Abstract
Cells have evolved a signaling pathway called the spindle assembly checkpoint (SAC) to increase the fidelity of chromosome segregation by generating a “wait anaphase” signal until all chromosomes are properly aligned within the mitotic spindle. It has been proposed that tension generated by the stretch of the centromeric chromatin of bioriented chromosomes stabilizes kinetochore microtubule attachments and turns off SAC activity. Although biorientation clearly causes stretching of the centromeric chromatin, it is unclear whether the kinetochore is also stretched. To test whether intrakinetochore stretch occurs and is involved in SAC regulation, we developed a Drosophila melanogaster S2 cell line expressing centromere identifier–mCherry and Ndc80–green fluorescent protein to mark the inner and outer kinetochore domains, respectively. We observed stretching within kinetochores of bioriented chromosomes by monitoring both inter- and intrakinetochore distances in live cell assays. This intrakinetochore stretch is largely independent of a 30-fold variation in centromere stretch. Furthermore, loss of intrakinetochore stretch is associated with enhancement of 3F3/2 phosphorylation and SAC activation.
Introduction
Equal partitioning of the replicated genome by the mitotic spindle is essential to avoiding aneuploidy. The spindle assembly checkpoint (SAC) improves the likelihood that each daughter cell receives a normal complement of chromosomes by ensuring that anaphase onset does not occur until every chromosome becomes bioriented at the spindle equator. The kinetochore, which is the structure on each sister chromatid where microtubules attach to the chromosome, is the site at which the localization and activities of SAC components are integrated into a “wait anaphase” signal when necessary (Musacchio and Salmon, 2007). Chromosome biorientation stretches the chromatin between kinetochores attached to microtubules emanating from opposite poles. Although it remains unclear whether tension signals directly to the SAC (McIntosh, 1991; Pinsky and Biggins, 2005), there is strong evidence that the generation of tension is a prerequisite for anaphase onset in normal mitoses (Li and Nicklas, 1995; Biggins and Murray, 2001; Stern and Murray, 2001). The traditional readout for tension is changes in interkinetochore distance, which is the distance between two sister centromeres (Waters et al., 1996). However, it is not clear whether chromosome biorientation also causes stretching of the core kinetochore structure and, if so, whether this physical change is detected by the SAC.
Results and discussion
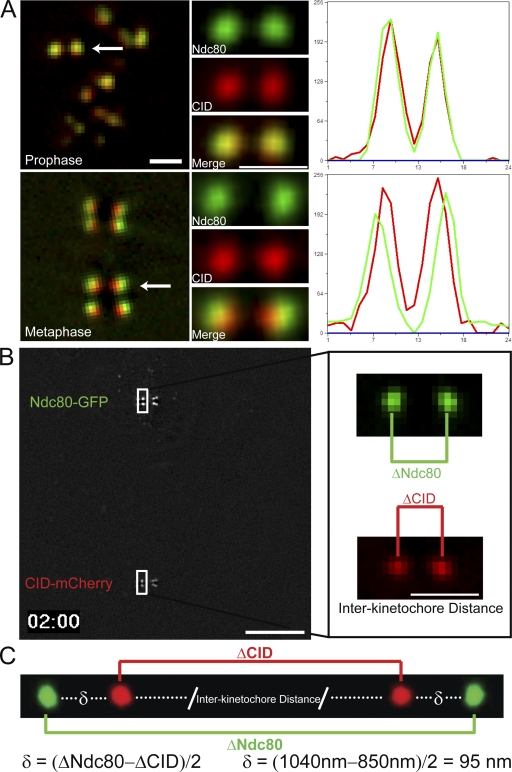
To investigate the physical changes that occur within the kinetochore structure itself, we generated a Drosophila melanogaster S2 cell line, which we have deemed K-Tensor (kinetochore tensometer and orientation) cells, that expresses both centromere identifier (CID)–mCherry and Ndc80-GFP (C-terminal label) to mark the inner and outer layers of the kinetochore, respectively (Fig. 1 A). The existence of movable elements within the kinetochore was first indicated by the fact that the distance between the inner and outer labels of the kinetochore changes between prophase and metaphase kinetochores (Fig. 1 A). Live imaging of K-Tensor cells (Fig. 1 B and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200808130/DC1) allowed us to monitor interkinetochore distance by measuring the distance between the peak CID intensities of a sister kinetochore pair and intrakinetochore distance (delta). Delta was calculated by measuring the difference in distance between the Ndc80 and CID signals for a pair of sister kinetochores divided by two (Fig. 1 C; Wordeman et al., 1991; Schittenhelm et al., 2007). This method corrects for errors caused by lateral chromatic aberration.
Figure 1.
