Abstract
Chromatin structure and transcription factor activity collaborate to set the transcription level of a gene. Our understanding of the relative contributions of each of these factors at a specific gene is limited. We studied the effects of an altered chromatin environment on the activity of the estrogen responsive pS2 promoter. We created stable cell lines with the pS2 promoter situated in an alternative chromatin site in addition to it being in its native site. Both promoters were estrogen responsive for estrogen receptor alpha (ERα) recruitment, but transcription was inducible only at the native site. At the recombinant site, transcription was high and constitutive. Higher histone H3 and H4 acetylation (acH3 and acH4), as well as trimethylated lysine 4 on histone H3 levels, were observed at the recombinant site compared to the native site in vehicle treated cells. Inhibition of histone deacetylases (HDACs) resulted in increased acH4, but only modest increases in acH3, ERα binding and basal transcription at the native pS2 site. Inhibiting HDACs had no effect on transcription from the recombinant site. These data suggest that highly active chromatin is not only permissive for transcription, but can override the requirement for the transcription factor at an inducible promoter.
Keywords: estrogen receptor, pS2, chromatin, histone acetylation, histone methylation, nuclear receptors, steroid receptors, flp recombination target
INTRODUCTION
Chromatin structure has been implicated as one modulator of gene expression status. For instance, the presence of nucleosomes has been linked to transcriptional repression [1–4]. Histone modifications are linked to the transcriptional activation or repression status of a gene, as well as marking chromosomal regions that constitute heterochromatin or euchromatin. Di- and tri-methylation of lysine 9 on histone H3 (H3K9me2 and H3K9me3) are usually associated with repressed genes or heterochromatin [5]. Histone hyperacetylation is usually linked to transcriptionally active genes and euchromatin [6–8]; while hypoacetylation is linked to less active genes and heterochromatin. Di-methylation of lysine 4 on histone H3 (H3K4me2) is associated with both active and inactive genes in euchromatin, but not heterochromatin [9]. On the other hand, tri-methylation of lysine 4 on histone H3 (H3K4me3) is associated with active genes [9, 10]. There are other histone modifications that can also influence gene expression, such as phosphorylation, ubiquitination and sumoylation [11, 12]. There are two types of mechanisms by which modification of histones are thought to regulate gene expression [13–16]. One is by directly altering the chromatin structure thus affecting the accessibility of transcription factors to their DNA binding sites. The second is by creating a new surface for the specific binding of proteins that regulate chromatin structure and transcription. These mechanisms are not necessarily mutually exclusive. Nucleosome assembly and histone modifications can act in concert with other factors such as transcription factor binding and DNA sequence to regulate gene expression. The association of transcriptional dysregulation with cancer underscores the importance of gene expression regulation. Epigenetic changes in the cell have been linked to a variety of malignancies [17–20]. For example, multiple cancers have been associated with alterations in histone deacetylase or histone acetyl transferase activity [19, 21]. Histone lysine methyl transferases are found to be dysregulated or overexpressed in hematopoietic malignancies and in breast cancer [19]. Another example is seen in the fact that EzH2, a methyltransferase for lysine27 on histone H3, which is involved in gene silencing, is known to be over-expressed in prostate cancer cells and in breast cancer [18, 22]. The presence and activation state of transcription factors are important in modulating the process of carcinogenesis. Many breast cancers contain estrogen receptors, which link hormonal treatment and gene expression regulation [23, 24]. The pS2 gene, which is the target of our study, is expressed in ERα positive breast cancer and is involved in tumor cell migration [25–28]. Expression of this gene has been investigated as a marker for breast cancer and is closely associated with ERα positive tumors [29].
