Abstract
Adhesion to epithelial cells1 and flagella-mediated motility are critical virulence traits for many Gram-negative pathogens, including enterotoxigenic Escherichia coli (ETEC)2, a major cause of diarrhoea in travellers and children in developing countries3,4. Many flagellated pathogens export putative adhesins belonging to the two-partner secretion (TPS) family5. However, the actual function of these adhesins remains largely undefined. Here we demonstrate that EtpA, a TPS exoprotein adhesin of enterotoxigenic Escherichia coli6, mimics and interacts with highly conserved regions of flagellin, the major subunit of flagella, and that these interactions are critical for adherence and intestinal colonization. Although conserved regions of flagellin are mostly buried in the flagellar shaft7, our results suggest that they are at least transiently exposed at the tips of flagella where they capture EtpA adhesin molecules for presentation to eukaryotic receptors. Similarity of EtpA to molecules encoded by other motile pathogens suggests a potential common paradigm for bacterial adhesion, while participation of conserved regions of flagellin in adherence has implications for development of vaccines for Gram-negative pathogens.
ETEC cause diarrhoea by delivery of heat labile and/or heat stable enterotoxins to the small intestine, a process that requires critical fimbrial bacterial adhesins known as colonization factors (CF)8,9. These essential proteinaceous finger-like projections are central to ETEC vaccines currently in development10. Generally, adhesion to intestinal epithelium by diarrhoeagenic E. coli is a very complex process that may involve a number of structures including flagella11. The role of flagella in ETEC pathogenesis has not been sufficiently explored12. Flagella are complex cylindrical structures assembled from approximately 20,000 flagellin (FliC) molecules that travel down the nascent flagellar cylinder to the distal tip where they are directed by cap proteins (FliD)13 into the growing flagellum. Flagellin has several major domains: the central domain projects on the surface of the flagellar shaft, accounting for antigenic variation used in E. coli “H” serotyping; conserved amino and carboxy regions interact with adjacent subunits, facing the inaccessible shaft core7.
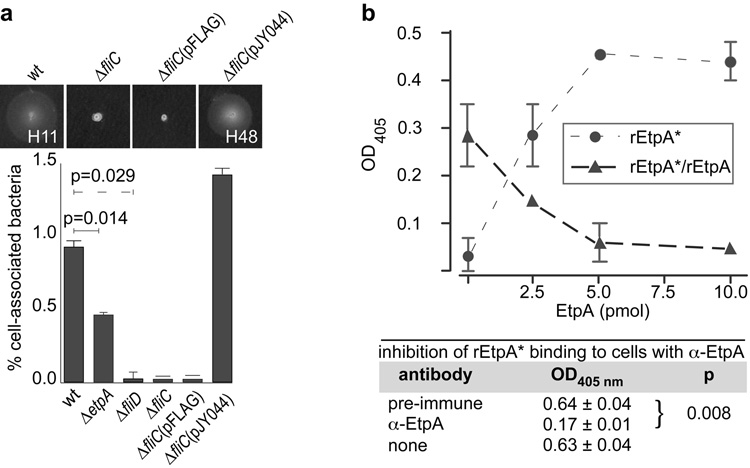
Efficient adherence of ETEC to intestinal cells required both intact flagella and etpA, however flagella-dependent adherence was independent of serotype, as complementation of a fliC (H11−) mutant with fliC (H48) restored both motility and adherence (Fig. 1a). Likewise, antibody generated against (H48) full-length flagellin inhibited adherence of ETEC (H11) (Supplementary Fig. 1a) in contrast to antibody against serotype-dependent regions of flagellin (Supplementary Fig. 1b). Similarly, anti-EtpA antibodies inhibited adherence by EtpA-producing ETEC of multiple (H11, H12, and H16) serotypes while amounts of endogenous (Supplementary Fig. 1c) or exogenously added EtpA (Supplementary Fig. 1d) paralleled the adherence phenotype.
Figure 1. EtpA and flagella contribute to ETEC adhesion.
a, Efficient ETEC adherence requires production of EtpA and intact flagella and is not dependent on flagellar serotype. wt = wild type ETEC stain H10407. Complementation of the fliC (H11−) isogenic deletion strain with pJY044 expressing fliC gene from MG1655 (E. coli K-12, flagellar serotype H48) restored motility and adherence phenotypes. p values (Mann-Whitney) generated by one-tailed test (etpA mutant and wt); others were two-tailed tests. (mean ± s.d. for n=4 replicates). b, EtpA binds specifically to target cells. Binding of purified biotinylated rEtpA (rEtpA*) to target Caco-2 epithelial cells was inhibited by unlabeled protein [mean values (n=3) ± s.e.m.]. Table below the graph demonstrates inhibition of EtpA* binding with EtpA antisera [mean values (n=3) ± s.e.m; p=0.0076: unpaired t test with Welch’s correction].
Recombinant EtpA labelled (Supplementary Fig. 2a) and bound specifically to the surface of intestinal cells, while antibodies against EtpA prevented this interaction (Fig. 1b). Interestingly, labelled EtpA localized to mucin-producing regions of small intestine (Supplementary Fig. 2b), suggesting that EtpA could promote ETEC interaction with intestinal mucosal surfaces.
