial dosing every hour, around the clock, is suggested. If worsening on initial therapy is noted, Candida may respond to the addition of amphotericin B 0.15% for combination therapy. Aspergillus may respond better by adding an azole, such as fluconazole, miconazole, or voriconazole, topically. Subconjunctival injection of antifungal medications is typically avoided because it results in extreme discomfort, with the exception of miconazole, and may be toxic. Voriconazole can also be used for intrastromal and intraocular injection. Cases of deep stromal involvement, scleritis, or endophthalmitis may benefit from systemic antifungal therapy with an azole, which demonstrates good corneal penetration. Ketoconazole is commonly used orally for this purpose, although fluconazole, voriconazole, and posaconazole may prove to have enhanced penetration and susceptibility profiles as further investigations are performed. Intravenous medical therapy can play a role in resilient, deeply infiltrating disease as well.
Once improvement of fungal ulcers with antifungal medications is noted, prolonged administration of therapy and careful observation are still needed. Typical fungal keratitis requires 30 to 40 days of therapy.1 Because of the persistence of fungal elements, the temptation to stop therapy quickly should be avoided, even in cases of presumed resolution. Adequate long-term administration of medication is essential to eradicate fungal pathogens. Given the extemporaneous nature of many of the agents used, toxicity is not uncommon and may mimic persistent disease. Therefore, cessation of therapy and careful observation may be necessary following 4 to 6 weeks of continuous treatment. Corticosteroids should be avoided until no infection activity has been noted for several weeks of observation off antifungal therapy.
As noted previously, corticosteroids have been implicated as a risk factor in the development of fungal keratitis, and their use should thus be generally avoided in this setting. Worsening of disease has been noted, despite their anti-inflammatory effects.1
Fungal ulcers remain an often virulent disease despite medical treatment, and surgical intervention may be warranted. Perforation may be treated with corneal glue to achieve tamponade continuing medical treatment in order to further eradicate the fungal organisms. Mycotic keratitis unresponsive to aggressive medical therapy deserves consideration of therapeutic penetrating keratoplasty, to aid in eliminating the fungal load. Keratoplasty should be pursued prior to scleral or vitreal involvement if possible, to avoid irreversible tissue invasion.1 As with other microbial keratitis, trephination should incorporate the entire involved area with an adjacent 1- to 1.5-mm clear zone to ensure adequate fungal eradication.21 Even with such wide excision, reinfection of the corneal graft has been reported in nearly 5% to 7% of cases.30 In cases of scleral involvement, cryotherapy applied to the sclera in a double freeze-thaw technique may be of benefit. Care in preserving lens integrity will reduce the likelihood of further posterior segment spread; however, lensectomy is recommended at the time of penetrating keratoplasty in cases of preexisting lenticular involvement.31 The surgical technique should involve avoiding recontamination of the new allograft tissue with instruments used to remove the infected tissue. As in the evaluation of corneal biopsy specimens, host corneal buttons harvested at the time of penetrating keratoplasty should be sent for both microbiologic and histopathologic examination to enhance fungal detection. Administration of intraocular antifungals should be done at the time of keratoplasty.
Following therapeutic penetrating keratoplasty, recurrence of infection is possible, and thus topical antifungal therapy should be maintained. Concomitant systemic antifungal therapy, if not already instituted, should be considered. Reinjection with intraocular antifungals should also be considered. Pathologic examination of the involved host corneal button that demonstrates margins clear of fungal organisms is reassuring, and typically antifungals can be stopped 2 weeks postoperatively in such cases. Evidence of intraocular or adjacent, unresected tissue spread is ominous and requires long-term therapy.
Controversy persists regarding the use of topical corticosteroids following therapeutic penetrating keratoplasty. Although maintenance of a clear graft postoperatively is beneficial, the reduction of recurrence is paramount. An optical regraft can be performed at a later date once the corneal condition is stabilized, and thus failure or rejection of the therapeutic graft is not catastrophic. Although use of corticosteroids may promote mycotic recurrence following penetrating keratoplasty,32 topical cyclosporin A has had more favorable outcomes in these fungal cases. In addition to its anti-inflammatory effects, topical cyclosporin A may exhibit direct fungicidal activity and a corneal allograft tolerance effect.33 As such, preference for topical cyclosporin A following penetrating keratoplasty for fungal keratitis should be considered over topical corticosteroid therapy. Once there is no clinical evidence of infection, usually after 4 weeks following the surgery, corticosteroids should be started to avoid a graft rejection.
CLASSIFICATION
Keratomycosis is a prevalent cause of ocular morbidity throughout the world.34 The number of patients affected has increased, and it has become a worldwide problem. The observed increased incidence may be due to the frequent use of topical corticosteroids and antibacterial agents, surgical procedures, inappropriate maintenance of contact lenses, trauma, chronic ocular diseases, corneal anesthetic abuse, and rise in the number of immunocompromised patients.13,35
The etiology of organisms causing keratitis is highly variable throughout the world, but fungal organisms are the etiologic agent in 1.2% to 62% of cases. Fungal keratitis is mostly caused by yeast and molds, molds being especially prevalent in subtropical and tropical regions.1,35 Among the molds, Fusarium and Aspergillus are the most common, with Fusarium representing 36% of infectious keratitis cases in South Florida.36
Classification of Fusarium at the subgenus level has been problematic, but increasing reports of Fusarium as a human pathogen in ocular infections has generated an interest in finding a more consistent basis for classification.37 Accurate identification of the etiologic agents causing keratomycosis is critical to rapid diagnosis. It will also help in advancing our understanding of each Fusarium species and may lead to a more rapid identification method, effective antifungal therapy, and improved clinical outcomes.38 This is particularly important for the Fusarium genus, for which minimum inhibitory concentrations (MICs) are higher and more variable than for other molds (eg, Aspergillus sp).39
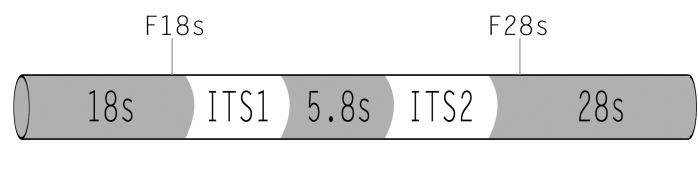
To reach these goals, we propose using traditional microscopic methods of species identification combined with DNA typing techniques: amplified fragment length polymorphism and DNA sequencing analysis of the internal transcribed spacer (ITS) and intergenic spacer (IGS) regions of the ribosomal DNA (rDNA) family. The ITS and IGS regions are the 2 most variable regions within the rDNA gene family, and both regions are widely used to elucidate variation in strain levels. These molecular techniques are extremely useful for tracing strains in epidemiologic studies by comparing genotypic patterns between the source and the infected individual. A clear consensus that has emerged from recent genotyping studies40–43 is that DNA sequence-based methods will be essential for species identification and subtyping of the Fusarium genus in clinical laboratories, especially within the pathogen-rich Fusarium solani and Fusarium oxysporum species complexes. The rDNA genes are good candidates to detect fungi from infection specimens, since they are multicopy genes, maximizing the sensitivity of the PCR. The transcriptional unit is composed of 18S, 5.8S, and 28S rDNA genes. Between the 18S and 5.8S and between 5.8S and 28S rDNA gene subunits are intergenic transcribed spacer regions (ITS 1 and 2) that are not translated into rRNA (Figure 1). Although rDNA genes are highly conserved, the ITS regions are variable and rich in informative sites, being good candidates for helping in the phylogenetic classification of Fusarium species.44–46
FIGURE 1.