Chromatin structure and transcription factor activity collaborate to set the transcription level of a gene. It is clear that chromatin structure dominates this collaboration in the extreme case of heterochromatin. In that case, the transcription factor binding sites in the DNA are inaccessible and transcription is not induced despite the presence of an appropriate transcription factor environment. Outside heterochromatin, chromatin can be more or less permissive for transcription and this correlates with levels of acH3 and H3K4me3 as shown in studies where viral promoter driven reporter constructs were inserted into different genomic sites [30]. In the case of inducible promoters in euchromatin sites, the transcription factors appear to dominate the chromatin structure. There are a number of examples of transcription factor binding to an inducible promoter recruiting chromatin modifying enzymes to the site resulting in increased levels of permissive histone modifications and increased transcription [31–33] This has become a model for the mechanism of transcriptional induction by steroid receptors. However, there are few mammalian systems where the effect of chromatin context on the function of a transcription factor within the same DNA sequence can be directly examined within the same cell.
This study focused on determining the relative importance of chromatin context and transcription factor (the estrogen receptor alpha (ERα)) binding in regulating the estrogen inducible pS2 gene. The pS2 promoter has an imperfect estrogen responsive element (ERE) located at 405 bp upstream of the transcriptional start site. ERα is recruited to the ERE following estradiol (E2) treatment which leads to induction of the pS2 gene [24]. We generated estrogen responsive stable cell lines that had the pS2 promoter situated both in its native chromatin site and in a second, recombinant, highly active chromatin site in the same cell. We observed that highly active chromatin structure could act as the direct opposite of heterochromatin. We see that the transcriptional activity of an inducible promoter in the highly active chromatin site was not simply permissive, but was independent of the ERα transcription factor required at the native site. We observed that even though ERα bound to the promoter at both sites in an estrogen dependent manner, the expression level of the pS2 gene in its native site was inducible with E2, whereas, it was highly constitutively active at the recombinant site. Chromatin immunoprecipitation (ChIP) analysis revealed substantially elevated levels of activating histone modifications on the promoter at the recombinant site compared to levels at the native site. Histone modification levels did not change following E2 induced ERα binding on the promoter at either site. We conclude that high levels of activating histone modifications override the function of the ERα transcription factor in regulating pS2 gene expression. This has important implications for breast cancer pathology in which the estrogen dependence of gene expression and proliferation is often lost. Accumulation of activating histone modifications in the genome, which dominate the transcription factor requirement for some genes, could contribute to pathways leading to the development of hormone-independent breast tumors.
MATERIALS AND METHODS
Cell Culture
The Flp-In™-293 parental cell line (Flp-In 293 or FlpIn 293) was purchased from Invitrogen Corp. (Carlsbad, CA). Cells were regularly maintained in phenol red-free Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 4 mM glutamine, and antibiotic-antimycotic liquid (Gibco or Cellgro) comprising 100 units/ml penicillin, 100 µg/ml streptomycin and 0.25 µg/ml amphotericin B. Other antibiotics were specific to the stable cell line under selection. Prior to performing hormone treatments, cells were maintained for 4 days (unless otherwise stated in the figure legends) in a steroid-free media containing phenol red-free DMEM supplemented with 5% dextran coated charcoal-stripped FBS, 4 mM glutamine, 1 mM pyruvate, 0.1 mM non-essential amino acids and antibiotic-antimycotic. In the steroid-free media, treatments included: 10 nM E2, 20 nM ICI 182780 (ICI), 200 nM Trichostatin A (TSA), or up to 0.1% (vol/vol) ethanol (EtOH) as a vehicle control, in various combinations. Stock solutions of E2, ICI and TSA were all in 100% EtOH. Final concentration of EtOH in cell culture was a maximum of 0.1% (vol/vol).
