Abstract
A quantitative trait locus (QTL) approach was used to define the genetic architecture underlying variation in systolic blood pressure (SBP) and heart rate (HR), measured indirectly on seven occasions by the tail cuff procedure. The tests were conducted in 395 F2 adult mice (197 males, 198 females) derived from a cross of the C57BL/6J (B6) and DBA/2J (D2) strains and in 22 BXD recombinant-inbred (RI) strains. Interval mapping of F2 data for the first 5 days of measurement nominated one statistically significant and one suggestive QTL for SBP on chromosomes (Chr) 4 and 14, respectively, and two statistically significant QTL for HR on Chr 1 (which was specific to female mice) and Chr 5. New suggestive QTL emerged for SBP on Chr 3 (female-specific) and 8 and for HR on Chr 11 for measurements recorded several weeks after mice had undergone stressful blood sampling procedures. The two statistically significant HR QTL were confirmed by analyses of BXD RI strain means. Male and female F2 mice did not differ in SBP or HR but RI strain analyses showed pronounced strain-by-sex interactions and a negative genetic correlation between the two measures in both sexes. Evidence for a role for mitochondrial DNA was found for both HR and SBP. QTL for HR and SBP may differ in males and females and may be sensitive to different environmental contexts.
Keywords: quantitative trait locus, sex specificity, stress
human essential hypertension, evidenced by persistent elevated systolic or diastolic blood pressure, is a highly prevalent complex disease (33) long known to be regulated by a polygenic system (20, 32). In the preclinical arena, Schlager and colleagues' (47–52) pioneering studies in laboratory mice amply confirmed the multigenic control of systolic blood pressure (SBP) and explored its genetic architecture.
Despite this background of quantitative genetic work on blood pressure (BP) in mice, attempts to describe the polygenic system underlying BP, identify its individual elements, via the quantitative trait locus (QTL) approach in animal models have mainly focused on crosses between several well-known hypertensive rat strains and normotensive controls (41). In part, the choice of species for these studies was directed by the extensive use of hypertensive rat strains in research before the beginning of the QTL era and the large body of research on these models. In any case, the research has been highly successful. All rat chromosomes have been associated with BP QTL (http://rgd.mcw.edu), and important progress has been made in fine-mapping of QTL regions (16, 24, 31, 36, 37, 46).
The genomic and technical resources available for study of the mouse genome still emphasize the great opportunities for cardiovascular research in this species, and an important beginning has been made with QTL studies. Crosses between inbred mouse strains have shown variation in SBP to be associated with Chr 1, 4, 5, 6, 7, 10, 11, 13, 15, and 16 (14, 60, 61, 74, 75). Strain variation in heart rate (HR) was reported many years ago (5), and QTL have also been identified that influence HR in mice, rat and human studies (23, 29, 56, 61).
The background of QTL research on the rat is a great advantage to analogous studies in mice. The degree of homology between rat and mouse genomes is the greatest among species commonly used in the laboratory (17, 28, 55), which permits information obtained from QTL studies in one species to inform research on the other. Examination of interspecific homologies is also likely to be helpful to attempts to map genes relevant to hypertension in humans where large-scale studies have often found it difficult to establish definitive linkage (22, 38, 40) and where factors such as antihypertensive therapy are suspected of interfering with the relationship between genotype and phenotype (12).
The present study, which is part of an analysis of a large number of behavioral, physiological, morphological, and pathological phenotypes, seeks to identify QTL affecting cardiovascular function in the C57BL/6J (B6) × DBA/2J (D2) lineage. Characterization of cardiovascular function in both a B6D2 intercross and BXD recombinant-inbred (RI) strains (66, 67) provided the opportunity for the genetic architecture underlying the various traits to be compared and contrasted in two different mapping populations with complementary strengths. The increasing sophistication of databases permitted comparison of genome sequence on both parental strains (59).
MATERIALS AND METHODS
Animals.
The animals used for SBP and HR measurements were subjects of a large cross-sectional study of biobehavioral, physiological, and morphological measures. All procedures were approved by the Pennsylvania State University Institutional Animal Care and Use Committee. C57BL/6J (B6) and DBA/2J (D2) mice and 23 of the derivative BXD RI strains were obtained from The Jackson Laboratory, derived by cesarean section into the barrier facility at the Center for Developmental and Health Genetics at Penn State University and raised by Swiss foster mothers. After establishment of the breeding colony, a 400-animal B6D2F2 intercross was generated in four birth cohorts of ∼100 mice each at 1-mo intervals. Each birth cohort of B6D2F2 was generated with an approximately equal number of four possible mating types of F1 mice i.e., B6D2×B6D2, B6D2×D2B6, D2B6×B6D2, and D2B6×D2B6 where the first animal in the mating combinations is a female: for example, D2B6 denotes an F1 animal derived from the mating of a D2 female and B6 male, and B6D2 is the offspring of a B6 female and a D2 male, etc. This mating plan permitted the origin of the Y chromosome (F2 males) and mitochondrial DNA to be specified. Four hundred F2 mice born to 50 primiparous females (range 8–13 pups per litter) were weaned for the study. A total of 198 females and 197 males were tested in the BP protocol. Mice of the BXD RI strains were weaned from both primiparous and multiparous females (range 2–12 pups/litter). The BXD study design called for 12 males and 12 females from each strain. Early death reduced the number of animals in some strains, or required additional breeding (BXD13). All animals were housed in groups of 4 of the same sex per cage at room temperature (20.5–22°C) on a 12/12 h light-dark cycle and fed ad libitum with sterilized Purina 5010 (designed to be nutritionally equivalent to Purina 5001 after sterilization).
BP schedule.
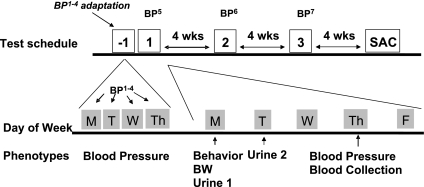
SBP and HR were measured from F2 animals on seven separate measurement occasions (four consecutive daily tests beginning at 115 days ± 2 wk, another test 1 wk later, then twice more at ∼4 wk intervals; Fig. 1). RI mice were measured on five separate occasions (four consecutive daily tests and once the following week). Test groups consisted of 12–16 animals equally divided by sex. The week after the first four BP measurements, a battery of behavioral and physiological measurements was initiated (noninvasive activity tests and urine collections preceded the fifth BP test by 48 h); 4–6 h after the BP measurement, animals were warmed and placed in a restrainer, and a blood sample was taken by tail-nick. This sequence of measurements was repeated at 4 wk intervals in tandem with BP measurements 6 and 7. Thus, BP and HR #1–5 preceded the first blood collection procedure, and BP #6 and 7 were each assessed 4 wk after a blood collection procedure. Figure 1 illustrates the study design.
Fig. 1.
Study design for F2 mice was initiated at 115 days ± 2 wk. Phenotypic measurements listed during week 1 (Behavior - Blood collection) were repeated during weeks when 6th and 7th blood pressure measurements (BP6 and BP7) were assessed.
BP.
