SUMMARY
Bacterial virulence mechanisms are attractive targets for antibiotic development, because they are required for the pathogenesis of numerous global infectious disease agents. The bacterial secretion systems used to assemble the surface structures that promote adherence and deliver protein virulence effectors to host cells could comprise one such therapeutic target. In this study, we developed and performed a high-throughput screen (HTS) of small molecule libraries and identified a small molecule, a 2-imino-5-arylidene thiazolidinone that blocked secretion and virulence functions of a wide array of animal and plant Gram-negative bacterial pathogens. This compound inhibited type III secretion-dependent functions, with the exception of flagellar motility, and Type II secretion-dependent functions, suggesting that the target of the compound could be an outer membrane component conserved between these two secretion systems. This work provides a proof of concept that compounds with a broad spectrum of activity against Gram-negative bacterial secretion systems could be developed to prevent and treat bacterial diseases.
INTRODUCTION
In the twentieth century the treatment of infectious diseases was revolutionized by the development of antibiotics (Morens et al., 2004). However, due to their widespread use resistance to antibiotics is increasing on a global scale, such that adequate therapies are lacking for both previously controlled and emerging bacterial diseases (Levy and Marshall, 2004; Marra, 2006; Morens et al., 2004). Moreover, the molecular targets and mechanisms of action of most newly developed antibiotics are similar to current ones (Levy and Marshall, 2004; Nathan, 2004), reducing their efficacy in the face of resistance. The effective treatment of infectious diseases in the face of increasing antibiotic resistance will likely require the development of pharmaceuticals that act upon previously unutilized conserved targets (Levy and Marshall, 2004). Recently, bacterial virulence properties have been proposed and explored as attractive targets for the development of new therapeutic agents (Marra, 2006). This strategy could decrease the likelihood for selection of resistance, because in contrast to currently available antibiotics these agents would presumably not require inhibition of general bacterial growth. Such compounds would likely have the advantage of sparing commensals, reducing the likelihood of side effects. A potential disadvantage of pathogenic mechanisms as therapeutic targets is that many are microbial specific, necessitating more rapid pathogen identification than currently is in clinical practice.
Gram-negative bacterial virulence secretion systems represent particularly appealing virulence factor targets; because they are essential for a wide array of animal and plant infectious diseases and have some functionally conserved components. Two prominent examples of Gram-negative bacterial virulence associated secretion systems, termed type II secretion (T2S) and type III secretion (T3S), are responsible for the pathogenesis of many infectious diseases including plague, gastroenteritis, Gram-negative pneumonia, dysentery, enteric fever, tularemia, trachoma, endometritis and a variety of plant diseases. T2S is also known as the terminal component of the sec-dependent or General Secretory Pathway (GSP), because substrates are secreted across the bacterial inner membrane by the GSP and subsequently transported across the outer membrane (Cianciotto, 2005). T2S systems secrete a variety of mammalian toxins as well as proteins, which degrade host cell components, including proteins, lipids and sugars of the extracellular matrix (Cianciotto, 2005). Interestingly, a number of the genes required for T2S are homologous to those required for type IV pilus (T4P) assembly on the cell surface of some bacteria (Mattick, 2002). T4P are required for twitching motility, a flagella-independent form of bacterial translocation, which plays a role in host colonization and biofilm formation in organisms, such as enterophathogenic E. coli (EPEC), Pseudomonas aeruginosa, Vibrio cholera and Nieserria gonorrhoea (Mattick, 2002). T3S systems are complex multi-protein organelles that assemble in the bacterial membrane of more than 25 Gram-negative animal and plant pathogens to deliver multiple virulence proteins, or effector proteins, directly from the bacterial cytosol into host cells. These secreted proteins influence host cell physiology by altering a variety of antibacterial functions with resultant disease (Cornelis and Van Gijsegem, 2000).
In recent years whole-cell based high-throughput screens have been performed to identify inhibitors of T3S systems (Gauthier et al., 2005; Kauppi et al., 2003; Pan et al., 2007). These screens have identified several classes of synthetic compounds and the natural product glycolipid caminosides, as active for inhibition of T3S in a broad range of Gram-negative bacterial pathogens, including Yersinia, Chlamydia and Salmonella (Gauthier et al., 2005; Kauppi et al., 2003; Linington et al., 2006; Linington et al., 2002; Negrea et al., 2007; Nordfelth et al., 2005; Wolf et al., 2006). The salicylanilides likely inhibit T3 gene transcription while the salicylideneacylhydrazides, sulfonylaminobenzanilides and caminosides have unknown targets. In this study, we designed and implemented a tractable high-throughput screen (HTS) for the identification of compounds that could function to inhibit T3S secretion systems and found a small molecule that broadly inhibits T3 and T2 bacterial secretion systems.
RESULTS
A high throughput screen using engineered S. typhimurium identifies a 2-imino-5-arylidene thiazolidinone as an inhibitor of T3S
To screen biological and chemical small molecule libraries for inhibitors of bacterial secretion; we designed and employed a whole-cell HTS for inhibitors of T3S. T3S systems, which are evolutionarily related to flagella, are complex multi-protein organelles that assemble in the bacterial membrane to deliver virulence proteins directly from the bacterial cytosol into host cells. A strain of S. typhimurium was constructed that secretes a phospholipase A2 reporter construct in a T3S-dependent manner. Specifically, this strain contains a protein fusion between the S. typhimurium T3S substrate SipA and the Yersinia enterocolitica T3 secreted substrate YplA. The first 59 amino acids were removed from YplA, because they contain the signal sequence for its secretion by Y. enterocolitica, but are not required for its enzymatic activity (Hatic et al., 2002; Warren and Young, 2005). Phospholipase activity is an efficient and well-recognized reporter in HT assays because of the availability of phospholipase substrates with a cleavage product that is fluorescent. Our assay was based on cleavage, by our phospholipase A2 reporter construct, of the substrate PED6: N-((6-(2,4-dinitro-phenyl)amino)-hexanoyl)-2-(4,4-difluro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine (Invitrogen, Carlsbad, CA). Such cleavage results in an increase in absorbance readily measured using a fluorometer. When PED6 is added directly to a culture of the engineered Salmonella strain, fluorescence is proportional to the amount of phospholipase reporter secreted by the T3S system.
Using this HTS, 92,000 small molecules from both natural and synthetic compound libraries were screened (NSRB screening facility, Harvard, MA) for compounds that resulted in a reduction in fluorescence (z-scores > −3.0), signifying a reduction in T3S similar to a T3S genetic mutant (ΔprgH–K). This screen yielded 89 putative T3S inhibitors that appeared to have no effect on bacterial growth (data not shown). Of these, 57 were judged to lack novelty or potential for drug development, seven compounds had an effect on bacterial growth upon further analysis (data not shown) and 25 were further studied. Because inhibition of a variety of general bacterial processes not specific to T3S would have been positive in the HTS, including gene transcription, protein translation, sec-dependent secretion, and disulfide bond isomerization, secondary assays were performed to define screening positives specific for secretion.