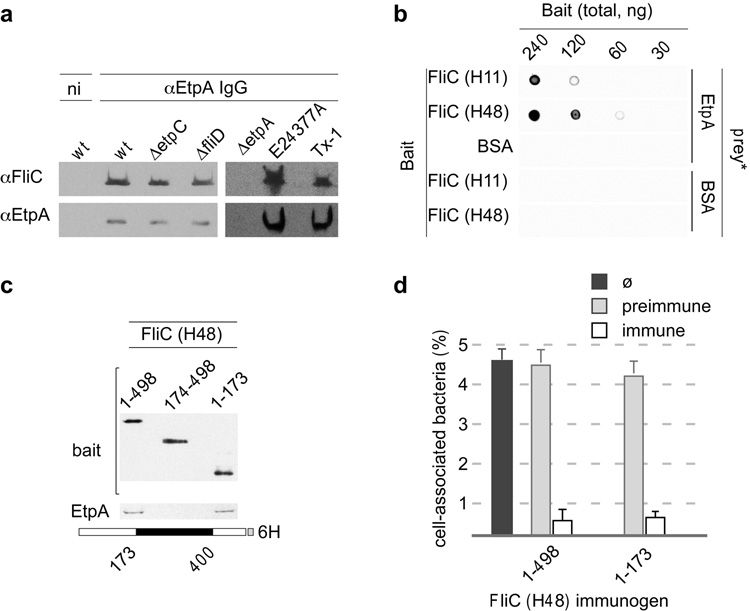
Theoretically, EtpA must maintain contact with ETEC to promote adherence. Attempts to purify recombinant EtpA from E. coli6 were confounded by co-isolation of another protein (≈50 kDa) despite attempted separation by column chromatography (Supplementary Fig. 3a), suggesting a potential protein-protein interaction14. MALDI-TOF definitively identified the co-purified protein as E. coli K-12 flagellin (H48, the same serotype as the recombinant used in the expression) (Supplementary Fig. 3b). Application of this purification strategy to ETEC strain H10407 (serotype H11) supernatants demonstrated that all EtpA-containing fractions also contained flagellin (Supplementary Fig. 3c), suggesting that EtpA interacts with multiple H serotypes of flagellin, and serendipitously alluding to a mechanism of action for EtpA. Co-immunoprecipitation of H10407 culture supernatants with anti-EtpA antibody6 confirmed that FliC (H11) and EtpA interact (Fig. 2a). FliC secreted by the fliD mutant, which cannot assemble monomers into intact flagella2, still interacted with EtpA, implying that EtpA can bind monomeric flagellin. Glycosylation of EtpA, a process dependent on the etpC gene6, also did not appear to be required. In co-IP experiments using additional motile ETEC strains, we could immunoprecipitate flagellin from EtpA-producing strains E24377A (H28) and TX-1 (H12), but not the etpA mutant control. Similarly, immobilized flagellins from different serotypes (FliCH11, and FliCH48) captured biotin-labelled EtpA (Fig. 2b) further demonstrating that these interactions are not dependent on flagellar serotype.
Figure 2. EtpA interacts with conserved regions of flagellins.
a, Co-immunoprecipitation of EtpA and flagellin from ETEC culture supernatants using anti-EtpA antibody. αEtpA IgG=immune antibody; ni=purified non-immune IgG (same rabbit). From left to right: wt, etpC mutant, fliD mutant; control isogenic etpA mutant (jf1668); other motile ETEC strains, E24377A (H28), TX-1 (H12). b, EtpA interacts with multiple flagellin serotypes. Membrane-immobilized FliCH11 and FliCH48 (bait) capture biotinylated rEtpA (“prey*”). c, EtpA interacts with a conserved region of flagellin. Immunoblots (anti-flagella, top; anti-EtpA, bottom) represent molecular pull-down experiments using recombinant polyhistidine-tagged FliC H48-based molecules as bait. Schematic at bottom of figure depicts full-length recombinant flagellin serotype H48. Conserved (white), serotype-dependent (black) regions are shown. d, Antibodies against conserved regions of flagellin (H48) block adherence of ETEC strain H10407 (H11) to Caco-2 epithelial cells. (n=3, error bars=s.e.m.)
These interactions of EtpA with FliC monomers of multiple serotypes suggest that EtpA engages highly conserved regions of these molecules. Interestingly, position-specific-iterative-BLAST (PSI-BLAST)15 searches identified not only putative bacterial adhesins, but subtle homology with amino and carboxy terminal regions of flagellins raising the possibility that EtpA interacts with multiple serotypes by mimicking these conserved domains. We next constructed recombinant (H48) flagellin proteins representing the full-length molecule including the unique serotype-determining region, as well as a truncated version consisting of amino acids 1–173 (the conserved N-terminal domain), and a third peptide containing amino acids 174–498. The conserved N-terminal domain was both necessary and sufficient to pull down EtpA (Fig. 2c). In addition, antibodies against either the full-length H48 flagellin or the conserved N-terminal (1–173) region significantly inhibited adherence of ETEC H10407 (H11) to Caco-2 cells (Fig. 2d). Together, these data implicate highly conserved regions of flagellin molecules in both EtpA binding and in adherence.