K-Tensor Drosophila S2 cells exhibit detectable changes in the distance between the inner and outer layers of the kinetochore. (A) Representative micrographs of prophase and metaphase K-Tensor cells expressing Ndc80-GFP (green) and CID-mCherry (red). Ndc80, CID, and merged images are shown for the highlighted kinetochore pairs (white arrows). Line scans for each of the kinetochore pairs are shown to the right of each pair, with the red line indicating CID-mCherry intensity and the green line reflecting Ndc80-GFP intensity. (B) A single frame from a dual-view imaged K-Tensor cell (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200808130/DC1) showing simultaneous imaging of Ndc80-GFP and CID-mCherry. The inset shows enlarged images of the Ndc80 (green) and CID (red) signals for the highlighted kinetochore pair (white boxes). The distances between the two brightest pixels for each pair are represented by ΔNdc80 and ΔCID. (C) To calculate delta (δ), which is the distance between Ndc80-GFP and CID-mCherry, ΔCID is subtracted from ΔNdc80 and divided by two. Bars: (A and B, inset) 1 µm; (B) 10 µm.
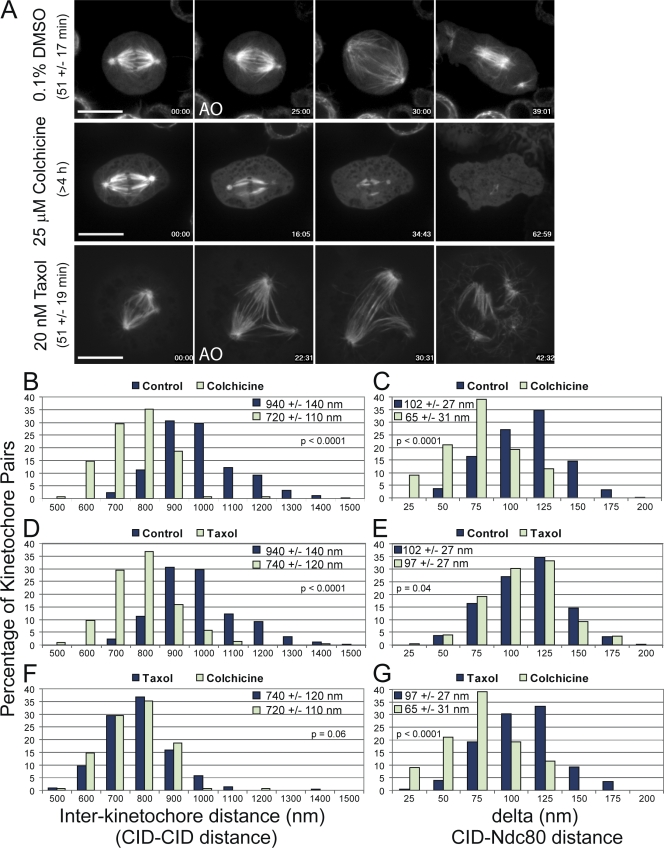
We first wanted to establish baseline measurements for mitotic progression, mean interkinetochore distance, and mean delta in K-Tensor cells without microtubules compared with metaphase cells under normal conditions. Spindle microtubules in mitotic S2 cells completely depolymerized within 1 h after the addition of 25 µM colchicine to the media (Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200808130/DC1). Most S2 cells are capable of arresting in mitosis for at least 4–5 h in the presence of colchicine (Fig. 2 A). The interkinetochore distance was 720 ± 110 nm and delta was 65 ± 31 nm for colchicine-treated cells. These values define the rest lengths for both interkinetochore distance and delta in the absence of attached kinetochore microtubules (Fig. 2, B and C). In comparison, spindles in control K-Tensor cells progressed through mitosis (defined as the time from the onset of chromosome condensation and anaphase) in 51 ± 19 min (Video 2). The interkinetochore distance for control cells at metaphase was 940 ± 130 nm, whereas delta was 102 ± 27 nm (Fig. 2, B–E). The mean centromere stretch, or increase in distance over rest length, was 220 nm between chromosomes with unattached kinetochore microtubules (colchicine treated) and bioriented chromosomes (control metaphase). The mean intrakinetochore stretch in control cells was 37 nm above the rest length distance of 65 nm in colchicine-treated cells.
Figure 2.
Microtubule poisons differentially affect interkinetochore distance and delta. (A) Selected frames from time-lapse imaging of GFP-tubulin–expressing S2 cells after treatment with DMSO, 25 µM colchicine, or 20 nM taxol. DMSO control cells progress through mitosis (as defined by chromosome condensation to anaphase onset [AO]) in 51 ± 17 min (n = 124 cells). Spindle microtubules completely depolymerize within 1 h after the addition of 25 µM colchicine, which causes cells to delay in mitosis for at least 4 h (n = 40 cells). Cells treated with 20 nM taxol progress through mitosis with the same kinetics as control cells (51 ± 19 min; n = 121 cells). (B–G) The distributions of interkinetochore distances (left) and delta (right) values are shown for all treatments. (B and C) Colchicine treatment causes reduction of both interkinetochore distance and delta, defining the rest lengths for each parameter (n = 156 kinetochore pairs). (D and E) Taxol treatment (n = 228 kinetochore pairs) causes a reduction in the interkinetochore distance; however, delta is not dramatically reduced relative to controls (n = 346 kinetochore pairs). (F and G) Comparing taxol and colchicine treatments highlights the fact that 20 nM taxol causes the interkinetochore distance to approach rest length without dramatically reducing delta. The mean values ± standard deviations and the two-tailed p-values for all conditions are shown in each graph. Bars, 10 µm.