The Visitech BP system (BP-2000, Visitech Systems, Apex, NC) was used to obtain measures of SBP and HR. Mice were guided into metal tunnels (the apparatus has four identical positions available) resting on a warmed surface. After we fitted an inflatable tail-cuff around its tail we restrained the mouse by taping its tail to the detector platform. We detected peripheral pulse by directing an infra-red light through the tail and measuring the changes resulting from the amount of blood between the light source and a photoelectric sensor. Mice were placed in the apparatus in groups of four animals of the same sex from a single cage. Cages of males and females were alternated throughout the test procedure.
On each measurement occasion, we placed the four animals in the restrainers at 37.8°C for 6 min before initiating the first measurement cycle. HR represented a 70-beat sample taken before the beginning of each cuff inflation cycle. SBP was the instantaneous reading of cuff pressure taken at the reappearance of the tail pulse during the deflation phase of each inflation-deflation cycle. Eight measurements of SBP and HR were then taken at 20- to 30-s intervals. The data were saved to a computer disk after each 8-trial block was completed. The 8-trial cycle was repeated twice more for a total of 24 measures per daily test. A complete set of measures for F2 mice was therefore 7 × 3 = 21, and for RIs 5 × 3 = 15, 8-trial blocks. Persistent struggling or escapes from the restrainer were recorded after each 8-trial block.
Molecular markers.
The F2 mice were genotyped with 96 microsatellite markers spaced at ∼15- to 20-cM intervals throughout the genome (68). We selected 622 microsatellite markers for use with the BXD RI (73).
Data analysis.
Means of SBP and HR were calculated for each 8-trial block for each mouse. If all readings were satisfactory (readings were discarded if animals struggled in the restrainer) there were means of SBP and HR available for the 21 8-trial blocks for each F2 animal and 15 8-trial blocks for each RI and parental strain mouse. Data from BXD13 were excluded because this strain was vulnerable to seizures. Standard data analyses were conducted using the SPSS 11.0 software package (SPSS, Chicago, IL). Unless otherwise stated, a two-tailed alpha of 0.05 was used to determine statistical significance. The results of analyses of variance are reported with F values, the associated degrees of freedom and the probability (nominally, P < 0.05, 0.01, 0.001).
Linkage analysis.
Interval mapping (IM) analysis and a genome-wide scan for epistatic interactions between all pairs of autosomal markers in the B6D2F2 intercross were carried out using the J/qtl and R/qtl packages (7). Sex was included as a covariate. Thresholds of statistical significance in IM were determined for each trait from 1,000 permutations (10). Significant epistatic interactions were identified if they exceeded thresholds of 9.1, 7.1, 6.3, 6.3, and 3.3 LOD for the full, conditional-interactive, interaction, additive, and conditional-additive models, respectively. These thresholds (http://www.rqtl.org/tutorials/new_summary_scantwo.pdf) have been obtained for an F2 intercross via 10,000 simulations by the developers of Rqtl (7).
Sex, suggestive, or significant QTL and epistatic interactions were included in a multiple-regression model for each trait. Individual terms were dropped by backward elimination until all terms of the model were significant at P < 0.05.
The effect of mitochondrial DNA was evaluated by analysis of variance using grandmaternal origin of F2 mice (i.e., originating in B6 or D2 females) as the group identifier.
To explore the genes in the QTL regions and polymorphisms between the C57BL/6J and DBA/2J strains and to assess mouse/rat homology, a variety of database tools was used (www.ncbi.nlm.nih.gov/, http://rgd.mcw.edu/, http://ratmap.org/, http://www.ensembl.org).
Single nucleotide polymorphism analysis.
Genes with nonsynonymous single nucleotide ploymorphisms (SNPs) were subjected to assessment of possible impact of amino acid substitution on the phenotype between B6 and D2 strains. Sequence of amino acids was obtained from Ensemble database (www.ensembl.org/Mus_musculus) and analyzed by PolyPhen software (http://genetics.bwh.harvard.edu/) (39). PolyPhen issues four predictions for the effects of nonsynonymous SNPs: 1) probably damaging (implicates high confidence for the variation to affect protein function), 2) possibly damaging (supposed to affect protein function or structure), 3) benign (most likely lacking any effect), and 4) unknown (typically due to the lack of data to make a prediction). The first three types of prediction have been encountered in our analysis. Genes falling within the first two categories of prediction have been further screened for their expression in heart, kidney, vascular/smooth muscle tissue by means of GEO database (www.ncbi.nlm.nih.gov/geo/).
RESULTS
Individual animal SBP and HR means were calculated for 1) the first 5 days of measurement (SBP1–5, HR1–5) before animals were exposed to the first blood collection procedure, and 2) the sixth and seventh days of measurement (SBP6,7, HR6,7; Fig. 1), which were each obtained ∼1 mo after blood collection after BP measures 5 and 6, respectively (see materials and methods and Fig. 1). Evidence that exposure to blood collection procedures influenced response to the BP restrainer among F2s was that struggling showed a significant increase in trials 6 and 7 compared with trials 1–5 (nonparametric Friedman's test, P < 0.001): mice were 3.5-fold more likely to struggle during trials 6 and 7 than during 1–5. HR for the combined group of males and females was normally distributed. SBP was positively skewed and was normalized with a logarithmic transformation.
SBP1–5 was higher in D2 mice (F 1,48 = 12.5, P < 0.001) largely due to the higher SBP of D2 males (sex-by-strain interaction, F 1,48 = 8.6, P < 0.005; Table 1). There was also a strain-by-sex interaction for HR1–5 (F 1,48 = 13.5, P < 0.001) with D2 males having higher HR than B6 males, the reverse being true for females.
Table 1.
Cardiovascular indexes of parental strains for measurements 1–5 and of B6/D2 F2 for measurements 1–5 and 6, 7
| Group | Sex | Mean HR | SD | Mean SBP | SD | n |
|---|---|---|---|---|---|---|
| B6 | F | 582.0 | 37.5 | 128.1 | 8.9 | 12 |
| M | 533.9 | 38.9 | 121.4 | 4.8 | 12 | |
| D2 | F | 537.0 | 34.5 | 129.6 | 11.9 | 16 |
| M | 561.7 | 30.4 | 137.4 | 6.9 | 12 | |
| F21–5 | F | 587.7 | 53.5 | 117.6 | 9.8 | 198 |
| F21–5 | M | 584.7 | 46.7 | 117.0 | 9.7 | 197 |
| F26,7 | F | 581.4 | 64.4 | 114.4 | 11.2 | 194 |
| F26,7 | M | 588.1 | 55.0 | 115.6 | 9.2 | 186 |
HR, heart rate; SBP, systolic blood pressure.
There was no sex difference in SBP or HR in the F2 data on either measurement occasion (F values <1). However, repeated-measures analysis found SBP6,7 was significantly lower than SBP1–5 (F 1,378 = 14.9, P < 0.001: Table 1). Body weight was ∼10 g heavier in males than females (39.0 ± 0.34 vs. 29.2 ± 0.34 g, P < 0.001). The small negative correlation between body weight and BP in males (r = −0.23, P < 0.001) was the only statistically significant relationship of cardiovascular function to body weight.
IM and epistatic interactions.