Constructing the pS2-SacII/FF and the Trn-pS2-sacII/FF reporters
The pcDNA5/FRT vector (Invitrogen) was modified to create the pS2-SacII/FF reporter as follows. The CMV promoter was eliminated by digesting with NheI and MluI, and blunt end ligation was performed to create pcDNA5/FRT/noCMV. A firefly (FF) luciferase coding sequence, two polyadenylation (pA) signals 5’ upstream of the luciferase and a multiple cloning site in between the luciferase sequence and the polyadenylation signal, were amplified by PCR from the plasmid pXP2 (ATCC) using platinum taq HiFi polymerase (Invitrogen). The product was subcloned into a pCR2.1 TA cloning vector (Invitrogen), transformed into INVαF’ cells (Invitrogen) and sequenced. The PCR product was recovered by digestion with ApaI and NheI, and then ligated into the pcDNA5/FRT/noCMV plasmid to obtain a pcDNA5/FRT/noCMV/2pA/FF construct. The pS2 promoter sequence was amplified by nested PCR of genomic DNA from MCF-7 cells to generate a 1.6 Kb product spanning −1587/+38 of the gene. The final PCR product was verified by sequencing. One mutation was detected and two additional mutations were introduced into the promoter to create a SacII site. However, none of these was in a region known to affect E2 dependent pS2 gene expression. The recombinant PCR product was recovered by digestion with HindIII and XhoI and then ligated into the pcDNA5/FRT/noCMV/2pA/FF construct to create the pS2-SacII/FF reporter. Generation of the Trn-pS2-sacII/FF reporter was similar to that of the pS2-SacII/FF reporter except in this case, the pS2 promoter was amplified using the pCR2.1 TA plasmid containing the 1.6kb pS2 promoter to give an ~0.6kb product from the region spanning −560/+38 of the pS2 gene. Plasmid preps were done using FastPlasmid mini kits (Eppendorf), and QIAfilter midi and maxi kits (Qiagen).
Generating stable ER27 and 27-9 cell lines
The pCLBabepuro/ERα plasmid, which encodes the human ERα gene regulated by an SV40 early promoter, as well as a puromycin resistance gene, was a gift from Dr. Elaine Alarid (University of Wisconsin). ER27 cells were generated by transfection of this construct into FlpIn 293 cells using lipofectamine 2000 (Invitrogen) and selection under puromycin and Zeocin (to ensure clones had both ERα and the FRT sequence stably integrated into the genome). Clones were also screened based on their LacZ expression status (to further confirm that the FRT was stably integrated into the genome), ERα expression status, and ability to induce E2 dependent pS2 expression from the endogenous gene at its native chromatin site. A total of 27 clones underwent some or all of the above mentioned screening process. The clone chosen as a host cell line to generate the 27-9 cell line was named the ER27 cell line. In addition to the ER27, four other clones namely, ER1, ER2, ER13 and ER26 were also used as host cell lines to generate other expression cell lines. 27-9 cells and other expression cell lines were generated by co-transfection of the flp recombinase expression vector, pOG44 (Invitrogen) together with the pS2 SacII/FF construct in a ratio of ~12.7 : 1 using the calcium phosphate transfection method. Site specific integration of the recombinant pS2 promoter regulating the firefly luciferase reporter into the FRT site of ER27 cells was promoted by expression of the flp recombinase. Correct site-specific integration into the FRT site places the hygromycin resistance gene downstream of a SV40 promoter and loss of lacZ expression. Thus, cells were selected with 50 µg/ml hygromycin. To further ensure that the pS2–SacII/FF plasmid had correctly integrated into the FRT site, a β–galactosidase assay was performed on hygromycin resistant clones to select for clones that had lost lacZ expression. A conventional PCR reaction using one primer complimentary to the integrating plasmid and the other to sequences at the FRT integration site was performed on select clones. Primers used were: F 5’ CGGATTACCAGGGA TTTCAG 3’; R 5’ GGATGGTTCGGATAATGC 3’. Candidate clones were also placed under puromycin selection to ensure maintenance of ERα expression. A total of about 29 clones were screened using some or all the above mentioned criteria. The selected cell line was named 27-9 and met all the criteria for integration into the FRT site as well as high ERα expression. The pS2 promoter in the FRT recombinant site was referred to as pS2-luc to distinguish it from the endogenous pS2 gene at its native chromatin site.
