Abstract
When yeast cells sense mating pheromone, they undergo a characteristic response involving changes in transcription, cell cycle arrest in early G1, and polarization along the pheromone gradient. Cells in G2/M respond to pheromone at the transcriptional level but do not polarize or mate until G1. Fus2p, a key regulator of cell fusion, localizes to the tip of the mating projection during pheromone-induced G1 arrest. Although Fus2p was expressed in G2/M cells after pheromone induction, it accumulated in the nucleus until after cell division. As cells arrested in G1, Fus2p was exported from the nucleus and localized to the nascent tip. Phosphorylation of Fus2p by Fus3p was required for Fus2p export; cyclin/Cdc28p-dependent inhibition of Fus3p during late G1 through S phase was sufficient to block exit. However, during G2/M, when Fus3p was activated by pheromone signaling, Cdc28p activity again blocked Fus2p export. Our results indicate a novel mechanism by which pheromone-induced proteins are regulated during the transition from mitosis to conjugation.
Introduction
Haploid yeast cells of opposite mating types undergo cell and nuclear fusion to form diploids. The haploid cell types (a and α) produce mating pheromones that bind to receptors on the plasma membrane of the opposite mating type, causing the cells to differentiate into mating cells or shmoos. Physiological changes associated with shmoo formation include transcriptional induction of genes required for mating, cell cycle arrest in G1, and polarization toward the source of pheromone. After the cells make contact, the cell walls between the mating partners are broken down, after which the plasma membrane and nuclear membranes can fuse (Ydenberg and Rose, 2008; for reviews see Marsh and Rose, 1997; White and Rose, 2001).
Interaction of α factor with its receptor stimulates guanine nucleotide exchange on the α subunit of a heterotrimeric G protein, causing it to release the βγ subunit (Gustin et al., 1998; Elion, 2000; Naider and Becker, 2004; Bardwell, 2005). Free Gβγ recruits the MAPK scaffold Ste5p to the plasma membrane, allowing phosphorylation and activation of the MAPK kinase kinase Ste11p (Pryciak and Huntress, 1998). After Ste11p activation of the MAPK kinase Ste7p, Ste7p phosphorylates and activates the MAPK Fus3p, which is required for many aspects of pheromone-induced differentiation. However, fus3 mutants are still capable of responding to pheromone as a result of the presence of the partially redundant Kss1 protein (Elion et al., 1991; Kusari et al., 2004). Fus3p and Kss1p both activate the transcription factor Ste12p and therefore induce an overlapping set of genes (Dolan et al., 1989; Madhani and Fink, 1997; Roberts et al., 2000). In addition, fus3 mutants are defective for G1 cell cycle arrest, but this phenotype can be suppressed by mutations in the G1 CLN3 (Elion et al., 1990). Fus3p also plays a key role in cell polarization that is not shared with Kss1p (Metodiev et al., 2002; Matheos et al., 2004).
Mating requires wholesale reorganization of the microtubule and actin cytoskeletons. In addition, many components of the mating and mitotic programs are shared but devoted to distinct tasks. For example, KAR3 encodes a microtubule motor protein with different roles in mating and mitosis (Meluh and Rose, 1990; Saunders and Hoyt, 1992; Page et al., 1994; Saunders et al., 1997). Kar3p is associated with cytoplasmic microtubules in shmoos and nuclear microtubules during mitosis. If cycling cells are treated with pheromone, what prevents morphological induction of the mating response before G1? Recent results indicate that during late G1 through S phase, the cyclin (Cln)-dependent kinase Cdc28p phosphorylates Ste5p, preventing its recruitment to the plasma membrane and thereby inhibiting activation of the pheromone response pathway (Strickfaden et al., 2007). However, cell synchrony experiments, genetic studies overexpressing the Clns, and in vitro kinase assays all show that Ste5p phosphorylation is specific to Cln1p/Cdc28p and Cln2p/Cdc28p and that pheromone-dependent transcription becomes fully active in G2/M (Oehlen and Cross, 1994, 1998; Wassmann and Ammerer, 1997; Strickfaden et al., 2007). Proteins promoting the mating response are therefore present in pheromone-treated cells in G2/M, but major effects such as cytoskeletal rearrangements do not take place.
Protein localization is an ideal readout for cell cycle–regulated signal transduction because it can be studied in unperturbed asynchronous cells. We previously found that the cell fusion protein Fus2p localizes to the shmoo tip after pheromone-induced cell cycle arrest but is nuclear when ectopically expressed in mitotic cells (Paterson et al., 2008). This change suggests that Fus2p localization is regulated by pheromone signaling, the cell cycle, or both. Fus2p interacts with an amphiphysin homologue, Rvs161p, which plays distinct roles in cell fusion and endocytosis (Munn et al., 1995; Brizzio et al., 1998). Although the biochemical function of Fus2p is unknown, the protein sequence contains a Dbl homology domain (Paterson et al., 2008), making it likely that Fus2p promotes cell fusion via Rho proteins, which are central regulators of cell polarity, exocytosis, and cell wall integrity during both mating and mitotic growth (Park and Bi, 2007). Regulation of Fus2p localization after induction may be critical to prevent interference with housekeeping functions.
In this study, we show that Fus2p expressed in G2/M cells in response to pheromone is sequestered in the nucleus until mitosis is completed, after which it exits and accumulates at the shmoo tip. Fus2p nuclear exit requires Fus3p-dependent pheromone signaling, and only the G1 phase of the cell cycle is permissive for exit. During the rest of the cell cycle, Cdc28p activity blocks Fus2p exit by at least two mechanisms; during late G1 through S phase, Cln/Cdc28 inhibits pheromone signaling and Fus3p activation, but during G2/M, Cdc28p blocks exit without inhibiting pheromone signaling. The antagonistic regulation of Fus2p nuclear exit by Fus3p and Cdc28p provides a novel pathway by which cells carefully control the transition from mitosis to mating.
Results
Fus2p exits the nucleus after mitosis in pheromone-treated cells
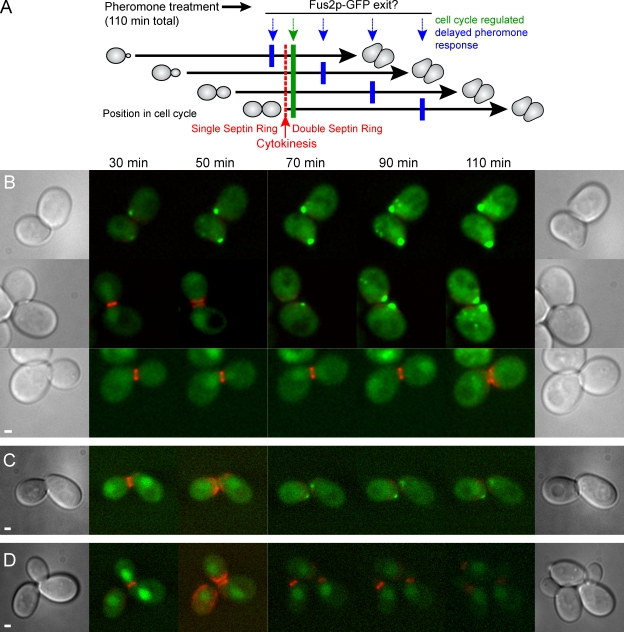
A previous study of the localization of Fus2p-GFP in pheromone-treated cells showed that it first appeared in the nucleus, after which it redistributed to the shmoo tip and cytoplasmic puncta (Paterson et al., 2008). Although Fus2p-GFP was concentrated in anaphase nuclei, it was apparently exported after anaphase because nuclear localization was not readily distinguishable in shmoos. We sought to determine whether the change in localization was caused by (a) the length of pheromone treatment, (b) increasing Fus2p levels during the pheromone treatment, or (c) a cell cycle–regulated switch. To do this, we treated asynchronous populations of cells with pheromone and examined the localization of Fus2p over time and with respect to cell cycle stage (Fig. 1 A). For these experiments, we used an internal GFP tag described previously (Paterson et al., 2008), which exhibits ∼90% of wild-type function. Cdc3p, a septin tagged with mCherry fluorescent protein (FP), served as a marker for the cell cycle. Septins form a single ring at the bud neck throughout G2 but split into two distinct rings during cytokinesis (Longtine and Bi, 2003), marking the time of cell division.
Figure 1.
Fus2p is expressed in pheromone-treated cells before cytokinesis but remains in the nucleus until after cell division. (A) Experiment schematic. An asynchronous cell population expressing Fus2p-GFP and Cdc3-mCherryFP, an indicator of cytokinesis (red), was treated with pheromone for 110 min (black arrows) and examined microscopically over time. If Fus2p-GFP nuclear exit is solely dependent on pheromone signaling, exit should occur at similar times after exposure for all cells (blue arrows). If Fus2p-GFP nuclear exit requires completion of the previous cell cycle, exit should occur soon after cytokinesis independent of the length of pheromone signaling (green arrow). (B) Fus2p-GFP expressed under its own promoter. Strain MY10176 containing FUS2∷GFP104 and CDC3-mCherry was pregrown in selective medium, and α factor was added at t = 0. Representative cells are shown in which cytokinesis occurred before 30 min, between 30 and 50 min, and between 90 and 110 min (top, middle, and bottom, respectively). (C) Fus2p-GFP expressed under the GAL1 promoter. Strain MY10177 containing PGAL1-FUS2∷GFP104 and CDC3-mCherry was pregrown in selective medium with galactose; α factor and glucose were added at t = 0. (D) Pheromone is required for Fus2p-GFP exit G1. MY10177 was examined as in C except that no α factor was added. Bars, 1 µm.