Ribosomal DNA genes, transcribed spacer regions (ITS 1 and ITS2), and primer locations.
Most clinical laboratories rely on methods that employ phenotypic characteristics that can be time-consuming and not very accurate. With rapid advances in molecular biology, as well as in-house fungal databases and the public accessibility of microbial sequence data, the development of a rapid and simple assay with a high-throughput capability to identify all the species within the genus Fusarium becomes clinically desirable. To achieve this, accurate DNA sequencing data from ocular isolates is necessary to develop a rapid identification system. A novel rapid identification system is the commercially available Luminex (Luminex Corporation, Austin, Texas). This method uses a novel flow cytometer, the Luminex 100. Luminex xMAP technology uses multiple color fluorescent microspheres by varying the proportion of red and infrared fluorescent dyes within microspheres to create an array of up to 100 separate bead classifications, each of which can represent a single species. Each dye, which is applied in different proportions, generates a 10×10 colored palette with a total of 100 sets of unique colors that are visible to a laser. The polystyrene microspheres are coated with carboxyl groups, which bind covalently to species-specific nucleic acid probes by carbodiimide coupling using 1-ethyl-3-(dimethylaminopropyl) carbodiimide hydrochloride. The capture probe microsphere-based hybridization assay uses PCR-biotinylated amplicon target DNA, which is inoculated into the microsphere bead mixture containing species-specific probes of interest. By adding a reporter molecule (streptavidin R-phycoerythrin), all hybridized species-specific amplicons captured by their complementary nucleotide sequence on the microsphere beads are recognized by the fluorescence of the reporter molecule. The median fluorescence intensity of the reporter molecule is then used to quantify the amount of DNA bound to the beads. This provides a sensitive molecular method that is rapid and simple to perform. This assay represents the validation of the technology with fungal cultures and details the methods for probe design and testing. This technology, which was adapted to identify species within the genus Fusarium, can be expanded to include other pathogenic fungal species. To our knowledge, this is the first application of ocular isolates of Fusarium to develop the assay necessary to use the Luminex technology for the detection of this organism as the causative agent in human ocular infections. Future studies will address the potential application of the assay with clinical specimens.
The purposes of this study are (1) to determine whether or not genotypic data support the morphologic classification of Fusarium species isolates; (2) to determine whether sequence analysis of ITS 1 and 2 and 5.8S rDNA can be used to detect and classify Fusarium from patients with ocular infections, using as the next step the Luminex system; and (3) to compare genotypic data with sensitivity data to antifungals.
METHODS
FUNGAL ISOLATES
A total of 58 fungal isolates from ocular sources of 52 patients were randomly selected. The isolates were obtained from patients presenting to Bascom Palmer Eye Institute with ocular infections during the period of April 2000 to May 2007. The isolates were obtained from 41 corneas, 4 aqueous humor samples, 1 vitreous sample, 8 contact lenses, and 4 contact lens cases. The isolates were retrieved from lyophilized storage state or from pure cultures in Sabouraud dextrose slants at the Bascom Palmer Eye Institute’s ocular microbiology laboratory (Miami, Florida) and grown in pure culture either in peptone-yeast-glucose (PYG) (Remel, Lenexa, Kansas) media or on Sabouraud dextrose agar plates. Species designation was determined by the microbiologist at the Bascom Palmer Eye Institute’s ocular microbiology laboratory and the Texas Fungus Testing Laboratory (San Antonio, Texas), a fungal reference laboratory.
DNA EXTRACTION
The fungi grown on Sabouraud dextrose agar plates for 7 days were scraped off the surface of the agar in phosphate-buffered saline (PBS) and transferred to 15-mL centrifuge tubes, whereas the fungi grown in PYG media for 7 days were poured directly into 15-mL centrifuge tubes. The tubes were centrifuged at 3000 rpm for 10 minutes, and the supernatant was removed. DNA isolation was performed using a modified Unset (lysis buffer) procedure as described by Hugo in 1992.47 One milliliter of the lysis buffer (urea 8M, sodium dodecyl sulfate (SDS) 2%, NaCl 0.15M, EDTA 0.001M, tris pH 7.5 0.1M) was added, and the fungal pellets were resuspended using a vortex mixer for 5 seconds. An equal volume of phenol-chloroform solution (phenol:chloroform:isoamyl alcohol 25:24:1) was added to the solution, along with 0.3 g of acid-washed glass beads, and the tubes were vortexed for 5 minutes. The samples were centrifuged for 5 minutes at 13,000 rpm, and the aqueous phase was transferred to new 1.5-mL Eppendorf tubes. The phenol-chloroform extraction procedure was repeated until satisfactory removal of the protein interface. A solution of 3M sodium acetate at 1/10 volume and isopropanol 1:1 was added and mixed. The tubes were spun for 10 minutes at 13,200 rpm at 4°C, the supernatant was decanted, and 1 mL 70% ethanol was added to each tube. The tubes were spun again for 10 minutes at 13,200 rpm. Next, the supernatant was decanted and residual ethanol pipetted off. The remaining ethanol was evaporated using a vacuum centrifuge at 45°C for 2 minutes. The resulting DNA pellet was resuspended in 30 μL of distilled water, and the genomic DNA was stored at −20°C.
The quality and concentration of the genomic DNA were assessed through spectrophotometric determination of the UV absorbance 260/280-nm ratio (Eppendorf BioPhotometer, Westbury, New York) and by gel electrophoresis of the sample with a 1 kilobase (kb) molecular weight ladder (Invitrogen, Carlsbad, California).