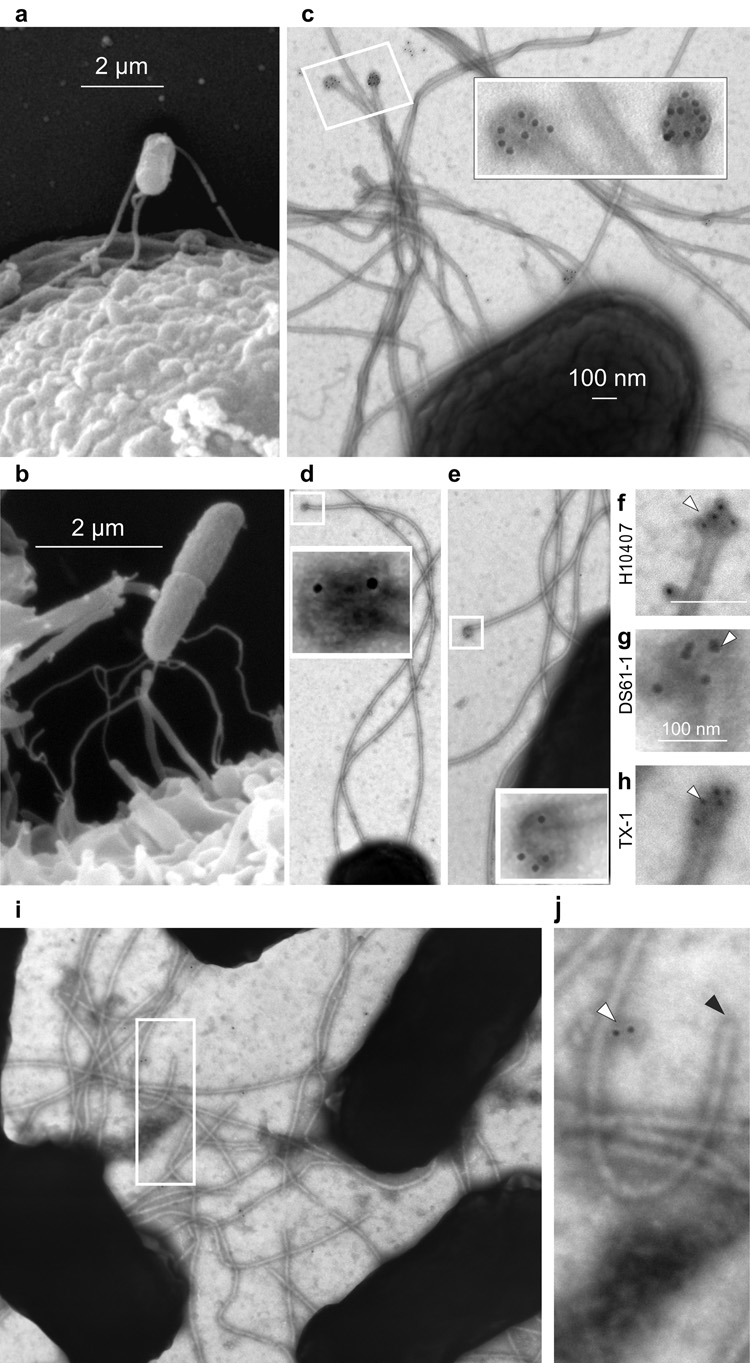
Intriguingly, following infection of host cells, ETEC were rapidly tethered to the surface via flagella (Fig. 3a–b). Although conserved regions (CR) of flagellin are largely buried within the flagellar shaft 7, we reasoned that EtpA might interact with CR of monomers available at the ends of flagella, particularly if the rotating FliD pentameric cap became dislodged from the tip. Accordingly, in situ immunogold labelling studies indicated that conserved FliC regions are available exclusively at the tips of flagella (Fig. 3c–e). Likewise, EtpA bound largely at tips of intact flagella from ETEC strains belonging to different serotypes (Fig. 3f–h). Conversely, fragmentation of flagella exposed additional EtpA binding sites supporting the concept that EtpA interacts with regions of flagellin typically buried within the flagellar shaft.
Figure 3. ETEC engage host cells via tips of flagella where EtpA interacts with highly conserved regions of flagellin.
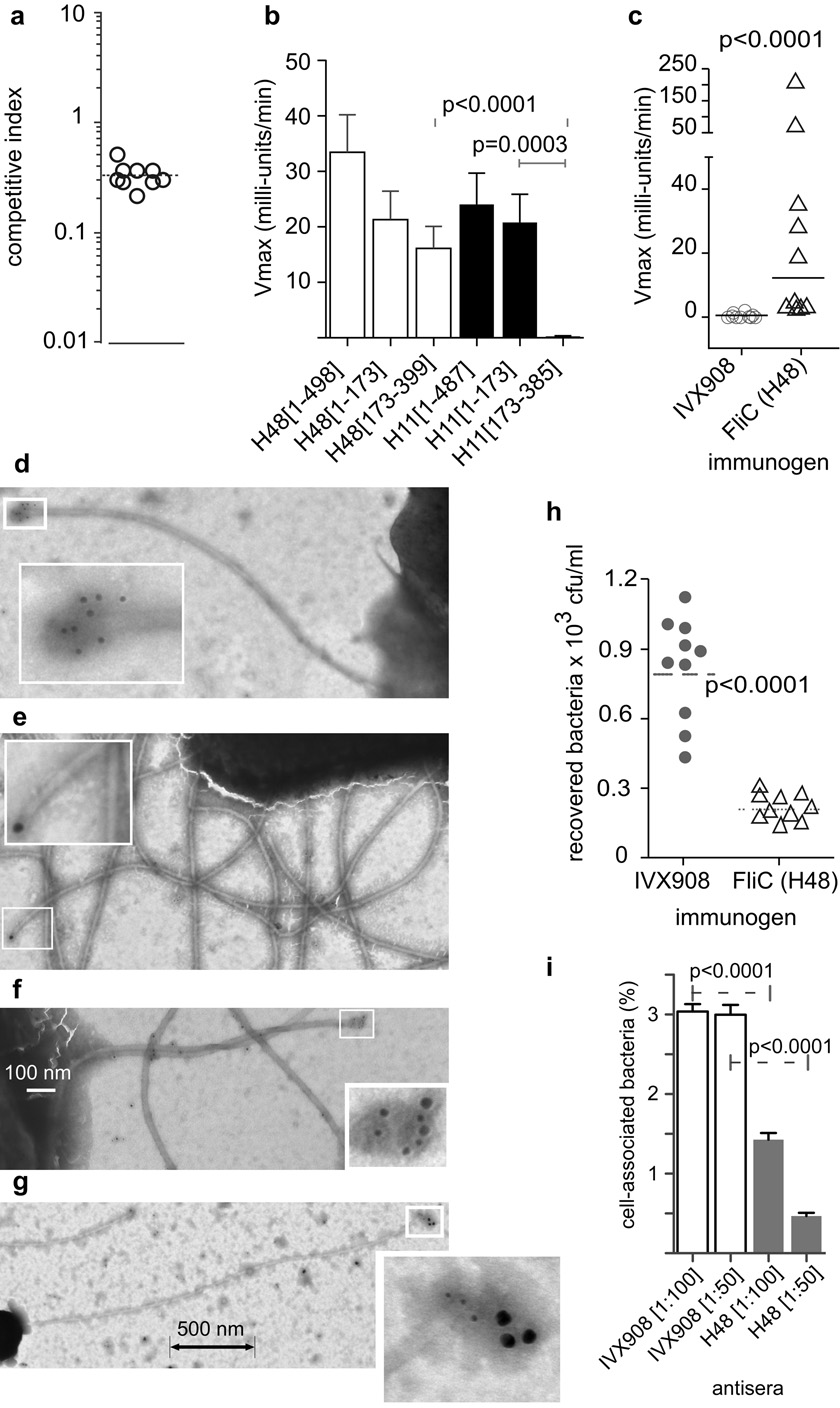
a–b, ETEC initially engage cells via the ends of their flagella. c–e, conserved regions of flagellin are exposed at the tips of H10407 (O78:H11) flagella shown by in situ immunolocalization using: c, anti-(H48)flagellar antibody affinity purified vs full-length (amino acids 1–498) rFliCH48; d, e: affinity-purified antisera against highly conserved N-terminal 173 amino acids of H48 flagellin. f–h, EtpA concentrates at flagellar tips (white arrowhead) of motile EtpA-producing ETEC strains: f, H10407 (O78:H11) g, DS61-1 (O6:H16), and h, TX-1 (O78:H12). i–j, Control labeling experiment with anti-EtpA antibody and secondary gold conjugate using an EtpA-negative strain (DS220-4) demonstrates minimal background (white arrow) with no labelling of flagellar tips (black arrow).
Because EtpA appeared to be concentrated at the tips of flagella, we also investigated whether rEtpA might also interact with the flagellar cap protein, FliD. However, we found no evidence for this in molecular pull-down experiments (Supplementary Fig. 4a). Additionally, we found that in vitro FliD inhibited interaction of EtpA with FliC (Supplementary Fig. 4b), suggesting that the FliD cap might inhibit binding of EtpA to (CR) of flagellin. Similarly, we could not demonstrate co-localization of either the CR or EtpA with the FliD protein (Supplementary Fig. 4c–i) arguing that when FliD is firmly attached, the CR are inaccessible to EtpA. Exogenously added rEtpA localized to the flagellar tips of the etpA mutant (Supplementary Fig. 4j–k) without compromising length, and deletion of etpA had no effect on flagellar length (Supplementary Fig. 5). Collectively, these data suggest that EtpA, binds opportunistically to conserved residues of flagellin at the tips of mature flagella when these domains are exposed by loss of the FliD cap protein complex7, or as these molecules leak16 from ends of flagella in the absence of an intact cap structure. We believe that our data, generated by in situ immunogold labelling17 of whole bacteria, are consistent with the highly dynamic rotating pentameric cap structure proposed previously based on elegant electron cryomicroscopic techniques13,18.
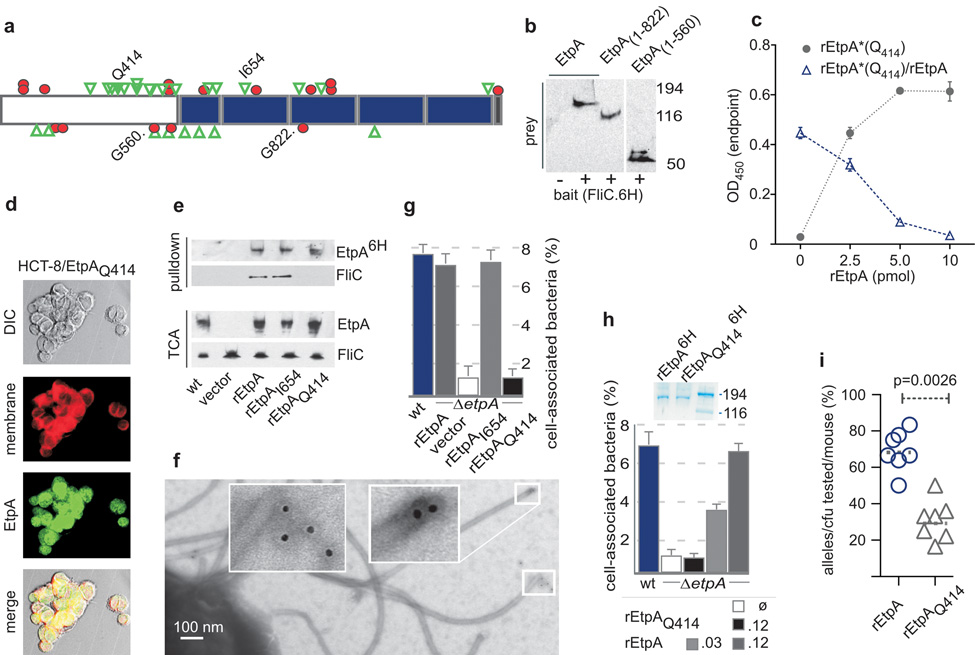
To investigate whether the EtpA-flagellin interaction was required for the adherence phenotype we performed linker scanning mutagenesis19 of etpA (Fig. 4a). In general, mutations within the repeat region of etpA did not interfere with secretion of EtpA or with binding to flagellin (Fig. 4b), suggesting that the 5’ end of etpA encodes domains essential for secretion (similar to other TPS molecules) and for flagellin binding. Further analysis of in-frame mutations mapping to this region lead to the identification of a secreted EtpA mutant molecule with an insertion beginning at residue Q414 of EtpA which retained its ability to bind to host cells (Fig. 4c–d) but that had lost the capacity to bind flagellin (Fig. 4e). Recombinant myc-tagged wt EtpA expressed in the etpA mutant localized to the tips of flagella (Fig. 4f) unlike the Q414 molecule. Compared to the wild type etpA gene, the mutant allele failed to complement the in vitro adherence defect in an isogenic etpA deletion strain (Fig. 4g), or when the mutated protein (EtpAQ414) was added exogenously to this mutant (Fig. 4h). EtpA is required for optimal colonization of the intestine by ETEC20. Likewise, bacteria expressing EtpAQ414 were not able to compete efficiently in intestinal colonization with those expressing the wt etpA gene (Fig. 4i), suggesting that the association of EtpA and flagellin is critical for efficient interaction of ETEC with target host cells, and for colonization of mucosal surfaces.