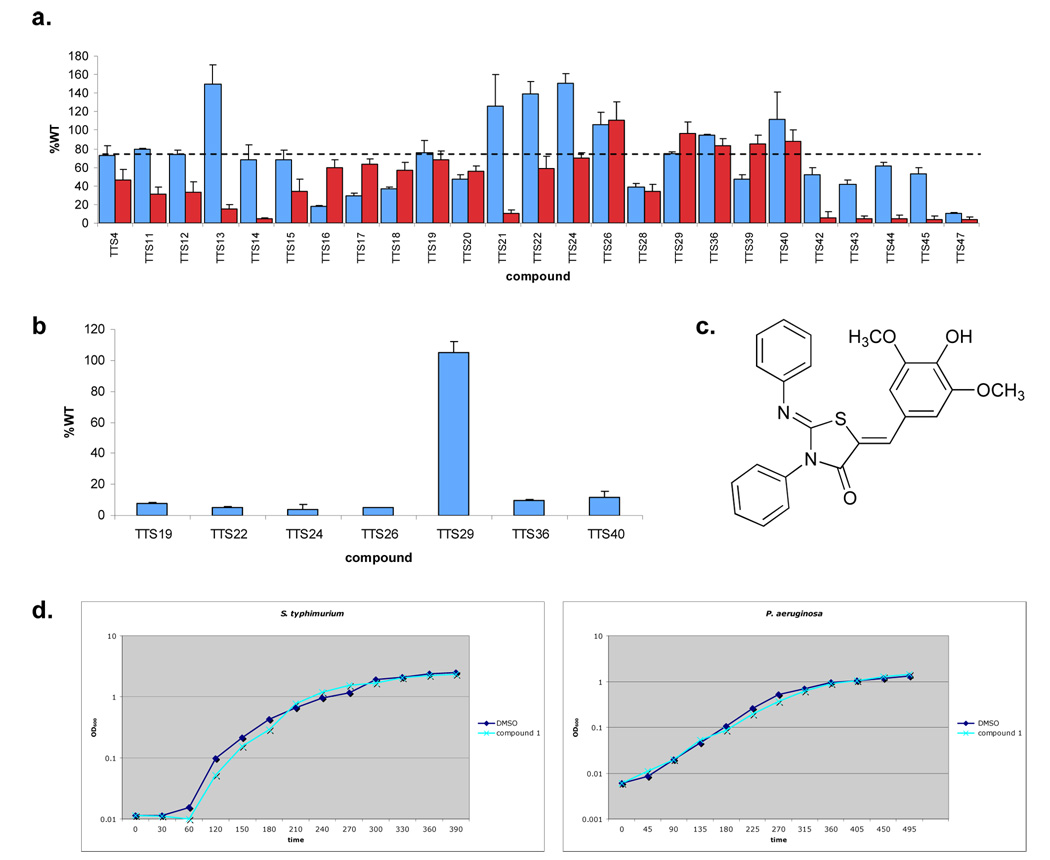
To identify non-specific inhibitors of transcription the expression of the flagellar regulatory gene (flhC) was measured using a transcriptional fusion to the lacZ gene. To eliminate compounds inhibiting bacterial translation, sec-dependent secretion, or disulfide bond isomerization, alkaline phosphatase activity of a PrgH’-‘PhoA protein fusion was measured in the presence of the various compounds. PrgH is an essential inner membrane component of the T3S apparatus secreted by the sec-dependent pathway (Kimbrough and Miller, 2000; Kubori et al., 1998), and alkaline phosphatase activity requires disulfide bond formation; thus, inhibition of fusion protein expression and/or its localization would result in decreased alkaline phosphatase activity. We found that while many of the identified compounds reduced β-galactosidase activity and/or alkaline phosphatase activity, cultures grown in the presence of seven (TTS19, 22, 24, 26, 29, 36 and 40) of the 25 compounds assayed had at least 60% of the activity observed for cells grown in the absence of compound on repeated measurement (Figure 1a). Thus, these seven compounds lacked a strong general effect on bacterial transcription, translation, sec-dependent secretion and/or bacterial growth.
Figure 1. Identification of an inhibitor of T3S in S. typhimurium.
a,β-galactosidase activity from an flhC-lacZ transcriptional fusion in S. typhimurium when grown in the presence of small molecules (~380 µm) identified in the HTS screen. Activity was recorded as a percent of WT (blue bars), i.e. the amount of β-galactosidase activity observed when bacteria are grown in the absence of compound. Three independent experiments were performed and the mean standard deviations are shown. Alkaline phosphotase activity of a PrgH-PhoA translational fusion in S. typhimurium when grown in the presence of small molecules (~380 m) identified in the HTS screen (red bars). The dashed line (---) marks 60% of WT activity, the level used to choose compounds for further study. b, β-galactosidase activity from an iagB-lacZ transcriptional fusion in S. typhimurium when grown in the presence of small molecules (380 µm) with levels of flhC-lacZ and PrgH-PhoA expression greater than 60%. As above, activity was recorded as a percent of WT. Three independent experiments were performed and the mean standard deviations are shown. c, Chemical structure of TTS29, renamed Compound 1. d, Growth curves for S. typhimurium and P. aeruginosa in LB medium with compound 1 or an equal volume of the solvent.
These compounds could still have had a specific transcriptional effect on T3S gene expression. Therefore, we measured the effect of these compounds on transcription of a T3S operon (the hilA operon) that encodes a transcriptional regulator for T3S structural components and secreted substrates by again using a transcriptional reporter fusion to lacZ. β-galactosidase activity was measured in the presence and absence of the compounds. We observed a greater than 85% reduction in β-galactosidase activity for six of these compounds when cultures were grown in the presence of compound, suggesting that they specifically inhibited T3S transcription (Figure 1b). In contrast, activity was similar in cultures grown in the presence or absence of one compound, TTS29, suggesting that this compound, of the 92,000 screened might specifically target the assembly or structure of the S. typhimurium T3S. This compound is a 2-imino-5-arylidene thiazolidinone and we labeled it compound 1 (Figure 1c). To corroborate our initial screening data that compound 1 does not affect bacterial growth; we assayed growth over time of S. typhimurium in growth medium with and without compound 1. No affect on bacterial growth was observed (Figure 1d). Bacterial colony forming units were also determined and no change was observed in the presence of compound 1 (data not shown).
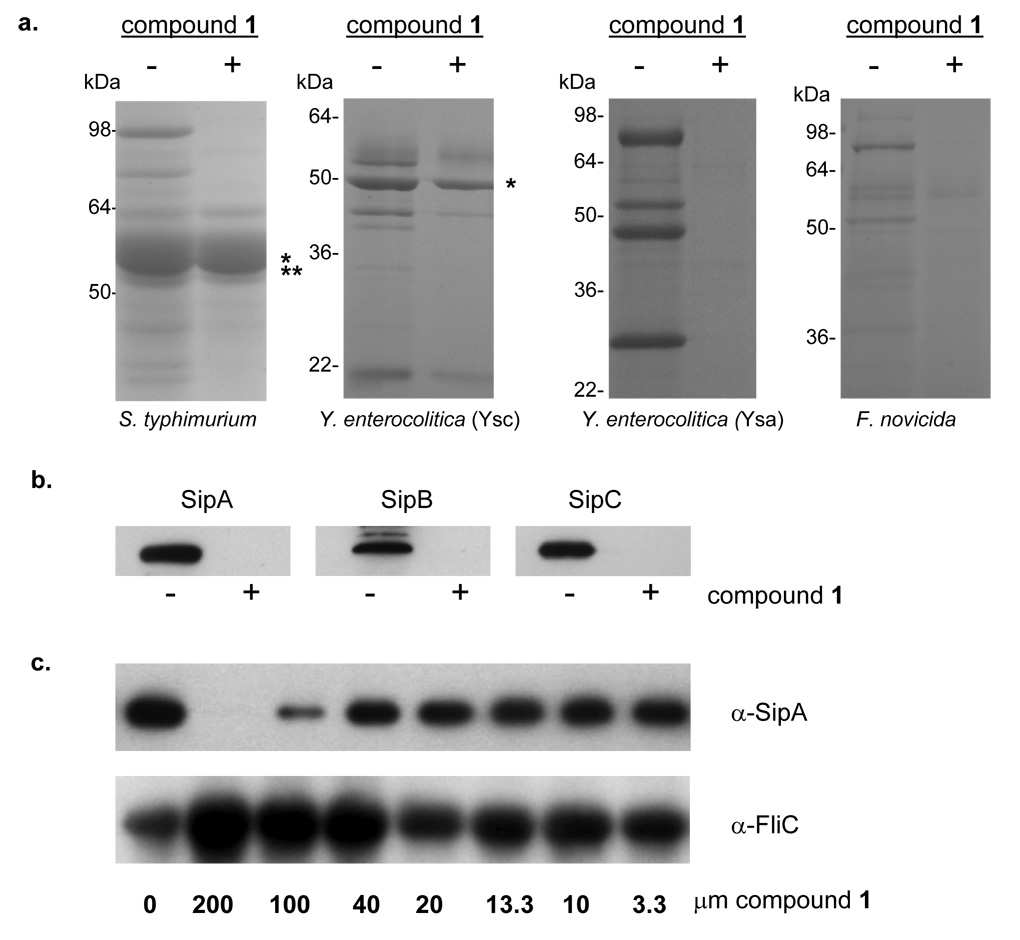
To demonstrate specific inhibition of T3S by this compound, we examined the culture supernatants of S. typhimurium for T3S secreted substrates when grown in the presence of compound 1 (380 µm) using Coomassie Blue-stained protein gels (Figure 2a) and Western Blots for the prominent T3S secreted virulence proteins, SipA, B and C (Figure 2b). We observed a marked decrease in the amount of the T3 secreted proteins when bacterial cultures were grown in the presence of compound 1. To clearly rule out a complex T3S-dependent transcriptional effect, secretion of two T3S substrates, SipA and SspH1, were expressed from the T3S-independent lac promoter and their secretion was examined by Western Blot. We found that neither of these proteins was secreted from bacteria grown in the presence of compound 1 (data not shown).