Although all cells were exposed to pheromone for the same amount of time, because the initial population was asynchronous, we observed two classes of cells, those in G1 at the start of the experiment and those that completed mitosis and entered G1 during the course of the experiment. The G1 cells formed shmoos immediately; the remaining cells were at various positions within the cell cycle and required different lengths of time to complete mitosis and form shmoos. In G1 cells, Fus2p-GFP was not observed in the nucleus at the first time point (30 min) but was already accumulated at the presumptive shmoo tip (Fig. 1 B, top; 16/16 cells). In cells that completed the cell cycle and underwent cytokinesis during imaging, Fus2p-GFP was initially nuclear. Nuclear exit was coincident with septin ring division in 23/38 cells or occurred within the time point immediately after septin ring division in 13/38 cells (Fig. 1 B, middle and bottom). In 2/38 cells, nuclear exit took ≥40 min after division. Therefore, Fus2p exit from the nucleus coincided with cell division and was not simply correlated with the amount of time the cells were treated with pheromone.
FUS2 transcription is strongly induced by pheromone (Elion et al., 1995), leading to an increase in total fluorescent signal during the experiment. However, because a low level of nuclear Fus2p-GFP was detected in untreated cells, we wanted to ensure that Fus2p-GFP expression was actually being induced by pheromone in G2 cells before shmoo formation. Accordingly, we compared GFP fluorescence levels in cells treated with pheromone for 30 min to untreated control cells. Examining only budded cells with fully formed single septin rings, we found that Fus2p-GFP levels were significantly higher in pheromone-treated cells than in untreated cells (103 ± 19, n = 30; vs. 28.6 ± 5.4, n = 25, respectively; P = 0.0018; numbers represent mean ± SEM). Together, these results show that FUS2 is induced before cell division but that exit only occurs after cell division.
To ensure that increasing Fus2p-GFP levels were not responsible for the observed change in localization, we expressed Fus2p-GFP under the GAL1 promoter. To examine the effect of pheromone on Fus2p localization independent of its role in promoting synthesis, cells were pregrown in galactose and transferred to glucose and pheromone simultaneously. Localization of GAL1-expressed Fus2p-GFP was similar to localization of Fus2p-GFP expressed under the native promoter. Fus2p-GFP remained nuclear until cell division and then exited in 22/22 cells in which septin ring division occurred during the experiment (Fig. 1 C). Therefore, Fus2p nuclear exit was not dependent on increasing Fus2p levels. If no pheromone was added, Fus2p-GFP remained nuclear regardless of cell cycle position (Fig. 1 D). We conclude that mating pheromone signaling and the cell cycle together regulate Fus2p localization.
Fus2p transiently exits in far1 mutants
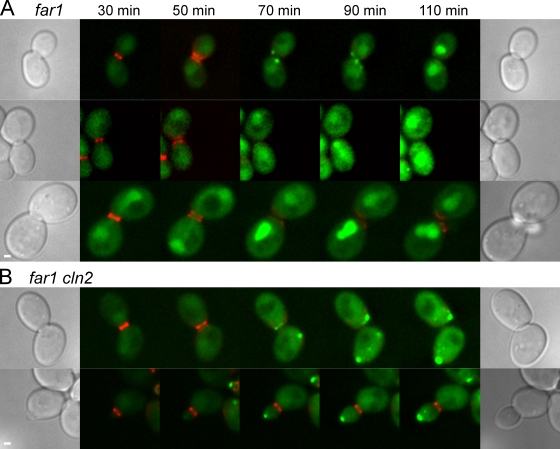
FAR1 encodes an inhibitor of Cln-dependent kinases that promotes pheromone-induced cell cycle arrest (Chang and Herskowitz, 1990; Peter et al., 1993). The far1 mutants respond to pheromone at the transcriptional level but fail to arrest in G1. Like wild-type cells, before cell division, Fus2p-GFP was nuclear in pheromone-treated far1Δ cells. After cytokinesis, Fus2p-GFP left the nucleus in one (8 of 21 cell pairs) or both (9 of 21 cell pairs) of the progeny cells (Fig. 2 A) for a total of 26 (8 + [9 × 2]) out of 42 cells. In seven of the eight asymmetrical cell pairs, the daughter cell exported Fus2p, whereas the mother retained it in the nucleus (Fig. 2 A, middle). In the remaining cell, it was not possible to identify the mother. Importantly, cytoplasmic localization of Fus2p-GFP in the far1Δ mutants was transient. In 9 of the 26 individual cells in which exit occurred, Fus2p returned to the nucleus during the time course (Fig. 2 A, top and middle), a phenotype was never observed in wild type (Fig. 1). It is likely that the asymmetry between mothers and daughters arises because G1 mother cells accumulate Clns faster than daughter cells and reenter the cell cycle earlier (Laabs et al., 2003). These results show that prolonged cell cycle arrest by itself is not required for Fus2p to exit the nucleus but rather suggest an inverse relationship between Cln levels and Fus2p-GFP export. Increased Cln/Cdc28p activity and reestablishment of the mitotic program in G1/S would coincide with Fus2p return to the nucleus.
Figure 2.
Fus2p transiently localizes to the cytoplasm in far1 mutants. (A) Fus2p-GFP localization was examined in a far1 mutant (MY10320) as described in Fig. 1 B. In representative images, Fus2p-GFP transiently localized to the cytoplasm in both cells (top), in the bud alone (middle), or in neither (bottom). (B) In medium- to large-budded far1 cln2 mutant cells (MY10321), Fus2p-GFP exited after cytokinesis, and cells arrested in G1 (top). In small-budded cells, Fus2p-GFP was present in the cytoplasm. Similar results were obtained when Fus2p-GFP was expressed from the GAL1 promoter. Bars, 1 µm.
The far1 defect in pheromone-induced cell cycle arrest can be suppressed by mutation of CLN2 (Chang and Herskowitz, 1990). To ensure that far1 was affecting Fus2p localization via its effect on the cell cycle, we examined Fus2p-GFP localization in far1Δ cln2 double mutants. Upon pheromone treatment, the far1Δ cln2 cells arrested at G1 and did not reenter the cell cycle. In cells with a medium to large bud at the start of the time course, Fus2p-GFP was exported only after cytokinesis and remained cytoplasmic (Fig. 2 B, top; 22/23 cells), indicating that cln2 suppressed the far1 defect. These data confirm that, with respect to Fus2p localization, Far1p is required only to prevent reentry into the cell cycle by inhibiting Cln levels. Remarkably, in four cells all with small buds, Fus2p-GFP was cytoplasmic and became localized at the bud tip (Fig. 2 B, bottom). Of these, Fus2p-GFP remained at the bud tip for the duration of the experiment in two cells, returned to the nucleus in one cell, and the remaining cell died. These data suggest that CLN2 is required to keep Fus2p in the nucleus specifically during G1/S.
Fus3p is required for Fus2p exit
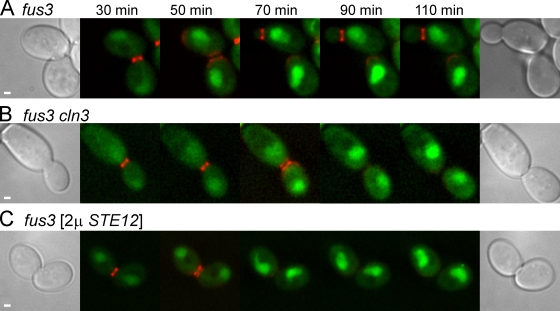
Previously, we found that MAPK Fus3p was required for Fus2p nuclear exit (Paterson et al., 2008). The failure of Fus2p to be exported in fus3 could be consequence of the inability of cells to arrest, reduced transcriptional activity, or loss of other Fus3p functions. The cell cycle arrest defect in fus3 can be suppressed by cln3 (Elion et al., 1991); we exploited this property to test the role of Fus3p in Fus2p exit.
In fus3Δ cells, Fus2p-GFP failed to exit the nucleus after cytokinesis as expected (Fig. 3 A; 33/33 cells). Fus2p-GFP also failed to exit in fus3Δ cln3 mutant cells, which remained arrested in G1 (Fig. 3 B; 13/13 cells). Therefore, the requirement for Fus3p in Fus2p's nuclear exit is independent of Fus3p's role in promoting cell cycle arrest.
Figure 3.
Fus3p is required for Fus2p nuclear exit. (A–C) Fus2p-GFP localization was examined in fus3 (MY10318; A), fus3 cln3 (MY10319; B), and fus3 [2µ STE12] (MY10322; C) as described in Fig. 1 B. Similar results were obtained when Fus2p-GFP was expressed from the GAL1 promoter. Bars, 1 µm.
The transcriptional response of fus3 mutants is partially defective, activating a fus1-LacZ reporter construct to ∼50% of wild-type levels (Elion et al., 1991) and showing an overall transcriptional profile consistent with compromised signaling (Roberts et al., 2000). Overexpression of Ste12p, the transcription factor downstream of Fus3p, increases transcription of pheromone-responsive genes. However, overexpression of Ste12p did not allow Fus2p to exit the nucleus (Fig. 3 C; 13/14 cells), indicating that the transcriptional defect in fus3 mutant cells was not responsible for the failure of Fus2p to exit the nucleus. These two lines of evidence suggest that Fus3p must regulate another pathway to promote Fus2p export.