Generating truncated pS2 cell lines and null-ERα cell lines
Generation of truncated pS2 cell lines was similar to that of the 27-9 cells. For these lines though, the Trn-pS2-sacII/FF reporter was used in place of the pS2-sacII/FF. The host cell lines used were ER27 and ER1. A total of 31 clones from the ER27 host cell line and two clones from the ER1 host cell line underwent some of the screening processes used for the 27-9 cells. Nine of the clones underwent more extensive screening and all met the criteria for integration into FRT site and ERα expression. Two out of the nine clones named Trn-21 and Trn-32 (both from the ER27 host cell line) were used for this study. Generation of Null-ERα cell lines was also similar to that of the 27-9 cells. The pS2-SacII/FF reporter, used in generating the 27-9 cells was used for the Null-ERα cell line generation. However, the host cell line used in this case was the ERα negative FlpIn 293 cells. Eighteen clones were screened and the two clones used in this study, met the criteria for integration in the FRT site.
Immunoblotting
Using standard procedures, samples from whole cell extracts were subjected to Western blot analysis. Primary antibodies were HC-20 against ERα (sc-543, Santa Cruz Biotechnology) at a 1:1000 fold dilution and C-11 against actin (sc-1615, Santa Cruz Biotechnology) at a 1:1000 fold dilution. Horseradish peroxidase-linked secondary antibodies (Amersham) were donkey anti-rabbit IgG and donkey anti-goat IgG at a 1:5000 fold dilution. Detection was by enhanced chemiluminescence (Amersham Biosciences).
RNA preparation
Total RNA was harvested from cells using the RNeasy mini kit and the QIAshreddar (Qiagen) according to manufacturer's protocol.
Quantitative RT-PCR and conventional RT-PCR
cDNA synthesis from RNA samples and quantitative PCR (qPCR) analysis of gene expression was as previously described [34] with a few modifications. Human ribosomal protein L19 gene was used as the internal control. Relative gene expression was determined using the 2−ΔΔCt method [35]. The following primers were used: L19: F: 5’ AGTATGCTCA GGCTT 3’, R: 5’ GGCGATTTCATTGGTCTC 3’; pS2: F: 5’ CCCAGCACGGTGATTAGTC 3’, R 5’GTCAAAGTCAGAGCAGTCAATC 3’; firefly luciferase: F:5’GCAGCCTACCGTAGTGTTTG 3’, R: 5’ CGACTGAAATCCCTGGTAATCC 3’; lacZ-Zeocin: F: 5’CGATTACCGTTGATGTTGAAGTG 3’, R: 5’AGTTTACCCGCTCTGCTACC3’. The following primers were used for conventional RT-PCR to span the pS2-luc gene and the lacZ-Zeocin gene: F:5’GCAGCCTACCGTAGTGTTTG 3’, R: 5’AGTTTACCCGCTCTGCTACC3’
Chromatin Immunoprecipitation (ChIP) analysis
A protocol originally adapted from Weinmann and Farnham [36] and Metivier et al [24], was used for ChIP analysis with a few modifications. Briefly, cells were crosslinked with 1% formaldehyde for 10 minutes and quenched with 0.125M glycine. Cells were lysed in cell lysis buffer (85 mM KCl, 0.5% NP40, 5 mM Pipes, 15 mM sodium butyrate). Then nuclei were lysed in nuclear lysis buffer (50 mM Tris-HCl [pH 8], 10 mM EDTA, 1% SDS, 15 mM sodium butyrate). Chromatin was sonicated to an average size of approximately 500 bp. Chromatin was diluted in IP dilution buffer (final concentration of buffer components after dilution: 0.92% TritonX-100, 0.008% SDS, 1 mM EDTA, 13.9 mM Tris-HCl [pH8], 13.9 mM NaCl, 12.5 mM sodium butyrate) and precleared with protein A or A/G-sepharose beads (Santa Cruz Biotechnology). Volumes of precleared chromatin equivalent to 10%, 8.3% and 36.7% respectively of a 100 mm plate of cells were used as total input (TI), for immunoprecipitation (IP) of modified histones, or