DNA AMPLIFICATION
A 0.2-μg sample of genomic DNA was used as a template for a PCR reaction. Two primers were developed to target Fusarium labeled F18S and F28S (Invitrogen, Carlsbad, California) (Figure 1). F18S (5'-GCGGAGGGATCATTACCGAGTT-3'), a Fusarium species-specific primer, is located at the end of the 18S rDNA. F28S (5'-CAGCGGGTATTCCTACCTGATC-3') hybridizes at the beginning of the 28S rDNA. F18S is located at position 41–62 and F28S at position 520–540 of reference strain F solani strain Fs-27, GenBank accession number EF432243. The targeted region amplifies the ITS region comprising ITS1 and ITS2 regions and the 5.8S rDNA gene. The PCR reaction used the Accuprime Taq DNA polymerase system: a 25-μL PCR mixture contained 1 μL (0.2 μg) of DNA template, 2.5 μL of buffer II solution (containing all the dNTPs and MgCl2), 1 μL of each 10-μm primer (F18S and F28S), 1μL of Taq DNA polymerase (Invitrogen, Carlsbad, California), and 18.5 μL of distilled water. PCR reactions were performed in a thermocycler (iCycler; Bio-Rad Laboratories Inc, Hercules, California) and involved 1 cycle at 95ºC for 3 minutes, followed by 45 cycles with a denaturation step at 95ºC for 30 seconds, an annealing step at 55ºC for 30 seconds, and an extension step at 68ºC for 2 minutes. A negative control was included in all experiments. The detection of amplified products was performed by running an aliquot of 5 μL of each amplicon in a 1% agarose gel with ethidium bromide 0.02% in 1× tris-acetate-EDTA (TAE) buffer electrophoresis for 20 minutes at 120V (PowerPac Basic; Bio-Rad Laboratories Inc, Hercules, California). The DNA bands were visualized under UV illumination (Universal Hood II; Bio-Rad Laboratories Inc, Hercules, California). A 1 kb molecular weight ladder was included in each run (Invitrogen, Carsbad, California). PCR products were purified using Geneclean-Spin kit (MP Biomedicals Inc, Solon, Ohio) according to the manufacturer’s instuctions.
DNA SEQUENCING
Direct sequencing of PCR products was performed by Genewiz Inc (South Plainfield, New Jersey) using Applied Biosystems BigDye version 3.1. The reactions were then run on Applied Biosystem’s 3730×1 DNA Analyzer. F18S and F28S were used as the sequencing primers. The PCR products were sequenced in duplicate or triplicate to ensure sequence fidelity.
PHYLOGENETIC ANALYSIS
Sequence alignments were performed in the Megalign program of the Lasergene package (DNAStar, Madison, Wisconsin). After alignment of the 58 sequences, nucleotide divergence and percent identity were determined. The MEGA 4.0 (Molecular Evolutionary Genetic Analysis software, version 4) phylogenetic program generated a distance tree. Lecanicillium lecanii was used as the outgroup sequence for comparison. The distance was produced in the program MEGA 4.0. Bootstrap values (a measure of similarity among different DNA sequences) for the maximum parsimony tree were obtained from a consensus tree based on 100 randomly generated data sets with jumbled sequence addition. All positions containing gaps were eliminated from the data set.
NUCLEOTIDE SEQUENCE ACCESSION NUMBERS
All DNA sequence data reported in this thesis have been deposited into GenBank under accession numbers EU721670 to EU721727.
ANTIFUNGAL AGENTS
The quality-control reference strains Candida albicans ATCC 90028, Candida parapsilosis ATCC 22019, Candida krusei ATCC 6258, and Aspergillus flavus ATCC 204304 were included as control isolates for both National Committee for Clinical Laboratory Standards (NCCLS) and E-test methods. As described in the Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) document M38-A, “Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi,” additive drug dilutions were prepared to yield twice the final strength required for the test. Stock solutions of these drugs at their final concentrations were frozen at −70°C until needed. The agents evaluated in this study have well-established microdilution MIC ranges for these QC strains. Twenty-seven isolates were sent to the University of Texas fungal testing laboratory for voriconazole susceptibilities to be tested (using the CLSI technique also). Stock inoculum suspensions were prepared from 7-day-old pure cultures grown on potato dextrose agar plates at 35°C for 3 days followed by 4 days at 25°C and adjusted to optical densities approximating McFarland 1.0.
A broth microdilution method adopted from document M38-A “Reference method for broth dilution antifungal susceptibility testing of filamentous fungi.” Amphotericin B and natamycin (US Pharmacopeia) were dissolved in dimethyl sulfoxide. Stock solutions were stored at −70°C. Ten twofold serial dilutions of the antifungal agents were prepared in microdilution 96-well plates at concentrations between 0.03125 and 16 μg/mL for amphotericin B and between 0.15 and 19.2 μg/mL for natamycin. The test medium used was RPMI-1640 (Sigma Chemical Co, St Louis, Missouri) with l-glutamine, but without sodium bicarbonate, and with 0.165M morpholinepropanesulfonic acid buffer. This medium was adjusted from pH 7.4 (originally from the manufacturer) to pH 7.0 through the addition of 105 μL of 12N HCl. The mold suspension to be tested was prepared from 7-day-old pure cultures grown on potato dextrose agar plates at 35°C for 3 days, followed by 4 days at 25°C, by covering the Sabouraud dextrose agar (SDA) slant culture with sterile 0.85% saline solution and gently probing the colonies with the tip of a Pasteur pipette. The clumped colonies were dispersed with a wooden stick and were homogenized in saline solution to obtain the harvest. The resulting mixture of conidia and hyphae was transferred to a sterile tube and was vortexed for 1 minute. Heavy particles of the suspension were allowed to settle for 3 to 5 minutes, and the upper homogeneous suspension was collected for inoculum adjustment. The inoculum density was adjusted with a spectrophotometer at a 530-nm wavelength. The target percentages of transmittance were 68% to 70% (concentration similar to McFarland 1.0) for Fusarium species. The adjusted inoculum was diluted 1:50 before it was dispensed and yielded a double-strength inoculum containing 0.4 to 5 106 conidia/mL. The inoculum size was verified by determination of the number of viable colony-forming units after plating of appropriate dilutions of the inoculum onto SDA. The plates were incubated at 35°C, and the results were read visually at 24, 48, and 72 hours. For amphotericin B and natamycin, the MIC was the lowest concentration of drug that produced an optically clear well.
RESULTS
A list of the 58 isolates examined in this study is shown in Table 1. Morphologic classification at the Bascom Palmer Eye Institute’s ocular microbiology laboratory identified 32 Fusarium species (without further identification), 16 F oxysporum, and 10 F solani. Thirty of the isolates were sent to the Texas Fungus Testing Laboratory (San Antonio, Texas) for morphologic classification. The isolates were identified as 17 F solani species complex (FSSC), 9 F oxysporum species complex (FOSC), and 4 Fusarium incarnatum-equiseti species complex (FIESC) (Table 2).