Figure 4. ETEC adherence to epithelial cells in vitro and small intestinal colonization require the interaction of EtpA and flagellin.
a, Linker scanning mutation map of EtpA (blue=repeats, red circles = stop codons, green triangles = in-frame insertions). b, Truncated N-terminal region of EtpA (M1-G560) is sufficient to interact with flagellin (H48) polyhistidine-tagged (FliC.6H) bait in molecular pull-downs. c, EtpAQ414 retains specific binding to Caco-2 cells. Circles=biotinylated (rEtpA*Q414). Triangles=competition with unlabelled rEtpA [mean (n=3) values ± s.e.m.]. d, EtpAQ414 retains binding to HCT-8 epithelial cells surfaces (confocal microscopy). e. EtpAQ414 does not interact with flagellin. etpA mutant (jf1668) was complemented with myc-polyhistidine-tagged EtpA variants as indicated prior to pull-down studies of supernatants with metal affinity resin. f, rEtpA-myc-6H identified with anti-myc 1° monoclonal antibody at flagellar tips of etpA mutant (jf1668) expressing pJY017. g, EtpAQ414 expression in isogenic etpA mutant (jf1668) fails to complement adherence (Caco-2) defect. h, Exogenous rEtpA, but not rEtpAQ414 complements the adherence defect in the etpA mutant. SDS-PAGE above the graph shows rEtpA(6H) and rEtpAQ414(6H) used in assays. (key = final [nM] concentrations). i, EtpAQ414 fails to complement colonization defect when expressed in isogenic etpA mutant. Graph shows the proportion of cfu bearing either the wild type allele (carried on pJL017) or mutant allele (carried on pJMF1087) recovered from each mouse (N=7) following simultaneous challenge with 1 × 104cfu of both strains. p=0.0026 (two-tailed Mann-Whitney test). An average of 15 colonies per mouse were analyzed by PCR and PmeI digestion.
Similar to other enteric pathogens21, we found that production of intact flagella was also required for efficient intestinal colonization by ETEC (Fig. 5a). Since EtpA interactions with CR of flagellin are critical to promoting colonization, and EtpA is a protective antigen in mice20, we examined whether immunization of mice with a single H serotype of flagellin could target shared regions of flagellins thereby affording heterologous protection against intestinal colonization. Immunization with full-length flagellin (H48) stimulated production of serum (Fig. 5b) and fecal22 (Fig. 5c) antibodies recognizing conserved regions of flagellin, accessible at the tips of H11 flagella (Fig. 5d–g) and dramatically impaired colonization by ETEC (Fig. 5h). These antibodies also inhibited adherence of ETEC expressing the heterologous flagellin H11 serotype (Fig. 5i). Collectively, these studies suggest that conserved residues of flagellin, implicated here in bacterial adherence and colonization, could also serve as viable vaccine targets.
Figure 5. Vaccination with flagellin inhibits ETEC infection in mice.
a, Motile ETEC bacteria out-compete fliC− mutants (geometric mean index≈0.33). b, Kinetic ELISA demonstrating that sera (1:128 dilution) from mice (n=10) immunized with H48 flagellin recognize full-length and N-terminal conserved regions of H48, and H11 molecules, serotype specific region of H48 (H48[173–399]), not serotype specific region of H11 (H11[174–385]). Values obtained with pooled sera from IVX908 control mice have been subtracted. [error bars=s.e.m.] c, Mice vaccinated with full-length H48 recombinant flagellin produce fecal antibodies against conserved regions of H11 flagellin. Shown are kinetic ELISA data from mice vaccinated intranasally with FliCH48/IVX908 (triangles) vs. IVX908 controls (circles). Horizontal bars = geometric mean values (p<0.0001, Mann-Whitney, 2-tailed) d, EtpA localized at tip of H10407 flagellum. e, antisera from mice immunized with H48 recognize the tips of ETEC H10407 (H11) flagella. f–g, EtpA and conserved regions of flagellin co-localize at tips of H10407 flagella. Primary antisera: (1) pooled from H48-immunized mice with (2) rabbit anti-EtpA; Secondary anti-mouse/anti-rabbit (IgG) gold conjugates: (10 nm/5 nm in f and 20 nm/10 nm in g). h, Vaccination with FliC (H48) flagellin protects against colonization with ETEC H10407 (H11). i, polyclonal anti-sera from mice immunized with recombinant H48 flagellin inhibits adherence of H10407 (H11) to Caco-2 cells in vitro. bars=mean ± s.e.m. of (n=8) wells. [all p values calculated using 2-tailed Mann-Whitney, except in i, (1-tailed test)].