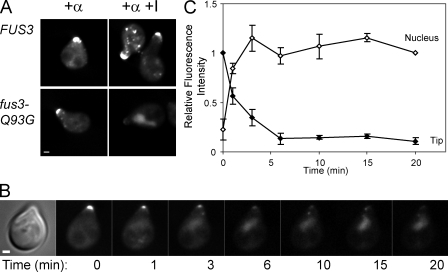
Fus2p rapidly returns to the nucleus when Fus3p is inhibited
Because Fus3p is localized in the nucleus and shmoo tip (van Drogen et al., 2001), Fus3p kinase activity could be required for Fus2p export, maintenance in the cytoplasm, or both. To determine whether Fus3p activity is required to maintain cytoplasmic Fus2p, we used an inhibitor-sensitive mutant, fus3-Q93G (Bishop et al., 2000). Cells grown in the presence of pheromone for 90 min were placed on slides with or without the inhibitor, and the location of Fus2p was examined. As shown in Fig. 4 A, in cells containing the fus3-Q93G mutation, Fus2p-GFP returned to the nucleus in response to inhibitor. Wild-type cells were unaffected, showing that the inhibitor is specific to Fus3p-Q93G. Therefore, Fus3p activity is required to maintain cytoplasmic Fus2p.
Figure 4.
Fus3p activity is required to maintain Fus2p in the cytoplasm. (A) FUS3 (MY10011) or FUS3-Q93G (MY10012) strains were pregrown in selective medium and induced with α factor for 90 min. Cells were placed on an agarose slide containing α factor with or without 1-NA-PP1, a selective inhibitor of Fus3p-Q93G (Matheos et al., 2004), and examined microscopically. (B) The fus3-Q93G strain (MY10012) was grown as in A and placed in a microscope flow cell chamber. Inhibitor was added at t = 0, and images were collected at 1-min intervals. (C) GFP fluorescence in the nucleus and at the shmoo tip was measured in 11 cells treated as in B and expressed as a fraction of t = 20 min (nucleus) or t = 0 (tip). Closed symbols, relative shmoo tip fluorescence; open symbols, relative nuclear fluorescence (see Materials and methods). Error bars represent ± SEM. Bars, 1 µm.
To examine the kinetics of the response, a similar experiment was performed using a flow cell to allow rapid image acquisition after addition of the inhibitor. Before inhibitor addition, Fus2p was concentrated at the shmoo tip as expected (Fig. 4 B). Within 1 min after inhibitor addition, Fus2p-GFP had begun to return to the nucleus. Approximately 90% of tip-localized Fus2p-GFP returned to the nucleus within 6 min with a t1/2 of ∼90 s (Fig. 4 C). The remaining ∼10% of Fus2p-GFP persisted at the shmoo tip for >20 min. These results show that constant Fus3p kinase activity is required to keep Fus2p in the cytoplasm and suggest that Fus3p might regulate Fus2p by direct phosphorylation.
Fus2p localization is controlled by Fus3p-dependent phosphorylation
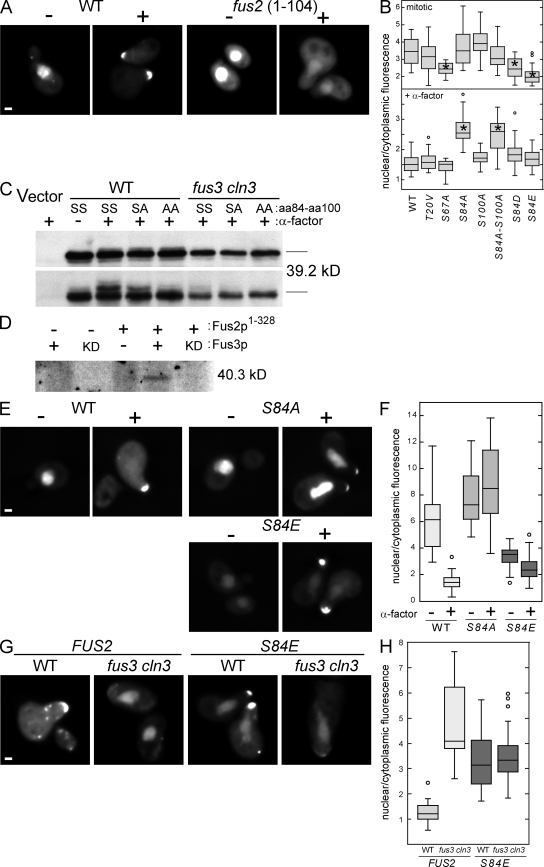
To identify the region of Fus2p responsible for pheromone-regulated localization, we generated a series of deletion mutations in Fus2p expressed under the GAL1 promoter (our unpublished data). A region encompassing the N-terminal 104 amino acids was both necessary and sufficient (Fig. 5 A) for localization of Fus2p-GFP to the nucleus in mitotic cells and will be referred to as the nuclear targeting domain (NTD). In α factor–treated cells, Fus2NTD-GFP became diffusely cytoplasmic but did not localize to the shmoo tip like full-length Fus2p (Fig. 5 A). Thus, NTD may regulate nuclear/cytoplasmic distribution but is not functional for cortical tip localization. Proteomic analysis identified four sites in NTD that are phosphorylated in pheromone-arrested cells (T20, S67, S84, and S100; Li et al., 2007). T20, S84, and S100 are followed by prolines and are good candidates for Fus3p phosphorylation sites. To identify the sites that are important for nuclear/cytoplasmic regulation, mutations in Fus2NTD-GFP were made, changing each phosphorylated serine or threonine residue to alanine or valine, respectively. The ratio of GFP fluorescence in the nucleus (N) relative to an equal area of the cytoplasm (C; N/C ratio) was measured in mitotic and pheromone-arrested cells expressing the mutant proteins (Fig. 5 B). The N/C ratio for wild-type Fus2NTD in mitotic cells was 3.6 ± 0.2 (SEM; n = 20), which was reduced to 1.55 ± 0.05 (SEM; n = 37) after pheromone treatment. Mutations T20V, S84A, and S100A had no significant effect on the N/C ratio in mitotic cells (3.1 ± 0.2, n = 26; 3.6 ± 0.2, n = 25; and 4.1 ± 0.2, n = 20, respectively). An S84 S100 double mutant also showed normal localization in mitotic cells (3.2 ± 0.2, n = 20). The mutation S67A significantly reduced localization to the nucleus under mitotic conditions (2.4 ± 0.1, n = 21), suggesting that this residue may be part of a nuclear localization sequence.
Figure 5.
Fus2p phosphorylation regulates localization. (A) The N-terminal domain of Fus2p is sufficient to localize GFP to the nucleus in mitotic cells and the cytoplasm in α factor–arrested cells. MY9181 containing full-length PGAL1-FUS2∷GFP104 (pMR5469) or the truncation mutant PGAL1-fus21-104-GFP (pMR5774) were pregrown in selective media with galactose; α factor was added 2 h before imaging. (B) Mutations at S84 affect the nuclear/cytoplasmic distribution of NTD. MY9181 containing PGAL1-fus21-104-GFP (pMR5774) with the indicated point mutations (Table II) were grown as in A. For each strain, 18 ≤ n ≤ 37. Asterisks indicate mutants significantly different from the wild-type control (P ≤ 0.0001 by one-way analysis of variance and Dunnett's multiple comparison). (C) NTD is phosphorylated in a Fus3p-dependent manner during pheromone signaling. MY9181 (wild type [WT]) or MY10273 (fus3 cln3) containing PGAL1 vector (YCpIF5), PGAL1-fus21-104-GFP (pMR5774), or pMR5774 with the indicated S84 and S100 mutations were grown in selective medium containing galactose and treated with α factor as indicated. Samples were run on standard gels (top) or gels containing 25 µM Phos-tag (bottom) to resolve phosphoisoforms. Proteins were detected by Western blot using α-GFP. SS, SA, and AA refer to the specific amino acids at residues 84 and 100. (D) Fus3p phosphorylates Fus2p in vitro. Flag-tagged Fus3p or kinase-dead (KD) Fus3pK42R was immunoprecipitated from α factor–treated yeast cultures and used to phosphorylate 6xHN-tagged Fus21–328 purified from Escherichia coli. (E) Mutations at S84 affect the nuclear/cytoplasmic distribution of full-length Fus2p-GFP. MY10174 containing PGAL1-FUS2∷GFP104 (pMR5469) or derivatives containing the indicated point mutations were grown and examined as in A. (F) Quantification of the experiment shown in E. For each strain/condition, 20 ≤ n ≤ 26. (G) S84E is insensitive to Fus3p for export. MY10174 or MY10273 containing either wild-type PGAL1-FUS2∷GFP104 (pMR5469) or the S84E mutant were grown, treated with α factor, and examined as in A. (H) Quantification of the experiment shown in G. For each strain, 22 ≤ n ≤ 25 cells. (B, F, and H) Box and whisker plots of the ratio of GFP fluorescence in the nucleus relative to an equal area of the cytoplasm are shown. Rectangles show the two inner quartiles separated by the median. Error bars indicate the entire range, and circles indicate outliers. Bars, 1 µm.