TABLE 1.
SUMMARY OF FUSARIUM SP ISOLATES EXAMINED
| PATIENT NUMBER | SOURCE | YEAR | BPEI LAB CLASSIFICATION | REFERENCE LAB CLASSIFICATION | GENOTYPE | HAPLOTYPE | GENBANK ACCESSION NUMBER |
|---|---|---|---|---|---|---|---|
| 1 | Cornea | 2005 | F solani | FSSC | FSSC1 | a | EU721672 |
| 2 | Contact lens case | 2006 | F sp | FSSC | FSSC1 | a | EU721721 |
| 3 | Cornea | 2006 | F oxysporum | FSSC | FSSC1 | b | EU721688 |
| 4 | Contact lens | 2003 | F sp | FSSC1 | a | EU721708 | |
| 5 | Cornea | 2006 | F sp | FSSC | FSSC1 | a | EU721704 |
| 6 | Contact lens | 2006 | F sp | FSSC | FSSC1 | b | EU721683 |
| 7 | Cornea | 2004 | F sp | FSSC1 | b | EU721701 | |
| 8 | Cornea | 2006 | F sp | FSSC1 | b | EU721671 | |
| 9 | Cornea | 2006 | F solani | FSSC1 | a | EU721677 | |
| 10 | Cornea | 2006 | F solani | FSSC2 | d | EU721693 | |
| 11 | Cornea | 2004 | F oxysporum | FSSC2 | c | EU721724 | |
| 12 | Cornea | 2006 | F sp | FSSC2 | c | EU721692 | |
| 13 | Cornea | 2007 | F solani | FSSC2 | e | EU721680 | |
| 14 | Cornea | 2007 | F oxysporum | FSSC2 | c | EU721686 | |
| 15 | Cornea | 2005 | F sp | FSSC | FSSC2 | f | EU721702 |
| 16 | Cornea | 2004 | F sp | FSSC2 | c | EU721722 | |
| 17 | Cornea | 2005 | F oxysporum | FSSC2 | c | EU721714 | |
| 18 | Vitreous | 2005 | F sp | FSSC2 | c | EU721678 | |
| 19 | Cornea | 2006 | F sp | FSSC | FSSC2 | c | EU721673 |
| 19 | Contact lens | 2006 | F sp | FSSC2 | c | EU721711 | |
| 20 | Cornea | 2005 | F oxysporum | FSSC2 | c | EU721696 | |
| 21 | Cornea | 2006 | F oxysporum | FSSC | FSSC2 | c | EU721717 |
| 22 | Contact lens | 2006 | F sp | FSSC | FSSC2 | c | EU721681 |
| 23 | Cornea | 2006 | F sp | FSSC2 | c | EU721703 | |
| 24 | Cornea | 2007 | F solani | FSSC3,4 | g | EU721679 | |
| 25 | Cornea | 2006 | F sp | FSSC | FSSC3,4 | g | EU721705 |
| 26 | Anterior chamber | 2006 | F solani | FSSC3,4 | h | EU721691 | |
| 27 | Cornea | 2004 | F solani | FSSC3,4 | g | EU721695 | |
| 28 | Cornea | 2000 | F oxysporum | FSSC | FSSC3,4 | g | EU721687 |
| 29 | Cornea | 2007 | F oxysporum | FSSC3,4 | g | EU721684 | |
| 30 | Cornea | 2007 | F oxysporum | FSSC3,4 | g | EU721699 | |
| 31 | Cornea | 2005 | F oxysporum | FSSC3,4 | g | EU721698 | |
| 32 | Cornea | 2007 | F oxysporum | FSSC | FSSC3,4 | g | EU721697 |
| 33 | Cornea | 2006 | F sp | FSSC3,4 | g | EU721675 | |
| 34 | Cornea | 2006 | F oxysporum | FSSC3,4 | g | EU721725 | |
| 35 | Cornea | 2006 | F oxysporum | FSSC3,4 | g | EU721719 | |
| 36 | Cornea | 2005 | F solani | FSSC | FSSC3,4 | g | EU721676 |
| 9 | Cornea | 2007 | F solani | FSSC5 | i | EU721674 | |
| 37 | Cornea | 2007 | F solani | FSSC | FSSC5 | j | EU721670 |
| 38 | Cornea | 2004 | F solani | FSSC5 | i | EU721694 | |
| 39 | Anterior chamber | 2004 | F oxysporum | FSSC | FSSC5 | i | EU721715 |
| 39 | Anterior chamber | 2006 | F oxysporum | FSSC | FSSC5 | i | EU721700 |
| 40 | Cornea | 2006 | F sp | FSSC5 | j | EU721727 | |
| 41 | Contact lens case | 2006 | F oxysporum | FOSC | k | EU721709 | |
| 41 | Contact lens | 2006 | F oxysporum | FOSC | FOSC | k | EU721718 |
| 42 | Cornea | 2006 | F sp | FOSC | FOSC | k | EU721712 |
| 43 | Contact lens case | 2005 | F sp | FOSC | FOSC | k | EU721710 |
| 43 | Contact lens | 2006 | F sp | FOSC | FOSC | k | EU721713 |
| 44 | Cornea | 2006 | F sp | FOSC | FOSC | k | EU721685 |
| 45 | Cornea | 2006 | F oxysporum | FOSC | FOSC | k | EU721682 |
| 46 | Contact lens | 2006 | F sp | FOSC | FOSC | l | EU721720 |
| 47 | Contact lens | 2006 | F sp | FOSC | FOSC | k | EU721723 |
| 48 | Contact lens | 2006 | F oxysporum | FIESC | FIESC | m | EU721716 |
| 49 | Contact lens | 2006 | F sp | FIESC | FIESC | m | EU721726 |
| 50 | Cornea | 2006 | F sp | FIESC | FIESC | m | EU721706 |
| 51 | Cornea | 2004 | F sp | FDSC | n | EU721707 | |
| 52 | Cornea | 2007 | F sp | FIESC | FDSC | o | EU721689 |
FDSC, Fusarium dimerum species complex; FIESC, Fusarium incarnatum-equiseti species complex; FOSC, Fusarium oxysporum species complex; FSSC, Fusarium solani species complex.
TABLE 2.