Next, we wanted to test how intrakinetochore stretch is related to centromere stretch. In S2 cells, spindles assembled with aligned and bioriented chromosomes in the presence of 20 nM taxol (Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200808130/DC1). Surprisingly, these cells exhibited the same kinetics of mitotic progression (51 ± 19 min) as control cells (Fig. 2 A). Thus, the SAC is satisfied in S2 cells in the presence of low doses of taxol. In the presence of 20 nM taxol, the mean interkinetochore distance was reduced to 740 ± 120 nm, which represents only 9% of control centromere stretch. Addition of 20 nM taxol slightly reduced delta compared with DMSO-treated cells to a mean value of 97 ± 27 nm (Fig. 2, D–G). The distribution of values for interkinetochore distance after taxol treatment was similar to the distribution for colchicine-treated cells, whereas the distribution of delta values for 20 nM taxol was very similar to control measurements (Fig. 2, D–F). These data indicate that the intra- and interkinetochore elements are not linear Hookean springs connected in series but rather exist as a more complex mechanical arrangement.
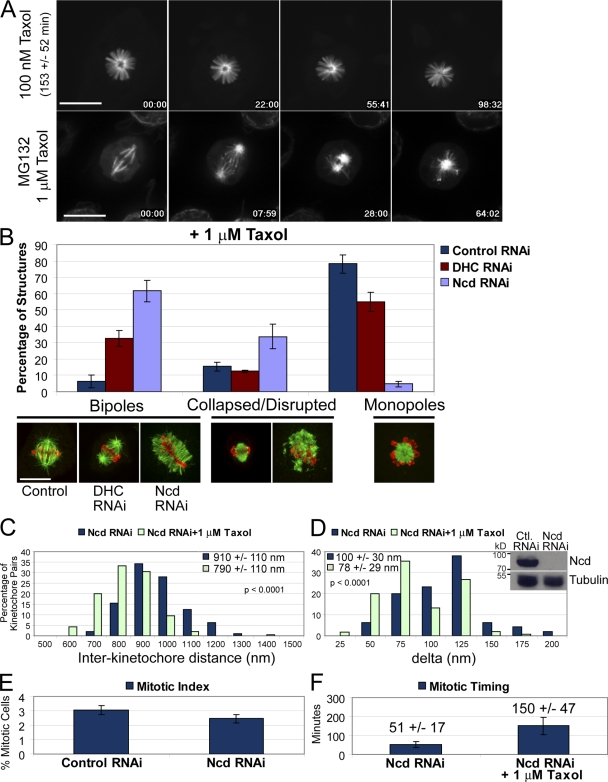
In many vertebrate cell types, addition of higher doses of taxol stabilizes microtubule dynamics and preserves kinetochore microtubules while inducing a near complete reduction of centromere stretch and activating the SAC. Addition of >20–50 nM taxol to S2 cells caused cells to arrest in mitosis for 153 ± 52 min with monopolar spindles (Fig. 3 A and Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200808130/DC1). These structures were not amenable to investigating delta because they lacked bioriented chromosomes. Time-lapse imaging of taxol-induced bipolar spindle collapse led us to hypothesize that this process required minus end–directed motor activity (Video 6). Indeed, we found that the percentage of taxol monopoles significantly decreased after RNAi of two different minus end–directed motors: dynein heavy chain (DHC) or nonclaret disjunction (Ncd; Fig. 3 B). The data indicate that DHC and Ncd share functional redundancy in the bipolar spindle collapse mechanism, albeit Ncd is the dominant minus end–directed motor activity involved in this process. DHC RNAi taxol bipoles maintained focused poles, whereas Ncd RNAi structures had unfocused kinetochore fibers. The Ncd RNAi structures that were classified as bipolar consisted of bioriented chromosomes largely aligned between unfocused kinetochore fiber arrays (Fig. 3 B).
Figure 3.
Treatment with 1 µM taxol reduces delta and activates the SAC. (A) Selected frames from time-lapse imaging of a preformed taxol monopole (top) and an MG132-treated bipolar spindle collapsing into a monopole after the addition of 1 µM taxol (bottom). Taxol monopoles delay in mitosis for 153 ± 52 min. (B) Taxol-induced spindle collapse is mediated by minus end–directed motor activity. The graph shows the percentage of each indicated structure with representative images after treatment with 1 µM taxol for 1 h. (C and D) Treatment of Ncd RNAi structures with 1 µM taxol (n = 240 kinetochore pairs) causes a reduction in both the interkinetochore distance and delta relative to Ncd RNAi alone (n = 95 kinetochore pairs). Ncd is undetectable by Western blotting (inset) after dsRNA treatment (96 h). (E) Ncd RNAi does not cause an increase in the number of mitotic cells in a cycling population. (F) Ncd RNAi cells divide with normal kinetics (51 ± 17 min; n = 100 cells), but addition of 1 µM taxol delays them in mitosis for 150 ± 47 min (n = 77 cells), indicating that the SAC is activated. The mean values ± standard deviations and the two-tailed p-values are shown in each graph. Error bars represent the standard deviations. Bars, 10 µm.