In α factor–arrested cells, the T20V, S67A, and S100A mutations had no significant effect on the cytoplasmic localization of Fus2NTD-GFP (N/C ratios of 1.6 ± 0.1, n = 18; 1.4 ± 0.1, n = 19; and 1.7 ± 0.1, n = 21, respectively). In contrast, in pheromone-treated cells, the S84A and S84A-S100A double mutant proteins remained nuclear, exhibiting high N/C ratios (2.6 ± 0.1, n = 23; and 2.5 ± 0.1, n = 23), indicating that phosphorylation of S84 is important for negatively regulating nuclear localization. In support of this, the phosphomimetic S84D and S84E alleles resulted in proteins that were significantly cytoplasmic in mitotic cells (2.5 ± 0.1, n = 21; and 2.3 ± 0.1, n = 20) and pheromone-arrested cells (1.8 ± 0.1, n = 21; and 1.7 ± 0.1, n = 22).
To determine whether NTD was phosphorylated in vivo, we expressed Fus2NTD-GFP in wild-type and fus3 cln3 mutant cells and looked for phosphoisoforms by SDS-PAGE and Western blot analysis (Fig. 5 C). Standard SDS-PAGE did not resolve different phosphorylated forms of Fus2NTD-GFP. However, electrophoresis using Phos-tag acrylamide, which reduces the mobility of phosphorylated proteins (Kinoshita-Kikuta et al., 2007), revealed two slower migrating species upon pheromone treatment (Fig. 5 C, bottom). The faint upper band was eliminated, and the major middle band was diminished in the S84A mutant. Both bands were eliminated in the S84A-S100A double mutant. Both bands were also eliminated when Fus2NTD-GFP was expressed in the fus3 cln3 mutant. Furthermore, a bacterially expressed fragment of Fus2p containing NTD was phosphorylated by Fus3p in vitro (Fig. 5 D). We conclude that S84 and S100 are phosphorylated by Fus3p and that phosphorylation at S84 controls the nuclear/cytoplasmic distribution of Fus2NTD.
To determine whether phosphorylation of S84 is relevant in the context of full-length Fus2p, we introduced S84A and S84E into Fus2p-GFP (Fig. 5, E and F). Expressed under the GAL1 promoter, in mitotic cells, full-length Fus2p-GFP was strongly nuclear localized, exhibiting a mean N/C ratio of 6.0 ± 0.4 (SEM; n = 24). Note that the N/C ratio measures only the enrichment of the nuclear signal relative to the cytoplasm and does not measure changes in the tip localization of Fus2p. The higher N/C ratio relative to NTD suggests that the full-length protein may contain additional determinants that affect localization. In pheromone-arrested cells, full-length Fus2p-GFP exhibited an N/C ratio of 1.5 ± 0.1 (SEM; n = 26), which is identical to NTD. In contrast, Fus2pS84A-GFP was significantly more nuclear localized in both mitotic- and pheromone-arrested cells (N/C ratios of 8.1 ± 0.6, n = 21; and 9.6 ± 1.1, n = 23, respectively) compared with the wild-type protein in mitotic cells (P = 0.01 and 0.001, respectively). Interestingly, the small quantity of Fus2pS84A-GFP in the cytoplasm became localized to the shmoo tip, indicating that S84 phosphorylation is not required for cortical localization. Nuclear localization of the phosphomimetic mutant Fus2pS84E-GFP was significantly reduced in mitotic cells relative to the wild-type protein, and a considerable amount was cytoplasmic (N/C = 3.4 ± 0.2, SEM, n = 21, P < 0.0001). After pheromone treatment, the relative fraction of nuclear Fus2pS84E-GFP was further reduced (N/C = 2.5 ± 0.2, SEM, n = 22), which may simply reflect localization of cytoplasmic protein to the shmoo tip. Therefore, mutations preventing phosphorylation or acting as a phosphomimetic at S84 affect nuclear localization in the context of full-length Fus2p just as they affect Fus2NTD.
We next sought to determine whether the phosphomimetic FUS2 mutant could bypass the loss of Fus3p for Fus2p nuclear exit. Although Fus2p-GFP remained largely nuclear in pheromone-treated fus3 cln3 cells, (N/C = 5.2 ± 0.5, SEM, n = 22; Fig. 5, G and H), Fus2pS84E-GFP localization to the nucleus was significantly reduced (N/C = 3.5 ± 0.2, SEM, n = 25, P = 0.0004). The N/C ratio of Fus2pS84E-GFP was not different between fus3 cln3 and wild type (3.3 ± 0.2, SEM, n = 23, P = 0.91), indicating that nuclear localization of the mutant was insensitive to Fus3p. Interestingly, tip localization of Fus2pS84E-GFP was partially defective, suggesting the existence of additional layers of Fus3p-dependent regulation.
Cdc28p inhibits Fus2p nuclear export in G1/S through down-regulation of Fus3p
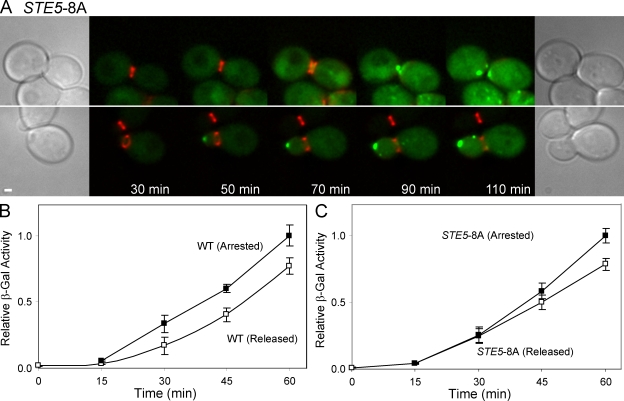
During the transition from G1 to S phase, Cln1p/Cdc28p and Cln2p/Cdc28p phosphorylate the MAPK scaffold Ste5p, modulating activation of the pheromone response pathway and Fus3p (Oehlen and Cross, 1994, 1998; Wassmann and Ammerer, 1997; Strickfaden et al., 2007). Because Fus3p activity is necessary for Fus2p export, this regulation alone may be sufficient to keep Fus2p in the nucleus during G1/S. Therefore, we examined the localization of Fus2p-GFP in STE5-8A, which abolishes Ste5p phosphorylation by Cdc28p (Strickfaden et al., 2007). In 10/23 STE5-8A cells, Fus2p-GFP accumulated in the nucleus before cell division, which is similar to wild type (Fig. 6 A, top). However, in the remaining 13 cells, Fus2p was observed outside the nucleus, usually at the bud tip, before cell division (Fig. 6 A, bottom). The cells in which Fus2p-GFP remained nuclear had larger buds at the start of the time course than those cells in which premature Fus2p-GFP exit was observed (ratio of bud diameter to mother diameter was 0.83 vs. 0.50, respectively; P = 0.005), which is consistent with STE5-8A playing a role primarily during G1/S. Interestingly, 8 of the 13 cells in which Fus2p-GFP exited prematurely also failed to reach cell division during the experiment. This phenotype was also observed by Strickfaden et al. (2007), who noted that many STE5-8A cells became arrested in G2 in response to pheromone. We conclude that the primary block to Fus2p export in G1/S is the inhibition of Fus3p activity via Ste5p phosphorylation.
Figure 6.
Cdc28p inhibits Fus2p nuclear export in G1/S through down-regulation of Fus3p. (A) Activation of Fus3p during G1/S allows Fus2p to exit before cell division. Fus2p-GFP localization was examined in STE5-8A as described in Fig. 1 B. Fus2p-GFP exited after cytokinesis in large-budded cells (top). Fus2p-GFP expressed from its own promoter was cytoplasmic and localized to the cortex in small-budded cells. Similar results were obtained when Fus2p-GFP was expressed from the GAL1 promoter. (B and C) Pheromone signaling in cells arrested at G2/M. cdc28-as1 (MY10451; B) or cdc28-as1 STE5-8A cells (MY10481; C) were arrested at G2/M by adding the Cdc28p-as1 inhibitor 1-NM-PP1 for 3 h. 85–92% of cells were budded under these conditions. After washing out inhibitor, the culture was split and treated with α factor alone (open symbols) or with Cdc28p-as1 inhibitor and α factor (closed symbols). Induction of pheromone-responsive genes was measured using fus1-LacZ expression (see Materials and methods). Activity is expressed relative to the 60-min time point of the arrested population. Error bars indicate SEM of three independent experiments. WT, wild type; β-Gal, β-galactosidase. Bar, 1 µm.
Given that the Clns can keep Fus2p in the nucleus by modulating Fus3p activity via Ste5p phosphorylation during G1/S, what is responsible for keeping Fus2p nuclear during other stages of the cell cycle? One possibility is that G2/M cells with active B-type Clns might also down modulate pheromone signaling. Previous studies (Oehlen and Cross, 1994, 1998; Wassmann and Ammerer, 1997; Strickfaden et al., 2007) have only observed repression during G1/S. However, these studies used short pheromone treatments in synchronized cells; sustained exposure to pheromone during G2/M might reveal significant cell cycle–dependent down modulation of pheromone signaling. Accordingly, inhibitor-sensitive cdc28-as1 cells were treated with 1-NM-PP1 3 h, after which >85% were arrested at the G2/M boundary. The culture was then split; one part was released from inhibitor and treated with α factor, whereas the other part was treated with both α factor and inhibitor to continue the arrest. If Cdc28p activity inhibited pheromone signaling during G2/M, we expect the culture with the inhibitor to show a substantially higher level of pheromone signaling. Allowing cells to reenter mitosis produced only a minor reduction in the rate of fus1-LacZ induction (Fig. 6 B). The difference was largely gone in the STE5-8A mutant (Fig. 6 C), suggesting that a low level of residual Cln/Cdc28p activity accounts for the delay. Regardless, these results confirm that pheromone signaling leads to robust induction during G2/M and that it is unlikely that the inhibition of Fus2p nuclear export in G2/M is regulated by global down modulation of pheromone signaling.