Figure 2. Phenotypic effects of compound 1 on type III secretion.
a, Bacterial cultures were grown in the absence (−) or presence (+) of 380 µM of compound 1. Secreted proteins were TCA precipitated and separated by 12.5% SDS-PAGE and stained with Coomaisse Blue. The flagellin proteins from S. typhimurium and Y. enterocolitica are marked (*) as well as the associated flagellar cap protein in S. typhimurium (**). b, Western Blots of the secreted proteins, SipA, B and C from S. typhimurium. c, Western Blots of secreted SipA protein as well as the flagellar filament protein, FliC in supernatants from S. typhimurium grown in LB alone or with decreasing concentrations of compound 1.
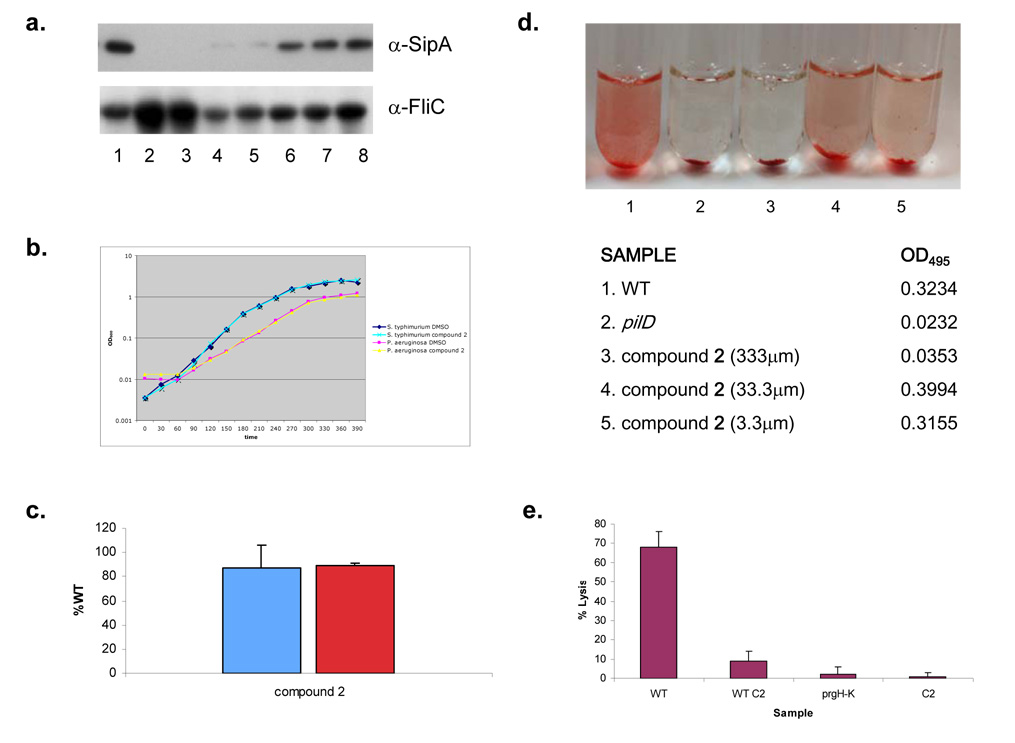
To support the hypothesis that compound 1 acted upon a specific biological mechanism for inhibition of T3S, we performed a dose-dependent curve with compound 1. We analyzed protein secretion by S. typhimurium in the presence of decreasing amounts (200, 100, 40, 20, 13.3, 10 and 3.3 µm) of compound 1. Secreted proteins were TCA precipitated and equal amounts of sample were loaded onto an SDS-PAGE, proteins were separated and Western Blotted with antibody to the SipA protein (Figure 2c). Inhibition of S. typhimurium protein secretion was greatest at 200µM and diminished incrementally with decreasing concentration of compound. To control for sample handling and loading of the supernatant proteins, we blotted with antibodies to FliC, the flagellar filament protein of S. typhimurium and a predominant protein found in culture supernatants. We did not observe a decrease, but actually observed an increase, in FliC protein levels in the presence of compound (Figure 2c).
Compound 1 inhibits S. typhimurium T3S needle complex assembly or stability, without altering secretin protein levels or membrane localization
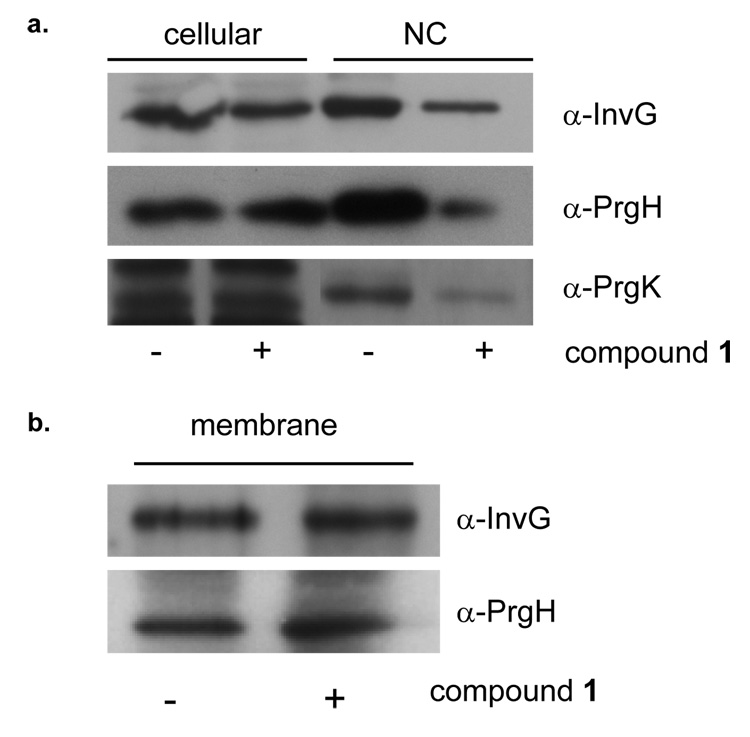
The highly conserved structure of the T3S apparatus includes a membrane-spanning complex associated with an extracellular needle, termed the needle complex (NC) (Kubori et al., 1998; Sekiya et al., 2001). In order to investigate the affect of compound 1 on the T3S apparatus, we purified NCs from S. typhimurium cells grown in the presence and absence of compound 1. The predominant NC proteins isolated in S. typhimurium are InvG, which forms the outer membrane ring or secretin complex, as well as PrgH and PrgK, which form the inner membrane ring (Kimbrough and Miller, 2000; Yip et al., 2005). Post CsCl-gradient fractions of NCs were separated by SDS-PAGE and analyzed by Western Blot. An overall reduction in NC proteins was observed when cells were grown in the presence of compound 1 (Figure 3a), suggesting that it may inhibit formation and/or destabilize NCs.
Figure 3. Phenotypic effects of compound 1 on type III secretion needle complex assembly.
a, Type III secretion needle complexes (NC) were purified from S. typhimurium grown in the absence (−) or presence (+) of 380 µM of compound 1. Proteins were separated by SDS-PAGE and Western Blotted with antibodies to InvG, PrgH and PrgK. b, Total membrane fractions were isolated from S. typhimurium grown in the absence (−) or presence (+) of 380 µM of compound 1. Proteins were separated by SDS-PAGE and Western Blotted with antibodies to InvG and PrgH.
To test whether the decrease in NCs actually resulted from a reduction in either cellular levels or localization of NC proteins to the bacterial membrane, we assayed cells grown in the presence and absence of compound 1 for the abundance and localization of InvG as well as PrgH. In the presence of compound 1, cellular and total membrane protein levels were unaltered suggesting localization was unchanged (Figure 3a & 3b). Given that stable and equal amounts of InvG and PrgH are present and localized to the membrane, these data indicate that NC formation or assembly is the likely target of compound 1.
