Summary
The control of mRNA stability is a complex biological process that involves numerous factors, including microRNA (miRNA) and short interfering RNA (siRNA). Here, we show that short interfering RNA (siRNA) and microRNA share some similarities in their response to cellular stress. miR16 expedites the degradation of mRNAs containing AU-rich elements (ARE) in their 3’ unltranslated region (UTR). si20 is an siRNA designed to target a non-ARE sequence in the TNF 3’UTR. We found that both si20 and miR16/ARE mediated degradation of mRNAs can be inhibited by stimulating cells with different stresses. By analyzing TNF-α stimulation-mediated stabilization of si20- and miR16-targeted mRNA, we show that this stabilization is not caused by modifying si20 and miR16 loading into Ago2 complexes, or mRNA targeting to Ago2, but by inhibiting mRNA deadenylation. This is the first report showing that a specific siRNA-mediated mRNA degradation can be regulated by inflammatory stimuli, and that deadenylation is involved in this siRNA-mediated mRNA decay.
Introduction
Gene expression is tightly controlled at various levels, from gene transcription to protein posttranslational modifications [1, 2, 3]. Deregulation of gene expression can result in dramatic phenotypes leading to pathological states [4, 5, 6, 7, 8]. The control of mRNA expression levels and translation have been highlighted as major control checkpoints for transiently expressed molecules like cytokines [9, 10, 11]. In cells of the innate immune system, inflammatory stimuli can be associated with an increase in mRNA translation and stability [12, 13, 14, 15]. AU-rich elements (ARE), present in the 3’ untranslated region (3’UTR) of many unstable mRNAs, control mRNA degradation through the recruitment of ARE binding proteins having various biological activities [14, 15]. Amongst ARE binding proteins that were studied and characterized at the functional level, some are destabilizing factors like tristetraprolin (TTP), some are stabilizing factors like the ELAV-related protein HuR [16–17]. Even more strikingly, the ARE and poly(A) binding protein AUF-1 has several isoforms: p37, p40, p42, and p45 that act in an antagonistic way in the control of ARE-mediated mRNA decay [17].
Research undertaken in our laboratory demonstrated that ARE-mediated mRNA degradation involves short non-coding endogenous RNA molecules called microRNAs (or miRNAs) [18]. In human cells, ARE-mediated mRNA degradation is triggered by the interaction of miR16 with a hexameric ribonucleotide sequence that exists in several ARE sequences. From a thermodynamic point of view, the interaction between miR16 and the hexamer is weak and unlikely to occur in vivo without the help of other factors. [18]. The biogenesis of miRNAs and short-interfering RNAs (siRNAs) share some common features in their maturation steps, processing by Dicer to mature 20–23 nucleotide miRNA/siRNAs and subsequent loading into Ago-containing complexes. Genes of miRNA are transcribed into large miRNA precursors, matured in the nucleus by Drosha to imperfect hairpin-like pre-miRNAs. These pre-miRNA precursors are then translocated to the cytosol by Exportin-5. Dicer further processes the hairpin structures to imperfect double-stranded RNA duplexes that are transferred to RISC [20, 21, 22, 23]. After loading into RISC, the passenger strand of the duplex RNA is destroyed by Ago2 as a consequence of the duplex RNA thermodynamic polarity, while the guide strand is retained into RISC to direct RNAi. The synthesis of siRNA from pSuper vectors is quite similar to the endogenous synthesis of miRNAs. Transcription of siRNA is performed by RNA polymerase III, and the perfectly matching hairpin is transferred into the cytosol where Dicer, present in the RISC-loading complex, generates a duplex RNA. This duplex is then transferred to RISC and the passenger strand is destroyed by Ago2 as a result of the same thermodynamic considerations as for the maturation of miRNAs [20, 21, 22, 23].
siRNA and miRNA are strikingly similar in structures and synthesis pathways; however, studies have also pointed out their different functions. It is well known that the ARE-mediated mRNA degradation, in which miR16 is involved, is subjected to regulation by inflammatory signals. Here we present evidence to demonstrate that siRNA-mediated mRNA decay is also subject to regulation by inflammatory signals. We show that TNF stimulation can inhibit siRNA-mediated RNA degradation by suppression of deadenylation.
Material and methods
Constructs
CMV-ARE and CMV-ΔARE were constructed by PCR amplification from the pBBB-TNF-3’UTR and pBBB-TNF-ΔARE vectors, as described earlier [19]. The forward primer contained a NotI restriction site whereas the reverse primer contained an XhoI restriction site. Both PCR amplification products were introduced into CMV-driven 3xFlag vectors by ligation into NotI and XhoI sites, generating the CMV-ARE and CMV-ΔARE plasmids. The Tet-ARE and Tet-ΔARE plasmids were generated by exchanging the CMV promoter for the Tetracycline responsive promoter from the CMV-ARE and CMV-ΔARE plasmids, respectively. Cloning of si20 and miR16 were performed by ligating duplex oligos into the BamHI and XhoI of pSuper vector in a similar way as was done by the Flemington lab, which is further described on their website (http://www.flemingtonlab.com). The sequence of si20 is ggttgcctctgtctcagaat. The sequence of miR16 is described in our previous work[18]. Flag-Ago2 was from Thomas Tuschl (Rockfeller Institute, New York, NY).
Cells and cell culture
HEK293T and HeLa Tet-off cells were grown at 37°C in a water-saturated atmosphere containing 5% CO2. Cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and further supplemented with 1% of a mixture of 100X-concentrated penicillin-streptomycin-glutamine solution (Invitrogen, Carlsbad, CA). Both cell lines were maintained in 10 cm dishes and passed at ~80% confluence. All transfections were performed using Lipofectamine 2000 (Invitrogen) following the procedure described by the manufacturer.
Quantitative RT-PCR
Total RNA was isolated from cells using Trizol reagent (Invitrogen) following the manufacturer’s instructions. Quantitative RT-PCR reactions were performed with the one step RT-PCR master mix reagents from Applied Biosystems (Foster City, CA) with the following primers: gtgacaagctgcacgtggat,; acagcacaataaccagcacgtt and Taqman® probe; FAM-ctgagaacttcagctcctgggc-Quantitative RT-PCR was normalized against GAPDH, as described in our previous work[18]. Detection was performed using an ABI 7900HT fast instrument (Applied Biosystems).
Northern-blots
The procedure to perform Northern blots is described elsewhere [24]. Briefly, Northern-blots were run on agarose gels containing formaldehyde. RNA was transferred to nitrocellulose membranes by capillary transfer. Hybridizations were performed with full-length RNA probes. Hybridizations were conducted at 70°C in a rotisserie oven for up to 18 hours. Membranes were washed until a specific signal appeared. Anti-sense full length RNA probes were internally labeled with [α-32P] UTP and generated with T7 polymerase (Roche Applied Science, Indianapolis, IN), following the manufacturer’s recommendations and as described in previous publications[24].
Primer extension
primer extensions were performed as in our previous work[18].
mRNA degradation experiments and TNF-α treatment
HeLa Tet-off cells were grown overnight in a 100 mm plate until reaching 80–85% confluence, and then they were transfected with 20 µg of Tet-ARE or Tet-ΔARE with pSuper-si20. Cells were then cultured for 15 additional hours, collected and redistributed in 12 different wells among two 6-well plates. Cells were grown for an additional 24h. In the first set of six wells, PBS was added 15 min. prior to doxycycline treatment (10 µg/ml), which was added for 0, 0.25, 0.5, 1, and 2 hours to block transcription. In a second set of six wells, 50 ng/ml of TNF-α were introduced 15 min. prior to doxycyclin addition (0, 0.25, 0.5, 1 and 2 hours) and total RNA was isolated with Trizol®.