Cdc28p inhibits Fus2p nuclear exit in G2/M
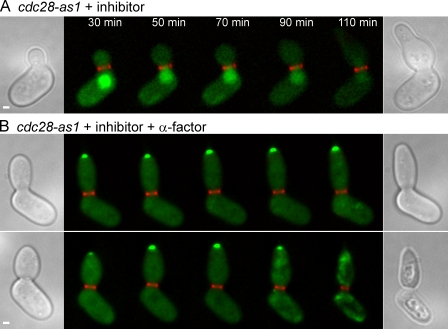
Given the lack of significant Cdc28p-dependent down modulation of Fus3p signaling in G2/M, we next asked whether Cdc28p plays any role in keeping Fus2p in the nucleus during this phase of the cell cycle. Fus2p-GFP was expressed under the GAL1 promoter in the cdc28-as1 background to compare pheromone-treated and untreated cells. When the cdc28-as1 mutant was treated with inhibitor alone, Fus2p-GFP remained in the nucleus, showing the requirement for pheromone signaling (Fig. 7 A; 29/29 cells). When cdc28-as1 cells were treated with inhibitor and α factor simultaneously, Fus2p-GFP exited the nucleus and accumulated at the tip of the bud (Fig. 7 B). Unlike STE5-8A, this effect was almost fully penetrant (34/36 cells), affecting cells at all stages of the cell cycle. Strikingly, 11/36 cells died during image acquisition (Fig. 7 B, bottom). In addition, in parallel experiments, cell viability was reduced by 43% (±4% SEM; n = 3) within 1 h after treatment. The addition of inhibitor or α factor alone did not lead to any loss of cell viability. These data suggest that premature loss of Cdc28p activity during α factor treatment induces a physiological state that is detrimental to cell survival. We conclude that active Cdc28p is required to keep Fus2p in the nucleus. Furthermore, the lack of a significant effect of Cdc28p activity on pheromone signaling during G2/M argues strongly that Cdc28p regulates Fus2p localization by a different mechanism during this part of the cell cycle.
Figure 7.
Cdc28p inhibits Fus2p-GFP nuclear exit in G2/M. Localization of GAL1-expressed Fus2p-GFP was examined as described in Fig. 1 D in cdc28-as1 (MY10324). (A) Cdc28p-as1 inhibitor was added without α factor. (B) Cdc28p-as1 inhibitor and α factor were added together. The images show cytoplasmic and cortical localization in large-budded cells before cytokinesis. The bottom panel shows a cell dying during the course of the experiment. Similar results were obtained when Fus2p-GFP was expressed under its own promoter. Bars, 1 µm.
Discussion
Fus2p is a key regulator of cell fusion, which localizes to the shmoo tip in pheromone-arrested cells. A previous study demonstrated that the localization of Fus2p-GFP is surprisingly dynamic (Paterson et al., 2008) and was altered in mutants affecting mating and cell fusion. Fus2p-GFP was found to be nuclear in fus3 mutants or when expressed under the GAL1 promoter in mitotic cells, i.e., when the pheromone pathway is compromised or not active. In this study, we dissect the conditions necessary for nuclear versus cytoplasmic localization and show that both the pheromone pathway and the cell cycle play a role in regulating Fus2p localization. Pheromone signaling dependent on Fus3p plays a positive regulatory role in Fus2p exit, whereas cell cycle signaling via Cdc28p plays a negative role.
When asynchronous cultures are exposed to pheromone, most cells will be post-START and must complete the current round of cell division before cell cycle arrest. Immediately after START, Clns associated with Cdc28p block pheromone signaling and downstream cellular responses (Strickfaden et al., 2007). However, as the cells enter G2, the pheromone signaling pathway becomes active, and the transcription of pheromone-regulated genes is allowed to proceed. Along with other pheromone-induced genes, Fus2p-GFP is expressed in G2 but localized to the nucleus until after mitosis. Because Fus2p is required for cell fusion and localizes to the site of cell fusion during conjugation, it is likely that the nuclear form is inactive with Fus2p sequestered away from its site of action. Although the specific function of Fus2p is not yet known, its homology with Rho–guanine exchange factor proteins suggests that it regulates a Rho-type G protein at the cell cortex. Because all of the Rho-type G proteins are constitutively expressed and play important roles during mitosis, premature activation by a pheromone-dependent regulatory protein would likely interfere with normal cell cycle progression. Thus, cells are faced with the problem of how to stockpile adequate levels of proteins required for mating while limiting their activity until after they have completed mitosis and are ready to mate.
We envision that four simple ways allow for delayed export and activation of Fus2p. First, Fus2p expression might be a delayed response to pheromone, perhaps by making expression dependent on an intermediate transcription factor like Kar4p (Lahav et al., 2007). Second, a protein required for Fus2p exit may be expressed in a delayed manner. Third, Fus2p exit might depend on its reaching a critical concentration that triggers export. Fourth, exit may be coupled to cell cycle–dependent regulation. Because the timing of Fus2p exit was correlated with the time of cell division in an asynchronous population and not with the length of time in pheromone, the first two scenarios are unlikely. Separating expression of Fus2p from pheromone signaling response using the GAL1 promoter also had no effect. Thus, export was not a consequence of increased Fus2p levels, making the third scenario unlikely. In contrast, the observed coupling of exit to posttranslational cell cycle regulation provides an elegant solution to the problem of transition between mitotic growth and cell cycle arrest. Proteins required for mating can be synthesized before they are needed and sequestered until their activation will not compromise cell cycle progression.
We do not know how general the mechanism of cell cycle–dependent nuclear localization is to regulate mating-specific proteins. Several proteins are known that show cell cycle–regulated (e.g., Swi5p and Cdc24p; Moll et al., 1991; Nern and Arkowitz, 2000) or signaling-regulated nuclear localization (e.g., Msn2p and Msn4p; Jacquet et al., 2003), and several proteins required for pheromone signaling shuttle between the nucleus and cytoplasm (e.g., Fus3p and Ste5p; Choi et al., 1999; Mahanty et al., 1999; van Drogen et al., 2001). To our knowledge, Fus2p is unique in being subject to two different pathways of regulation. Two other proteins with different roles in mitosis and mating, Kar3p and Far1p, also show mating-regulated localization patterns being largely nuclear in mitotic cells but cytoplasmic in mating cells (Meluh and Rose, 1990; Nern and Arkowitz, 1999). It is not yet known whether the transition between these patterns is restricted during the cell cycle.
Cell cycle requirement for exit
Prolonged arrest in the cell cycle by itself is not required for Fus2p exit. Instead, it appears that Fus2p exit is negatively correlated with Cln levels in G1. First, far1 mutants do not arrest in the presence of pheromone, yet many far1 cells showed transient Fus2p exit as they progressed from cytokinesis into G1. Fus2p usually returned to the nucleus as the cells progressed into the cell cycle when Cln levels would be rising. Second, we observed frequent mother–daughter asymmetry in far1 mutants, with daughters showing exit more often than mothers. This is most likely because mothers have higher Cln levels immediately after cell division and progress into the cell cycle before daughter cells (Laabs et al., 2003). Third, introduction of the cln2 deletion, which removes a G1 Cln, allowed cell cycle arrest and Fus2p exit in all far1 mutant cells. Moreover, in the far1 cln2 mutant, we observed Fus2p at the bud tip only in small-budded cells when the G1 Cln would be expected to be active. Fourth, inhibition of Cdc28p allowed Fus2p export in all stages of the cell cycle but only in the presence of pheromone. Thus, high Cdc28p activity is correlated with Fus2p nuclear localization, and low Cdc28p activity is permissive for Fus2p nuclear exit.
Pheromone requirement for exit
Previous results showed that Fus2p does not exit the nucleus in fus3 mutants (Paterson et al., 2008). This is not simply a result of the inability of fus3 mutants to cell cycle arrest because the cln3 mutation suppressed arrest but not Fus2p exit. Moreover, it is not likely that Fus3p is only required for the transcriptional induction of a protein required for Fus2p exit. First, the fus3 mutant was not suppressed by high copy STE12. Second, Fus2p that had already localized to the shmoo tip returned very rapidly to the nucleus when Fus3p kinase activity was chemically inhibited. It is much more likely that Fus2p is regulated directly by Fus3p. First, the Fus2NTD fragment is phosphorylated at S84 in vivo in response to pheromone. Second, the phosphorylation is dependent on Fus3p, and Fus3p can phosphorylate Fus2p in vitro. Third, mutation of Ser84 to alanine prevented export of both Fus2NTD and full-length Fus2p. Conversely, mutation of Ser84 to glutamate or aspartate (negatively charged residues that mimic a phosphoryl group) allowed constitutive export of Fus2p-GFP in mitotic cells. Collectively, these results are most consistent with a direct requirement for Fus3p activity for Fus2p exit.