The presence of bioriented kinetochores in the Ncd and DHC RNAi conditions allowed us to measure interkinetochore distance and delta under high taxol conditions in which kinetochore–microtubule attachment was not evidently compromised (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200808130/DC1). RNAi of DHC or Ncd alone had minor effects on metaphase interkinetochore distance and delta (Fig. 3, C and D; and Fig. S2). In contrast, addition of 1 µM taxol to Ncd and DHC RNAi cells reduced both interkinetochore distance and delta (Fig. 3, C and D; and Fig. S2). Note that for both DHC RNAi and Ncd RNAi, taxol treatment reduced delta without reducing interkinetochore distance below the 20-nM taxol level (Fig. 2, D and F). This is further evidence that the mechanical linkages between centromeric DNA and structures within sister kinetochores are not a simple arrangement of linear Hookean springs.
In the absence of taxol, DHC RNAi induces a mitotic delay (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200808130/DC1) because Mad2 is retained at attached kinetochores (Griffis et al., 2007); however, Ncd RNAi does not cause an increase in the mitotic index or slow mitotic progression (Fig. 3, E and F). Importantly, addition of 1 µM taxol delayed Ncd RNAi cells in mitosis for 150 ± 47 min, indicating that the SAC was active. Thus, suppression of microtubule dynamics in Ncd RNAi cells with 1 µM taxol reduced delta and activated the SAC without clearly disrupting microtubule attachment.
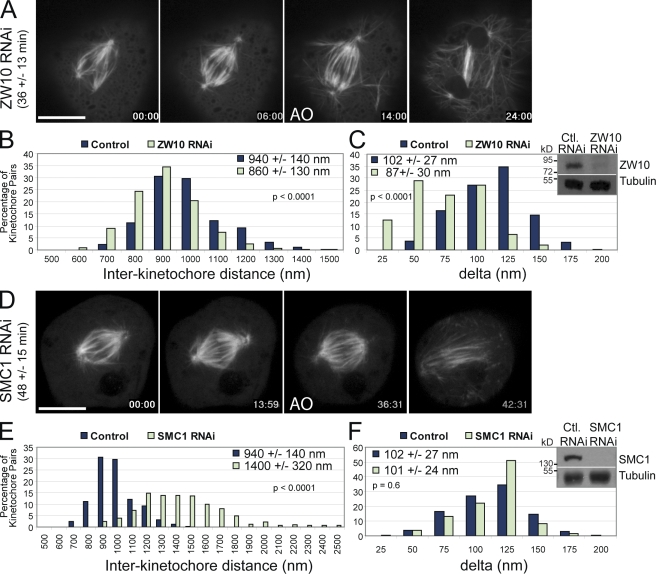
To further understand how interkinetochore distance and delta are related, we used molecular perturbations to alter interkinetochore distances and assayed how delta was affected. The Rod–ZW10–Zwilch complex member ZW10 is a bona fide SAC regulator that is required to target dynein and Mad1/Mad2 to kinetochores (Starr et al., 1998; Basto et al., 2000; Buffin et al., 2005). In addition, ZW10 has been implicated in regulating centromere stretch, kinetochore–microtubule attachment stability, and tension sensing (Famulski and Chan, 2007; Yang et al., 2007). In agreement with previous findings (Basto et al., 2000; Kops et al., 2005; Famulski et al., 2008), ZW10 RNAi in S2 cells led to loss of SAC activity, as the mitotic index of ZW10 RNAi cells treated for 18 h with colchicine was fourfold lower than control cells (unpublished data). Furthermore, ZW10 knockdown caused cells to progress through mitosis faster than control cells (36 ± 13 min vs. 51 ± 17 min; Fig. 4 A and Video 7, available at http://www.jcb.org/cgi/content/full/jcb.200808130/DC1). We found that the mean interkinetochore distance was reduced from 940 ± 130 nm in controls to 860 ± 130 nm after ZW10 RNAi (Fig. 4 B). This represents an ∼40% reduction in centromere stretch as previously observed (Yang et al., 2007). ZW10 RNAi yielded a broad distribution of delta distances with a mean delta of 87 ± 30 nm (Fig. 4 C). Because ZW10 targets a population of dynein to the kinetochore, we hypothesized that dynein could be responsible for generating delta. However, DHC RNAi produced a smaller reduction in delta than ZW10 RNAi and did not reduce the mean interkinetochore distance (Fig. S3). Thus, although we cannot rule out some contribution of dynein to intrakinetochore stretch, the effects of ZW10 RNAi cannot be explained solely by the reduction of dynein levels at the kinetochore.