Immunohystochemistry
HeLa cells were seeded onto glass cover slips at the bottom of the 6-well plates. Fixation and staining procedures were done as described elsewhere [24]. M2 anti-Flag monoclonal antibodies were from Sigma, anti-TIA-1 goat polyclonal antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA), anti-Dcp1a rabbit polyclonal antibodies were obtained elsewhere [25]. Fluorescent-conjugated secondary antibodies were from Molecular Probes (Invitrogen, Carlsbad, CA). Fluorescence signal was detected with a CCD camera and an Axiovert 200 from Carl Zeiss Inc (Thornwood, NY).
Co-immunoprecipitations
Cells (HEK-293T cells or HeLa Tet-off cells) were rinsed with PBS and lysed on ice with lysis buffer (20 mM Tris pH 7.5, 0.9% NaCl, 0.5% Triton X-100, and 0.5 mM PMSF). Cell particles were spun down at 10,000 rpm at 4°C for 10 min., and M2 anti-Flag agarose conjugated beads (Sigma, St Louis, MA) were added to the supernatant and incubated at 4°C for 2–4h under gentle agitation. Beads were then washed with lysis buffer 5 times. To extract RNA from the beads, Trizol® was directly added to the beads.
Anti-Cap-immunoprecipitation
Trizol®-extracted total RNA was utilized for immunoprecipitations. K121 anti-2,2,7-trimethylguanosine agarose-conjugated mouse monoclonal antibodies (Calbiochem, Darmstadt, Germany) were pre-blocked with 100 µg/ml tRNA for 2 hours at 4°C under gentle agitiation. Immunoprecipitation was performed in a lysis buffer (20 mM Tris pH 7.5, 0.9% NaCl, 0.5% Triton X-100) with 100 µg/ml tRNA and K121 beads for 5 hours at 4°C under gentle agitation.
Poly(A) tail analysis
Poly(A) tail experiments were performed following Lykke-Andersen’s protocol [26]. Trizol®-extracted total RNA was mixed with oligonucleotides complimentary to the 3’UTR of ARE and ΔARE mRNAs, heated to 80°C for 2 min., and cooled down to room temperature to allow annealing of the oligonucleotides with complimentary mRNA sequences. RNAse H reaction buffer and RNAse H were then added to the mixture following the manufacturer’s recommendation (Roche Applied Science). RNAse H digestion was then performed at 37°C for 1 hour, and total RNA was extracted using Trizol® and then resuspended in Northern-blot loading buffer [26].
Results
Stabilization of siRNA-targeted mRNA by extracellular stimuli
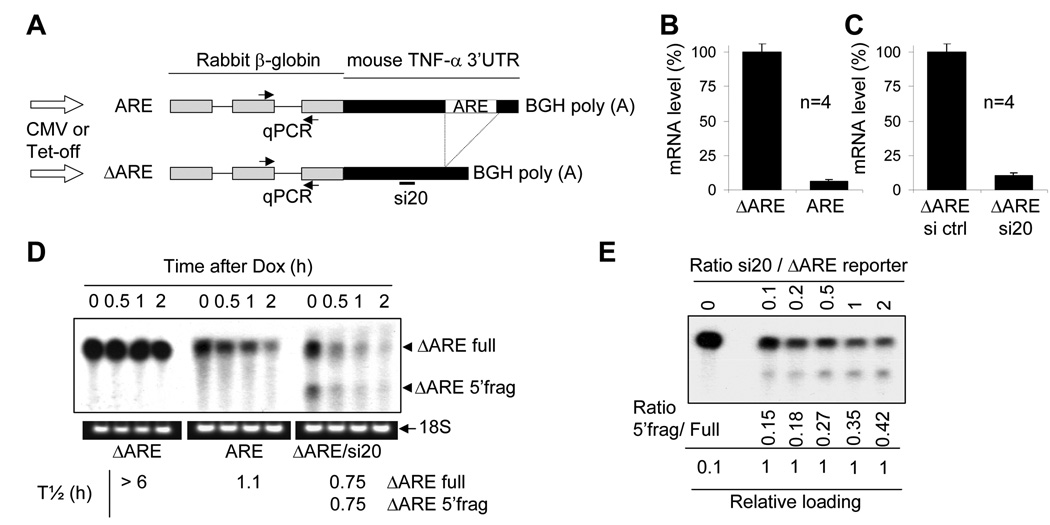
To study the effect of extracellular stimulation on siRNA-mediated mRNA decay, we established reporter mRNA systems. As shown in Figure 1A, reporters of siRNA-dependant as well as miR16/ARE-dependant mRNA degradations were constructed. Both a constitutive promoter (CMV promoter) and an inducible promoter (Tet promoter) were used in these reporters. Rabbit β-globin (β-globin) gene was used as a reporter, and either the full length (ARE constructs) or a part (ΔARE constructs) of the 3’ untranslated region (3’UTR) of the mouse tumor necrosis factor-α (TNF-α) were added downstream of the globin gene. The ΔARE constructs contain all of the TNF 3’UTR except the AU-rich elements. Figure 1A also shows the location of the quantitative RT-PCR (qPCR) primers (spanning intron 2 of the rabbit β-globin gene), as well as the site targeted by the siRNA si20.
Figure 1. ARE and ΔARE reporter constructs and their expression in mammalian cells.
A)Schematic representation of the ARE and ΔARE constructs. The full length mouse TNF-α 3’UTR was added downstream of the rabbit β-globin gene (ARE reporter). The AU-rich elements in the TNF-α 3’UTR were deleted to generate the ΔARE reporter. Both the ARE and ΔARE reporters were constructed under the control of the cytomegalovirus (CMV) or Tet promoters. The quantitative RT-PCR detection system spans intron 2. si20-targeting site and AU-rich elements (ARE) are localized in the 3’UTR of the constructs. Bovine growth hormone poly (A) signal (BGH poly (A)) was used. B) HEK-293T cells were transfected with CMV–ΔARE (ΔARE) or CMV-ARE (ARE) plasmids and cultured for 40 hours. Total RNA was extracted with Trizol® reagent. Quantitative one-step RT-PCR assays were then run to measure ARE and ΔARE mRNA. ARE and ΔARE mRNA levels were normalized against GAPDH mRNA levels. Results are expressed as the mean of 4 independent experiments, the error bar represents the standard deviation. C) HEK-293T cells were cotransfected with CMV-ΔARE and pSuper-si20 (ΔARE si20), or with CMV-ΔARE and pSuper-si ctrl (ΔARE si ctrl). Samples were analyzed as in B). D) HeLa Tet-off cells were transfected with pcDNA3 and Tet-ΔARE (ΔARE); pcDNA3; Tet-ARE (ARE); or pSuper si20 and Tet-ΔARE (ΔARE/si20) and cultured for 40 additional hours. Doxycycline was then added to block transcription, and total RNA was isolated with Trizol. Reporter mRNA degradation was analyzed by Northern-Blotting and 18S ribosomal RNA was utilized to control RNA loading. Half-lives were analyzed using the phosphoimager. E) HeLa Tet-off cells were transfected with a total of 5 µg of plasmid DNA. The mass ratio of pSuper-si20 and Tet-ΔARE plasmids were 0, 0.1, 0.2, 0.5, 1, and 2. pcDNA3 was utilized to normalize a total of 5 µg plasmid per transfection. Cells were then cultured for 40 additional hours before total RNA was isolated using Trizol. Reporter mRNA was analyzed by Northern-Blotting and the ratios of 5’frag/Full were analyzed using the phosphoimager.