QTL affecting SBP and HR traits are summarized in Table 2. There was no effect of the Y chromosome (see Animals, above) on either SBP or HR. Combined genetic effects accounted for 6.9% of phenotypic variance of SBP1–5, 13.6% of SBP6,7, 17.6% of HR1–5, and 15.1% of HR6,7. Two of the QTL were female-specific (Table 2). It is noteworthy that the three QTL that were statistically significant during the first five measurements retained their significance during the sixth and seventh measures. However, this was not true of the QTL that satisfied only the suggestive level of significance during the first five measures. During the sixth and seventh measurements, which were obtained in each case ∼4 wk after invasive procedures for blood-sampling, three suggestive QTL (two for SBP, one for HR) were found that had not been seen in analyses of the first five measurements. In addition to evidence from frequency of struggling (see above) that the aversiveness of the procedure may have increased, “r” between measurements 1–5 and 6 and 7 of both SBP and HR were modest (SBP 0.247, HR 0.465; both Ps <0.001) consistent with the idea that control of the two variables differed across measurement occasions. Additional evidence that the two HR measures were differentially controlled was the finding that B6 mitochondrial DNA was associated with increased HR6,7 but not with HR1–5 (mitochondrial DNA by measurement occasion, F 1,342 = 17.6, P < 0.001): mice with B6 mitochondrial DNA had faster HR6,7 (means ± SE = 594.3, ± 4.4) than D2 (575.3 ± 4.6), compared with the lack of a significant difference for HR1–5 (B6, 584.4 ± 3.7; D2, 590.6 ± 3.8, not significant) The opposite finding (between measurements 1–5 and 6 & 7) was noted for the same interaction term for SBP (F 1,370 = 4.2, P < 0.04) where mice with B6 mitochondrial DNA had higher SBP1–5 (B6, 118.9 ± 0.7; D2, 115.7 ± 0.7) with no difference between the two groups on SBP6,7.(B6, 115.3 ± 0.76; D2, 114.7 ± 0.75).
Table 2.
Genetic architecture of SBP and HR measured during adaptation (BP/HR1–5) or ∼1 mo after stressful blood-collection procedures (BP/HR6,7) in B6/D2 F2s
| Trait | Chr | Peak, cM | Confidence Interval,a cM | Peak,b LOD | Marker Near QTL Peak | Variance,c % | Increasing Allele | Moded | Locus |
|---|---|---|---|---|---|---|---|---|---|
| BP6,7 | 3F | 58.8 | 49–65 | 3.5 | D3Mit42 | 4.5 | B6 | DomF | Bpq13 |
| BP1–5; BP6,7 | 4 | 9.9 | 0–37 | 5.8* | D4Mit104 | 4.7 | B6 | Add/Rec | Bpq10 |
| BP6,7 | 8 | 37 | 27–46 | 4.2 | D8Mit249 | 4.0 | D2 | Dom | Bpq12 |
| BP1–5 | 14 | 31 | 10–42 | 3.4 | D14Mit203 | 1.9 | B6 | Rec | Bpq11 |
| HR1–5; HR6,7 | 1F | 72 | 48–86 | 7.9† | D1Mit87 | 8.5 | B6 | RecF | Hrq4 |
| HR1–5 | 4 | 1.9 | 0–12 | 3.2 | D4Mit104 | — | B6 | Add/Rec | |
| HR1–5; HR6,7 | 5 | 54 | 45–64 | 8.5† | D5Mit10 | 9.6 | D2 | Dom | Hrq5 |
| HR6,7 | 11 | 2 | 0–24 | 3.1 | D11Mit227 | 2.7 | B6 | Dom | Hrq6 |
1.5 Logarithm of the odds ratio (LOD) drop-off on either side of the quantitative trait locus (QTL) peak;
genome-wide threshold was determined by 1,000 permutations;
Significant (P < 0.05) and
highly significant (P < 0.01) QTL, otherwise suggestive. Where QTL were statistically significant in both 1–5 and 6,7 measures, the measure with the highest level of significance is entered in the table.
Percentage of phenotypic variance accounted for by QTL was obtained for the terms that were retained in the regression model.
Mode of the QTL effect was determined via t-test between marker classes: Dom, homozygous for the increasing allele significantly greater than homozygous for the decreasing allele but not different from heterozygous; Rec, homozygous for the increasing allele significantly greater than heterozygous and homozygous for the decreasing allele; Add/Rec, homozygous for the increasing allele significantly greater than heterozygous or homozygous for the decreasing allele. In case of female-specific QTL
, mode of effect was examined in females only.
Search for epistatic interactions identified an interaction between D1Mit87 and D5Mit10 affecting HR1–5 and an interaction between D10Mit117 and D13Mit230 affecting HR6,7. Neither of these interactions, however, were retained in the regression model, suggesting their limited influence on the phenotypes.
Verification in BXD RIs.
The RI analyses did not reveal any QTL that satisfied the threshold for a whole genome scan. Statistically significant QTL identified in the F2 intercross (Bpq10, Hrq4, and Hrq5) were submitted to the RI panel for confirmation in the following manner. Using the F2 QTL plots, we identified RI markers closest to the F2 QTL peaks. Single marker regression analyses were then conducted for each phenotype in males and females. In each case, a directional prediction from the F2 data regarding the allele associated with increased SBP or HR pointed to the appropriateness of a one-tailed test. In this manner the Hrq5 locus (D5Mit10; P < 0.0243 and P < 0.0198) was verified in females and males, respectively. For the Hrq4 locus marker D1Mit106 was within a 1.5-LOD drop-off interval and was significant at P = 0.021 in females. It has to be noted that these levels of significance do not match criteria of confirmation described by Lander and Kruglyak (27).
SNP analysis.
Extensive SNP analysis was applied to the Bpq10 locus on Chr 4, which was found in the present study and is concordant with the previously identified blood pressure loci Bpq3 and Abbp2 in the A/B6 lineage (60, 74). The B6 allele conferred higher BP at all three loci. Concordance of the loci and allelic effect suggest involvement of the same gene. We used the mouse SNP database (www.ncbi.nlm.gov) to search for nonsynonymous SNPs between the B6 and D2 / A/J strains within the 1.5-LOD support interval (∼1–73,000,000 bp) on Chr 4. There were 33 SNPs found in 20 genes of the region with alleles shared between the A/J and D2 strains but distinct in the B6 strain (Supplemental Fig. S1).1 Variants of the following genes were predicted (see materials and methods) to have probably damaging effects: E130016E03Rik, expressed in heart (GDS2614), embryonic kidney (GDS1583), smooth muscle cells (GDS799); B230312A22Rik, expressed in embryonic kidney (GDS1583), heart (GDS2304), aortic smooth muscle cells (GDS2704); Nipsnap3a, expressed in heart (GDS1228), aortic smooth muscle cells (GDS2704), kidney (GDS1583). The following variants are predicted to be possibly damaging: Tln1, expressed in heart (GDS2614, GDS1228), embryonic kidney (GDS1748), aortic smooth muscle (GDS2704); Mdn1, expressed in heart (GDS1228), embryonic kidney (GDS1583), smooth muscle cells (GDS799); Ltb4dh, expressed in heart (GDS1080, GDS2727), embryonic kidney (GDS1583), aortic smooth muscle (GDS2704). For the rest of the genes variation was predicted to be benign.