ACCURACY OF MORPHOLOGIC COMPARED TO GENOTYPIC CLASSIFICATION
| SPECIES (n = 52) | GENOTYPIC IDENTIFICATION | GENOTYPE/PHENOTYPE CONCORDANCE AT BPEI’S MICROBIOLOGY LABORATORY (n = 52) | GENOTYPE/PHENOTYPE CONCORDANCE AT A FUNGAL REFERENCE LABORATORY (n = 30) |
|---|---|---|---|
| FSSC | 40 | 23% | 100% |
| FOSC | 7 | 14% | 100% |
| FIESC | 3 | 67% | 100% |
| FDSC | 2 | 50% | Misclassified |
| Total | 52 | 25% (average) | 97% (average) |
FDSC, Fusarium dimerum species complex; FIESC, Fusarium incarnatum-equiseti species complex; FOSC, Fusarium oxysporum species complex; FSSC, Fusarium solani species complex.
The PCR amplicons of the ITS region ranged in size from 457 to 482 base pairs. Sequencing identified 15 unique genotypes, 8 of which comprised multiple isolates. These 8 sequences represented 51 of the 58 isolates. Sequence analysis placed the isolates into 4 major clades: FSSC 76%, FOSC 16%, FIESC 5%, and Fusarium dimerum species complex (FDSC) 3%. The FSSC group, representing the majority of the isolates, could be further divided into 5 subclades (Table 3). Sequence variation ranged from 100% to 84% identity within and between species.
TABLE 3.
GENETIC IDENTIFICATION BREAKDOWN OF THE 58 ISOLATES AND COMPARISON TO THE REFERENCE CLASSIFICATION FOR OCULAR ISOLATES
| SPECIES COMPLEX | NO ISOLATES | CLADES | REFERENCE CLASSIFICATION43 |
|---|---|---|---|
| FSSC | 44 (76%) | FSSC 1 = 9 (16%) | 62% (FSSC clades 1 to 6) |
| FSSC 2 = 16 (28% | |||
| FSSC 3,4 = 13 (22%) | |||
| FSSC 5 = 6 (10%) | |||
| FOSC | 9 (16%) | 29% | |
| FIESC | 3 (5%) | 2% | |
| FDSC | 2 (3%) | 1% | |
| FCSC | — | — | |
| GFSC | — | 6% |
FCSC, Fusarium chlamydosporum species complex; FDSC, Fusarium dimerum species complex; FIESC, Fusarium incarnatum-equiseti species complex; FOSC, Fusarium oxysporum species complex; FSSC, Fusariumsolani species complex; GFSC, Gibberella fujikuroi species complex.
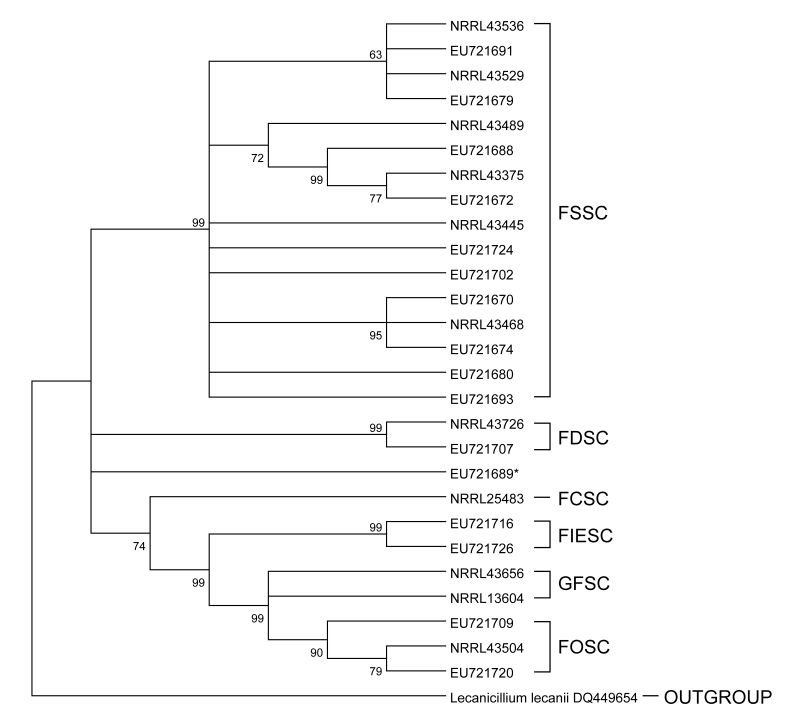
The phylogenetic relationships between the 58 isolates were examined with distance analyses (Figure 2). The closely related L lecanii was used to root the tree. The FSSC clade was well supported by a terminal bootstrap value of 99%. The distance-based phylogenetic reconstruction also supported this grouping based on the small evolutionary distances within the FSSC clade. Isolates within this clade had differences ranging from 0 to 4% and evolutionary distance less than 3.8%. EU721706, EU721716, and EU721726, which were found to have identical sequences and were morphologically classified as F incarnatum-equiseti, differed from members of the FOSC clade by less than 5%. This clade was also supported by a terminal bootstrap value of 99%. EU721689, also morphologically classified as F incarnatum-equiseti and differing from EU721706, EU721716, EU721726, and members of the FOSC clade by more than 13%, may be representative of this clade. The branching order for EU721689 and EU721707 was not well supported, as they both differ from other Fusarium isolates and each other by greater than 12%, illustrated by the large evolutionary distance observed in the distance tree. A similar difference was observed when comparing the FSSC and FOSC clades to each other.
FIGURE 2.
Distance tree phylogenetic relationships between the 58 isolates examined with parsimony and neighbor-joining analyses. FCSC, Fusarium chlamydosporum species complex; FDSC, Fusarium dimerum species complex; FIESC, Fusarium incarnatum-equiseti species complex; FOSC, Fusarium oxysporum species complex; FSSC, Fusarium solani species complex; GFSC, Gibberella fujikuroi species complex.
The susceptibilities to 3 different antifungals are shown at Table 4. There was only one isolate resistant to natamycin (>4.8) and amphotericin B (>2), belonging to the clade FSSC3,4.
TABLE 4.
COMPARISON OF SUSCEPTIBILITIES TO VORICONAZOLE, AMPHOTERICIN B, AND NATAMYCIN
| ISOLATE | VORICONAZOLE MIC90 | VORICONAZOLE MFC90 | AMPHOTERICIN B MIC90 | AMPHOTERICIN B MFC90 | NATAMYCIN MIC90 | NATAMYCIN MFC90 |
|---|---|---|---|---|---|---|
| FSSC | 16 | — | 2 | 2 | 4.8 | 19.2 |
| FNSSC | 4 | — | 2 | 4 | 4.8 | 19.2 |
FNSSC, Fusarium non-solani species complex; FSSC, Fusarium solani species complex; MIC, minimum inhibitory concentration; MFC, minimum fungicidal concentration.
Fifteen of 16 tested FSSCs were resistant (MIC90 > 4 μg/mL) to voriconazole, whereas only 1 in 14 FNSSCs was resistant.