Figure 1B shows a measurement by quantitative RT-PCR of the cellular mRNA levels in HEK-293T cells transfected with CMV-ΔARE or CMV-ARE. The presence of the AU-rich elements in the 3’UTR of the ARE mRNA results in low mRNA levels, as compared to ΔARE mRNA expression. Figure 1C shows mRNA levels of the reporter in HEK-293T cells cotransfected with the CMV-ΔARE and pSuper expression vectors of si20 or control siRNA. Cotransfecting pSuper-si20 strongly downregulates the cellular levels of the ΔARE mRNA when compared to cotransfection with a control siRNA (pSuper-si ctrl). Therefore, CMV-driven reporters can be used to study ARE- and si20-mediated mRNA decay.
The addition of tetracyclin derivatives (doxycyclin) to the Tet-off system results in quick and complete transcription blocking of the tetracycline responsive promoter. Doxycyclin addition to HeLa Tet-off cells results in very specific, quick, and strong transcription suppression that does not affect many other genes’ expression or cell viability, as compared to other drugs like Actinomycin D. HeLa Tet-off cells were transfected with Tet-ΔARE, Tet-ARE, or Tet-ΔARE and pSuper-si20, as detailed in the legend of figure 1D. Using Tet-off cells, our Northern-blots revealed that the ΔARE mRNA was quite stable (half-life greater than 6h). Coexpression with si20 resulted in a strong downregulation of ΔARE mRNA that was attributable to ΔARE mRNA instability (half-life of 0.75 h). Moreover, an endonucleolytic fragment (ΔARE 5’frag) can be observed, and oligo-RNAse H mapping (results not shown) revealed that this mRNA fragment was the 5’ fragment upstream of the si20 targeting site. It was quite surprising that the 5’ fragment was not degraded immediately after its generation. By contrast, the ARE mRNA does not display any endonucleolytic fragments but is similarly unstable (half-life of 1.1 h).
A range of pSuper-si20/Tet-ΔARE ratios were used to transfect HeLa Tet-off cells to investigate whether the level of si20 plays a role in the presence of the 5’ fragment (Figure 1E). It appears that diminishing the amount of pSuper-si20 for transfection resulted in a higher expression level of the full length ΔARE mRNA and a gradual decrease in the amount of the 5’ fragment. Since the level of the 5’ fragment is determined by the speed of endo-cutting of full-length reporter mRNA and the rate of 5’ fragment degradation, the speed of endo-cutting is apparently faster than the decay of the 5’ fragment, even when si20 levels are low.
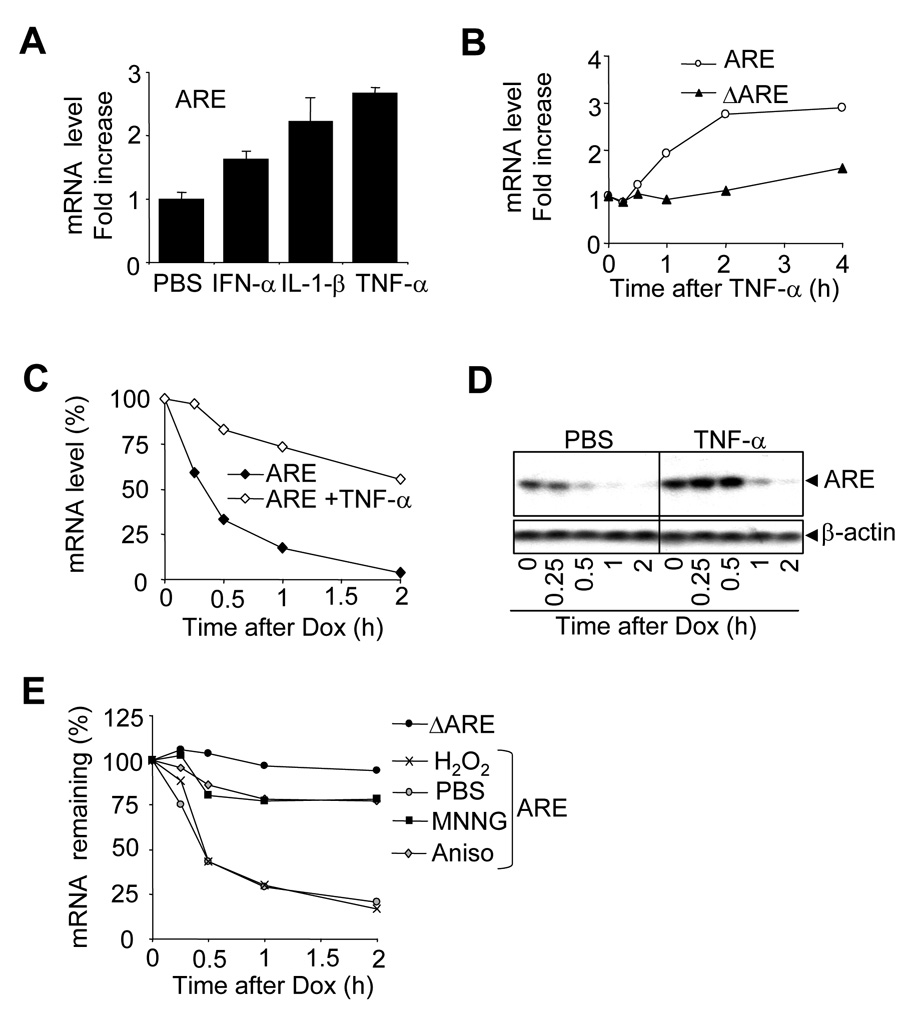
It has been well described that cell stimulation can result in the stabilization of some unstable mRNA molecules. Some mRNAs are of low abundance when cells are in a resting stage, whereas inflammatory stimulation results in transcriptional activation that is accompanied by mRNA stabilization, further enhancing the efficiency of the transcriptional pulse. The treatment of CMV-ARE-transfected HEK-293T cells with cytokines results in a general upregulation of ARE mRNA cellular levels, as compared to PBS-treated cells (Figure 2A). To determine whether this increase in ARE reporter expression occurred at transcriptional or post-transcriptional levels, we compared CMV-riven ARE and ΔARE reporters. HEK-293T cells were transfected with the CMV-ARE or the CMV-ΔARE, and were treated with TNF-α for up to 4 hours (Figure 2B). It appears that even though both constructs are under the control of the exact same CMV promoter, only the ARE mRNA is upregulated by TNF-α treatment, suggesting that TNF-α treatment regulates the stability of the ARE mRNA.
Figure 2. Inflammatory stimuli and other stresses interfere with ARE mRNA degradation.