These studies offer a novel paradigm for bacterial adhesion as they demonstrate for the first time that a secreted virulence protein interacts with highly conserved regions of flagellin exposed at the tips of flagella, exploiting these long (≈10–15 µm) appendages to tether EtpA adhesins that anchor bacteria on initial engagement of host cells. The precise contributions of EtpA and flagella relative to the established critical role of colonization factors in ETEC adherence remain undefined. However, preliminary data suggest that these are early transient interactions acting in concert to facilitate resilient pathogen-host interactions mediated by shorter (0.5–1µm) spring-like23 colonization factors.
EtpA or EtpA-like molecules are present in many ETEC strains from multiple flagellar serotypes6, suggesting that interactions between EtpA and flagellin are not limited to a small subset of ETEC strains. Likewise, homology searches of EtpA revealed the presence of many potential EtpA-like molecules in other motile Gram-negative pathogens (Supplementary Fig. 6) raising the possibility that these could function in a similar fashion. Interestingly, another TPS exoprotein, HMW1 (Haemophilus influenzae), employs disulfide bonds between carboxy-terminal cysteine residues to link fibrillar adhesin structures to the bacterial surface24. Absence of similar residues in EtpA and EtpA-like TPS proteins identified in genomes of other flagellated pathogens5 may permit free secretion of these molecules for interactions with flagella.
Flagellar organelles have previously been dismissed in development of ETEC vaccines due to substantial variation in H serotypes and the assumption that only variant FliC regions were exposed12. The highly conserved flagellin domains implicated in the present studies, potent stimuli of innate immunity25, might also be exposed on commensal organisms. However, dependence of ETEC flagella-mediated adhesion on TPS, a unique adaptation of pathogenic bacteria, is likely one critical step in a process involving several adhesins including well-defined ETEC fimbrial colonization factors8. Although the precise mechanism linking EtpA and flagella is not entirely clear, our data do suggest that EtpA and similar molecules as well as highly conserved flagellin ligands could serve as viable antigenic targets in design of novel vaccines to prevent infections caused by ETEC and other important motile pathogens.
Methods Summary
Detailed methods are contained within the Supplementary Information (SI). A description of bacterial strains and plasmids used is given in Supplementary Table 1. Primers used are detailed in Supplementary Table 2. Adherence6 and motility assays2 were done as previously described. Mouse intestinal colonization studies followed established protocols20,26. Antisera against recombinant EtpA, FliC, and FliD were generated in rabbits6 and mouse polyclonal antibodies were recovered as previously outlined20. For in situ gold labelling17, bacteria were fixed on carbon-coated nickel grids and probed with anti-EtpA, -FliC or -FliD antibodies followed by immunodetection using secondary antibodies coupled to gold particles of various diameters.
Cloning, protein expression and purification techniques are described in detail in the Methods section of SI. Beads pre-charged with Co2+ (Talon® Clontech) were used to pull down polyhistidine-tagged proteins in protein interaction studies. Co-immunoprecipitation was performed using EtpA antibody bound to solid support gel matrix (Pierce).
Comparisons of data were performed using the nonparametric Mann-Whitney (two-tailed) test unless otherwise specified. Statistical calculations were performed using Prism v4.0c and InStat v3.0 (GraphPad Software).
Full Methods and associated references are available in the online version of the paper at http://www.nature.com/nature.
Methods
Binding of EtpA to the surface of intestinal epithelial cells
Concentrated recombinant EtpA (rEtpA) was added to live HCT-8 or Caco-2 cells at a final concentration of 12.5 µg/ml in PBS. After incubation with the cells at 4°C for one hour, cells were washed, and fixed with methanol. Bound EtpA was detected by immunofluorescence using anti-EtpA primary antibodies6, and secondary anti-rabbit Alexa Fluor 488-conjugated IgG. During subsequent washes 4’,6-diamidino-2-phenylindole, dihydrochloride (DAPI) was added to stain nuclei. Plasma membranes were stained with CellMask (Deep Red, Molecular Probes) at final concentration of 5µg/ml. Confocal immunofluorescence images acquired on a BioRad MRC 1024 imaging system is equipped with a Krypton/Argon laser were coloured in ImageJ (http://rsb.info.nih.gov/ij/).
rEtpA, biotinylated with biotin LC hydrazide, and dialyzed vs PBS to remove excess biotin, was 1st added to target monolayers (paraformaldehyde-fixed, 1 % BSA blocked) at final concentrations of 0.05, 0.1, and 0.2 µM, and incubated at 37°C for 1 hour. After washing with PBS, bound label was detected by using streptavidin-HRP (1:10000) and tetramethylbenzidine/H2O2 (TMB) peroxidase substrate. Reactions were stopped by addition of 1M H2SO4, and the OD405 determined. Experiments were then conducted using biotinylated rEtpA at a final concentration of 0.05 µM combined with increasing amounts of unlabeled EtpA.
To visualize EtpA binding to intestine, frozen sections of mouse ileum were blocked, incubated with biotinylated rEtpA, washed, and bound rEtpA was detected with streptavidin-coated quantum dots (Qdot525 ITK SA). Both CellMask membrane stain and DAPI were incorporated into final washes.