Compound 1 inhibits both type III secretion systems in Yersinia spp
Phylogenetic analyses of conserved T3S systems indicate that these structures have evolved into different families typified by the Ysc system of Yersinia spp., the Inv/Spa system of S. typhimurium and Shigella flexneri, and the Esc system of enterohaemorrhagic E. coli (EHEC) and the Hrp/Hrc system characteristic of plant pathogens such as Pseudomonas syringae (Cornelis, 2006). To examine the potential inhibitory effect of our compound on other classes of T3S systems, we analyzed the secretion profiles for the T3S systems of Yersinia enterocolitica grown in the presence of compound 1. The plasmid-encoded Ysc T3S system of Yersinia pestis, Y. enterocolitica and Yersinia pseudotuberculosis represents a well characterized T3S system that delivers a set of bacterial effector proteins, termed Yops (Yersinia outer proteins) into the lumen of the target host cell resulting in inhibition of the innate immune response (Viboud and Bliska, 2005). In contrast, the chromosomally encoded Ysa T3S system of Y. enterocolitica secretes a set of proteins termed Ysps (Yersinia secreted proteins) (Matsumoto and Young, 2006). Although translocation of these proteins into host cells has not been clearly demonstrated, it is believed to contribute to the gastrointestinal stage of infection. We found that both Yop and the Ysp proteins were absent in supernatants from Y. enterocolitca cultures grown in the presence of compound 1 (Figure 2a), indicating that these Yersinia T3S systems were also inhibited by this compound. Similar to S. typhimurium no difference in growth was observed for Y. enterocolitica grown in the presence of compound 1 (Supplemental Figure 1). Thus compound 1 appears to be a relatively broad inhibitor of T3S systems.
Compound 1 does not inhibit the flagellar-specific T3S system in S. typhimurium
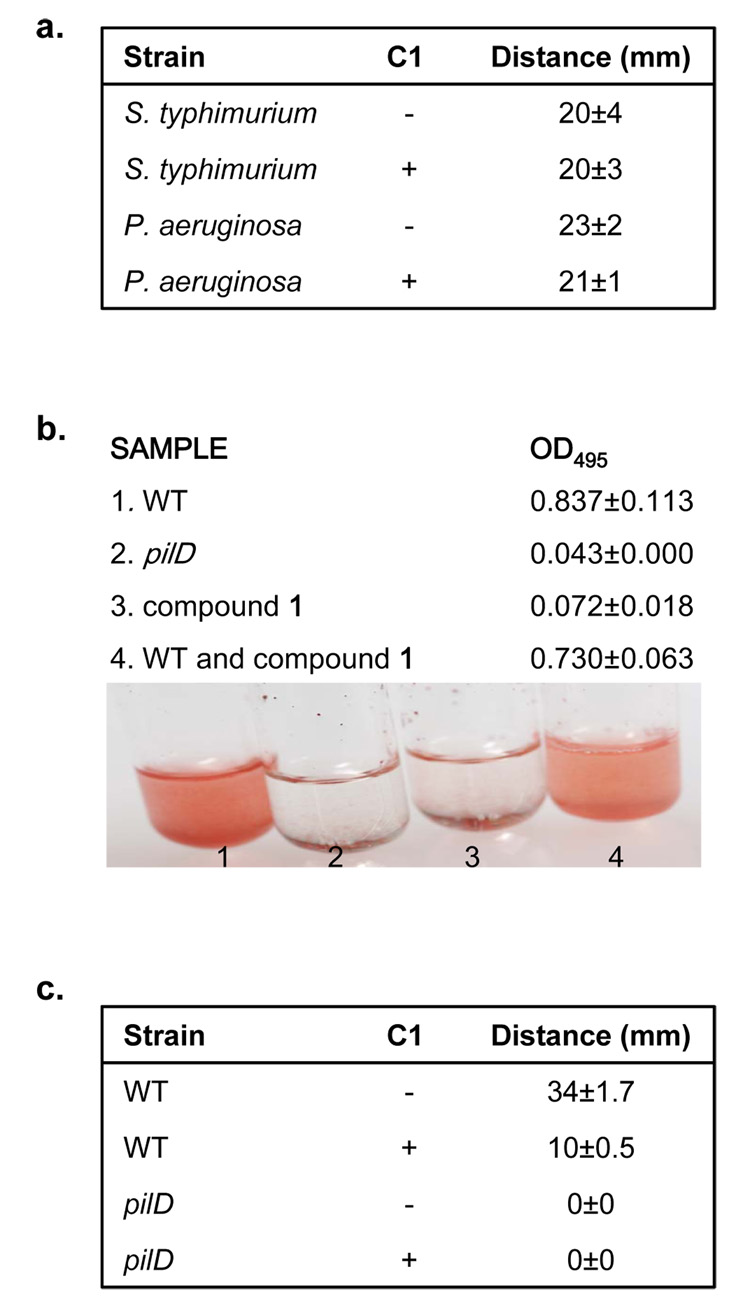
Virulence T3S systems are evolutionarily related to the flagellar-specific T3S system and therefore it was plausible that compound 1 inhibited flagellar secretion and mobility. Specifically, a core of eight conserved proteins assemble the foundation of T3S apparatus’ as well as the base of the bacterial flagella, but the flagellar system does not utilize the outer membrane component of the NC the secretin protein (Cornelis, 2006). Interestingly, secretion of flagellin, the predominant substrate of this system observable in the culture supernatant, was unaltered by the presence of compound 1 (Figure 2a) suggesting that compound 1 does not inhibit flagellar-specific T3S. These data were corroborated by Western Blot of the S. typhimurium supernatant proteins with antibody to the flagellar filament protein, FliC (Figure 2c). To further determine the effect of compound 1 on flagellar secretion and hence function, we measured bacterial motility of S. typhimurium in motility plates containing compound 1 and compared it to motility in plates lacking compound 1. Motility was measured after incubation at 37°C for 6 hours. We found that motility of S. typhimurium was similar in plates with and without compound 1 (Figure 4a). These data suggested that compound 1 may target a component of T3S systems not conserved with the flagellar-specific T3S system.
Figure 4. Phenotypic effects of compound 1 on alternate secretion systems.
a, Motility assays of S. typhimurium and P. aeruginosa in the absence (−) or presence (+) of 380 µM of compound 1. Bacteria were stabbed into motility plates and incubated at 37°C for 6 hours. The diameter of the ring was measured for three independent replicates and the mean standard deviations are shown. b, Elastolytic activity for 18 hour culture supernatants of P. aeruginosa grown in the absence (WT; 1) or presence of 380 µM of compound 1 (compound 1; 3) was determined using elastin Congo Red as a substrate. Elastase activity was measured as a change in OD495 and can be observed as an increase in the red color of the sample. As a negative control elastase activity was determined for the culture supernatant of a P. aeruginosa T2S mutant (pilD; 2). As a control for inhibition of elastase activity by the compound, compound 1 was added directly to culture supernatants from P. aeruginosa grown in the absence of compound and assayed for elastolytic activity (WT and compound 1; 4). Pictures and the corresponding OD495 of samples are shown. c, Twitching assays of P. aeruginosa in either the absence (−) or presence (+) of 380 µM of compound 1. Bacteria were stabbed into LB plates with 1.0% agar and incubated for 2 days at room temperature. Growth media was removed and the bacteria were stained with crystal violet for better visualization.
Compound 1 inhibits other secretin containing secretion systems of Gram-negative bacteria
The specificity of compound 1 for other T3S systems, but not the flagellar-specific T3S system as well as a decrease in NC formation in the presence of compound suggested that the outer membrane secretin protein may be the target of this small molecule. If this were the case other secretion systems containing a secretin component would possibly be inhibited, such as T2S and T4P assembly. Bacterial T2S systems transport many substrates from the periplasm across the outer membrane, including a variety of mammalian toxins, as well as other proteins, which degrade host cell components, such as proteins, lipids and sugars of the extracellular matrix (Cianciotto, 2005). An important virulence factor of the opportunistic pathogen P. aeruginosa is the T2 secreted extracellular enzyme elastase, which is required for the ability of this organism to produce corneal ulcers, skin infections and pneumonia (Wretlind and Pavlovskis, 1983). To determine if compound 1 inhibits the T2-dependent secretion of elastase, we measured elastase in culture supernatants from bacteria grown in the presence and absence of compound 1 by monitoring its enzymatic activity. Elastase activity can be measured by adding the substrate elastin conjugated to Congo Red to culture supernatants. Cleavage of this substrate by elastase results in a detectable increase in absorbance at OD495 (McIver et al., 2004; Ohman et al., 1980). An isogenic strain that lacks the prepillin peptidase PilD, which is an essential component of the Pseudomonas T2S system was used as a negative control. We observed a reduction in supernatant elastolytic activity in culture supernatants from bacteria grown in the presence of compound 1, similar to the pilD mutant, as indicated by a decrease in the OD495, when bacteria were grown in the presence of compound 1 (Figure 4b) as opposed to solvent alone suggesting an inhibition of the P. aeruginosa T2S system by this molecule. As a control for the potential inhibition of the compound on the enzymatic activity of elastase, compound 1 was added directly to culture supernatants from P. aeruginosa grown in the absence of compound 1 and assayed for elastolytic activity. We did not observe a decrease in the OD495 indicating that compound 1 does not affect the enzymatic activity of elastase. To eliminate the possibility that this affect is due to a growth defect of P. aeruginosa in the presence of the compound, growth curves were performed for P. aeruginosa and no difference in growth for P. aeruginosa was observed in the presence of compound 1 (Figure 1d).