A) HEK-293T cells were transfected with CMV-ARE (ARE) and cultured for 40h. One hour before total RNA isolation, cells were treated with PBS, 50 ng/ml IFN-α, 50 ng/ml Il-1β, or with 50 ng/ml TNF-α. Quantitative one-step RT-PCR assays were used to measure ARE mRNA levels. ARE mRNA levels were normalized against GAPDH mRNA levels and increases were calculated using PBS-treated sample controls. Results are expressed as the mean of 3 independent experiments, with the error bar representing the standard deviation. B) HEK-293T cells were transfected with either CMV-ARE (ARE) or CMV-ΔARE (ΔARE) and cultured for 40 additional hours. Prior to RNA isolation, cells were treated with 50ng/ml TNF-α for 0.25, 0.5, 1, 2, and 4 hours. Quantitative one-step RT-PCR assays were performed and ARE and ΔARE mRNA increases were calculated using the time 0 point as 1. C) HeLa Tet-off cells were transfected with Tet-ARE and cultured for 40 additional hours. Then cells were treated with either PBS or 50 ng/ml TNF-α for 15 min. before doxycycline was added to block transcription. Samples were analyzed as in B. ARE mRNA levels were expressed as a percentage of the amount existing at time 0. D) As C, but the presence of ARE mRNA in the cells was analyzed by Northern-blotting. β-actin was used to ensure equal loading. E) As C, but HeLa Tet-off cells were pretreated with PBS (PBS), hydrogen peroxide (H2O2), MNNG, or anisomycin (Aniso) 30 min. before addition of doxycycline. Samples were analyzed as in B.
To further exclude any promoter activation issue, the ARE mRNA was expressed in HeLa Tet-off cells under the control of the tetracyclin-sensitive promoter (Figure 2C and D). HeLa Tet-off cells expressing the ARE mRNA were stimulated with TNF-α prior to transcriptional shut down by doxycyclin treatment (Figure 2C). Quantitative RT-PCR showed a strong stabilization of the ARE mRNA when cells were pre-treated TNF-α. In another set of experiments, this was further confirmed by Northern-blotting analysis, but in a more transient way, as ARE mRNA expression dropped after one hour (Figure 2D). One might notice that the TNF-mediated stabilization that is observed in the northern-blot experiments (Figure 2D) is less pronounced that the one observed in the quantitative RT-PCR experiments (Figure 2C). This is most likely due to variations between experiments. To investigate whether other stress-related stimuli are able to stabilize ARE mRNA, HeLa Tet-off cells transfected with Tet-ARE were pre-stimulated with chemicals known to induce strong cellular stresses, like oxidative stress (H2O2), DNA damage (MNNG), or inhibition of protein synthesis (anisomycin) (Figure 2E). It appears that MNNG and anisomycin strongly induce ARE mRNA stabilization, whereas oxidative stress had no obvious effect on ARE mRNA stability.
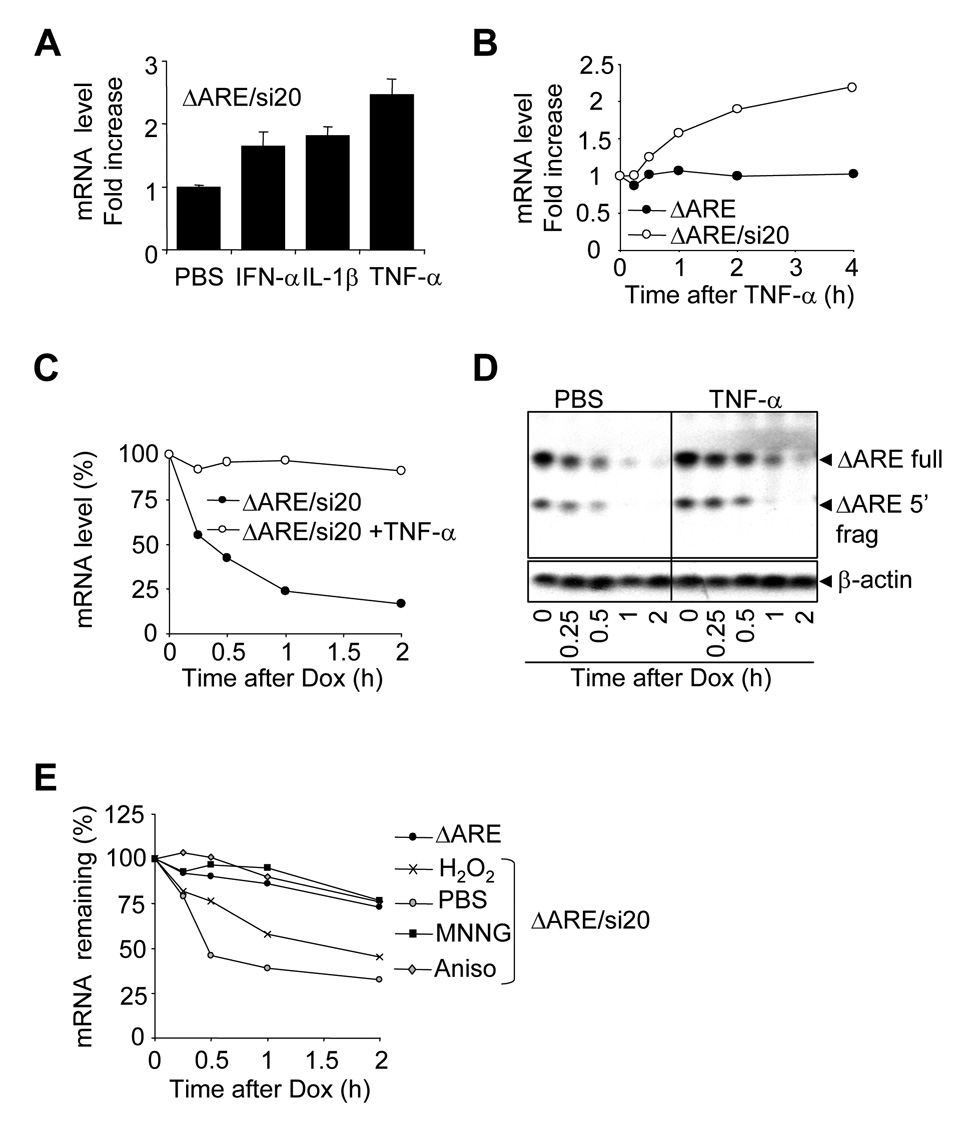
Because miRNA is involved in ARE-mediated mRNA degradation, and because certain similarities exist between miRNA and siRNA, we thought to test whether siRNA-mediated mRNA decay is also subject to regulation by extracellular stress stimuli. HEK-293T cells were transfected with Tet-ΔARE and pSuper-si20, and were then exposed to PBS, IFN-α, Il-1β, or TNF-α for 1 hour (Figure 3A). TNF-α stimulation resulted in a strong upregulation of ΔARE mRNA, as compared to PBS. IFN-α and Il-1β also resulted in ΔARE mRNA upregulation, but to lesser extent. To rule out that cytokines treatment stimulates the CMV promoter, HEK-293T cells were transfected with CMV-ΔARE alone or CMV-ΔARE plus pSuper-si20. The cells were treated with TNF-α for up to 4 hours, and ΔARE mRNA levels were analyzed (Figure 3B). Although the basal level of the CMV-ΔARE reporter mRNA was reduced by expressing si20 (Figure 1C), TNF-α stimulation led to an increase in ΔARE mRNA in the presence of si20 (Figure 3B). In contrast, TNF-α stimulation did not affect the level of ΔARE mRNA in the absence of si20. This suggests that si20-mediated mRNA degradation is impaired by TNF-α in a similar way as ARE mRNA. To further analyze the effect of TNF-α stimulation on si20-mediated mRNA degradation, HeLa Tet-off cells were transfected with Tet-ΔARE or Tet-ΔARE plus pSuper-si20. The cells were pretreated with or without TNF-α and then the Tet promoter was switched off by the addition of doxycycline for up to 2 hours (Figure 3C). ΔARE mRNA is very unstable when co-expressed with si20, and TNF-α treatment resulted in strong ΔARE mRNA stabilization (Figure 3C). To investigate whether TNF-α stimulation could affect siRNA-mediated endonucleolytic cleavage, HeLa Tet-off cells were transfected with Tet-ΔARE and pSuper-si20 and then pretreated with or without TNF-α before doxycycline addition. Northern-blots revealed that TNF-α pretreatment interfered with si20-mediated ΔARE mRNA degradation but that the presence of the 5’ endonucleolytic fragment in the cell lysates was not affected (Figure 3D), indicating that the si20-mediated endonucleolytic activity is not blocked by the TNF-α treatment. This data suggests that in addition to endo-cleavage, other degradation mechanisms may be involved in si20-mediated mRNA decay.