Figure 4.
Interkinetochore distance and delta can be experimentally uncoupled. (A) Selected frames from time-lapse imaging of GFP-tubulin cells after ZW10 RNAi treatment. Cells progress through mitosis in 36 ± 13 min (n = 141 cells) in the absence of ZW10, which is faster than control cells (51 ± 17 min). (B and C) Reduced levels of ZW10 cause interkinetochore distances and delta (n = 777 kinetochore pairs) to be reduced relative to controls. Western blot analysis (inset) reveals efficient knockdown of ZW10 by RNAi (96 h). (D) Frames from time-lapse imaging of GFP-tubulin cells after SMC1 RNAi. Similar to control cells (51 ± 17 min), SMC1 dsRNA–treated cells progress through mitosis in 48 ± 15 min (n = 222 cells). (E and F) SMC1 RNAi increases the interkinetochore distance without affecting delta (n = 451 kinetochore pairs). SMC1 is nearly undetectable by Western blotting (inset) after dsRNA treatments (96 h). The mean values ± standard deviations and the two-tailed p-values for all conditions are shown in each graph. AO, anaphase onset. Bars, 10 µm.
SMC1 is a component of the cohesin complex, which helps to physically link sister chromatids until anaphase onset (Nasmyth et al., 2000). We found that SMC1 RNAi caused an increase in the interkinetochore distance to 1,400 ± 320 nm (Fig. 4 E), a threefold increase in centromere stretch over control metaphase cells, without significantly perturbing kinetochore structure or mitotic progression (Fig. 4 D and Video 8, available at http://www.jcb.org/cgi/content/full/jcb.200808130/DC1). However, the “hyperstretched” centromeres of SMC1 RNAi cells did not impact the delta distances (101 ± 24 nm), which were indistinguishable from control cells (Fig. 4 F). These data show that normal delta is produced even when the physical linkages between kinetochores have been altered by loss of SMC1.
The 3F3/2 phosphoepitope serves as a molecular readout for kinetochore tension during mitosis whereby low-tension conditions exhibit high levels of kinetochore 3F3/2 staining that become reduced upon biorientation (Gorbsky and Ricketts, 1993; Nicklas et al., 1995). Because we could control delta and interkinetochore distance experimentally, we next set out to determine which of these parameters correlated with 3F3/2 staining (Fig. 5). Microtubule depolymerization with colchicine, which reduced both interkinetochore distance and delta, induced the highest levels of 3F3/2 staining. In control metaphase cells, 3F3/2 levels dropped two- to fourfold (a mean of 2.4-fold) relative to colchicine-treated cells. Experimental conditions that yielded delta near control levels on bioriented chromosomes (20 nM taxol and SMC1, DHC, and Ncd RNAi) all had 3F3/2 staining near control levels, and, with the exception of DHC RNAi, which fails to remove Mad1/Mad2 from kinetochores, all satisfied the SAC even though interkinetochore distances varied 30-fold. However, conditions that yielded >40% reduction in intrakinetochore stretch (colchicine, Ncd RNAi + 1 µM taxol, DHC RNAi + 1 µM taxol, and ZW10 RNAi) had elevated levels of 3F3/2 and, with the exception of ZW10, which lacks a functional checkpoint, activated the SAC (Fig. 5 B and Figs. S2 and S3). Collectively, these data demonstrate that levels of the tension-sensitive 3F3/2 phosphoepitope correlates inversely with delta (high delta = low 3F3/2 and low delta = high 3F3/2) rather than simply centromere stretch (Fig. 5 B). Furthermore, SAC activation correlated with a reduction in delta rather than centromere stretch.
Figure 5.
Generation of the tension-sensitive phosphoepitope 3F3/2 correlates with changes in delta. (A) Representative micrographs from each of the indicated conditions. In the merged images, DNA is blue, CID is red, and 3F3/2 is green. (B) The ratio of fluorescent intensities for 3F3/2-CID signals versus centromere stretch, intrakinetochore stretch, and mitotic progression is shown for each experimental condition. The 3F3/2 levels (n = 373 pairs from seven experiments), centromere stretch, kinetochore stretch, and mitotic progression for control + DMSO cells were each assigned a value of 100%, and all other experimental conditions were normalized accordingly. Colchicine treatment results in a 2.4-fold increase in 3F3/2 levels (n = 522 kinetochore pairs from nine experiments) and a reduction of kinetochore and centromere stretches to their minima (rest lengths) as well as a mitotic delay. 20 nM taxol does not cause a mitotic delay or elevated levels of 3F3/2 (n = 102 pairs from three experiments) despite an ∼90% reduction in centromere stretch. Ncd RNAi + DMSO (3F3/2 measurements: n = 154 pairs from three experiments) behave similar to control cells; however, addition of 1 µM taxol to Ncd RNAi cells causes a 2.4-fold increase in 3F3/2 levels (n = 161 pairs from three experiments), a 65% reduction in centromere and kinetochore stretches, and a mitotic delay. ZW10 RNAi cells (n = 189 pairs from three experiments) have 1.8-fold higher levels of 3F3/2 and an ∼40% reduction in both centromere stretch and kinetochore stretch but progress through mitosis faster than control cells. SMC1 RNAi cells have levels of 3F3/2 staining similar to controls (n = 144 pairs from three experiments), hyperstretched centromeres, normal kinetochore stretch, and normal mitotic progression. The error bars represent the standard deviations. Bar, 10 µm.