DISCUSSION
Keratomycosis is a prevalent cause of ocular morbidity throughout the world.1 Molds are more likely to be ocular pathogens than yeasts.48 Fusarium species are a common pathogen in ocular infections and have an increased incidence over the last decade.2,49,50 Keratitis caused by these pathogens can cause severe visual compromise.6 Diagnosis and treatment are often delayed owing to lack of sensitive and rapid diagnostic tests.51
This study examined the usefulness of PCR amplification and analysis of the ITS regions (ITS1 and ITS2) in detecting and differentiating Fusarium species from ocular infections and to assessed the correlation between the genotypic data and the morphologic classification.
In the first objective of this study, primers capable of amplifying Fusarium in a genus-specific manner were successfully designed to target the ribosomal ITS region.
Clearly, morphologic (or phenotypical) identification performed at a general ocular microbiology laboratory (Bascom Palmer Eye Institute) appears to be reliable to the genus level, with accordance to the genotypes in 100% of the 58 cases. However, identification at such a laboratory may be unreliable at the species level for the Fusarium genus, with accurate species identification in only 25% of the cases. Classification performed by a fungal reference laboratory (Texas Fungus Testing Laboratory, San Antonio, Texas), on the other hand, was very accurate at both the genus and species level, with 100% correspondence at the genus level and 97% at the species level.
The majority of the sequences fell into the FSSC followed by the FOSC. Phylogenetic analysis separated the F solani and F oxysporum clades with high terminal bootstrap values and showed evolutionary distances of greater than 12%. Phylogenetic analysis also suggested that the FIESC is polyphiletic or the 3 isolates, EU721706, EU721716, and EU721726, were misclassified, the latter being more likely.
Zhang and associates43 genotyped 191 isolates, including isolates obtained from human and environmental sources, from the Centers for Disease Control and Prevention (CDC) investigation. They broke down the isolates in 5 different species complexes (Table 3) and the FSSC into 6 clades. A sixth species complex, called Fusarium chlamydosporum species complex, was present only in environmental samples in the study. The main species complex found in their study was the FSSC, with 62%. Similarly, Godoy and associates52 found 91% of the isolates to lie within the FSSC. These studies correlate with our data, where 76% of the isolates were found to be from the FSSC. In the present study, although the ITS region was not effective at resolving branching orders of clades within the FSSC, it did allow successful and accurate speciation.
This study is based on correlations of genotypes with clinical outcomes and antifungal susceptibilities, which showed that in a clinical perspective, the most relevant differentiation is between FSSC and Fusarium non-solani species complexes (FNSSCs), which have shown different susceptibility and pathogenicity profiles (Oechsler RA et al, Invest Ophthalmol Vis Sci 2008;49:E-abstract 2495). The in vitro susceptibility testing, when coupled with the genotypic findings, provides more accurate information for the selection of an antifungal. All 50 isolates tested exhibited an MIC90 that was resistant to caspofungin, fluconazole, and moxifloxacin. All isolates except FNSSCs, which included FOSC, FIESC, and FDSC, exhibited sensitivity to posaconazole and voriconazole but resistance to all others, including amphotericin B. Of significance, FSSC clades 1, 2, and 6 were resistant to voriconazole. This genotypic classification, when correlated with in vitro microbiologic sensitivity data, will provide the clinician rapid information to select the most appropriate antifungal.53
Such DNA-based diagnostic tests show significant promise in allowing precise and rapid diagnosis of fungal ocular infections, and their design and implementation will certainly hasten the initiation of appropriate antimicrobial therapy and decrease the ocular morbidity resulting from infections caused by Fusarium.
On the basis of this and previously published data,44–46 my colleagues and I propose that the ITS regions provide a sufficient genetic scaffolding for DNA-based diagnostic tests to detect and differentiate Fusarium isolates causing ocular infections to a clinically relevant level.
Furthermore, the ITS region is attractive to be used in diagnostic tests, because it is a multicopy gene, and a lower number of pathogens would be necessary to yield sufficient DNA for a positive recognition of the pathogen. This is principally important in the ophthalmic field, where extremely small volume samples are usually available for the diagnostic workup.
In conclusion, the sequence variation within the ITS provides a sufficient quantitative basis for the development of a molecular diagnostic approach for the Fusarium species isolated from ocular infections. Morphology based on microscopic and macroscopic observations yields inconsistent results, particularly at nonreference laboratories, emphasizing the need for a more reproducible test with less user-dependent variability. F solanis tend to be more resistant to certain antifungals (azoles).
Further modification of the probes will allow identification of species complexes. In the future, the use of probes in a high-throughput multiplex assay (eg, Luminex) will allow rapid identification of clinical samples.53–57 The Luminex xMap system uses a flow cytometer with 2 laser detectors that can analyze up to 100 different fluorescent color beads covalently bound to species-specific probes. The hybridized target DNA carries a fluorescent dye, which is detected and quantified by one of the dual laser systems. The assay, which was designed for clinical use, is rapid: the tests are run in 96-well plates with the capability of processing up to 100 different analyses at a rate of 0.47 minute per well. In contrast to gene chips, the method is inexpensive and versatile, as a variety of probes can be added or subtracted to create different platform arrays that can be employed in a wide variety of applications, such as ribosomal and functional genes. This thesis provides the genotypic probes for Fusarium that will allow multiplex analysis of ocular samples for rapid identification of pathogenic species in ocular tissues.
ACKNOWLEDGMENTS
Funding/Support: This work is funded in part by institutional grants from Research to Prevent Blindnes Inc, the National Eye Institute, and the Coulter Center at the University of Miami. Funding to the Coulter Center is provided by a matching grant from Bausch & Lomb and Alcon Laboratory.
Financial Disclosures: Dr Alfonso has received financial support in the form of honoraria and research support from Bausch & Lomb and Alcon Laboratory.
Conformity With Author Information: Institutional review board review and approval was not obtained, since this study does not pertain to human subjects.
Other Acknowledgments: I gratefully acknowledge the support and assistance provided by my colleagues Rafael A. Oechsler, MD, Michael R. Feilmeier, MD, Dolena L. Leedee, PhD, Darlene Miller, DSc, Jack Fell, PhD, Mara J. Diaz, PhD, M. Elizabeth M. Fini, PhD, and Alfonso Iovieno, MD in the conduct and design of this study.