Figure 3. Inflammatory stimuli and other stresses interfere with si20-mediated mRNA degradation.
A) HEK-293T cells were transfected with pSuper-si20 and CMV-ΔARE and cultured for 40 h. One hour before total RNA isolation, cells were treated with PBS, 50ng/ml IFN-α, 50ng/ml Il-1β or 50ng/ml TNF-α. Samples were analyzed as in 2A B) HEK-293T cells were transfected and cultured as in A. Prior to RNA isolation, cells were treated with 50ng/ml TNF-α for 0.25, 0.5, 1, 2, and 4 hours. Quantitative one-step RT-PCR assays were run to measure ΔARE mRNA levels in HEK-293T cells. ΔARE mRNA levels were normalized against GAPDH mRNA levels. ΔARE mRNA increases were calculated by using the time 0 point as 1. C) HeLa Tet-off cells were transfected with Tet-ΔARE and pSuper-si20 and cultured for 40h. Then cells were treated PBS or 50 ng/ml of TNF-α 15 min. prior to doxycycline treatment. Samples were analyzed as in B. ΔARE mRNA levels were expressed as a percentage of the amount existing at time 0. D) As C, but the presence of ΔARE mRNA in the cells was analyzed by Northern-blotting. Re-probing β-actin was used to ensure equal loading. E) As C, but HeLa Tet-off cells were pretreated with PBS (PBS), hydrogen peroxide (H2O2), MNNG, or anisomycin (Aniso) 30 min. before the addition of doxycycline. Samples were analyzed as in B. ΔARE mRNA levels were normalized against GAPDH mRNA levels. ΔARE mRNA levels were expressed as a percentage of the amount existing at time 0.
Next, we investigate whether other stress-related stimuli are able to stabilize siRNA-targeted mRNA. HeLa Tet-off cells cotransfected with Tet-ΔARE and pSuper-si20 were pre-stimulated with H2O2, MNNG, or anisomycin, and the mRNA stabilities were measured (Figure 3E). Similar to what we observed with the ARE reporter, MNNG and anisomycin strongly induce ARE mRNA stabilization whereas oxidative stress only had a modest effect.
TNF-α induced stabilization of siRNA-targeted mRNA is not related to siRNA loading or to mRNA targeting to the Ago2 complex
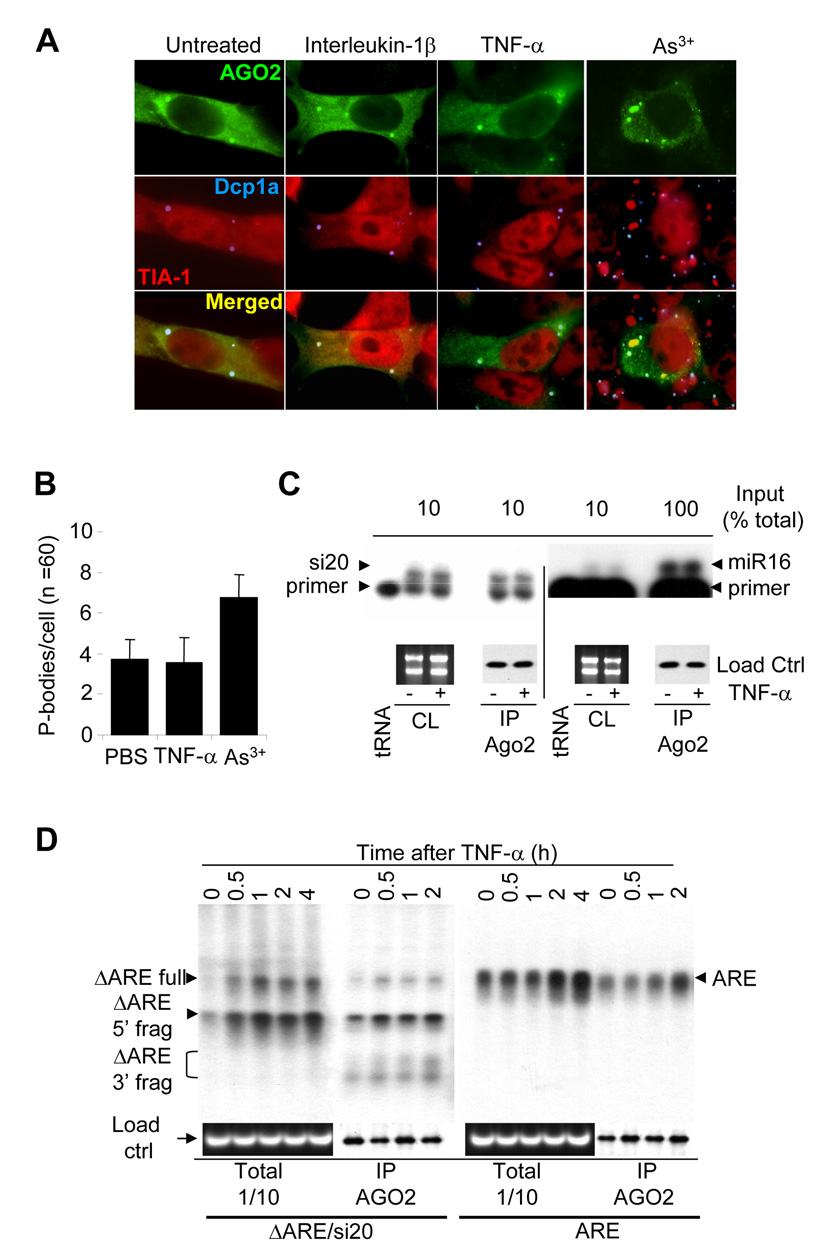
In mammalian cells, the Ago2 complex directs siRNA-dependant mRNA degradation, and therefore TNF-α could interfere with this process at several levels. Since Ago2 is localized in cytoplasmic foci called p-bodies [25], we investigated whether TNF-α stimulation could interfere with the p-body localization of Ago2. HeLa cells were transfected with Flag-Ago2 and stimulated for 1h with IL-1β, TNF-α, or As3+. Ago2 subcellular localization and p-bodies morphology were then analyzed by immuno-histochemistry with anti-Flag (green), anti-Dcp1a (blue; p-body marker) or anti-TIA-1 (red; stress granule marker) (Figure 4A). As expected from other group’s data, stimulating HeLa cells with As3+ resulted in the docking of p-bodies to newly formed stress granules, whereas the average number of p-bodies per cell increased significantly (Figure 4B). This was served as a positive control of our experiments to examine p-body dynamism. Compared to the untreated control, stimulating HeLa cells with IL-1β or TNF-α did not result in any changes in Ago2 p-body localization, p-body morphology (Figure 4A), and the average number of p-bodies per cell (Figure 4B), suggesting that TNF-α-induced stabilization of siRNA-targeted mRNA is not due to changes of Ago2 subcellular localization.