SNP analysis of Hrq4 and Hrq5 was focused on promising candidates among the genes exhibiting SNPs between B6 and D2 in the relevant genomic regions (Hrq4 region, 122/674 genes of which 82 were annotated; Hrq5 region, 48/420, of which 27 were annotated). For Hrq4, SNPs in the Chrng gene (nicotinic cholinergic receptor may modify autonomic regulation of heart rate), Mybph (myosin binding protein, expressed in the chicken cardiac conduction system) (1), and Cav1.1 (L-type calcium channel gene, potentially involved in cardiac excitation contraction coupling and discussed later in relation to its potential role in sex differences in cardiac function) were examined. One SNP in the Chrng gene was predicted to be possibly damaging. Those in other genes were considered benign. For Hrq5, an interesting candidate is Adrbk2 (β-adrenergic receptor kinase 2 with potential influence on HR) (6). It has three nonsynonymous SNPs between the B6 and D2 strains. All were predicted benign. Aside from alterations in protein structure and function, variation at QTL could be related to differences in gene expression between B6 and D2 variants. Exploration of potential differences in tissue expression in the many genes within the various QTL intervals were beyond the scope of the present investigation but could be profitably explored in future studies.
Cross-species comparison of cardiovascular QTL and candidate genes.
Each of the statistically significant SBP and HR QTL found in the present experiment was compared with previous studies in the laboratory rat (http://rgd.mcw.edu/). Overlaps between QTL in the present study, previously identified mouse QTL and QTL in homologous regions of the rat and human genome may be inspected in Table 3. There was overlap with QTL in homologous regions of the rat and human genomes for the Chr 4 SBP QTL but for neither of the HR QTL identified in this study. The QTL confidence intervals in all three species are large and do not appreciably reduce the genomic regions (and therefore the number of genes) for scrutiny.
Table 3.
Relationship of statistically significant cardiovascular QTL in the present study to previously identified mouse QTL and homologous QTL in humans and the laboratory rat
|
Mouse |
Rat
|
Human
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| “New” QTL | Chr | cM | Overlaps with* | Overlaps with* | Chr | Mb | Overlaps with* | Chr | Mb |
| Bpq10 | 4 | 5–26 | Bpq3, Abbp2 | Bp7,49,119 | 5 | 62.5–168.3, 62.5–103.2, 2.4–32.4 | BP49_H | 8 | 82.26–118.15 |
| Hrq4 | 1 | 45–91 | Hrq3† | ||||||
| Hrq5 | 5 | 37–66 | |||||||
Overlaps identified from QTL confidence intervals in http://rgd.mcw.edu/.
Demonstration of Hrq3 depends on interaction with Hrq1 on mouse Chr 2 (61). In some studies, BP of mice and rats were measured after animals had been exposed to a high-salt diet.
For the eight annotated genes identified as potential candidates (see above) for Bpq10 and Hrq4 and 5, search was made for their role as candidates in human and rat hypertension and heart rate control (http://rgd.mcw.edu/). None of the genes has been proposed as candidates in these two species, although in two instances (Mybpc3 and Adrbk1), genes in related functional pathways have been implicated (8, 35).
Analysis of RI strain data.
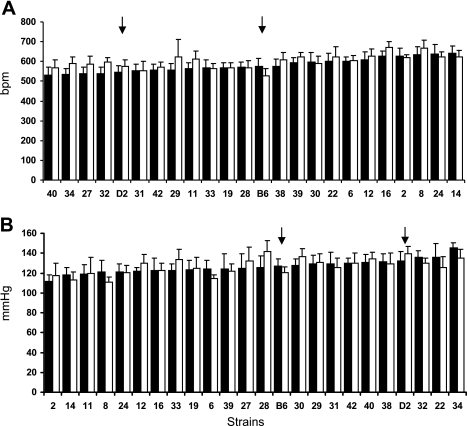
HR1–5 was normally distributed across RI strains. SBP1–5 was positively but modestly skewed. Analysis of variance of SBP1–5 (log transformed) revealed a statistically significant effect of strain (F, 21,470 = 7.7, P < 0.001), no sex difference, but a statistically significant strain by sex interaction (F, 21,470 = 2.7, P < 0.001). SBP1–5 ranged from ∼115 mmHg (BXD2, 8, 14) to 140 mmHg (BXD34). Males and females had similar SBP levels in most strains but differed in specific strains in different directions (BXD28 males, 18 mmHg higher than females; BXD22 females, 12 mmHg higher than males; Fig. 2A).
Fig. 2.
Strain means distribution of heart rate (A), systolic blood pressure (B) in females (dark bars) and males (open bars) of 22 recombinant-inbred, B6 and D2 (arrows) strains. Traits sorted by ascending order of female means.
For HR1–5, there were statistically significant effects of strain (F, 21, 468 = 15.6, P < 0.0001), sex (F, 1,468 = 72.7, P < 0.0001), and a statistically significant interaction between strain and sex (F, 21, 468 = 4.1, P < 0.0001). BXD8 and 16 had HR approaching 640 beats per minute (bpm), while BXD19 and 40 had HR that was ∼100 bpm slower. Although males had faster HR overall the sex difference was strain dependent. The sex-by-strain interactions are depicted in Fig. 2B. Approximately eight strains showed no tendency to sex differences in HR, but the remaining strains tended to exhibit higher HR in males with the sex difference reaching from 60 to 100 bpm in specific strains (e.g., BXD29, 32).
Analysis of variance was used to estimate narrow sense heritability of cardiovascular function from between strain (RI) and total sum of squares (Table 4). The results suggest that the proportion of total variance in cardiovascular function accounted for by between-strain variation in the BXD RIs was substantial in both males and females. Estimates for HR1–5 were somewhat higher than those for SBP1–5.
Table 4.
Narrow sense heritabilities for cardiovascular function
| SBP1–5 | HR1–5 | |
|---|---|---|
| Males | 0.34 | 0.45 |
| Females | 0.29 | 0.48 |
*Calculated from analyses of variance of BXD recombinant inbreds (RIs).
Phenotypic correlations between SBP1–5 and HR1–5 for male and female RI mice were −0.259 and −0.254, respectively (P < 0.001). Genetic correlations (4) were also calculated from strain means: a significant negative genetic correlation between mean HR and mean SBP was found within both males and females (Table 5). Male and female means were moderately positively correlated for HR and SBP, indicating the existence of common genetic factors for males and females.
Table 5.
Genetic correlations (Pearson's r) for mean HR and BP calculated separately for males and females from 22 BXD RI strain means
| fMEAN HR1–5 | fMEAN SBP1–5 | mMEAN HR1–5 | mMEAN SBP1–5 | |
|---|---|---|---|---|
| fMEAN HR1–5 | −0.562 | 0.653 | −0.582 | |
| fMEAN SBP1–5 | −0.25 | 0.502 | ||
| mMEAN HR1–5 | −0.481 | |||
| mMEAN SBP1–5 |
The raw data for this correlation matrix are means of male (prefix “m”) or female (prefix “f”) groups for 22 BXD RI strains. HR in beats per min.1P < 0.05, 2P < 0.01, 3P < 0.001.