Twitching motility, which is a flagellar-independent form of bacterial motility, is mediated by T4P(Mattick, 2002). As a number of the genes required for T4P assembly are homologous to those required for T2S, we also examined twitching motility in P. aeruginosa in the presence and absence of compound 1 (Mattick, 2002). After incubation for two days at 30°C we measured the diameter of the twitching ring for three independent biological replicates. We found that the spread of bacteria from the point of inoculation was reduced in the plates containing compound 1 (Figure 4c). In contrast, the flagellar mediated swimming motility of P. aeruginosa was unaffected by the presence of compound 1 in motility media, further indicating that flagellar-specific T3S systems are unaffected by compound 1 (Figure 4a).
Recently, Francisella novicida, a subspecies of F. tularensis the causative agent of the zoonotic disease tularemia, was demonstrated to secrete a number of virulence factors through an atypical secretion system related to T4P secretion systems (Hager et al., 2006). Francisella secretion was shown to require a protein with amino acid similarity to secretins, and thus to further examine the potential broad spectrum nature of 2-imino-5-arylidene thiazolidinone inhibition we analyzed secretion of virulence proteins by F. novicida when grown in the presence of compound 1 using Coomaisse Blue-stained protein gels (Figure 2a). Compound 1 inhibited secretion of the known Francisella virulence proteins and as the secretin is the only shared component with T3S systems, these data suggest that compound 1 might block secretion by targeting the outer membrane ring proteins of these multi-protein systems. The growth of F. novicida was unaltered by the presence of the compound as determined by OD600, 1.0385 (DMSO) vs. 0.9604 (compound 1), prior to removal of the supernatant.
Compound 1 blocked secretion and virulence functions in both animal and plant models of infection
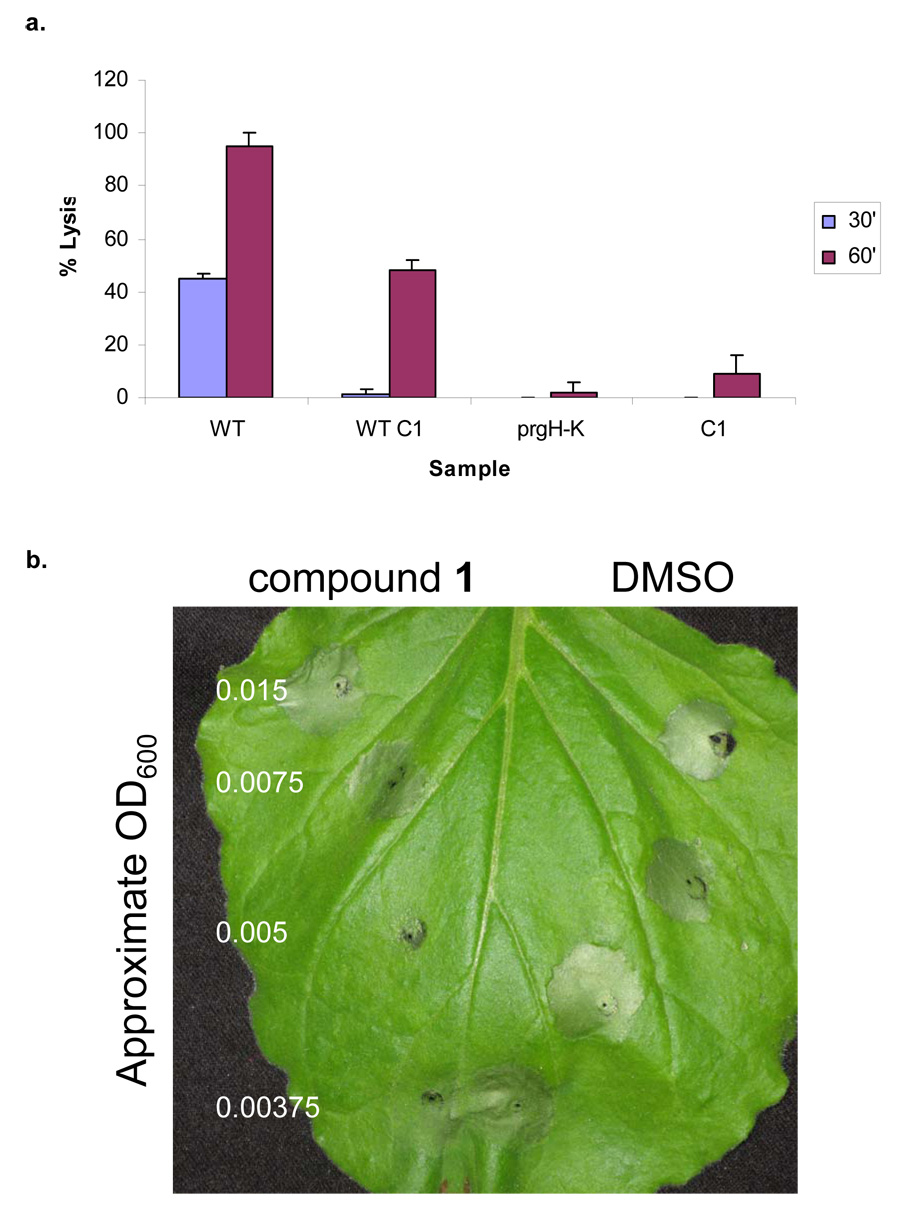
Although we showed that compound 1 inhibited in vitro virulence protein secretion, we wanted to test the impact of the inhibition of secretion on eukaryotic cells. To determine whether compound 1 blocked delivery of virulence determinants into host cells and thus inhibit microbial virulence function, we measured cytotoxicity of bone marrow macrophages by S. typhimurium in the presence of compound 1, which is T3S-dependent. The enteric pathogens Salmonella and Shigella stimulate caspase-1 mediated cell death as a result of T3S-mediated translocation (Miao et al., 2006). S. typhimurium cytotoxicity of macrophages was measured by monitoring the release of lactate dehydrogenase, a stable cytosolic enzyme that is released upon macrophage lysis. Bacteria were grown in the presence or absence of compound 1 under T3S-inducing conditions and added to macrophages at an MOI of 40. Infections were allowed to proceed for 30 minutes or 60 minutes at 37°C and in an atmosphere of 5% CO2. A decrease in caspase-1 dependent cell death of macrophages, 40% to 0.5% (30 minute infection) or 96% to 48% (60 minute infection), was observed for bacteria grown in the presence of compound 1, consistent with the protection of mammalian cells from bacterial virulence in a tissue culture model of infection (Figure 5a). Therefore compound 1 appears to block the T3-dependent cytotoxicity of S. typhimurium for eukaryotic cells. In addition, compound 1 was added directly to macrophages at a concentration equal to the sample with bacteria and compound 1. Macrophages exposed to compound 1 did not lyse indicating that compound 1 was not overtly cytotoxic to mammalian cells. Cytotoxicity of macrophages was not observed with the S. typhimurium T3S genetic mutant (ΔprgH–K), as previously observed.
Figure 5. Inhibition of bacterial virulence phenotypes by compound 1.
a, Mouse bone marrow macrophages (BMM) from Balb/c mice were infected with WT S. typhimurium grown in the absence (WT) or presence of 380 µM of compound 1 (WT C1) to SPI1 inducing conditions. A T3S genetic mutant (prgH–K) was used as a negative control. To confirm that compound 1 was not itself cytotoxic to the macrophages, compound without bacteria (C1) was added at a concentration equal to the experimental samples. Bacteria were added at an MOI of 40 and cytotoxicity was assessed by LDH release at 30 minutes and 60 minutes after infection. Assays were performed in quadruplicate and mean standard deviations are shown. b, P. syringae and compound 1 were co-inoculated on non-host tobacco plants and monitored for HR. Varying concentrations of bacteria were added, while the amount of compound or solvent (v/v) remained constant. Four independent experiments were performed and a representative experiment is shown. Tissue collapse near the point of inoculation for either compound 1 or solvent alone is due to tissue damage caused during injection of compounds by a blunt-ended syringe.