Figure 4. TNF-α stimulation does not affect Ago2’s localization to p-bodies and has no effect on siRNA/miRNA and mRNA loading into Ago2 complexes.
A) HeLa cells transfected with Flag-Ago2 were treated with interleukin-1β, TNF-α or As3+ for 1 hour. Cells were fixed, permeabilized, and stained for Flag-Ago2 (green), TIA-1 (red), or Dcp1a (blue). Panels show merged pictures of TIA-1 and Dcp1a (2nd row) and merged pictures of Flag-Ago2, TIA-1, and Dcp1a (third row). B) Average p-body counts per cell (N = 60) from A). C) HEK-293T cells were cotransfected with Flag-Ago2 and pSupermiR16, or Flag-Ago2 and pSuper-si20. Cells were stimulated for 30 min. with or without TNF-α, lysed and subjected to anti-Flag immunoprecipitations. Total RNAs were extracted from the immunoprecipitates and subjected to primer extension to detect si20 and miR16 in Ago2 complexes, whereas aliquots were conserved for protein analysis by anti-Flag western-blots. Ribosomal RNAs were used as cell lysate loading controls, and were visualized on agarose gels by ethidium bromide staining. D) HEK-293T cells were cotransfected with Flag-Ago2 and CMV-ARE, or CMV-ΔARE and pSuper-si20. Cells were stimulated with TNF-α (for 0, 0.5, 1, 2, or 4 hours), lysed, and then subjected to anti-Flag immunoprecipitations. Total RNAs were extracted and subjected to Northern-blot analysis, and aliquots of the samples were conserved for protein analysis by anti-Flag western-blotting. Ribosomal RNAs were used as cell lysate loading control, and were visualized on agarose gels by ethidium bromide staining.
TNF-α and other inflammatory stimuli could also block miRNA and siRNA loading to the Ago2 complex, therefore interfering with their biological functions. HEK-293T cells were transfected with Flag-Ago2 and pSuper-si20, or with pSuper-miR16. Cells were treated with or without TNF-α and subjected to anti-Flag immunoprecipitation (Figure 4C). Primer extension analyses for the presence of miR16 and si20 revealed that the levels of mature miR16 and si20 in the Ago2 immuno-complex and in total cell lysates were not changed by TNF-α stimulation (Figure 4C). Therefore, TNF-α-induced stabilization of ARE-mRNA and si20-targeted mRNA is not controlled by the loading of siRNAs and miRNAs to AGO-2 complexes.
Another possible explanation for TNF-α-mediated inhibition of ARE/miR16- and si20-mediated mRNA degradation is that the targeting of mRNA to Ago2 complexes is controlled by TNF-α stimulation. To investigate this possibility, HEK-293T cells were transfected with Flag-Ago2 in addition to CMV-ARE (ARE), or with CMV-ΔARE plus pSuper-si20 (ΔARE/si20). Cells were stimulated with TNF-α for 2 hours and subjected to anti-Flag immunoprecipitation. Northern-blotting analyses were then performed on total RNA extracted from the cell lysates or from the Ago2 immuno-complex (Figure 4D). As expected, TNF-α treatment resulted in the up-regulation of the cellular levels of ARE mRNA, as well as the ΔARE mRNA and ΔARE mRNA 5’ fragments. TNF-α stimulation does not block ARE mRNA or si20-targeted ΔARE mRNA targeting to Ago2 (Figure 4D). Surprisingly, the ΔARE mRNA 3’ fragment, that has never been detected in the cell lysates, is detectable in the Flag-Ago2 immunoprecipitates. RNAse H treatment with oligo(dT)20 demonstrated that the 3’ fragment was polyadenylated (data not shown). This data indicates that Ago2 does not require mRNA deadenylation prior to cleaving siRNA-targeted mRNA. TNF-α treatment results in a general uploading in Ago2 complexes of the full length ΔARE mRNA, as well as the 3’ and 5’ fragments. Even if TNF-α appears to upregulate the general level of the ΔARE mRNA targeted by si20, TNF-α does not impair si20-mediated ΔARE mRNA cleavage. This suggests that TNF-α stabilizing activity occurs as an event that is independent from Ago2 endonucleolytic activity.
Deadenylation of siRNA-targeted mRNA is subjected to regulation by TNF-α stimulation
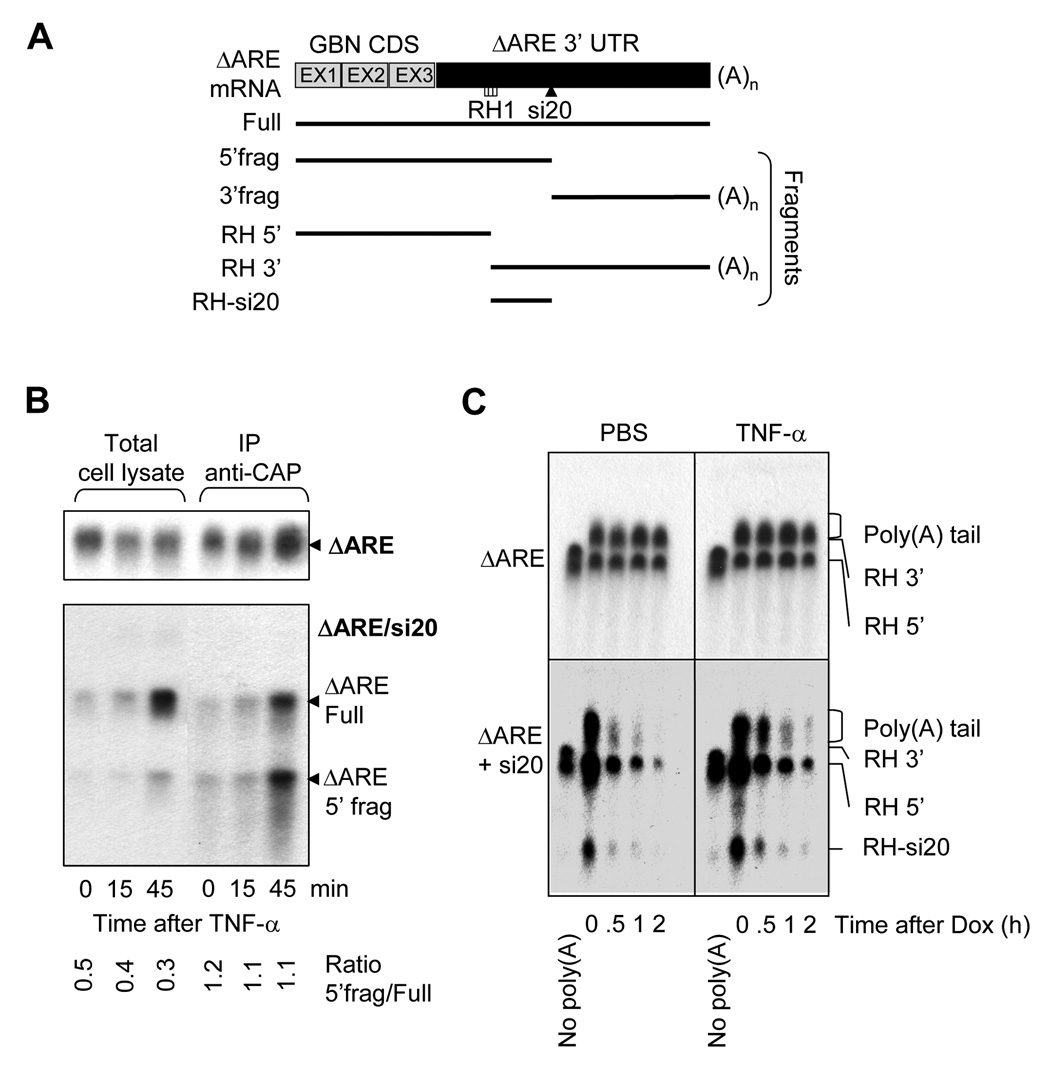
Since decapping and deadenylation are known to be involved in ARE-mediated mRNA degradation, we next examined whether decapping and deadenylation are involved in si20-mediated mRNA decay. Anti-Cap mRNA immunoprecipitations were performed to investigate whether TNF-α stimulation is involved in deccaping inhibition, or whether decapping occurs prior to siRNA-mediated mRNA endonucleolytic degradation (Figure 5B). We showed earlier that TNF-α stimulation resulted in the induction of cellular ARE and si20-targeted ΔARE mRNA, but not ΔARE mRNA that was expressed alone. When performing the anti-Cap immunoprecipitations on RNA extracted from cells expressing the ΔARE mRNA alone, ΔARE mRNA capping was not significantly affected by TNF-α treatment. When si20 was co-expressed, ΔARE mRNA was induced by the TNF-α stimulation. Accordingly, cap-containing ΔARE mRNA was increased after TNF-α treatment (Figure 5B). It appears that TNF-α treatment did not affect capping and decapping of si20-targeted mRNA. The ratio between the 5’fragment and the full length ΔARE mRNA (ratio 5’frag/full ΔARE) in cell lysates was around 0.4 and did not change much with TNF-α treatment. This ratio is about 1.1 in anti-Cap immunoprecipitates, and also did not change by TNF-α stimulation. Since the 5’ fragment contains the cap, the 5’ fragment should be generated from a capped ΔARE mRNA, indicating that decapping is not required for si20-mediated cleavage.
Figure 5. Capping and Poly(A) tail of si20 targeted ΔARE mRNAs after TNF-α stimulation.