DISCUSSION
The present study has identified five chromosomal regions containing allelic variants that influence mean SBP and HR during the first five measurement sessions. The two HR QTL (with the largest effect sizes of all QTL: Table 2) were verified by BXD RI-QTL analyses. The relatively small number of RI strains used (22) limited power for verification of QTL with small effect. In addition, QTL that reflect the action of more than one gene (in F2 analyses) may be disrupted in RI studies because of the multiple opportunities for recombination that occur during inbreeding of RIs. Nonsyntenic association (73) among different regions of the genome, which contain QTL, may also interfere with detection, a problem that is likely to be pronounced when only a small number of RI strains are studied. In any case, the proportion of QTL verified by RI analysis in this study approximates previous F2/RI simulations and empirical comparisons (2, 65).
In addition to the QTL that contributed to variation in SBP and HR during the first five measurement sessions, three other suggestive QTL were discovered (SBP, Chr 3 and 8; HR, Chr 11) to influence the sixth and seventh measurements. Incidence of struggling during BP measurement increased significantly on the sixth and seventh measurement occasions, compared with the level observed during the first five measurements, a finding consistent with the hypothesis that mice had learned to associate handling and restraint with the invasive procedure of blood collection that had taken place 1 mo before both measurement occasions (Fig. 1). Mitochondrial DNA also influenced both SBP and HR differentially in a measurement-specific manner (1–5 vs. 6 and 7). We hypothesize that associations with these invasive procedures activated stress-sensitive biological systems with the capacity to influence cardiovascular function and that allelic differences between B6 and D2 contributed to their variability. It cannot be ruled out, however, that the altered genetic architecture could also be related to age-related changes and/or repeated cardiovascular measurements on the same animals over a 2 mo period. In general, these findings draw attention to the potential of QTL studies of cardiovascular function to incorporate manipulations, which may reveal the influence of novel variants (58). The widely held belief that stress contributes to cardiovascular disease in humans is a powerful reason to carry out additional QTL studies of cardiovascular function incorporating controlled stressors.
Bpq10 on proximal mouse Chr 4 is in the same chromosomal region as Bpq3 previously reported by Sugiyama et al. (60) and Abbp2 by Woo and Kurtz (74) in crosses between A/J and B6 strains. In all three studies, the B6 allele was associated with higher SBP. The Sugiyama et al. study was restricted to male offspring that had been exposed to 1% NaCl for 2 wk. However, the study by Woo and Kurtz (74) and the present study used both male and female subjects, which received no addition of NaCl to their drinking water and ate standard diets. If there is a gene in common to these different reports, it exerts its influence on both males and females and does not require excess NaCl to exert its influence. In the Sugiyama et al. study, D4Mit214 (17.9 cM) and D4Mit164 (28.6 cM) were associated with QTL peaks and D4Mit164 was found to be the best predictor of SBP. However, the more proximal marker, D4Mit214, is close to Bpq10 in the present study and D4Mit164 was outside our 1-LOD drop-off interval. Thus, the present data and those of Woo and Kurtz (74) support a more proximal chromosomal location for Bpq3 than that suggested by the analyses of Sugiyama et al. In any case, the possibility of multiple loci contributing to BP variation in this region of mouse Chr 4 cannot be dismissed (60).
We used similar methods to those employed by Sugiyama et al., but Woo and Kurtz used a different protocol that only required warming of animals to 31°C. Thus, verification of this QTL in different laboratories and under different experimental conditions in our laboratory (see earlier discussion of stress during SBP6,7) speaks to the stability of its influence on cardiovascular function.
Sex differences and SBP and HR QTL.
In previous research with rats, sex specificity of BP QTL has been a pronounced feature of the genetic architecture (30). In the present experiment, Bpq13, the suggestive QTL on Chr 3, was female specific, but the other BP QTL were independent of sex. However, B6 females (but not males) had faster HR than D2s (Table 1), RI analyses revealed pronounced strain by sex interactions (Fig. 2), and Hrq4 was specific to females with B6 as the increasing allele (Table 2). Similar sex-specific findings are evident in studies of human BP. Both systolic and diastolic BP have been shown to be strongly sexually dimorphic yet only SBP manifested a significant sex interaction in heritability (72).
As noted, analyses of RI data showed statistically significant strain by sex interactions for both cardiovascular indexes (Fig. 2). Nevertheless, the finding that genetic correlations calculated across male/female strain means approximated r = 0.6 for cardiovascular indices (Table 5) indicated that there was substantial communality of genes influencing cardiovascular function in males and females. RI data also revealed a significant negative genetic correlation between HR and SBP in both male and female mice. It is accepted that HR and cardiac output are not linearly related because stroke volume decreases with increasing HR, so a positive relationship between SBP and HR would not be expected. Nevertheless, the significant negative relationship between the two measures is intriguing and worthy of further dissection in attempts to understand genetic mechanisms of cardiovascular regulation (9).
QTL with sex-specific effects could be the result of hormonal effects on gene expression and regulation or other nongenetic factors associated with sex differences, providing additional criteria with which to evaluate candidate genes.
Two genes within the Hrq4 confidence interval (Cav1.1 and Rgs2) are worthy of consideration. Cav1.1 encodes an L-type calcium channel, may be expressed in cardiac tissues (54, 63), and has the potential to be involved in cardiac excitation-contraction coupling (3). Estrogen is known to affect cardiac function (15, 57) possibly via its effect on L-type calcium channels (13, 15, 25). Rgs2 (regulator of G protein signaling 2) has been linked to several cardiac phenotypes (18, 21, 44, 62, 64). Perhaps relevant to the specific situation in which our measurements were obtained, Gross et al. (18) suggested that HR in Rgs2 knockout mice may be more sensitive to environmental stimuli, such as handling. Rgs2 also appears to be sensitive to sex-related hormones such as gonadatropin releasing hormone and oxytocin (34, 76). While these background data enhance the plausibility of these two genes as candidates, the large number of genes within the interval needs to be remembered when considering their feasibility as realistic candidates.
Homology mapping.
The identification of mouse QTL for BP parallels similar efforts in other organisms, particularly in the laboratory rat and in humans. One general goal of using the mouse as a model is that the genes underlying the QTL will correspond to homologous genes that also regulate BP in humans. This goal is shared by other investigators studying complex traits (45) and QTL mapping to homologous chromosomal regions in human and mouse have been found for diabetes (53), HDL cholesterol (70), LDL cholesterol and triglycerides (71), bone mineral density (26), and atherosclerosis (69). Such concordance has also been reported for hypertension (59, 60). One murine SBP QTL identified here did show substantial overlap with a homologous region in a rat BP QTL, as well as with several human BP/hypertension QTL. The Chr 3 “suggestive” QTL (Bpq13) overlaps with several QTL on the homologous region of human Chr 3 (19, 43). Bpq10 on Chr 4 at 5–26 cM overlaps with previously identified mouse QTL (Bpq3 and Abbp2), as well as with rat QTL Bp7,49,119 on rat Chr 5 and with a QTL reported for human systolic blood pressure on Chr 8 (42). In previous comparative mapping, five out of six of the then known QTLs for human hypertension were correctly predicted based upon the location of rat QTLs (59). However, a limitation of this approach is the imprecision of the location of the QTLs identified using low resolution genome scans. Further fine mapping of these QTL regions should allow for a more precise physical map location and improved homology mapping. In addition, genome-wide linkage studies of high BP and BP-related phenotypes in humans have now identified well over 100 hypertension-related QTLs across the genome (11), thus the large number of loci may increase the likelihood of chance overlap with mouse QTL found here.