Expression and purification of recombinant EtpA
Following induction of LMG194(pJY019) 6 with arabinose, supernatant was sterile-filtered (0.22 µm), and concentrated (≈150X) via 30,000 MWCO filtration (YM30, Millipore). Retentate was desalted with 50 mM sodium phosphate, 150 mM NaCl, pH 7.2, concentrated via 100,000 MWCO filter, and loaded onto a Sephacryl S-300 column. Elution of protein (flow rate of 0.5–1.0 ml/minute) was monitored by absorbance at 280 nm. 1 ml fractions were then collected and stored at 4°C.
Identification by Mass Spectrometry
Bands of interest excised from Coomassie-stained polyacrylamide were prepared27 and the mass spectra of the resulting peptides were recorded on a Bruker Ultraflex MALDI-TOF/TOF reflecting time-of-flight mass spectrometer (Bruker Daltonics, Bremen, Germany). A non-redundant protein sequence collection (NCBI) was then searched with peptide mass data using ProFound28 software at an error tolerance of 100ppm within the E. coli taxonomy.
Cloning, expression, and purification of recombinant flagellin molecules
To clone the full-length fliC (H48) gene, primers jf030106.1 and jf030106.2 were used to amplify fliC from MG1655. The product was ligated into pTrcHisB in-frame with the polyhistidine encoding region to produce pKR001b. Primers jf050206.1 and jf030106.2 were used to amplify and clone the region encoding amino acids 174–498 [FliC174–498] to produce pKR005b. Finally, the region encoding residues 1–173 of FliC was amplified using primers jf030106.1 and jf050206.2 and cloned into pTrcHisB to produce pKR007b. These plasmids, were introduced into Top10 which does not produce native flagellin 29. Recombinant flagellins with myc and polyhistidine tags at their carboxy termini were purified by nickel-affinity chromatography.
Membrane-based protein interaction studies
Varying amounts of target (bait) proteins were absorbed onto nitrocellulose, followed by addition of biotinylated prey proteins (10 µg/ml) and incubation (1 hour) at room temperature. Strips were washed to remove unbound prey protein. Bound biotinylated prey proteins were detected with streptavidin-HRP and chemiluminescent substrate.
Co-immunoprecipitation
Anti-EtpA IgG polyclonal antibodies were first purified from highly cross-absorbed anti-EtpA antisera 6 by protein G affinity column chromatography (HiTrap ProteinG, Amersham Biosciences). Preimmune sera from the same rabbit were treated in an identical fashion. The resulting antibodies (200 µg) were coupled to solid support gel matrix. Concentrated supernatants from ETEC were incubated overnight at 4°C with matrix containing either pre-immune IgG or anti-EtpA (IgG) antibodies. After washing of the matrix proteins were eluted (ImmunoPure, IgG Elution Buffer, pH 2.8, Pierce), separated by SDS-PAGE (10 %), and transferred to nitrocellulose for subsequent immunoblotting.
Molecular pull-down assays
Polyhistidine-tagged flagellin molecules and rEtpA were added together in solution in a molar ratio of 1:1 (approximately 150 pmol each). After incubation for 1 hour at 4°C polyhistidine flagellins and interacting EtpA were pulled down by the addition of 100 µl of Co2+ metal affinity bead suspension in PBS. Bound proteins were released by incubation in SDS-PAGE sample buffer and used for immunoblotting.
Transmission and scanning electron microscopy
Immunogold labelling of EtpA and conserved regions of flagellin in situ 17 used suspensions of live bacteria grown on the surface of UV-sterilized grids and fixed with formaldehyde and glutaraldehyde. Immunogold detection was then carried out using highly cross-absorbed anti-EtpA polyclonal rabbit antisera or affinity purified anti-flagellin antibodies followed by anti-rabbit IgG gold (10 nm) conjugate. Negative staining was then carried out in 1 % phosphotungstic acid, pH 6.5. Scanning images were acquired on a Philips XL30 ESEM instrument.
Linker scanning mutagenesis of etpA
pJL017 was the target for in vitro Tn7-based mutagenesis19. Plasmids with etpA-mapped, non-redundant insertions were digested with PmeI, and religated to remove the majority of the transprimers. This yielded etpA mutants with 15 bp scar sequences that (along with pJL030) were used in transformation of Top10 to AmpR/CmR. Plasmids encoding effectively secreted EtpA mutants in Top10 were introduced into isogenic etpA mutant jf1668 for subsequent adherence assays.
Intestinal competition
The previously described model of murine intestinal colonization 26 was used in competition between etpA mutant jf1668 complemented with EtpA expression plasmid pJL017, or the same mutant complemented with pJMF1087 which expresses EtpA that has lost the ability to bind to FliC following introduction of a linker mutation. Each mouse received 1 ×104 cfu of jf1668(pJL017) and 1 ×104 cfu of jf1668(pJMF1087) (together by gavage). Following challenge, water supply for the mice included ampicillin (50 µg/ml) and arabinose (0.0002%), respectively. 24 hours later, intestinal lysates26 were plated onto Luria agar plates containing chloramphenicol and ampicillin. AmpR/CmR colonies were subjected to PCR with primers jf122205.1 and jf092605.3 to generate a ≈1690 bp amplicon that was digested with PmeI to distinguish the WT allele from pJL017 from the mutant containing a PmeI site (pJMF1087).