REFERENCES
- 1.Rosa RH, Jr, Miller D, Alfonso EC. The changing spectrum of fungal keratitis in south Florida. Ophthalmology. 1994;101:1005–1013. doi: 10.1016/s0161-6420(94)31225-5. [DOI] [PubMed] [Google Scholar]
- 2.Alfonso EC, Cantu-Dibildox J, Munir WM, et al. Insurgence of Fusarium keratitis associated with contact lens wear. Arch Ophthalmol. 2006;124:941–947. doi: 10.1001/archopht.124.7.ecs60039. [DOI] [PubMed] [Google Scholar]
- 3.Marangon FB, Miller D, Giaconi JA, Alfonso EC. In vitro investigation of voriconazole susceptibility for keratitis and endophthalmitis fungal pathogens. Am J Ophthalmol. 2004;137:820–825. doi: 10.1016/j.ajo.2003.11.078. [DOI] [PubMed] [Google Scholar]
- 4.Tu EY, McCartney DL, Beatty RF, et al. Successful treatment of resistant ocular fusariosis with posaconazole (SCH-56592) Am J Ophthalmol. 2007;143:222–227. doi: 10.1016/j.ajo.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 5.Ando N, Takayori K. Fungal flora of the conjunctival sac. Am J Ophthalmol. 1982;94:67–74. doi: 10.1016/0002-9394(82)90193-3. [DOI] [PubMed] [Google Scholar]
- 6.Dursun D, Fernandez V, Miller D, Alfonso EC. Advanced Fusarium keratitis progressing to endophthalmitis. Cornea. 2003;22:300–303. doi: 10.1097/00003226-200305000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Kuriakose T, Thomas PA. Keratomycotic malignant glaucoma. Indian J Ophthalmol. 1991;39:118–121. [PubMed] [Google Scholar]
- 8.Vemuganti G, Garg KP, Gopinathan V, et al. Evaluation of agent and host factors in progression of mycotic keratitis. A histologic and microbiologic study of 167 corneal buttons. Ophthalmology. 2002;109:1538–1546. doi: 10.1016/s0161-6420(02)01088-6. [DOI] [PubMed] [Google Scholar]
- 9.Fong LP, Ormerod LD, Kenyon KR, Foster CS. Microbial keratitis complicating penetrating keratoplasty. Ophthalmology. 1988;94:269–1275. doi: 10.1016/s0161-6420(88)33036-8. [DOI] [PubMed] [Google Scholar]
- 10.Karp CL, Tuli SS, Yoo SH, et al. Infectious keratitis after LASIK. Ophthalmology. 2003;110:503–510. doi: 10.1016/S0161-6420(02)01760-8. [DOI] [PubMed] [Google Scholar]
- 11.Alfonso E, Mandelbaum S, Fox MJ, et al. Ulcerative keratitis associated with contact lens wear. Am J Ophthalmol. 1986;101:429–433. doi: 10.1016/0002-9394(86)90641-0. [DOI] [PubMed] [Google Scholar]
- 12.Rosenthal RA, Dasanayake NL, Schlitzer RL, et al. Biocide uptake in contact lenses and loss of fungicidal activity during storage of contact lenses. Eye Contact Lens. 2006;32:262–266. doi: 10.1097/ICL.0b013e31802b413f. [DOI] [PubMed] [Google Scholar]
- 13.Tanure MA, Cohen EJ, Sudesh S, et al. Spectrum of fungal keratitis at Wills Eye Hospital, Philadelphia, Pennsylvania. Cornea. 2000;3:307–312. doi: 10.1097/00003226-200005000-00010. [DOI] [PubMed] [Google Scholar]
- 14.Mselle J. Fungal keratitis as an indicator of HIV infection in Africa. Trop Doct. 1999;29:133–135. doi: 10.1177/004947559902900303. [DOI] [PubMed] [Google Scholar]
- 15.Berson EL, Kobayashi GS, Becker B, Rosenbaum L. Topical corticosteroids and fungal keratitis. Invest Ophthalmol. 1967;6:512–517. [PubMed] [Google Scholar]
- 16.Stern GA, Buttross M. Use of corticosteroids in combination with antimicrobial drugs in the treatment of infectious corneal disease. Ophthalmology. 1991;98:847–853. doi: 10.1016/s0161-6420(91)32211-5. [DOI] [PubMed] [Google Scholar]
- 17.Thygeson P, Okumoto M. Keratomycosis: a preventable disease. Trans Am Acad Ophthalmol Otolaryngol. 1974;78:433–439. [PubMed] [Google Scholar]
- 18.Sevel D, Kassar B. Suppurative keratitis and fungal keratitis. Trans Ophthalmol Soc N Z. 1973;25:228–232. [PubMed] [Google Scholar]
- 19.Jones DB, Wilson L, Sexton R, et al. Early diagnosis of mycotic keratitis. Trans Ophthalmol Soc U K. 1970;89:805–813. [PubMed] [Google Scholar]
- 20.Bharathi MJ, Ramakrishnan R, Vasu S, et al. Epidemiological characteristics and laboratory diagnosis of fungal keratitis: a three year study. Ind J Ophthalmol. 2003;51:315–321. [PubMed] [Google Scholar]
- 21.Ishibashi Y, Hommura S, Matsumoto Y. Direct examination vs culture of biopsy specimens for the diagnosis of keratomycosis. Am J Ophthalmol. 1987;103:636–640. doi: 10.1016/s0002-9394(14)74322-3. [DOI] [PubMed] [Google Scholar]
- 22.Kaufman SC, Musch DC, Belin MW, et al. Confocal microscopy: a report by the American Academy of Ophthalmology. Ophthalmology. 2004;111:396–406. doi: 10.1016/j.ophtha.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 23.O’Brien TP. Therapy of ocular fungal infections. Ophthalmol Clin North Am. 1999;12:33–50. [Google Scholar]
- 24.Mauger TF, Craig EL. Havener’s ocular pharmacology. 9th ed. St Louis, MO: CV Mosby; 1994. [Google Scholar]
- 25.O’Day DM. Selection of appropriate antifungal therapy. Cornea. 1987;6:238–245. doi: 10.1097/00003226-198706040-00002. [DOI] [PubMed] [Google Scholar]
- 26.Foster CS. Miconazole therapy for keratomycosis. Am J Ophthalmol. 1981;91:622–629. doi: 10.1016/0002-9394(81)90063-5. [DOI] [PubMed] [Google Scholar]
- 27.O’Day DM, Foulds G, Williams TE, et al. Ocular uptake of fluconazole after oral administration. Arch Ophthalmol. 1990;108:1006–1008. doi: 10.1001/archopht.1990.01070090108050. [DOI] [PubMed] [Google Scholar]
- 28.Yee RW, Cheng CJ, Meenakshi S, et al. Ocular penetration and pharmacokinetics of topical fluconazole. Cornea. 1997;16:64–71. [PubMed] [Google Scholar]
- 29.Goldblum D, Frueh BE, Sarra GM, et al. Topical caspofungin for treatment of keratitis caused by Candida albicans in a rabbit model. Antimicrob Agents Chemother. 2005;49:1359–1363. doi: 10.1128/AAC.49.4.1359-1363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cristol SM, Alfonso EC, Guildford JH, et al. Results of large penetrating keratoplasty in microbial keratitis. Cornea. 1996;15:571–576. [PubMed] [Google Scholar]