The mechanical elements responsible for intrakinetochore stretch in control metaphase cells exhibited a much different behavior than those for centromere stretch. The latter exhibited a wide range of length changes dependent on biorientation, sister chromatid linkages (SMC1 depletion), and suppression of microtubule dynamics with taxol. In contrast, intrakinetochore stretch did not behave as a linearly elastic element. Thus, it is possible that the kinetochore contains a molecular switch with an “on” conformation for distances between 65 and 87 nm and an “off” conformation for deltas near 100 nm; “on” corresponds to high levels of 3F3/2 phosphorylation and an active SAC. Strong yet flexible filaments of finite lengths that connect the inner and outer kinetochore may limit intrakinetochore stretch to ∼100 nm as the centromere becomes stretched above its rest length.
Changes in delta could partly be explained by structural rearrangements within the kinetochore after microtubule attachment, as previously observed by electron tomography (Dong et al., 2007). Because microtubule depolymerization with colchicine yielded a mean delta of 65 nm, whereas 1-µM taxol treatment of Ncd RNAi cells had a mean delta of 78 nm, microtubule attachment alone may account for ∼10–15 nm of intrakinetochore stretch at metaphase. How is the remaining 20 nm of intrakinetochore stretch generated? 1-µM taxol treatment reduced the majority of both centromere stretch and delta. In contrast, 20 nm taxol, which also greatly reduced centromere stretch, did not dramatically reduce delta. This indicates that force generation, driven primarily by plus end dynamics of kinetochore microtubules, generates the necessary intrakinetochore stretch that contributes to 3F3/2 phosphorylation and SAC activity. Furthermore, this stretch is likely to require low levels of force that can still be generated by the significantly dampened microtubule dynamics in 20 nM taxol that we believe are completely suppressed by 1-µM taxol treatment. Our data do not exclude the ability for tension generated by centromere stretch to produce intrakinetochore stretch; however, it does show that intrakinetochore stretch occurs at very low levels of centromere stretch and requires dynamic microtubules.
Our data also show that changes in delta rather than centromere stretch correlate with 3F3/2 phosphorylation. Importantly, a reduction in delta also correlates with SAC activation when a functional checkpoint pathway is present. Furthermore, we believe that any SAC mechanism that is regulated by intrakinetochore stretch must be upstream of kinetochore-bound Mad1/Mad2 for two reasons: (1) ZW10 RNAi, which prevents Mad1/Mad2 from binding kinetochores (Buffin et al., 2005), fails to arrest cells even with reduced delta and elevated levels of 3F3/2, and (2) DHC RNAi, which prevents stripping of Mad1/Mad2 from kinetochores (Griffis et al., 2007), causes a mitotic delay even with high delta and reduced levels of 3F3/2.
In conclusion, we postulate that intrakinetochore stretch is generated by the translocation of protein elements in response to the attachment of dynamic microtubules. Furthermore, this movement may behave like a low-tension “mechanical switch” that controls SAC activity by changing the relative proximity and localization of regulatory factors within the kinetochore. Future investigation of this hypothesis will have significant implications for kinetochore structure, mechanics, and SAC function.
Materials and methods
Cell culture and RNAi
Drosophila S2 cells were cultured at 24°C in Schneider’s medium supplemented with 10% heat-inactivated FBS (Invitrogen) and 0.5× antibiotic–antimycotic cocktail (Invitrogen). For RNAi experiments, media was aspirated off semiadhered cells at 25% confluence in 6-well plates and replaced with 1 ml of serum-free Schneider’s medium containing ∼20 µg double-stranded RNA (dsRNA). After 1 h, 1 ml of fresh Schneider’s medium + FBS was added to the wells, and they were incubated for 4 d at 24°C.