The bacterial plant pathogen Pseudomonas syringae pv tomato DC3000 requires T3S of virulence determinants to cause disease. The hypersensitivity response (HR) elicited by P. syringae in non-host tobacco plants depends upon a functional T3S system and is an established virulence model for this pathogen (Huang et al., 1988). To determine the effect of compound 1 on P. syringae HR we co-inoculated P. syringae with compound 1 or solvent (v/v) onto the leaves of Tobacco plants. The bacteria were added at decreasing dilutions, while the concentration of the compound remained constant. HR was measured by tissue collapse of plants cells surrounding the point of inoculation. In the presence of compound 1, P. syringae exhibited a reduced ability to elicit HR in the presence of compound 1 compared with solvent alone (Figure 5b). Tissue collapse near the point of inoculation for compound 1 or solvent alone is tissue damage caused during injection of compounds by a blunt-ended syringe. Therefore this compound can block disease in an important model of plant pathogenesis.
Structure activity relationships established features of the thiazolidinone chemotype effective for T3S inhibition
Thiazolidinones are a structural class associated with a variety of biological effects, that include antibacterial, antiviral, cardiotonic and anti-inflammatory activities (Pulici and Quartieri, 2005). The fact that diverse and specific activities have been demonstrated for individual compounds from this family, which has a pluripotent common scaffold, suggests that the substituents are responsible for target selectivity. Using compounds obtained both commercially and from our own synthetic work, we performed a preliminary structure activity relationship (SAR) study of thiazolidinone analogs. We tested 40 compounds structurally related to compound 1 for inhibition of S. typhimurium SipA secretion by Western Blot. We found that while most of the 40 related compounds tested had minimal inhibitory activity on T3S (Supplemental Table 1), six compounds inhibited greater than 90% of SipA secretion as determined by Western Blot of supernatant proteins (Table 1 and Supplementary Table 1). These data allowed us to draw the following preliminary conclusions: First, these results are consistent with requirements for the imino nitrogen and the aryl group for inhibition of T3S (compounds 5 & 6). Second, the 5-substituent likely must be an appropriately substituted arylidene group as reduction (compound 9), removal (compound 8) or stripping the substituents from (compound 7) this ring significantly diminishes activity. Finally, the amido nitrogen is fairly permissive (compounds 2, 3 & 4) and thus allows for the directed evolution of increasingly active thiazolidinone analogs from substitution on this nitrogen. In addition, a few of these compounds fell out of the mainstream of the SAR and may represent a new chemotype.
TABLE 1.
Effect of a sample of thiazolidinone anologs on type III secretion of the SipA protein in S. typhimurium. The amount of SipA protein in culture supernatants was determined by Western Blot for three independent replicates and standard deviations are shown. Compounds were tested at a final concentration of 380 µm.
| Compound | Structure | % secretion | Compound | Structure | % secretion |
|---|---|---|---|---|---|
| 1 | 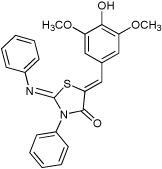 |
0 ± 0 | 5 | 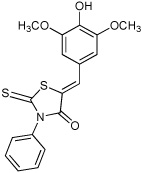 |
81 ± 15 |
| 2 | 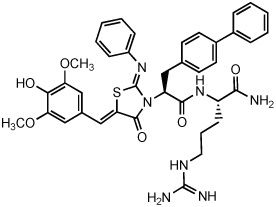 |
0 ± 0 | 6 | 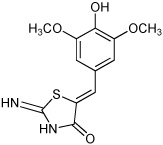 |
64 ± 4 |
| 3 | 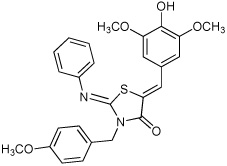 |
0 ± 0 | 7 | 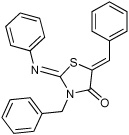 |
90 ± 18 |
| 4 | 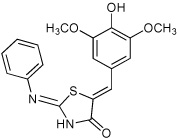 |
9 ± 6 | 8 | 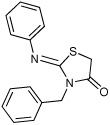 |
72 ± 0 |
| 9 | 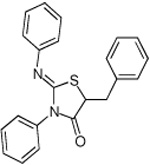 |
62 ± 3 |
Synthesis of an analog of compound 1 with increased solubility and activity
Because it was possible that the poor solubility of compound 1 contributed to its requirement for a high micromolar concentration for full activity, we synthesized an analog (compound 2) that was more hydrophilic and contained protonatable groups that would enhance solubility. We then performed dose-dependent curves. Again, secreted proteins from S. typhimurium grown in the presence or absence of this compound were TCA precipitated, separated by SDS-PAGE and Western Blotted with anti-SipA antibody. In contrast to compound 1, the analog appears to inhibit most of the detectable SipA at 100 µM. Minimal SipA protein was detected in the supernatant at 40 and 20 µM (Figure 6a), suggesting that compound 2 is approximately 10-fold more potent than compound 1. It was possible that the increased activity of compound 2 was due to an acquired affect on the growth of S. typhimuirum, and therefore we measured growth of S. typhimurium and P. aeruginosa in the presence and absence of compound 2. We found no affect of compound 2 on growth (Figure 6b). We also confirmed that compound 2, like compound 1, did not have an affect on type III gene transcription by measuring β-galactosidase activity of the iagB-lacZ transcriptional fusion for bacteria grown in the presence and absence of compound 2. Similar levels of β-galactosidase activity were observed under both conditions (Figure 6c), suggesting that compound 2 does not have an affect on T3S gene expression. We also measured expression of our PrgH’-‘PhoA fusion in the presence of compound 2. As with compound 1, we observed similar levels of alkaline phosphatase activity when bacteria grown in the absence or presence of compound 2 (Figure 6c). To determine if compound 2 maintained the broad spectrum of activity, we measured T2-dependent secretion of elastase into the supernatant by P. aeruginosa. Similar to compound 1, compound 2 demonstrated an ability to inhibit elastase secretion of P. aeruginosa as measured by cleavage of Congo Red conjugated to elastin (Figure 6d), providing evidence that both compounds are able to inhibit T3 and T2S systems. Finally, compound 1 and 2 were found to have no effect on bacterial membrane permeability, as determined using 1-N-phenyl-naphthylamine and ethidium bromide uptake assays, indicating that they were not having a non-specific effect on the outer membrane (data not shown) and providing further evidence that these compounds have a specific mechanism of action.
Figure 6. Characterization of secretion inhibition by compound 2.
a, Western Blots of secreted SipA protein as well as the flagellar filament protein, FliC in supernatants from S. typhimurium grown in LB alone or with decreasing concentrations of compound 2. b, Growth curves for S. typhimurium and P. aeruginosa in LB medium with compound 2 or an equal volume of solvent. c, β-galactosidase activity (blue bar) from an iagB-lacZ transcriptional fusion in S. typhimurium or the amount of alkaline phosphotase activity (red bar) observed from a PrgH-PhoA translational fusion in S. typhimurium when grown in the presence of compound 2 (100 µm). Enzymatic activity was recorded as a percent of WT, i.e. the amount of activity observed in the absence of compound. Three independent experiments were performed and the mean standard deviations are shown. d, Elastolytic activity for 18 hour culture supernatants of P. aeruginosa grown in the absence (1) or presence of compound 2 at varying concentrations (3, 4 & 5) was determined using elastin Congo Red as a substrate. Elastase activity was measured as a change in OD495 and can be observed as an increase in the red color of the sample. As a negative control elastase activity was determined for the culture supernatant of a P. aeruginosa T2S mutant (pilD; 2). Pictures and the corresponding OD495 of samples are shown. e, Mouse BMMs were infected with WT S. typhimurium grown in the absence (WT) or presence of 100 µm of compound 2 (WT C2), a T3S mutant (prgH–K) or compound 2 without bacteria (C2). Bacteria were added at an MOI of 40 and cytotoxicity was assessed by LDH release at 60 minutes. Assays were performed in quadruplicate and mean standard deviations are shown.
To determine if compound 2 was more effective at protecting mammalian cells from bacterial virulence in a tissue culture model of infection, we measured cytotoxicity of bone marrow macrophages by S. typhimurium in the presence of compound 2. Bacteria were grown in the presence or absence of compound 2 under T3S-inducing conditions and added to macrophages at an MOI of 40. Infections were allowed to proceed for 60 minutes at 37°C and in an atmosphere of 5% CO2. A decrease in caspase-1 dependent cell death of macrophages, from 68% to 9%, was observed for bacteria grown in the presence of compound 2 (100µm) (Figure 6e). This is greater than the protection observed with compound 1 (380µm) at 60 minutes, 95% to 48%, suggesting that compound 2 is more effective than compound 1 at protecting mammalian cells from bacterial virulence. Again, compound was added directly to macrophages at a concentration equal to the sample with bacteria and compound. Macrophages exposed to compound 2 exhibited minimal LDH release suggesting that compound 2 is also not cytotoxic to mammalian cells. These data indicate compounds of higher activity can be generated without sacrificing features important for potential bioavailability as well as the broad spectrum of activity.