Conclusions
The QTL yield in the present study has been promising. Bpq10 on Chr 4 is concordant with Bpq3 previously identified by Sugiyama et al. (60). A distinction has been made among the other suggestive and statistically significant QTL: several influenced SBP (Bpq10) and HR (Hrq4 and 5) throughout the seven measurement occasions, while others (Bpq 12 and 13) and Hrq7 were only identified after animals had been exposed to invasive stressors several weeks before measurement. We raise the possibility that stress-sensitive processes capable of influencing cardiovascular function may be affected by variation in relevant genes. Two of the QTL (Bpq13 and Hrq4) are demonstrable in females and not in males, and we briefly discuss two candidates within the Hrq4 locus with the potential to play a role in cardiac function and which may be plausibly influenced by hormones in a sex-specific manner.
GRANTS
This work was supported by National Institute on Aging Grants P01 AG-14731 and T32 AG-00276.
Supplementary Material
Acknowledgments
We thank David A. Bienus, Susan E. Lingenfelter, Terri Rothermel, and Kimberley A. Seymour of the barrier facility in the Center for Developmental and Health Genetics for excellent technical assistance.
Address for reprint requests and other correspondence: D. A. Blizard, Center for Developmental and Health Genetics, 201, Research Bldg. D, Pennsylvania State Univ., University Park, PA 16802 (e-mail: dab22@psu.edu).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Alyonycheva T, Cohen-Gould L, Siewert C, Fischman DA, Mikawa T. Skeletal muscle-specific myosin binding protein-H is expressed in Purkinje fibers of the cardiac conduction system. Circ Res 80: 665–672, 1997. [DOI] [PubMed] [Google Scholar]
- 2.Belknap JK, Mitchell SR, O'Toole LA, Helms ML, Crabbe JC. Type I and type II error rates for quantitiative trait loci (QTL) mapping studies using recombinant-inbred mouse strains. Behav Genet 26: 149–160, 1996. [DOI] [PubMed] [Google Scholar]
- 3.Bers DM Cardiac excitation-contraction coupling. Nature 415: 198–205, 2002. [DOI] [PubMed] [Google Scholar]
- 4.Blizard DA, Bailey DW. Genetic correlation between open-field activity and defecation: analysis with CXB recombinant-inbred strains. Behav Genet 9: 349–357, 1979. [DOI] [PubMed] [Google Scholar]
- 5.Blizard DA, Welty R. Cardiac activity in the mouse: strain differences. J Comp Physiol Psychol 77: 337–344, 1971. [DOI] [PubMed] [Google Scholar]
- 6.Brodde OE, Bruck H, Leineweber K. Cardiac adrenoceptors: physiological and pathophysiological relevance. J Pharm Sci 100: 323–337, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Broman KW Mapping quantitative trait loci in the case of a spike in the phenotype distribution. Genetics 163: 1169–1175, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrier L, Bonne G, Bahrend E, Yu B, Richard P, Niel F, Hainque B, Cruaud C, Gary F, Labeit S, Bouhour JB, Dubourg O, Desnos M, Hagege AA, Trent RJ, Komajda M, Fiszman M, Schwartz K. Organization, and sequence of human cardiac myosin binding protein C gene (MYBPC3) and identification of mutations predicted to produce truncated proteins in familial hypertrophic cardiomyopathy. Circ Res 80: 427–434, 1997. [PubMed] [Google Scholar]
- 9.Churchill GA Recombinant inbred strain panels: a tool for systems genetics. Physiol Genomics 31: 174–175, 2007. [DOI] [PubMed] [Google Scholar]
- 10.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowley AW Jr. The genetic dissection of essential hypertension. Nat Rev Genet 7: 829–840, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Cui JS, Hopper JL, Harrap SB. Antihypertensive treatments obscure familial contributions to blood pressure variation. Hypertension 41: 207–210, 2003. [DOI] [PubMed] [Google Scholar]
- 13.Curl CL, Wendt IR, Canny BJ, Kotsanas G. Effects of ovariectomy and 17 beta-oestradiol replacement on Ca2+(i) in female rat cardiac myocytes. Clin Exp Pharm Physiol 30: 489–494, 2003. [DOI] [PubMed] [Google Scholar]
- 14.DiPetrillo K, Tsaih SW, Sheehan S, Johns C, Kelmenson P, Gavras H, Churchill GA, Paigen B. Genetic analysis of blood pressure in C3H/HeJ and SWR/J mice. Physiol Genomics 17: 215–220, 2004. [DOI] [PubMed] [Google Scholar]
- 15.Du XJ, Fang L, Kiriazis H. Sex dimorphism in cardiac pathophysiology: experimental findings, hormonal mechanisms, and molecular mechanisms. Pharm Ther 111: 434–475, 2006. [DOI] [PubMed] [Google Scholar]
- 16.Garrett MR, Rapp JP. Defining the blood pressure QTL on chromosome 7 in Dahl rats by a 177-kb congenic segment containing Cyp11b1. Mamm Genome 14: 268–273, 2003. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Fabre PM, Helou K, Stahl F. Predictions based on the rat-mouse comparative map provide mapping information on over 6000 new rat genes. Mamm Genome 13: 189–193, 2002. [DOI] [PubMed] [Google Scholar]
- 18.Gross V, Tank J, Obst M, Plehm R, Blumer KJ, Diedrich A, Jordan J, Luft FC. Autonomic nervous system and blood pressure regulation in RGS2-deficient mice. Am J Physiol Regul Integr Comp Physiol 288: R1134–R1142, 2005. [DOI] [PubMed] [Google Scholar]
- 19.Hamet P, Merlo E, Seda O, Broeckel U, Tremblay J, Kaldunski M, Gaudet D, Bouchard G, Deslauriers B, Gagnon F, Antoniol G, Pausová Z, Labuda M, Jomphe M, Gossard F, Tremblay G, Kirova R, Tonellato P, Orlov SN, Pintos J, Platko J, Hudson TJ, Rioux JD, Kotchen TA, Cowley AW Jr. Quantitative founder-effect analysis of French Canadian families identifies specific loci contributing to metabolic phenotypes of hypertension. Am J Hum Genet 76: 815–32, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton M, Pickering GW, Roberts JAF, Sowry GSC. The aetiology of hypertension. 4 The role of inheritance. Clin Sci 13: 273, 1954. [PubMed] [Google Scholar]
- 21.Heximer SP, Knutsen RH, Sun XG, Kaltenbronn KM, Rhee MH, Peng N, Oliveira-dos-Santos A, Penninger JM, Muslin AJ, Steinberg TH, Wyss JM, Mecham RP, Blumer KJ. Hypertension and prolonged vasoconstrictor signaling in RGS2-deficient mice. J Clin Invest 111: 1259–1259, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt SC, Ellison RC, Atwood LD, Pankow JS, Province MA, Leppert MF. Genome scans for blood pressure, and hypertension: the National Heart, Lung, and Blood Institute Family Heart Study. Hypertension 40: 1–6, 2002. [DOI] [PubMed] [Google Scholar]