A)Schematic representation of the fragments generated by RNAse H during ΔARE mRNA poly(A) tail analysis. B) HEK-293T cells were transfected with CMV-31 ΔARE (ΔARE), or CMV-ΔARE plus pSuper-si20 (ΔARE/si20). Cells were treated with TNF-α for 0, 15, or 45 min. and total RNA was isolated. Cap-containing mRNAs were immunoprecipitated with K121 anti-2,2,7-trimethylguanosine agarose-conjugated mouse monoclonal antibodies. Northern-blotting was done with RNA extracted from cell lysates and after K121 immunoprecipitation. C) HeLa-Tet-off cells were transfected with Tet-ΔARE (ΔARE), or Tet-ΔARE plus pSuper-si20 (ΔARE/si20). Cells were treated for 15 min. with PBS or TNF-α prior to doxycycline addition for 0, 0.5, 1, and 2 hours. RNA was extracted and subjected to RNAse H treatment to visualize poly(A) tail length. As a control of deadenylation, oligo d(T)20 was added to remove the poly(A) tail. Northern-blots were used to analyze poly (A) tail distributions.
To investigate whether the mRNA poly(A) tail deadenylation was affected by TNF-α stimulation, we designed oligos that can hybridize to sequences of ΔARE mRNAs for RNAse H digestion. The localization of RH1 oligo on the ΔARE mRNA and the length of digesting products are schematized on Figure 5A. Oligos d(T)20 were utilized to determine the size of the deadenylated fragments. HeLa Tet-off cells were chosen to perform the experiments to exclude transcription from the assay. Cells were pretreated with PBS or TNF-α for 15 min., and doxycycline was added for 0, 0.5, 1, or 2 hours to block transcription. RNAse H treatments were undertaken to shorten mRNA molecules for a better visualization of the poly(A) tail distribution. The distribution of the ΔARE mRNA poly(A) tail (top panel) did not change after doxycycline addition, regardless PBS or TNF-α treatment (Figure 5C, top panels). When co-expressing si20, the addition of doxycycline resulted in a quick ΔARE mRNA decay that was accompanied by a quick deadenylation, characterized by a mean poly(A) tail distribution that got shorter with time (Figure 5C, left-low panel). When TNF-α was added 15 min. prior to doxycycline treatment, the degradation of the ΔARE mRNA in presence of si20 was slower than the PBS-treated control (Figure 5C, right-low panel). Additionally, TNF-α stimulation resulted in the stabilization of the poly(A) tail, as the average size of the poly(A) tail remained the same after doxycycline treatment, even though the total amount of ΔARE mRNA decreased after doxycycline addition (Figure 5C, right-low panel). These data indicate that deadenylation is involved in si20-mediated degradation of ΔARE mRNA, and TNF-α treatment inhibits deadenylation of si20-targeted ΔARE mRNA.
Discussion
miRNA and siRNA, even if they share common features, have been reported to repress gene expression through different mechanisms, as miRNAs act mainly through translational control mechanisms, whereas siRNAs induce mRNA degradation [27, 28, 29, 30]. The mechanism by which siRNA mediates mRNA degradation is partly uncovered by our work, as Ago2 was demonstrated to direct an endonucleolytic cleavage at the siRNA targeting sites [31]. On the other hand, the mechanism by which miRNA mediates translational initiation suppression is still unclear, and it is only recently that Ago2 was shown to repress translation through a cap binding-like motif within Ago2 [29]. Additionally, Mathonnet et al. reported that miRNA inhibition of translation initiation occurrs in vitro by targeting the cap-binding complex EIF4F [30]. Despite the well accepted translational suppressing function of miRNA, recent work from our and other laboratories has shown that miRNA can also mediate mRNA degradation [18, 34]. Our work described in this report demonstrates that at least some siRNA-mediated mRNA degradation is subjected to regulation by extracellular stimulation, revealing an additional feature that is similar between miRNA- and siRNA-mediated mRNA decay.
mRNA instability is a common phenomenon that is associated with mRNAs that are transient, inducible, or tissue-specific. Amongst factors controlling mRNA stability, AU-rich elements (ARE) present in the 3’UTR of many mRNAs have been shown to control their fate by recruiting ARE-binding proteins to the 3’UTR, and by promoting or blocking their degradation [9, 14, 15, 16, 17, 19]. Recently, miRNA was found to be involved in the degradation of ARE-containing mRNAs [18]. Considering that ARE-mediated mRNA degradation is a regulated process, and that miR16 controls ARE-mediated mRNA decay [18], it is highly probable that miR16 is involved in the regulation of the degradation of ARE-containing mRNAs. Wu et al. showed recently that miRNAs direct the deadenylation of targeted mRNAs [34], suggesting that the regulation may occur at this step.
siRNA-mediated mRNA degradation is believed to be initiated by cleaving the mRNA target sites complementary to siRNA, followed by 3’→5’ and 5’→3’ exonucleolytic digestions [33]. We observed both 5’ and 3’ fragments in the Ago2 complex (Figure 4D), but only 5’ fragments in the total cell lysate (Figure 1D, 1E, Figure 3D, Figure 4D), suggesting that 3’→5’ exonucleolytic digestion is a speed-limiting factor for siRNA-mediated degradation, and that the 3’ fragment is quickly degraded after release from Ago2. Since RNAi is not a natural phenomenon in mammalian cells, the physiological relevance of the different speeds in degradation of 5’ and 3’ fragments is unclear.