DISCUSSION
The increase in knowledge of bacterial virulence properties has led investigators to consider them as targets for the development of therapeutic agents (Marra, 2006). Such agents would possibly block disease without killing the pathogen or commensal bacteria. In this study, we designed and implemented a tractable high-throughput screen (HTS) that identified small molecule inhibitors of Gram-negative bacterial virulence associated secretion systems, required for the pathogenesis of numerous global infectious agents. We identified a 2-imino-5-arylidene thiazolidinone that inhibits bacterial secretion systems in a wide array of animal and plant pathogens without altering bacterial growth. A variety of genetic and biochemical studies suggest that compound 1 and 2 act by inhibiting the conserved components of secretion systems not utilized by the flagellar secretion system. The active compound was further demonstrated to inhibit T3S-dependent S. typhimurium cytotoxicity for macrophages as well as the pathology of the plant pathogen, P. syringae on Tobacco. Finally, by testing for activity of a broad array of purchased and synthesized analogs, a defined secretion inhibitor chemotype was determined and an analog with increased solubility and activity was identified.
Thiazolidinones appear in a diverse array of drug discovery programs and substituted thiazolidinones are under investigation as anticancer antagonists, antischistosomal agents and general antifungal/antimicrobial agents (Dayam et al., 2006; Khare et al., 1995; Manhi and Mahoud, 2007; Merja et al., 2004; Qin et al., 2006; Rajanarendar et al., 2004). Moreover, many thiazolidinones are highly potent and target specific, suggesting that the substituents impart a high degree of selectivity to the common core of these small molecules. Our initial SAR studies suggest that the 2-imino-5-arylidene thiazolidinone mediating our effect is highly sensitive to the substituents at the imino N-2, amido N-3, and 5-arylidene groups. This sensitivity supports the hypothesis that specific molecular interactions between the small molecule ligand and the cognate protein(s) are required for inhibiting bacterial secretion. In addition, we were able to rationally design and synthesize compound 2 (Kline et al., unpublished data), an analog of compound 1 that demonstrated increased activity and enhanced solubility. The reappearance of the thiazolidinone motif in a wide variety of research studies indicates that compounds developed from this scaffold may have the potential to be developed into therapeutic agents with low toxicity and the required pharmacokinetic and bioavailability properties to be useful clinically.
As T2 and T3 bacterial secretion systems have a single shared protein component, our data suggest that compound 1 is mediating its broad spectrum of activity by targeting this protein – the secretin (Yip and Strynadka, 2006). Secretins are outer membrane-spanning proteins that are synthesized in the bacterial cytoplasm and subsequently exported across the inner membrane by the sec-dependent pathway. These proteins have a similar overall structure, i.e. secretin proteins have two major domains, which are approximately equal in length. The C-terminal domain is well conserved and believed to anchor the protein in the membrane by 10–14 transmembrane amphipathic β strains characteristic of other outer membrane proteins (Guilvout et al., 1999). In contrast, the N-terminal domain is much less conserved and believed to facilitate recognition of substrates and confer secretion specificity. These proteins associate into a highly stable oligomeric complex of 12–14 subunits in the bacterial outer membrane, which functions as an export channel for substrate secretion across this barrier, by both T2 and T3 bacterial secretion systems (Bitter, 2003). Here we present data that the amount of assembled NCs, the multi-protein secretin containing transmembrane complex of the T3S system of S. typhimuirum, in the bacterial membrane is reduced when bacterial cultures are grown in the presence of compound 1, but the amount of secretin protein, InvG, in the bacterial membrane is unaltered. These data suggest that compound 1 may mediate its effect by targeting the seretin protein, but the molecular mechanism of action of these compounds needs to be further examined.
Since the discovery of the secretion of virulence factors, investigators studying bacterial pathogenesis have investigated the feasibility of developing broad spectrum antimicrobials with the ability to specifically inhibit toxin secretion. The diversity of delivery systems for bacterial toxins, as well as the vast homo- and heterooligomeric architecture of these structures has made the practicality of this concept a daunting endeavor. It was impossible to predict that 2-imino-5-arylidene thiazolidinones could be developed that would have broad activity, but in this work a combination of an HTS screen, secondary assays and hit to lead chemistry identified these small molecules as potential broad spectrum anti-virulence agents for Gram-negative bacteria by inhibiting a diversity of bacterial secretion systems.
MATERIALS AND METHODS
Strain construction and bacterial growth conditions
Salmonella, Yersinia and Pseudomonas strains were grown in Luria-Bertani (LB) medium, while Francisella was grown in Tryptic Soy Broth supplemented with 0.1% cysteine. Motility media was made as previously described (Maloy et al., 1996; Semmler et al., 1999). For cytotoxicity assays, Salmonella strains were grown overnight and diluted 1:40 (v/v) and were grown for 3 h to induce T3S gene expression. For all experiments, unless indicated otherwise, compound 1 was used at a concentration of 380µM and compound 2 was used at a concentration of 100µm. Corresponding cultures were grown in the presence of an equal concentration of the solvent (v/v) without compound. Bacterial strains were constructed using P22HT int transduction and the λ-RED system (Datsenko and Wanner, 2000; Maloy et al., 1996). Strains used in these studies are listed in the Supplemental Material (Supplemental Table 2).
Screening Screening
Enzymatic assays
β-galactosidase and alkaline phosphatase assays were performed following protocols previously described (Maloy et al., 1996). Elastase assays were performed as described (McIver et al., 2004; Ohman et al., 1980).
Compounds
Compounds were purchased from Chem Div (compounds TTSS4, TTSS11-22, TTSS24), Biomol (compounds TTSS26, TTSS28-29), Maybridge (compounds TTSS36, TTSS39-40 and TTSS42-43), IF Labs (TTSS44-45) and Bionet (TTSS47). Compounds were checked for purity by HPLC and found to be > 90% pure. Compounds for the SAR were purchased from Akos, Aurora, Chembridge, ChemDiv, Interchim AnaLogix, Princeton Bio and Ryan as well as synthesized at the University of Washington, Seattle, Washington (Kline et al., unpublished data).
Protein chemistry
Secreted proteins were prepared as previously described for S. typhimurium (Kimbrough and Miller, 2000), Y. enterocolitica (Young and Young, 2002) and F. novicida (Hager et al., 2006). Cellular samples were derived from lysed whole cells, while total membrane fractions resulted from ultracentrifugation of this lysate to pellet the membrane. T3S NCs were isolated from S. typhimurium as previously described (Kimbrough and Miller, 2000). SDS/PAGE and Western Blot techniques were performed as described (Pegues et al., 1995).