Because general inhibition of the mating signaling pathway is known to occur in late G1 through S phase, down-regulation of Fus3p would account for inhibition of Fus2p exit during this part of the cell cycle. Consistent with this, Fus2p was frequently found at the bud tip of small-budded STE5-8A cells, which are defective in the G1/S inhibition of Fus3p. Inhibition of Fus3p most likely accounts for the return of Fus2p to the nucleus observed in far1 mutant cells; as Cln levels accumulate at the end of G1, the mating pathway would be shut off and Fus3p made inactive.
Interestingly, pheromone regulation plays a separate role in the recruitment of cytoplasmic Fus2p to the plasma membrane because Fus2pS84E is diffusely cytoplasmic in mitotic cells, whereas it is tip localized in shmoos. Moreover, cytoplasmic Fus2pS84E also fails to localize to the cortex in the fus3 cln3 mutant treated with pheromone, suggesting that the defect is not simply a result of the absence of pheromone-induced proteins.
Mechanism of cell cycle regulation in late G2
Inhibition of Fus3p activation via Ste5p is limited to G1/S. Even upon extended pheromone treatment, active Cdc28p has, at best, a very minor effect on pheromone-dependent transcription during G2, arguing that Fus3p is fully active during G2. Accordingly, we favor a model in which the pheromone pathway and Cdc28p converge to regulate Fus2p localization.
How could such regulation work mechanistically? We envisage two general classes of models that differ in whether Cdc28p acts directly or indirectly. As an example of direct regulation, Cdc28p might phosphorylate Fus2p, causing nuclear import (or blocking export) independent of Fus3p phosphorylation. Alternatively, Cdc28p phosphorylation of Fus2p might block phosphorylation by Fus3p. If Fus2p is phosphorylated by Cdc28p, it is unlikely to occur within the NTD based on the mobility of the mitotically expressed protein in Phos-tag polyacrylamide gels. Arguing against the first model is the fact that the S84E mutant has constitutively greater cytoplasmic distribution than the wild-type protein, making it unlikely that Cdc28p phosphorylation can override S84 phosphorylation.
Examples of indirect regulation include the possibility that Cdc28p-dependent regulation controls the specificity of nuclear import or export receptors, thereby controlling Fus2p localization independent of Fus3p regulation. Alternatively, Cdc28p might regulate Fus3p substrate specificity in such a way that targets required for the transcriptional response to pheromone (Dig1p and Dig2p) can be phosphorylated, but Fus2p and possibly other targets required for later events in mating are not.
Ultimately, phosphorylation at S84 impacts the nuclear/cytoplasmic distribution of the protein. Phosphorylation is known to regulate the nuclear import of many proteins. For example, in Msn2p, phosphorylation by cAMP-dependent protein kinase interferes with NLS function (Gorner et al., 2002). The S84 residue in Fus2p does not lie in a predicted NLS; indeed, there is no predicted NLS within the Fus2NTD fragment that is both necessary and sufficient for nuclear localization. Elucidation of the mechanism of Fus2p localization will require identification of the true NLS and nuclear export signal and determining the impact of S84 phosphorylation and possibly phosphorylation at other sites.
We have described a novel mechanism regulating mating-dependent processes in yeast. Before pheromone-treated cells complete mitosis, FUS2 becomes active at the transcriptional level, but Fus2p is negatively regulated at the level of localization. As cells enter G1, two antagonistic signaling pathways converge to regulate the export Fus2p to the cytoplasm: positive regulation by the pheromone response pathway and relief of negative regulation by the cell cycle. We suggest that similar patterns of regulation impact other genes involved in the mating pathway as well as other differentiated states of the cell. Elucidation of the mechanism of Fus2p control will help us further understand the intricacies of this highly regulated biological process.
Materials and methods
Strains and general yeast methods
Yeast media, general methods, and transformations were performed as described previously (Adams et al., 1997). Yeast strains are listed in Table I. MY10011 and MY10012 were derived from MY9211 (provided by J. Paterson, Princeton University, Princeton, NJ). CDC3 was tagged with mCherry using the method described by Longtine et al. (1998) using pMR5598 as a template. FUS3 and FAR1 deletion constructs were created by amplifying natMX6 with primers containing homology to regions just outside the coding sequence of each gene. CLN2 and CLN3 disruption constructs were derived by amplifying these loci from strains already containing the disruptions (Epstein and Cross, 1992). The cln3∷URA3 disruption was replaced with cln3∷ura3∷LEU2 using the method of Cross (1997). In all cases, deletions were confirmed by PCR.
Table I.
Strains used in this study
| Strain | Genotype | Source |
| BY4741a | MATa ura3Δ0 his3Δ200 leu2Δ0 met15Δ0 | Brachmann et al., 1998 |
| MY9181 | fus2Δ∷HIS3 | Paterson et al., 2008 |
| MY10011 | fus2Δ∷HIS3 fus3Δ∷kanMX pMR5642 [FUS2∷GFP104 LEU2 CEN] pMR4937 [FUS3 CEN URA3] | This study |
| MY10012 | fus2Δ∷HIS3 fus3Δ∷kanMX pMR5642 [FUS2∷GFP104 LEU2 CEN] pMR4938 [fus3-Q93G CEN URA3] | This study |
| MY10174 | fus2Δ∷HIS3 CDC3-mCherry∷kanMX | This study |
| MY10176 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX pMR5482 [FUS2∷GFP104 URA3 CEN] | This study |
| MY10177 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX pMR5469 [PGAL1-FUS2∷GFP104 URA3 CEN] | This study |
| MY10273 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX fus3Δ∷natMX cln3∷ura3∷LEU2 | This study |
| MY10318 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX fus3Δ∷natMX pMR5482 | This study |
| MY10319 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX fus3Δ∷natMX cln3∷ura3∷LEU2 pMR5482 | This study |
| MY10320 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX far1Δ∷natMX pMR5482 | This study |
| MY10321 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX far1Δ∷natMX cln2∷LEU2 pMR5482 | This study |
| MY10322 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX fus3Δ∷natMX pMR5482 pSY1 [2µ LEU2 STE12] | This study |
| MY10324 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX cdc28-as1 pMR5469 | This study |
| MY10329 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX STE5-8A pMR5482 | This study |
| MY10451 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX cdc28-as1 pSB231 [CEN URA3 fus1-LacZ] | This study |
| MY10481 | fus2Δ∷HIS3 CDC3- mCherry∷kanMX cdc28-as1 STE5-8A pSB231 | This study |
| EY700 | MATa fus3-6Δ∷LEU2 ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 can1-100 | G. Finkb |
| YM1933 | Σ2000 ura3Δ0 leu2Δ0 trp1Δ∷hisG his2ΔhisG fus3K42R-3×FLAG∷kanMX | Bao et al., 2004 |
All strains created for this study were derived from BY4741.
Massachusetts Institute of Technology, Cambridge, MA.
The STE5-8A mutation was integrated by the loop-in/loop-out procedure using XbaI-cut pPP2330 (Strickfaden et al., 2007). Loop outs were screened via a SmaI digest of a PCR product spanning the mutated region and confirmed by sequencing this same PCR product. The cdc28-as1 mutation was integrated by the loop-in/loop-out procedure using AflII-cut pJAU01 (Bishop et al., 2000). Loop outs were confirmed by cell cycle arrest in the presence of 500 nM 1-NM-PP1.
Plasmids used in this study are listed in Table II. To create pMR5642, FUS2∷GFP104 was cut out of pMR5482 with XhoI and SpeI and cloned into the XhoI-SpeI sites of pRS415 (Sikorski and Hieter, 1989). To create pMR5774, a stop codon was introduced into pMR5469 at the 3′ end of GFP by dut ung mutagenesis (Kunkel, 1985). Point mutations were created in pMR5469 and pMR5774 by dut ung mutagenesis. To create pMR5630, a fragment of FUS2 comprising codons 1–328 was amplified using primers containing XhoI and PstI sites. This fragment was cloned into the SalI and PstI sites of pPROTet.E133 (Clontech Laboratories, Inc.).
Table II.
Plasmids used in this study
| Plasmid | Markers | Source |
| pRS413 | CEN HIS3 | Sikorski and Hieter, 1989 |
| pSB231 | CEN URA3 fus1-LacZ | Trueheart et al., 1987 |
| pJAU01 | URA3 cdc28-as 1 | Bishop et al., 2000 |
| pSY1 | 2µ LEU2 STE12 | Dolan et al., 1989 |
| pPP2330 | URA3 STE5-8A | Strickfaden et al., 2007 |
| YCpIF5 | CEN URA3 PGAL1 | Foreman and Davis, 1994 |
| pPROTet.E133 | Ptet-6xHN Cmr | Clontech Laboratories, Inc. |
| pMR4937 | CEN URA3 FLAG-FUS3 | Matheos et al., 2004 |
| pMR4938 | CEN URA3 FLAG-fus3Q93G | Matheos et al., 2004 |
| pMR5048 | CEN HIS3 FLAG-FUS3 | D. Matheosc |
| pMR5469a | CEN URA3 PGAL1-FUS2∷GFP104 | Paterson et al., 2008 |
| pMR5482 | CEN URA3 FUS2∷GFP104 | Paterson et al., 2008 |
| pMR5598 | pFa6a-mCherry-kanMX6 | S. Clarkc |
| pMR5630 | Ptet-6xHN-fus21–328 Cmr | This study |
| pMR5642 | CEN LEU2 FUS2∷GFP104 | This study |
| pMR5774b | CEN URA3 PGAL1-fus21–104-GFP | This study |
Point mutations in FUS2∷GFP used in Fig. 5 D were created for this study and are derived from pMR5469.