By comparing ARE/miR16- and si20-mediated mRNA degradations, it was quite unexpected to have discovered that the si20-mediated RNAi is regulated by inflammatory mediators (Figure 3). To our knowledge, this is the first report mentioning that a particular RNAi efficiency is controlled by cell stimuli, and specifically by inflammatory signals like TNF-α. This finding is of major importance as it raises questions about the reliability and efficiency of RNAi and knock-down procedures when experiments are undertaken in the context of cells under stress. While attempting to determine the mechanism of siRNA-mediated mRNA suppression, we found that TNF-α has no direct or indirect effect on si20 levels, its loading to Ago2 complexes, or the efficiency of si20-targeting mRNA (Figure 4). TNF treatment has no global effect on p-body number or Ago2’s p-body localization. It appears that TNF-α regulation targets an event that is upstream of or independent from Ago2-mediated endo-cleavage of mRNA (Figure 4). Further analysis revealed that deadenylation is a regulatory event in si20-mediated mRNA degradation (Figure 5) in a manner similar to miRNA-mediated mRNA degradation [34]. Our data demonstrate that there are more commonalities in the manner by which miRNA and siRNA execute their respective functions than was previously believed.
Acknowledgements
We thank J. Lykke-Andersen (University of Colorado at Boulder) for providing the anti- Dcp1a antibodies. This work was supported by grants from National Institutes of Health grants AI41637 and AI54696.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that aply to the journal pertain.
References
- 1.Khabar KS, Young HA. Post-transcriptional control of the interferon system. Biochimie. 2007;89:761–769. doi: 10.1016/j.biochi.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang J, Fillebeen C, Chen G, Biederbick A, Lill R, Pantopoulos K. Iron-dependant degradation of apo-IRP1 by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 2007;27:2423–2430. doi: 10.1128/MCB.01111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quattrone A, Pascale A, Nogues X, Zhao W, Gusev P, Pacini A, Alkon DL. Postranscriptional regulation of gene expression in learning by the neuronal ELAV-like mRNA-stabilizing proteins. Proc. Natl. Acad. Sci. USA. 2001;98:11668–116673. doi: 10.1073/pnas.191388398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verrechia F, Mauviel A. Transforming growth factor-beta and fibrosis. World J. Gastroenterol. 2007;13:3056–3062. doi: 10.3748/wjg.v13.i22.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lubberts E, Koenders MI, Van den Berg WB. The role of T-cell interleukin-17 in conducting destructive arthritis: lessons from animal models. Arthritis Res. Ther. 2005;7:29–37. doi: 10.1186/ar1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salahshor S, Goncalves J, Chetty R, Gallinger S, Woodgett JR. Differential gene expression profile reveals deregulation of pregnancy specific beta1 glycoprotein 9 early during colorectal carcinogenesis. BMC Cancer. 2005;5:66. doi: 10.1186/1471-2407-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang HF, Murphy TF, Shu P, Barton AB, Barton BE. Stable expression of constitutively-activated STAT3 in benign prostatic epithelial cells changes their phenotype to that resembling malignant cells. Mol. Cancer. 2005;4:2. doi: 10.1186/1476-4598-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrick DM, Lai WS, Blackshear PJ. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res. Ther. 2004;6:248–264. doi: 10.1186/ar1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao W, Liu M, Kirkwood KL. p38alpha stabilizes interleukin-6 mRNA via multiple AU-rich elements. J. Biol. Chem. 2007 doi: 10.1074/jbc.M707573200. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao C, Hamilton T. Introns regulate the raye of unstable mRNA decay. J. Biol. Chem. 2007;282:20230–20237. doi: 10.1074/jbc.M700180200. [DOI] [PubMed] [Google Scholar]
- 11.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan J, Heller NM, Gorospe M, Atasoy U, Stellato C. The role of post-transcriptional regulation in chemokine gene expression in inflammation and allergy. Eur. Respir. J. 2005;26:933–947. doi: 10.1183/09031936.05.00120204. [DOI] [PubMed] [Google Scholar]
- 13.Wang G, Guo X, Floros J. Differences in the translation efficiency and mRNA stability mediated by 5'-UTR splice variants of human SP-A1 and SP-A2 genes. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005;289:497–508. doi: 10.1152/ajplung.00100.2005. [DOI] [PubMed] [Google Scholar]
- 14.Dean JL, Sarsfield SJ, Tsounakou E, Saklatvala J. p38 Mitogen-activated protein kinase stabilizes mRNAs that contain cyclooxygenase-2 and tumor necrosis factor AU-rich elements by inhibiting deadenylation. J. Biol. Chem. 2003;278:39470–39476. doi: 10.1074/jbc.M306345200. [DOI] [PubMed] [Google Scholar]
- 15.Kontoyiannis D, Kotlyarov A, Carballo E, Alexopoulou L, Blackshear PJ, Gaestel M, Davis R, Flavell R, Kollias G. Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology. EMBO J. 2001;20:3760–3770. doi: 10.1093/emboj/20.14.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai WS, Parker J, Grissom SF, Stumpo DJ, Blackshear PJ. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell. Biol. 2006;26:9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dean JL, Sul G, Clark AR, Saklatvala J. The involvement of AU-rich elementbinding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilization, Cell. Signal. 2004;16:1113–1121. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 19.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetrarolin: 14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Kim K, Lee YS, Carthew RW. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA. 2007;13:22–29. doi: 10.1261/rna.283207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strnad cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzye complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian p-bodies. Nat. Cell. Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fenger-Gron M, Fillman C, Norrild B, Lykke-andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 27.Valencia Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 28.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 29.Kiriakidou M, Tan GS, Lamprinaki S, De Plnell-Saguer M, Nelson PT, Mourelatos Z. An mRNA M7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, Fillipowicz W, Duchaine TF, Sonenberg N. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 31.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Jiang F, Kalida S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514–1520. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orban TI, Izzauralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan SP, Slack FJ. microRNA-mediated silencing inside p-bodies. RNA Biol. 2006;3:97–100. doi: 10.4161/rna.3.3.3499. [DOI] [PubMed] [Google Scholar]
- 36.Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EK. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eulalio A, Behl-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kedersha N, Stoecklin G, Avodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell. Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilks N, Kedersha N, Avodeles M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cramer P, Srebrow A, Kadener S, Werbajh S, de la Mata M, Melen G, Noquès G, Kornblihtt AR. Coordination between transcription and pre-mRNA processing. FEBS Lett. 2001;498:179–182. doi: 10.1016/s0014-5793(01)02485-1. [DOI] [PubMed] [Google Scholar]
- 42.Read RL, Norbury CJ. Roles for cytoplasmic polyadenylation in cell cycle regulation. J. Cell Biochem. 2002;87:258–265. doi: 10.1002/jcb.10300. [DOI] [PubMed] [Google Scholar]
- 43.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. In situ fluorescence analysis demonstrates active siRNA exclusion from the nucleus by Exportin 5. Nucleic Acids Res. 2006;34:1369–1380. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]