Virulence studies
For cytotoxicity assays, bone marrow macrophages (BMMs) were plated and infected the following day. S. typhimurium was added at a multiplicity of infection (MOI) of 40, diluted in 100µl media and added to BMMs. Infections were allowed to proceed for 30 or 60 minutes at 37°C in an atmosphere of 5% CO2. Lactate dehydrogenase (LDH) activity was measured on 50µl of supernatant using the CytoTox 96 assay (Promega). Each sample was done in quadruplicate and the average values are shown. For the HR assays, P. syringae DC3000 was resuspended from plates in 100mM sucrose and 10mM MgCl2 to a final OD600 of 0.3 and diluted 1:20, 1:40, 1:60 and 1:80. Bacteria were incubated with DMSO or compound for 30 minutes at RT in the dark. Samples were injected into Nicotiana benthamiana plants and pictures taken 36 hrs post-inoculation.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank L. Hoffman and S. Sanowar for critical reading of the manuscript and the staff at the NSRB for their technical assistance with the HTS. E. E. Galyov (Division of Microbiology, Institute for Animal Health, UK) generously provided the SipA, B and C antibodies. V. Koronakis (Department of Pathology, University of Cambridge) generously provided the InvG antibodies. We would like to thank K. T. Hughes (Department of Biology, University of Utah) for providing the flhC transcriptional reporter fusion and the flgC::Tn10 insertion. This work was funded by the NIH and NIAID (U54 A105714) and the investigators are members of the Northwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bitter W. Secretins of Pseudomonas aeruginosa: large holes in the outer membrane. Arch Microbiol. 2003;179:307–314. doi: 10.1007/s00203-003-0541-8. [DOI] [PubMed] [Google Scholar]
- Cianciotto NP. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 2005;13:581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayam R, Aiello F, Deng J, Wu Y, Garofalo A, Chen X, Neamati N. Discovery of small molecule integrin alphavbeta3 antagonists as novel anticancer agents. J Med Chem. 2006;13:4243–4252. doi: 10.1021/jm051296s. [DOI] [PubMed] [Google Scholar]
- Gauthier A, Robertson ML, Lowden M, Ibarra JA, Puente JL, Finlay BB. Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrob Agents Chemother. 2005;2005:4101–4109. doi: 10.1128/AAC.49.10.4101-4109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilvout I, Hardie KR, Sauvonnet N, Pugsley AP. Genetic dissection of the outer membrane secretin PulD: are there distinct domains for multimerization and secretion specificity? J Bacteriol. 1999;181:7212–7220. doi: 10.1128/jb.181.23.7212-7220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager AJ, Bolton DL, Pelletier MR, Brittnacher MJ, Gallagher LA, Kaul R, Skerrett SJ, Miller SI, Guina T. Type IV pili-mediated secretion modulates Francisella virulence. Mol Microbiol. 2006;62:227–237. doi: 10.1111/j.1365-2958.2006.05365.x. [DOI] [PubMed] [Google Scholar]
- Hatic SO, Picking WL, Young BM, Young GM, Picking WD. Purification and characterization of two active derivatives of recombinant yplA, a secreted phospholipase from Yersinia entercolitica. Biochem Biophys Res Commun. 2002;292:463–467. doi: 10.1006/bbrc.2002.6690. [DOI] [PubMed] [Google Scholar]
- Huang HC, Schuurink R, Denny TP, Atkinson MM, Baker CJ, Yucel I, Hutcheson SW, Collmer A. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitivity response in tobacco plants. J Bacteriol. 1988;170:4748–4756. doi: 10.1128/jb.170.10.4748-4756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi AM, Nordfelth R, Uvell H, Wolf-Watz H, Elofsson M. Targetingbacterial virulence: inhibitors of type III secretion in Yersinia. Chem Biol. 2003;10:241–249. doi: 10.1016/s1074-5521(03)00046-2. [DOI] [PubMed] [Google Scholar]
- Khare RK, Srivastava MK, Singh H. Synthesis and fungicidal activity of some 5-methylene-2-[5'aryl-1',3',4'-oxa(thia)-diazol-[2'-yl]amino-4-thiazolones. Indian Journal of Chemistry. 1995;34B:168–173. [Google Scholar]
- Kimbrough TG, Miller SI. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa S. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- Levy SB, Marshall B. Antibiotic resistance worldwide: causes, challenges and responses. Nat Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- Linington RG, Robertson M, Gauthier A, Finlay BB, MacMillan JB, Molinkski TF, van Soest R, Andersen RJ. Caminosides B-D, antimicrobial glycolipids isolated from the marine sponge Caminus sphaeroconia. J Nat Prod. 2006;69:173–177. doi: 10.1021/np050192h. [DOI] [PubMed] [Google Scholar]
- Linington RG, Robertson M, Gauthier A, Finlay BB, van Soest R, Andersen RJ. Caminoside A, an antimicrobial glycolipid isolated from the marine sponge Caminus sphaeroconia. Org Lett. 2002;4:4089–4092. doi: 10.1021/ol0268337. [DOI] [PubMed] [Google Scholar]
- Maloy SR, Stewart VJ, Taylor RK. Genetic analysis of pathogenic bacteria: a laboratory manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- Manhi F, Mahoud MR. Studies on the reactivity of fused tiazole toward nucleophilic reagents; synthesis of new thiazolo-derivatives of potential antischistomsomal activity. Heteroatom Chem. 2007;16:121–131. [Google Scholar]
- Marra A. Targeting virulence for antibacterial chemotherapy. Drugs R D. 2006;7:1–16. doi: 10.2165/00126839-200607010-00001. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Young GM. Proteomic and functional analysis of the suite of Ysp proteins exported by the Ysa type III secretion system of Yersinia enterocolitica. Biovar 1B. Mol Microbiol. 2006;59:689–706. doi: 10.1111/j.1365-2958.2005.04973.x. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- McIver KS, Kessler E, Ohman DE. Identification of residues in the Pseudomonas aeruginosa elastase propeptide required for chaperone and secretion activities. Microbiology. 2004;150:3969–3977. doi: 10.1099/mic.0.27340-0. [DOI] [PubMed] [Google Scholar]
- Merja BC, Joshi AM, Parikh KA, Parikh AR. Synthesis and biological evaluation of pyrido-[1,2-1]pyrimidie an disoxazoline derivatives. Indian Journal of Chemistry. 2004;43B:909–912. [Google Scholar]
- Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. Cytoplasmic flagellin activates caspase 1-and secretion of interleukin 1 beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- Morens DM, Folkers GK, Fauci AS. The challenge of emerging and reemerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C. Antibiotics at the crossroads. Nature. 2004;431:899–902. doi: 10.1038/431899a. [DOI] [PubMed] [Google Scholar]
- Negrea A, Bjur E, Ygberg SE, Elofsson M, Wolf-Watz H, Rhen M. Salicylidene acylhydrazides that affect type III protein secretion in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2007;51:2867–2876. doi: 10.1128/AAC.00223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordfelth R, Kauppi AM, Norberg HA, Wolf-Watz H, Elofsson M. Small-molecule inhibitors specifically targeting type III secretion. Infect Immun. 2005;73:3104–3114. doi: 10.1128/IAI.73.5.3104-3114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman DE, Cryz SJ, Iglewski BH. Isolation and characterization of Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol. 1980;142:836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Lee C, Goguen J. High throughput screening for small-molecule inhibitors of type III secretion in Yersinia pestis. Exp Med Biol. 2007;603:367–375. doi: 10.1007/978-0-387-72124-8_34. [DOI] [PubMed] [Google Scholar]
- Pegues DA, Hantman MJ, Behlau I, Miller SI. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- Pulici M, Quartieri F. Traceless solid-phase synthesis of 2-amino-5-alkylidine-tiazol-4-ones. Tetrahedron Letters. 2005;46:2387–2391. [Google Scholar]
- Qin Z, Zhang J, Xu B, Chen L, Wu Y, Yang X, Shen X, Molin S, Danchin A, Jiang H, Qu D. Structure-based discovery of inhibitors of the YycG histidine kinase: new chemical leads to combat Staphylococcus epidermidis infections. BMC Microbiol. 2006;6:96. doi: 10.1186/1471-2180-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanarendar E, Afzal M, Karunakar D. Synthesis of isoxazolylpryrazolo[3,4,-d]thiazoles and isoxazolylthiazoles and their antibacterial and antifungal activity. Indian Journal of Chemistry. 2004;43B:168–173. [Google Scholar]
- Sekiya K, Ohishi M, Ogino T, Tamano K, Sasakawa C, Abe A. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc Natl Acad Sci. 2001;98:11638–11643. doi: 10.1073/pnas.191378598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmler ABT, Whitchurch CB, Mattick JS. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology. 1999;145 doi: 10.1099/00221287-145-10-2863. [DOI] [PubMed] [Google Scholar]
- Viboud GI, Bliska JB. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu Rev Microbiol. 2005;59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- Warren SM, Young GM. An amino-terminal secretion signal is required for YplA export by the Ysa, Ysc and flagellar type III secretion systems of Yersinia enterocolitica biovar 1B. J Bacteriol. 2005;187:6075–6083. doi: 10.1128/JB.187.17.6075-6083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Betts HJ, Chellas-Gery B, Hower S, Linton CN, Fields KA. Treatment of Chlamydia trachomatis with a small molecule inhibitor of Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol Microbiol. 2006;61:1543–1555. doi: 10.1111/j.1365-2958.2006.05347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wretlind B, Pavlovskis OR. Pseudomonas aeruginosa elastase and its role in pseudomonas infections. Rev Infect Dis. 1983;5:S998–S1004. doi: 10.1093/clinids/5.supplement_5.s998. [DOI] [PubMed] [Google Scholar]
- Yip CK, Kimbrough TG, Felise HB, Vuckovic M, Thomas NA, Pfuetzner RA, Frey EA, Finlay BB, Miller SI, Strynadka NCJ. Structural characterization of the molecular platform for type III secretion system assembly. Nature. 2005;435:702–707. doi: 10.1038/nature03554. [DOI] [PubMed] [Google Scholar]
- Yip CK, Strynadka NCJ. New structural insights into the bacterial type III secretion system. Trends Biochem Sci. 2006;31:223–230. doi: 10.1016/j.tibs.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Young BM, Young GM. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems in Yersinia enterocolitica. J Bacteriol. 2002;184:1324–1334. doi: 10.1128/JB.184.5.1324-1334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.