- 31.Panda A, Vajpayee RB, Kumar TS. Critical evaluation of therapeutic keratoplasty in cases of keratomycosis. Ann Ophthalmol. 1991;23:373–376. [PubMed] [Google Scholar]
- 32.Stern GA, Buttross A. Use of corticosteroids in combination with antimicrobial drugs in the treatment of infectious corneal disease. Ophthalmology. 1991;98:847–853. doi: 10.1016/s0161-6420(91)32211-5. [DOI] [PubMed] [Google Scholar]
- 33.Perry HD, Doshi SJ, Donnenfeld ED, et al. Topical cyclosporin A in the management of therapeutic keratoplasty for mycotic keratitis. Cornea. 2002;21:161–163. doi: 10.1097/00003226-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 34.Whitcher JP, Srinivasan M, Upadhyay MP. Corneal blindness: a global perspective. Bull World Health Organ. 2001;79:214–221. [PMC free article] [PubMed] [Google Scholar]
- 35.Chowdhary A, Singh K. Spectrum of fungal keratitis in North India. Cornea. 2005;24:8–15. doi: 10.1097/01.ico.0000126435.25751.20. [DOI] [PubMed] [Google Scholar]
- 36.Liesegang TJ, Forster RK. Spectrum of fungal keratitis in South Florida. Am J Ophthalmol. 1980;90:38–47. doi: 10.1016/s0002-9394(14)75075-5. [DOI] [PubMed] [Google Scholar]
- 37.O’Donnell K, Sarver BAJ, Brandt M, et al. Phylogenetic diversity and microsphere array-based genotyping of human pathogenic Fusaria, including isolates from the multistate contact lens-associated US keratitis outbreaks of 2005 and 2006. J Clin Microbiol. 2007;45:2235–2248. doi: 10.1128/JCM.00533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas PA, Leck AK, Myatt M. Characteristic clinical features as an aid to the diagnosis of suppurative keratitis caused by filamentous fungi. Br J Ophthalmol. 2005;89:1554–1558. doi: 10.1136/bjo.2005.076315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azor M, Gene J, Cano J, et al. Universal in vitro antifungal resistance of genetic clades of the Fusarium solani species complex. Antimicrob Agents Chemother. 2007;51:1500–1503. doi: 10.1128/AAC.01618-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Donnell K, Kistler HC, Tacke BK, et al. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc Natl Acad Sci U S A. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Donnell K, Ward TJ, Geiser DM, et al. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet Biol. 2004;41:600–623. doi: 10.1016/j.fgb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Summerbell RC, Lévesque CA, Seifert KA, et al. Microcoding: the second step in DNA barcoding. Philos Trans R Soc Lond B Biol Sci. 2005;360:1897–1903. doi: 10.1098/rstb.2005.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang N, O’Donnell K, Sutton DA, et al. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J Clin Microbiol. 2006;44:2186–2190. doi: 10.1128/JCM.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fell JW. Rapid identification of yeast species using three primers in a polymerase chain reaction. Mol Mar Biol Biotechnol. 1993;2:174–180. [PubMed] [Google Scholar]
- 45.Fell JW. rDNA targeted oligonucleotide primers for the identification of pathogenic yeasts in a polymerase chain reaction. J Ind Microbiol. 1995;14:475–477. doi: 10.1007/BF01573961. [DOI] [PubMed] [Google Scholar]
- 46.Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- 47.Espinel-Ingroff A, Boyle K, Sheehan DJ. In vitro antifungal activities of voriconazole and reference agents as determined by NCCLS methods: review of the literature. Mycopathologia. 2001;150:101–150. doi: 10.1023/a:1010954803886. [DOI] [PubMed] [Google Scholar]
- 48.Rosenberg KD, Flynn HW, Alfonso EC, et al. Fusarium endophthalmitis following keratitis associated with contact lenses. Ophthalmic Surg Lasers Imaging. 2006;37:310–313. doi: 10.3928/15428877-20060701-08. [DOI] [PubMed] [Google Scholar]
- 49.Alfonso EC, Miller D, Cantu-Dibildox J, et al. Fungal keratitis associated with non-therapeutic soft contact lenses. Am J Ophthalmol. 2006;142:154–155. doi: 10.1016/j.ajo.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 50.Chang DC, Grant GB, O’Donnell K, et al. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA. 2006;296:953–963. doi: 10.1001/jama.296.8.953. [DOI] [PubMed] [Google Scholar]
- 51.Höfling-Lima AL, Forseto A, Duprat JP, et al. Laboratory study of the mycotic infectious eye diseases and factors associated with keratitis. Arq Bras Oftalmol. 2005;68:21–27. doi: 10.1590/s0004-27492005000100005. [DOI] [PubMed] [Google Scholar]
- 52.Godoy P, Cano J, Gené J, et al. Genotyping of 44 isolates of Fusarium solani, the main agent of fungal keratitis in Brazil. J Clin Microbiol. 2004;42:4494–4497. doi: 10.1128/JCM.42.10.4494-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye F, Li MS, Taylor JD, et al. Fluorescent microsphere-based readout technology for multiplexed human single nucleotide polymorphism analysis and bacterial identification. Hum Mutat. 2001;17:305–316. doi: 10.1002/humu.28. [DOI] [PubMed] [Google Scholar]
- 54.Dunbar SA, Vander Zee CA, Oliver KG, et al. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex LabMAP system. J Microbiol Methods. 2003;53:245–252. doi: 10.1016/s0167-7012(03)00028-9. [DOI] [PubMed] [Google Scholar]
- 55.Diaz MR, Boekhout T, Kiesling T, et al. Comparative analysis of the intergenic spacer regions and population structure of the species complex of the pathogenic yeast Cryptococcus neoformans. FEMS Yeast Res. 2005;5:1129–1140. doi: 10.1016/j.femsyr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 56.Diaz MR, Fell JW. Use of a suspension array for rapid identification of the varieties and genotypes of the Cryptococcus neoformans species complex. J Clin Microbiol. 2005;43:3662–3672. doi: 10.1128/JCM.43.8.3662-3672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diaz MR, Fell JW. High-throughput detection of pathogenic yeasts of the genus Trichosporon. J Clin Microbiol. 2004;42:3696–3706. doi: 10.1128/JCM.42.8.3696-3706.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]