Live cell imaging
Cells were seeded onto Con A (Sigma-Aldrich)–treated acid-washed coverslips (Corning) for 1 h. The coverslips were assembled into rose chambers containing media and imaged at room temperature. GFP-tubulin cells (provided by G. Goshima and R. Vale, University of California, San Francisco, San Francisco, CA) were imaged on an inverted microscope stand (TE300; Nikon) equipped with a Yokogawa spinning disk confocal head (CSU10; PerkinElmer) attached to a cooled charge-coupled device camera (Orca ER; Hamamatsu Photonics) using a 100× 1.4 NA plan-Apochromatic differential interference contrast objective (Nikon). Most K-Tensor data were acquired using a 100× 1.4 NA plan-Apochromatic differential interference contrast objective on an inverted microscope stand (TE2000-U; Nikon) with the 1.5× Optivar in place and equipped with a beamsplitter (Dual-View; Photometrics) attached to a camera (iXON EMCCD; Andor Technology) to simultaneously image mCherry and GFP. Some K-Tensor cell data were obtained by acquiring near simultaneous confocal fluorescence images at 488 and 568 nm. This technique yielded results similar to the Dual-View imaging technique. Images were processed in Photoshop CS3 (Adobe).
Interkinetochore distances and delta measurements
K-Tensor cells were analyzed using MetaMorph version 6.3 software (MDS Analytical Technologies). The single line region tool was used to draw a straight line between the brightest pixels of each kinetochore pair for the GFP (ΔNdc80) and mCherry (ΔCID) spots. The length of each line was then calibrated based on a units/pixel value assigned by imaging a calibration micrometer. The delta value (delta = [ΔNdc80 − ΔCID]/2) was calculated for ∼200–800 kinetochore pairs per condition. Because of pixelation, we believe that the measurements for each condition may overestimate the actual distance between fluorophores by ≤10%. The mCherry-CID construct was provided by S. Moutinho-Pereira and H. Maiato (Instituto de Biologia Molecular e Celular, Porto, Portugal). Genomic DNA for amplifying the Ndc80 gene was provided by G. Rogers and S. Rogers (University of North Carolina at Chapel Hill, Chapel Hill, NC).
Immunofluorescence
Coverslips were briefly washed in BRB80 and fixed in 100% methanol at −20°C for 10 min. The coverslips were then incubated in PBS + 1% Triton X-100 for 10 min. Next, they were washed briefly three times with PBS + 0.1% Triton X-100 before blocking with 5% boiled donkey serum for 30–60 min. Coverslips were incubated in primary antibodies diluted in 5% boiled donkey serum (1:100 CID [Abcam], 1:500 3F3/2 [Boston Biologicals], and 1:150 DHC) overnight at 4°C. The next day, coverslips were washed three times for 5 min in PBS + 0.1% Triton X-100 before incubating them in secondary antibodies (Jackson ImmunoResearch Laboratories) diluted 1:200 in 5% boiled donkey serum containing 0.1 mg/ml DAPI. The coverslips were then washed three times for 5 min in PBS + 0.1% Triton X-100 and mounted in a solution of 90% glycerol and 0.5% N-propyl gallate. For 3F3/2 staining, 10 µM microcystin lysine-arginine (Sigma-Aldrich) was included in all buffers. Aliquots of 10-mM microcystin stock were stored at −80°C and added fresh to each of the buffers. The DHC antibody was provided by T. Hays (University of Minnesota, Minneapolis, MN). The ZW10 antibody was provided by M. Goldberg (Cornell University, Ithaca, NY). The SMC1 antibody was provided by S. Bickel (Dartmouth College, Hanover, NH). The Ncd antibody was provided by J. Scholey (University of California, Davis, Davis, CA).
Fluorescence quantification for 3F3/2 and DHC was performed similarly to previous work (Hoffman et al., 2001); however, the larger and smaller regions were drawn manually in MetaMorph, and all measurements were normalized to CID staining. The following equations were used: background signal = (integrated fluorescence intensitybig area − integrated fluorescence intensitysmall area)/(areabig − areasmall) and total intensity = integrated fluorescence intensitysmall area × (background signal × small area).
dsRNAs
dsRNAs were synthesized from DNA templates using the T7 RiboMAX Express Large Scale RNA Production System (Promega). Templates were generated from pQE30 (QIAGEN) or the cDNA constructs for ZW10 (CG9900), SMC1 (CG6057), DHC (CG7507-RA), or Ncd (CG7831). The following sequences were placed after the T7 promoter sequence (5′-TAATACGACTCACTATAGGG-3′): control forward, 5′-AACTCCATCTGGATTTGTTC-3′; control reverse, 5′-GTTGTCCATATTGGCCACGT-3′; ZW10 forward, 5′-ATGGAGGAAGAGGCGCCGCG-3′; ZW10 reverse, 5′-CTCCTGGAGGCTCTGCTGCA-3′; SMC1 forward, 5′-CACATATCGGATGCCATTGA-3′; SMC1 reverse, 5′-ACGTCTACACATTTACTGGC-3′; DHC forward, 5′-TGCCCAGGCGAATAGTTGGT-3′; DHC reverse, 5′-CAAGTTTAAAGTATTTCATT-3′; Ncd forward, 5′-ATGGAATCCCGGCTACCGAA-3′; and Ncd reverse, 5′-GTGGAAGCGGGCCTTGAAGT-3′.