Supplementary Material
Supplementary Information is linked to the online version of the paper at http://www.nature.com/nature.
acknowledgements
The authors wish to thank Benita Westerlund-Wikström for supplying anti-flagellar antisera, and Katherine Troughton of the Department of Anatomy and Neurobiology for her assistance with TEM and Lou Boykins of the Integrated Microscopy Center at the University of Memphis for her assistance with SEM. We thank Gerry Byrne, Harry Courtney, James Dale, Sam Dagogo-Jack, and Ted Strom for their critical reading of the manuscript. This research was supported by grants from the National Institutes of Health (NCRR) RR16190-05, the Department of Veterans Affairs, and funds from the University of Tennessee Microbial Pathogenesis Research Center.
Footnotes
competing financial interests University of Tennessee Research Foundation has applied for a patent related to studies included here.
References
- 1.Torres AG, Zhou X, Kaper JB. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect Immun. 2005;73(1):18. doi: 10.1128/IAI.73.1.18-29.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorsey FC, Fischer JF, Fleckenstein JM. Directed delivery of heat-labile enterotoxin by enterotoxigenic Escherichia coli. Cellular Microbiology. 2006;8(9):1516. doi: 10.1111/j.1462-5822.2006.00736.x. [DOI] [PubMed] [Google Scholar]
- 3.WHO. Future directions for research on enterotoxigenic Escherichia coli vaccines for developing countries. Wkly Epidemiol Rec. 2006;81(11):97. [PubMed] [Google Scholar]
- 4.Qadri F, Svennerholm AM, Faruque AS, et al. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev. 2005;18(3):465. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob-Dubuisson F, Locht C, Antoine R. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol Microbiol. 2001;40(2):306. doi: 10.1046/j.1365-2958.2001.02278.x. [DOI] [PubMed] [Google Scholar]
- 6.Fleckenstein JM, Roy K, Fischer JF, et al. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect Immun. 2006;74(4):2245. doi: 10.1128/IAI.74.4.2245-2258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424(6949):643. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 8.Evans DG, Silver RP, Evans DJ, Jr, et al. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect Immun. 1975;12(3):656. doi: 10.1128/iai.12.3.656-667.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaastra W, Svennerholm AM. Colonization factors of human enterotoxigenic Escherichia coli (ETEC) Trends Microbiol. 1996;4(11):444. doi: 10.1016/0966-842x(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 10.Svennerholm AM, Tobias J. Vaccines against enterotoxigenic Escherichia coli. Expert review of vaccines. 2008;7(6):795. doi: 10.1586/14760584.7.6.795. [DOI] [PubMed] [Google Scholar]
- 11.Giron JA, Torres AG, Freer E, et al. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol Microbiol. 2002;44(2):361. doi: 10.1046/j.1365-2958.2002.02899.x. [DOI] [PubMed] [Google Scholar]
- 12.Wolf MK. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin Microbiol Rev. 1997;10(4):569. doi: 10.1128/cmr.10.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yonekura K, Maki S, Morgan DG, et al. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science. 2000;290(5499):2148. doi: 10.1126/science.290.5499.2148. [DOI] [PubMed] [Google Scholar]
- 14.Golemis E, Adams PD. Protein-protein interactions : a molecular cloning manual. 2nd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 15.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komoriya K, Shibano N, Higano T, et al. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol Microbiol. 1999;34(4):767. doi: 10.1046/j.1365-2958.1999.01639.x. [DOI] [PubMed] [Google Scholar]
- 17.Jin Q, He SY. Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae. Science. 2001;294(5551):2556. doi: 10.1126/science.1066397. [DOI] [PubMed] [Google Scholar]
- 18.Maki-Yonekura S, Yonekura K, Namba K. Domain movements of HAP2 in the cap-filament complex formation and growth process of the bacterial flagellum. Proc Natl Acad Sci U S A. 2003;100(26):15528. doi: 10.1073/pnas.2534343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goff SP, Prasad VR. Linker insertion mutagenesis as probe of structure-function relationships. Methods Enzymol. 1991;208:586. doi: 10.1016/0076-6879(91)08030-l. [DOI] [PubMed] [Google Scholar]
- 20.Roy K, Hamilton D, Allen KP, et al. The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect Immun. 2008;76(5):2106. doi: 10.1128/IAI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ottemann KM, Miller JF. Roles for motility in bacterial-host interactions. Mol Microbiol. 1997;24(6):1109. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 22.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47(3):401. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 23.Mu XQ, Savarino SJ, Bullitt E. The three-dimensional structure of CFA/I adhesion pili: traveler's diarrhea bacteria hang on by a spring. J Mol Biol. 2008;376(3):614. doi: 10.1016/j.jmb.2007.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buscher AZ, Grass S, Heuser J, et al. Surface anchoring of a bacterial adhesion secreted by the two-partner secretion pathway. Mol Microbiol. 2006 doi: 10.1111/j.1365-2958.2006.05236.x. [DOI] [PubMed] [Google Scholar]
- 25.Smith KD, Andersen-Nissen E, Hayashi F, et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4(12):1247. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 26.Allen KP, Randolph MM, Fleckenstein JM. Importance of heat-labile enterotoxin in colonization of the adult mouse small intestine by human enterotoxigenic Escherichia coli strains. Infect Immun. 2006;74(2):869. doi: 10.1128/IAI.74.2.869-875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cummings ED, Brown JM, Sarva ST, et al. High-throughput proteomics processing of proteins in polyacrylamide in a multiwell format. Journal of proteome research. 2007;6(4):1603. doi: 10.1021/pr060472y. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Chait BT. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal Chem. 2000;72(11):2482. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi F, Smith KD, Ozinsky A, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410(6832):1099. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at http://www.nature.com/nature.