Point mutations in fus21–104-GFP used in Fig. 5 (B and C) were created for this study and are derived from pMR5774.
Princeton University, Princeton, NJ.
Live cell imaging
For time-course experiments, cells were grown to early log phase in selective medium at 30°C. At t = 0, 6 µM α factor (Princeton University Molecular Biology Syn/Seq facility), 2% glucose, and 500 nM of the cdc28-as1 inhibitor 1-NM-PP1 was added (provided by K. Shokat, University of California, San Francisco, San Francisco, CA) as indicated. In most experiments, the first image was acquired at 30 min, and subsequent images were acquired every 20 min. Differential interference contrast images were taken before and after the first and last fluorescent image. For experiments in which FUS2∷GFP104 was expressed under the native FUS2 promoter, the culture was shifted to 23°C at t = 0 to allow GFP to fold. After 15 min, 5 µl of cells was placed on a 2% agarose pad containing the same media, which was mounted on a microscope slide.
For experiments using the inhibitor-sensitive fus3-Q93G allele, cells were grown to early log phase in selective medium at 30°C and treated with 6 µM α factor for 90 min. For still images, 5 µl of cells was placed on agarose pads with or without the inhibitor (1-NA-PP1; final concentration 100 µM; provided by K. Shokat). Experiments showing addition of the inhibitor during acquisition were conducted in a similar manner except using a homemade flow cell. Cells in selective medium containing α factor were immobilized on coverslips using concanavalin A. The coverslip was affixed to Scotch tape walls overlaid on a glass slide floor to create an ∼0.1-mm-thick chamber. To remove unbound cells, the flow cell was first flushed three times with selective medium containing α factor by pipetting from one end and inserting the corner of a tissue paper at the other. After the first images were acquired, this medium was exchanged for medium containing α factor and 1-NA-PP1 by the same procedure.
Images were acquired using a microscopy system (DeltaVision; Applied Precision, LLC) using an inverted microscope (TE200; Nikon), a charge-coupled device camera (CoolSNAP HQ; Roper Scientific), and a 100× objective with a 1.4 NA. Time course images (Fus2p-GFP and Cdc3-mCherryFP) are composed of either six z sections spaced 0.8 µm apart with 2 × 2 binning or 10 z sections spaced 0.45 µm apart without binning. Rhodamine and FITC filter sets were used to visualize Cdc3p-mCherryFP and Fus2p-GFP, respectively. All other images are composed of 24 z sections spaced 0.2 µm apart using the FITC filter set. All images were deconvolved using the softWoRx program (Applied Precision, LLC), and projections were made using a summing algorithm.
Figures were prepared for publication using Photoshop and Illustrator (Adobe). For presentation purposes, pixel density was increased using bicubic resampling where necessary. The brightness of the red channel showing Cdc3p-mCherryFP was increased in some panels to allow the separation of the septin rings to be seen clearly.
Time courses on each strain were collected during at least two different imaging sessions. In some cases, data collected using Fus2p-GFP expressed from both the FUS2 promoter and the GAL1 promoter were pooled. Except where indicated, time courses were counted only if a cytokinesis event occurred during imaging.
Image analysis
Image quantification was performed using softWoRx and ImageJ (National Institutes of Health). For experiments using the inhibitor-sensitive fus3-Q93G allele, fluorescence intensity was measured at three regions: overlapping the shmoo tip, inside the nucleus, and in the cytoplasm. The background intensity was subtracted from the tip intensity, and this adjusted tip intensity was normalized for each cell, taking the value at the 0-min time point to be 1. Normalized intensities were averaged between all cells in the dataset for each time point. A similar method was used to calculate nuclear intensities except that normalization was done such that the value at the final time point was 1.
For experiments comparing the nuclear/cytoplasmic distribution of the NTD point mutants, total fluorescence intensity was measured on image projections in two equal areas inside the nucleus and inside the cytoplasm. Background autofluorescence was measured in a separate population of cells not expressing GFP that were imaged on the same day. After subtracting the mean autofluorescence value from each measurement, the ratio of cytoplasmic to nuclear fluorescence was calculated for each cell. Note that because the projected image contains fluorescence from the entire volume, the N/C ratio underestimates the concentration of GFP in the nucleus. Statistical analysis was performed using one-way analysis of variance followed by Dunnett's multiple comparison test or the Wilcoxon-Mann-Whitney rank sum test for non-normally distributed data.
To quantify total Fus2p-GFP levels in G2 cells, we measured total fluorescence intensity in a box encompassing the whole cell and subtracted the background intensity of a congruent box in an empty field. This protocol was used to measure the mean autofluorescence in MY10174, which does not contain GFP. We measured MY10176 treated with pheromone as in the time-course experiments for 30 min or left untreated. Only budded cells with fully formed septin rings were selected for analysis. The mean autofluorescence intensity was subtracted from each cell, and the populations were compared with Student's t test.
Western blotting
Proteins were prepared for SDS-PAGE by TCA precipitation, resolved on a 10% polyacrylamide gel with or without 25 µM Phos-tag acrylamide (FMS Laboratory). Proteins were transferred to polyvinylidene difluoride and detected using monoclonal mouse anti-GFP (Roche) at 1:1,000 followed by incubation with a secondary antibody and standard chemiluminescence.
In vitro kinase assay
In vitro kinase assays were performed essentially as described by Bao et al. (2004) using bacterially expressed Fus2p fragment. BL21PRO cells containing pMR5630 expressing 6xHN-tagged Fus21–328 or a vector control were grown in 100 ml LB medium, cooled to room temperature, induced with 100 ng/ml anhydrotetracycline for 4 h, collected by centrifugation, and flash frozen. Cells were lysed in 1 ml TPB (50 mM Tris-HCl, pH 8.0, 1 mM MgCl2, 10 mM β-mercaptoethanol, and protease inhibitor cocktail [Roche]) by incubation with 1 mg/ml lysozyme followed by sonication. The lysate was clarified by centrifugation at 11,000 rpm for 20 min in a tabletop centrifuge. Proteins were bound to 375 µl nickel-nitrilotriacetic acid Superflow beads (QIAGEN) and prewashed in TPB in batch format for 60–90 min. The beads were washed in column format with 3 ml TPB containing 250 mM NaCl and 10 mM imidazole. The column was eluted using a step gradient of imidazole in TPB, and fractions containing Fus21–328 were identified by SDS-PAGE followed by Coomassie blue staining.
Flag-tagged Fus3p was immunoprecipitated from yeast extracts made from 200-ml cultures of EY700 containing pRS413 (vector control), pMR5048 (Fus3p), or YM1933 (Fus3pK42R) induced with 6 µM α factor for 90 min. Cells were collected by centrifugation and flash frozen. Cells were thawed in 1 ml of TENNI buffer (50 mM Tris-HCl, pH 7.4, 250 mM NaCl, 50 mM NaF, 5 mM EDTA, 0.1% NP-40, 1 mM PMSF, protease inhibitor cocktail, and phosphatase inhibitor cocktail [Roche]) and lysed using glass beads. The lysate was centrifuged for 10 min at 3,000 rpm, and the supernatant was transferred to a new tube and centrifuged again for 15 min at 11,000 rpm. 30 µl anti-Flag affinity gel (Sigma-Aldrich) prewashed in TENNI buffer was added to the extract, and proteins were adsorbed for 60 min at 4°C. The beads were washed four times in TENNI buffer, two times in kinase buffer (50 mM Tris, pH 7.4, 20 mM MgCl2, 1 mM DTT, protease inhibitor cocktail, and phosphatase inhibitor cocktail), and resuspended in 40 µl of kinase buffer. 20 µl slurry was combined with ∼200 ng Fus21–328 eluate in a total volume of 60 µl. After 5 min, 5 µl of reaction buffer (2 µCi/µl γ-[32P]ATP and 10 µM ATP in kinase buffer) was added. Reactions were continued for 15 min at 30°C and stopped by addition of electrophoresis buffer and boiling. Proteins were resolved on a 12% polyacrylamide gel and processed for autoradiography.
Cell cycle arrest and β-galactosidase assays
cdc28-as1 cells were arrested at the G2/M transition by treating with 500 nM 1-NM-PP1 for 3 h (Bishop et al., 2000). The inhibitor was washed out by vacuum filtration. The cells were washed with 3 vol of fresh medium and resuspended in 1 vol of fresh medium. The culture was split and treated with either DMSO + 10 µg/ml α factor or 500 nM 1-NM-PP1 (in DMSO) + 10 µg/ml α factor. At 15-min intervals, aliquots were taken for analysis of β-galactosidase activity. Activity was measured by the permeabilized cell method of Adams et al. (1997) with minor modifications and is expressed relative to the 60-min time point of the arrested population in each experiment.
Acknowledgments
We thank Peter Pryciak, David Morgan, and Kevan Shokat for providing reagents and materials, Sean Clark for creation of the mCherryFP integration cassette, and Peter Houston for help with the flow cell experiments. We thank Rebecca Davis for excellent help with media and materials.