Online supplemental material
Fig. S1 shows that kinetochore fibers are present in 1-µM taxol conditions. Fig. S2 characterizes the effects of 1 µM taxol on 3F3/2 levels, interkinetochore distance, and delta in DHC RNAi bipolar spindles. Fig. S3 shows a comparison of ZW10 and DHC RNAi conditions. Video 1 shows time-lapse imaging of Drosophila K-Tensor cells with both the GFP and mCherry signals imaged simultaneously. Videos 2–8 show time-lapse imaging of GFP-tubulin–expressing S2 cells after various chemical and molecular perturbations. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200808130/DC1.
Acknowledgments
We are grateful to all members of the Salmon laboratory, especially to Dr. J. Gatlin for sharing many insightful scientific discussions of this work. We thank the members of the Cell Division Group at Marine Biological Laboratories (MBL), especially Dr. A. Groen for advice, and we are grateful to Dr. C. Waterman-Storer for providing lab space at MBL. We also thank Drs. G. Rogers and S. Rogers for sharing their expertise and reagents, Dr. H. Maiato and S. Moutinho-Pereira for the pMT–CID-mCherry construct, and Drs. S. Bickel, M. Goldberg, T. Hays, J. Scholey, G. Goshima, and R. Vale for reagents.
This work was supported by the American Cancer Society (grant PF0711401 to T.J. Maresca) and the National Institutes of Health (grant GM24364 to E.D. Salmon).
Footnotes
Abbreviations used in this paper: CID, centromere identifier; DHC, dynein heavy chain; dsRNA, double-stranded RNA; Ncd, nonclaret disjunction; SAC, spindle assembly checkpoint.
References
- Basto R., Gomes R., Karess R.E. 2000. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila.Nat. Cell Biol. 2:939–943 [DOI] [PubMed] [Google Scholar]
- Biggins S., Murray A.W. 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint.Genes Dev. 15:3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M.E., Karess R.E. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex.Curr. Biol. 15:856–861 [DOI] [PubMed] [Google Scholar]
- Dong Y., Vanden Beldt K.J., Meng X., Khodjakov A., McEwen B.F. 2007. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions.Nat. Cell Biol. 9:516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulski J.K., Chan G.K. 2007. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores.Curr. Biol. 17:2143–2149 [DOI] [PubMed] [Google Scholar]
- Famulski J.K., Vos L., Sun X., Chan G. 2008. Stable hZW10 kinetochore residency, mediated by hZwint-1 interaction, is essential for the mitotic checkpoint.J. Cell Biol. 180:507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky G.J., Ricketts W.A. 1993. Differential expression of a phosphoepitope at the kinetochores of moving chromosomes.J. Cell Biol. 122:1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Stuurman N., Vale R.D. 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore.J. Cell Biol. 177:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., Salmon E.D. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores.Mol. Biol. Cell. 12:1995–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G.J., Kim Y., Weaver B.A., Mao Y., McLeod I., Yates J.R., III, Tagaya M., Cleveland D.W. 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore.J. Cell Biol. 169:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Nicklas R.B. 1995. Mitotic forces control a cell-cycle checkpoint.Nature. 373:630–632 [DOI] [PubMed] [Google Scholar]
- McIntosh J.R. 1991. Structural and mechanical control of mitotic progression.Cold Spring Harb. Symp. Quant. Biol. 56:613–619 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time.Nat. Rev. Mol. Cell Biol. 8:379–393 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Peters J.M., Uhlmann F. 2000. Splitting the chromosome: cutting the ties that bind sister chromatids.Science. 288:1379–1385 [DOI] [PubMed] [Google Scholar]
- Nicklas R.B., Ward S.C., Gorbsky G.J. 1995. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint.J. Cell Biol. 130:929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B.A., Biggins S. 2005. The spindle checkpoint: tension versus attachment.Trends Cell Biol. 15:486–493 [DOI] [PubMed] [Google Scholar]
- Schittenhelm R.B., Heeger S., Althoff F., Walter A., Heidmann S., Mechtler K., Lehner C.F. 2007. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes.Chromosoma. 116:385–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Williams B.C., Hays T.S., Goldberg M.L. 1998. ZW10 helps recruit dynactin and dynein to the kinetochore.J. Cell Biol. 142:763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern B.M., Murray A.W. 2001. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast.Curr. Biol. 11:1462–1467 [DOI] [PubMed] [Google Scholar]
- Waters J.C., Skibbens R.V., Salmon E.D. 1996. Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push.J. Cell Sci. 109:2823–2831 [DOI] [PubMed] [Google Scholar]
- Wordeman L., Steuer E.R., Sheetz M.P., Mitchison T. 1991. Chemical subdomains within the kinetochore domain of isolated CHO mitotic chromosomes.J. Cell Biol. 114:285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tulu U.S., Wadsworth P., Rieder C.L. 2007. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint.Curr. Biol. 17:973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]