- 23.Jaworski R, Jirout M, Closson S, Breen L, Flodman PL, Spence MA, Kren V, Krenova D, Pravenec M, Printz MP. Heart rate and blood pressure quantitative trait loci for the airpuff startle reaction. Hypertension 39: 348–352, 2002. [DOI] [PubMed] [Google Scholar]
- 24.Joe B, Garrett MR, Dene H, Rapp JP. Substitution mapping of a blood pressure quantitative trait locus to a 2.73 Mb region on rat chromosome 1. J Hypertens 21: 2077–2084, 2003. [DOI] [PubMed] [Google Scholar]
- 25.Johnson BD, Zheng W, Korach KS, Scheuer T, Catterall WA, Rubanyi GM. Increased expression of the cardiac L.-type calcium channel in estrogen receptor-deficient mice. J Gen Physiol 110: 135–140, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein RF Genetic regulation of bone mineral density in mice. J Musculoskelet Neuronal Interact 2: 232–236, 2002. [PubMed] [Google Scholar]
- 27.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Genetics 11: 241–247, 1995. [DOI] [PubMed] [Google Scholar]
- 28.Levan G, Szpirer J, Szpirer C, Klinga K, Hanson C, Islam MQ. The gene map of the Norway rat (Rattus norvegicus) and comparative mapping with mouse and man. Genomics 10: 699–718, 1991. [DOI] [PubMed] [Google Scholar]
- 29.Martin LJ, Comuzzie AG, Sonnenberg GE, Myklebust J, James R, Marks J, Blangero J, Kissebah AH. Major quantitative trait locus for resting heart rate maps to a region on chromosome 4. Hypertension 43: 1146–1151, 2004. [DOI] [PubMed] [Google Scholar]
- 30.McBride MW, Carswell HV, Graham D, Carr FJ, Charchar FJ, Macrae IM, Dominiczak AFD. Genetic and gender determinants of cerebrovascular disease. Sem Nephrol 22: 127–134, 2002. [DOI] [PubMed] [Google Scholar]
- 31.Meng H, Garrett MR, Dene H, Rapp JP. Localization of a blood pressure QTL to a 2.4-cM interval on rat chromosome 9 using congenic strains. Genomics 81: 210–220, 2003. [DOI] [PubMed] [Google Scholar]
- 32.Miall WE, Oldham PD. A study of arterial blood pressure and its inheritance in a sample of the general population. Clin Sci (Lond) 14: 459–488, 1955. [PubMed] [Google Scholar]
- 33.Ostchega Y, Dillon CF, Hughes JP, Carroll M, Yoon S. Trends in hypertension prevalence, awareness, treatment, and control in older U.S. adults: data from the National Health and Nutrition Examination Survey 1988 to 2004. J Am Geriatr Soc 55: 1056–1065, 2007. [DOI] [PubMed] [Google Scholar]
- 34.Park ES, Echetebu CO, Soloff S, Soloff MS. Oxytocin stimulation of RGS2 mRNA expression in cultured human myometrial cells. Am J Physiol Endocrinol Metab 282: E580–E584, 2002. [DOI] [PubMed] [Google Scholar]
- 35.Park SJ, Choi DJ, Kim CW. Hypertensive left ventricular hypertrophy: relation to beta-adrenergic receptor kinase-1 (betaARK1) in peripheral lymphocytes. J Hypertens 22: 1025–32, 2004. [DOI] [PubMed] [Google Scholar]
- 36.Pravenec M, Zidek V, Musilova A, Vorlicek J, Kren V, St Lezin E, Kurtz TW. Genetic isolation of a blood pressure quantitative trait locus on chromosome 2 in the spontaneously hypertensive rat. J Hypertens 19: 1061–1064, 2001. [DOI] [PubMed] [Google Scholar]
- 37.Pravenec M, Kren V, Krenova D, Zidek V, Simakova M, Musilova A, Vorlicek J, Lezin ES, Kurtz TW. Genetic isolation of quantitative trait loci for blood pressure development, and renal mass on chromosome 5 in the spontaneously hypertensive rat. Physiol Res 52: 285–289, 2003. [PubMed] [Google Scholar]
- 38.Province MA, Kardia SLR, Ranade K, Rao DC, Thiel BA, Cooper RS, Risch N, Turner ST, Cox DR, Hunt SC, Weder AB, Boerwinkle EA. Meta-analysis of genome-wide linkage scans for hypertension: the National Heart Lung and Blood Institute Family Blood Pressure Program. Am J Hypertens 16: 144–147, 2003. [DOI] [PubMed] [Google Scholar]
- 39.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 30: 3894–3900, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao DC, Province MA, Leppert MF, Oberman A, Heiss G, Ellison RC, Arnett DK, Eckfeldt JH, Schwander K, Mockrin SC, Hunt SC. A genome-wide affected sibpair linkage analysis of hypertension: the HyperGEN Network. Am J Hypertens 16: 148–150, 2003. [DOI] [PubMed] [Google Scholar]
- 41.Rapp JP Genetic analysis of inherited hypertension in the rat. Physiol Res 80: 135–172, 2000. [DOI] [PubMed] [Google Scholar]
- 42.Rice T, Rankinen T, Province MA, Chagnon YC, Pérusse L, Borecki IB, Bouchard C, Rao DC. Genome-wide linkage analysis of systolic and diastolic blood pressure: the Québec Family Study. Circulation 102: 1956–1963, 2000. [DOI] [PubMed] [Google Scholar]
- 43.Rice T, Rankinen T, Chagnon YC, Province MA, Pérusse L, Leon AS, Skinner JS, Wilmore JH, Bouchard C, Rao DC. Genome-wide linkage scan of resting blood pressure: HERITAGE Family Study. Health, risk factors, exercise training, and genetics. Hypertension 39: 1037–1043, 2002. [DOI] [PubMed] [Google Scholar]
- 44.Riddle EL, Rana BK, Murthy KK, Rao FW, Eskin E, O'Connor DT, Insel PA. Polymorphisms and haplotypes of the regulator of G protein signaling-2 gene in normotensives and hypertensives. Hypertension 47: 415–420, 2006. [DOI] [PubMed] [Google Scholar]
- 45.Rollins J, Chen Y, Paigen B, Wang X. In search of new targets for plasma high-density lipoprotein cholesterol levels: promise of human-mouse comparative genomics. Trends Cardiovasc Med 16, 220–34, 2006. [DOI] [PubMed] [Google Scholar]
- 46.Saad Y, Garrett MR, Manickavasagam E, Yerga-Woolwine S, Farms P, Radecki T, Joe B. Fine-mapping and comprehensive transcript analysis reveals nonsynonymous variants within a novel 1.17 Mb blood pressure QTL region on rat chromosome 10. Genomics 89: 343–353, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlager G Heritability of blood pressure in mice. J Hered 56: 278–284, 1965. [DOI] [PubMed] [Google Scholar]
- 48.Schlager G Genetic and physiological studies of blood pressure in mice. I. Crosses between A/J, SWR/J and their hybrids. Can J Genet Cytol 10: 853–864, 1968. [DOI] [PubMed] [Google Scholar]
- 49.Schlager G Selection for blood pressure levels in mice. Genetics 76: 537–549, 1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlager G Biometrical genetic analysis of blood pressure levels in the genetically hypertensive mouse. Clin Exp Hypertens 16: 809–824, 1994. [DOI] [PubMed] [Google Scholar]
- 51.Schlager G, El Seoudy AA. Genetic analysis of blood pressure levels in the genetically hypertensive mouse. In: Hypertensive mechanisms–The Spontaneously Hypertensive Rat as a Model to Study Human Hypertension edited by Rascher W, Clough D, Ganten D. Stuttgart: Schattauer Verlag, 1982, p. 147–150.