This work was made possible by the National Institutes of Health (grant GM37739 to M.D. Rose).
Footnotes
Abbreviations used in this paper: Cln, cyclin; FP, fluorescent protein; NTD, nuclear targeting domain.
References
- Adams A., Gottschling D.E., Kaiser C.A., Stearns T. 1997. Methods in Yeast Genetics : a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 177 pp [Google Scholar]
- Bao M.Z., Schwartz M.A., Cantin G.T., Yates J.R., III, Madhani H.D. 2004. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast.Cell. 119:991–1000 [DOI] [PubMed] [Google Scholar]
- Bardwell L. 2005. A walk-through of the yeast mating pheromone response pathway.Peptides. 26:339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A.C., Ubersax J.A., Petsch D.T., Matheos D.P., Gray N.S., Blethrow J., Shimizu E., Tsien J.Z., Schultz P.G., Rose M.D., et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase.Nature. 407:395–401 [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications.Yeast. 14:115–132 [DOI] [PubMed] [Google Scholar]
- Brizzio V., Gammie A.E., Rose M.D. 1998. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae.J. Cell Biol. 141:567–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Herskowitz I. 1990. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2.Cell. 63:999–1011 [DOI] [PubMed] [Google Scholar]
- Choi K.Y., Kranz J.E., Mahanty S.K., Park K.S., Elion E.A. 1999. Characterization of Fus3 localization: active Fus3 localizes in complexes of varying size and specific activity.Mol. Biol. Cell. 10:1553–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F.R. 1997. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics.Yeast. 13:647–653 [DOI] [PubMed] [Google Scholar]
- Dolan J.W., Kirkman C., Fields S. 1989. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction.Proc. Natl. Acad. Sci. USA. 86:5703–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E.A. 2000. Pheromone response, mating and cell biology.Curr. Opin. Microbiol. 3:573–581 [DOI] [PubMed] [Google Scholar]
- Elion E.A., Grisafi P.L., Fink G.R. 1990. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation.Cell. 60:649–664 [DOI] [PubMed] [Google Scholar]
- Elion E.A., Brill J.A., Fink G.R. 1991. FUS3 represses CLN1 and CLN2 and in concert with KSS1 promotes signal transduction.Proc. Natl. Acad. Sci. USA. 88:9392–9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E.A., Trueheart J., Fink G.R. 1995. Fus2 localizes near the site of cell fusion and is required for both cell fusion and nuclear alignment during zygote formation.J. Cell Biol. 130:1283–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein C.B., Cross F.R. 1992. CLB5: a novel B cyclin from budding yeast with a role in S phase.Genes Dev. 6:1695–1706 [DOI] [PubMed] [Google Scholar]
- Foreman P.K., Davis R.W. 1994. Cloning vectors for the synthesis of epitope-tagged, truncated and chimeric proteins in Saccharomyces cerevisiae.Gene. 144:63–68 [DOI] [PubMed] [Google Scholar]
- Gorner W., Durchschlag E., Wolf J., Brown E.L., Ammerer G., Ruis H., Schuller C. 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor.EMBO J. 21:135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin M.C., Albertyn J., Alexander M., Davenport K. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae.Microbiol. Mol. Biol. Rev. 62:1264–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet M., Renault G., Lallet S., De Mey J., Goldbeter A. 2003. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae.J. Cell Biol. 161:497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita-Kikuta E., Aoki Y., Kinoshita E., Koike T. 2007. Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis.Mol. Cell. Proteomics. 6:356–366 [DOI] [PubMed] [Google Scholar]
- Kunkel T.A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection.Proc. Natl. Acad. Sci. USA. 82:488–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusari A.B., Molina D.M., Sabbagh W., Jr, Lau C.S., Bardwell L. 2004. A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1.J. Cell Biol. 164:267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laabs T.L., Markwardt D.D., Slattery M.G., Newcomb L.L., Stillman D.J., Heideman W. 2003. ACE2 is required for daughter cell-specific G1 delay in Saccharomyces cerevisiae.Proc. Natl. Acad. Sci. USA. 100:10275–10280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav R., Gammie A., Tavazoie S., Rose M.D. 2007. Role of transcription factor Kar4 in regulating downstream events in the Saccharomyces cerevisiae pheromone response pathway.Mol. Cell. Biol. 27:818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Gerber S.A., Rudner A.D., Beausoleil S.A., Haas W., Villen J., Elias J.E., Gygi S.P. 2007. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae.J. Proteome Res. 6:1190–1197 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., Bi E. 2003. Regulation of septin organization and function in yeast.Trends Cell Biol. 13:403–409 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae.Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Madhani H.D., Fink G.R. 1997. Combinatorial control required for the specificity of yeast MAPK signaling.Science. 275:1314–1317 [DOI] [PubMed] [Google Scholar]
- Mahanty S.K., Wang Y., Farley F.W., Elion E.A. 1999. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade.Cell. 98:501–512 [DOI] [PubMed] [Google Scholar]
- Marsh L., Rose M.D. 1997. The pathway of cell and nuclear fusion during mating in S. cerevisiae. In The Molecular and Cellular Biology of the Yeast Saccharomyces. Vol. 3 Broach J.R., Pringle J.R., Jones E.W.editors Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 827–888 [Google Scholar]
- Matheos D., Metodiev M., Muller E., Stone D., Rose M.D. 2004. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p.J. Cell Biol. 165:99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Rose M.D. 1990. KAR3, a kinesin-related gene required for yeast nuclear fusion.Cell. 60:1029–1041 [DOI] [PubMed] [Google Scholar]
- Metodiev M.V., Matheos D., Rose M.D., Stone D.E. 2002. Regulation of MAPK function by direct interaction with the mating-specific Galpha in yeast.Science. 296:1483–1486 [DOI] [PubMed] [Google Scholar]
- Moll T., Tebb G., Surana U., Robitsch H., Nasmyth K. 1991. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5.Cell. 66:743–758 [DOI] [PubMed] [Google Scholar]
- Munn A.L., Stevenson B.J., Geli M.I., Riezman H. 1995. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae.Mol. Biol. Cell. 6:1721–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naider F., Becker J.M. 2004. The alpha-factor mating pheromone of Saccharomyces cerevisiae: a model for studying the interaction of peptide hormones and G protein-coupled receptors.Peptides. 25:1441–1463 [DOI] [PubMed] [Google Scholar]
- Nern A., Arkowitz R.A. 1999. A Cdc24p-Far1p-Gβγ protein complex required for yeast orientation during mating.J. Cell Biol. 144:1187–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., Arkowitz R.A. 2000. Nucleocytoplasmic shuttling of the Cdc42p exchange factor Cdc24p.J. Cell Biol. 148:1115–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlen L.J., Cross F.R. 1994. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle.Genes Dev. 8:1058–1070 [DOI] [PubMed] [Google Scholar]
- Oehlen L.J., Cross F.R. 1998. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases.J. Biol. Chem. 273:25089–25097 [DOI] [PubMed] [Google Scholar]
- Page B.D., Satterwhite L.L., Rose M.D., Snyder M. 1994. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein.J. Cell Biol. 124:507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.O., Bi E. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond.Microbiol. Mol. Biol. Rev. 71:48–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson J.M., Ydenberg C.A., Rose M.D. 2008. Dynamic localization of yeast Fus2p to an expanding ring at the cell fusion junction during mating.J. Cell Biol. 181:697–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M., Gartner A., Horecka J., Ammerer G., Herskowitz I. 1993. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast.Cell. 73:747–760 [DOI] [PubMed] [Google Scholar]
- Pryciak P.M., Huntress F.A. 1998. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway.Genes Dev. 12:2684–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C.J., Nelson B., Marton M.J., Stoughton R., Meyer M.R., Bennett H.A., He Y.D., Dai H., Walker W.L., Hughes T.R., et al. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles.Science. 287:873–880 [DOI] [PubMed] [Google Scholar]
- Saunders W., Lengyel V., Hoyt M.A. 1997. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors.Mol. Biol. Cell. 8:1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W.S., Hoyt M.A. 1992. Kinesin-related proteins required for structural integrity of the mitotic spindle.Cell. 70:451–458 [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae.Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden S.C., Winters M.J., Ben-Ari G., Lamson R.E., Tyers M., Pryciak P.M. 2007. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway.Cell. 128:519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart J., Boeke J.D., Fink G.R. 1987. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein.Mol. Cell. Biol. 7:2316–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F., Stucke V.M., Jorritsma G., Peter M. 2001. MAP kinase dynamics in response to pheromones in budding yeast.Nat. Cell Biol. 3:1051–1059 [DOI] [PubMed] [Google Scholar]
- Wassmann K., Ammerer G. 1997. Overexpression of the G1-cyclin gene CLN2 represses the mating pathway in Saccharomyces cerevisiae at the level of the MEKK Ste11.J. Biol. Chem. 272:13180–13188 [DOI] [PubMed] [Google Scholar]
- White J.M., Rose M.D. 2001. Yeast mating: getting close to membrane merger.Curr. Biol. 11:R16–R20 [DOI] [PubMed] [Google Scholar]
- Ydenberg C.A., Rose M.D. 2008. Yeast mating: a model system for studying cell and nuclear fusion.Methods Mol. Biol. 475:3–20 [DOI] [PubMed] [Google Scholar]