- 52.Schlager G, Weibust RS. Genetic control of blood pressure in mice. Genetics 55: 497–506, 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt C, Gonzaludo NP, Strunk S, Dahm S, Schuchhardt J, Kleinjung F, Wuschke S, Joost HG, Al-Hasani H. A meta-analysis of QTL for diabetes-related traits in rodents. Physiol Genomics 34: 42–53, 2008. [DOI] [PubMed] [Google Scholar]
- 54.Seisenberger C, Specht V, Welling A, Platzer J, Pfeifer A, Kühbandner S, Striessnig J, Klugbauer N, Feil R, Hofmann F. Functional embryonic cardiomyocytes after disruption of the L-type alpha(1C) [Ca(v)1.2] calcium channel gene in the mouse. J Biol Chem 275: 39193–39199, 2000. [DOI] [PubMed] [Google Scholar]
- 55.Serikawa T, Cui Z, Yokoi N, Kuramoto T, Kondo Y, Kitada K, Guenet JL. A comparative genetic map of rat, mouse and human genomes. Exp Anim 47: 1–9, 1998. [DOI] [PubMed] [Google Scholar]
- 56.Silva GJJ, Pereira AC, Krieger EM, Krieger JE. Genetic mapping of a new heart rate QTL on chromosome 8 of spontaneously hypertensive rats. BMC Med Genet 8: 17, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stevenson JC Various actions of oestrogens on the vascular system. Maturitas 30: 5–9, 1998. [DOI] [PubMed] [Google Scholar]
- 58.Stoll M, Cowley AW Jr, Tonellato PJ, Greene AS, Kaldunski ML, Roman RJ, Dumas P, Schork NJ, Wang Z, Jacob HJ. A genomic-systems biology map for cardiovascular function. Science 294: 1723–1726, 2001. [DOI] [PubMed] [Google Scholar]
- 59.Stoll M, Kwitek-Black AE, Cowley AW Jr, Harris EL, Harrap SB, Krieger JE, Printz MP, Provoost AP, Sassard J, Jacob HJ. New target regions for human hypertension via comparative genomics. Genome Res 10: 473–482, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugiyama F, Churchill GA, Higgins DC, Johns C, Makaritsis KP, Haralambos G, Paigen B. Concordance of murine quantitative QTL for salt-induced hypertension with rat and human loci. Genomics 71: 70–77, 2001. [DOI] [PubMed] [Google Scholar]
- 61.Sugiyama F, Churchill GA, Li R, Libby LJ, Carver T, Yagami K, John SW, Paigen B. QTL associated with blood pressure, heart rate and heart weight in CBA/CaJ and BALB/cJ mice. Physiol Genomics 10: 5–12, 2002. [DOI] [PubMed] [Google Scholar]
- 62.Sun XG, Kaltenbronn KM, Steinberg TH, Blumer KJ. RGS2 is a mediator of nitric oxide action on blood pressure and vasoconstrictor signaling. Mol Pharmacol 67: 631–639, 2005. [DOI] [PubMed] [Google Scholar]
- 63.Takemura H, Yasui K, Opthof T, Niwa N, Horiba M, Shimizu A, Lee JK, Honjo H, Kamiya K, Ueda Y, Kodama I. Subtype switching of L-Type Ca2+ channel from Cav1.3 to Cav12 in embryonic murine ventricle. Circ J 69: 1405–1411, 2005. [DOI] [PubMed] [Google Scholar]
- 64.Tang M, Wang G, Lu P, Karas RH, Aronovitz M, Heximer SP, Kaltenbronn KM, Blumer KJ, Siderovski DP, Zhu Y, Mendelsohn ME. Regulator of G-protein signaling-2 mediates vascular smooth muscle relaxation and blood pressure. Nat Med 9: 1506–1512, 2003. [DOI] [PubMed] [Google Scholar]
- 65.Tarantino LM, McClearn GE, Rodriguez LA, Plomin R. Confirmation of quantitative ttrait loci for alcohol preference in mice Alc. Clin Exp Res 22: 1099–1105, 1998. [PubMed] [Google Scholar]
- 66.Taylor BA Recombinant-inbred strains: use in gene-mapping. In: Origins of Inbred Mice, edited by Morse H. New York: Academic, 1978, 423–438.
- 67.Taylor BA, Wnek C, Kotlus BS, Roemer N, MacTaggart T, Phillips SJ. Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm Genome 10: 335–348, 1999. [DOI] [PubMed] [Google Scholar]
- 68.Vandenbergh DJ, Heron K, Peterson R, Shpargel KB, Woodroffe A, Blizard DA, McClearn GE, Vogler GP. Simple tests to detect errors in high-throughput genotype data in the molecular laboratory. J Biomol Techn 14: 9–16, 2003. [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X, Ishimori N, Korstanje R, Rollins J, Paigen B. Identifying novel genes for atherosclerosis through mouse-human comparative genetics. Am J Hum Genet 77: 1–15, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, Paigen B. Quantitative trait loci and candidate genes regulating HDL cholesterol: a murine chromosome map. Arterioscler Thromb Vasc Biol 22: 1390–1401, 2002. [DOI] [PubMed] [Google Scholar]
- 71.Wang X, Paigen B. Genome-wide search for new genes controlling plasma lipid concentrations in mice and humans. Curr Opin Lipidol 16: 127–137, 2005. [DOI] [PubMed] [Google Scholar]
- 72.Weiss LA, Pan L, Abney M, Ober C. The sex-specific genetic architecture of quantitative traits in humans. Nat Genet 38: 218–222, 2006. [DOI] [PubMed] [Google Scholar]
- 73.Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol 2: 11, RESEARCH0046, 2001. [DOI] [PMC free article] [PubMed]
- 74.Woo DL, Kurtz I. Mapping blood pressure loci in (A/J × B6)F2 mice. Physiol Genomics 15: 236–242, 2003. [DOI] [PubMed] [Google Scholar]
- 75.Wright FA, O'Connor DT, Roberts E, Kutey G, Berry CC, Yoneda LU, Timberlake D, Schlager G. Genome scan for blood pressure loci in mice. Hypertension 34: 625–630, 1999. [DOI] [PubMed] [Google Scholar]
- 76.Wurmbach E, Yuen T, Ebersole BJ, Sealfon SC. Gonadotropin-releasing hormone receptor-coupled gene network organization. J Biol Chem 276: 47195–47201, 2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.