for IP of ERα. Overnight immunoprecipitations were performed followed by an additional 1.5 hour incubation with protein A/G-sepharose beads. Antibodies used were against ERα (HC-20, sc-543, Santa Cruz Biotechnology), histone H3 (ab7191, Abcam), acH3 (06599, Upstate), acH4 (06-866, Upstate), H3K4me3 (Ab-8580, Abcam), H3K27me3 (Upstate), H3K9me3 (upstate) or Pol II (H-224, sc-9001, SantaCruz Biotechnology). Following washes, samples were incubated at 67°C for approximately 4 hrs to reverse formaldehyde crosslinks. DNA was purified using Qiaquick PCR purification kit. Primers used for qPCR analysis around the ERE region on recovered DNA were: pS2: F: 5’GCCATCTCTCACTATGAATCAC3’, R: 5’CGCAGATCACCTTGTTCTC3’; pS2-luc: F: 5’GCCATCTCTCACTATGAATC AC3’, R: 5’CGGAATGCCAAGCTCAGATC3’. Primers for pS2 spanned −354/+70 of the native gene and primers for pS2-luc spanned –354/+62 of the recombinant gene. Specificity of the two sets of primers were verified by PCR using genomic DNA from a pS2-luc positive cell line (27-9) and a pS2-luc negative cell line (ER27). Primers used for qPCR analysis of the 3’distal region were: pS2: F: 5’GGTTTG GTTTCCTGTGGCATTTC3’, R: 5’CCTTGGTGAGAGCGGTTGTTC3’; pS2luc: F: 5’GGCG ATTA CCGTTGATGTTGAAG3’, R: 5’CCCTAATCCGAGCCAGTTTACC3’.
ChART PCR assay
ChART (chromatin accessibility by real time) PCR was performed [37, 38] with the following modifications. About 18 million cells were harvested and washed with PBS. To isolate nuclei, cells were resuspended in buffer FG1 from the FlexiGene DNA kit (Qiagen) supplemented with 0.15 M sucrose. Cells were incubated on ice for ~5 minutes and spun at 2500 rpm for 5 minutes to pellet nuclei. Buffer FG1 was removed and nuclei resuspended in 441 µl buffer D (150 mM sucrose, 50 mM Tris-HCl [pH7.6] and 50 mM KCl ) containing no CaCl2 as adapted from a paper by Okino and Whitlock [39]. 49 µl of the nuclei suspension was used for each Mnase digestion condition to which 1 µl of 0.05 M CaCl2 solution was added, and nuclei prewarmed at 37°C for 1.5 minutes. Then 50 µl of each micrococcal nuclease (Mnase) solution was added and incubated at 37°C for 5 minutes Final Mnase concentrations in the reactions were: 2, 16, 32, 64, 128, 256, and 512 Units/ml. The reaction was stopped by the addition of 100 µl buffer C [39] (20 mM Tris-HCl [pH8.0], 20 mM EDTA, 20 mM NaCl, 600 µg/ml proteinase K, 1% sodium dodecyl sulfate), and incubated at 37°C for at least 3 hours. DNA was purified using the QIAquick PCR purification kit (Qiagen). 3.8 % of the recovered DNA was resolved on a 1% agarose gel stained with Vistra green (Amersham), and visualized under UV light. 1.5% of the recovered DNA was used for qPCR analysis. Primers were the same as for ChIP analysis and displayed similar efficiencies. Percent residual DNA for a specific chromatin region (pS2-luc ERE or pS2 ERE) in a single experiment was determined using the following formula: 2−ΔCt × 100 where ΔCt = Average Ctdigested DNA-Average Ct2 U/ml digested DNA. We found that the qPCR amplification efficiency of control, undigested genomic DNA was much lower than that of slightly digested DNA recovered from nuclease treated nuclei. We therefore defined results from the 2 U/ml digested DNA as 100% residual DNA. Note that an equal number of nuclei were initially used for the digestion and equal percentages of the total purified DNA were subjected to amplification.
RESULTS
Generation of a stable cell line that expresses ERα and has a recombinant pS2 promoter regulating a luciferase reporter construct integrated into a unique FRT site in chromatin
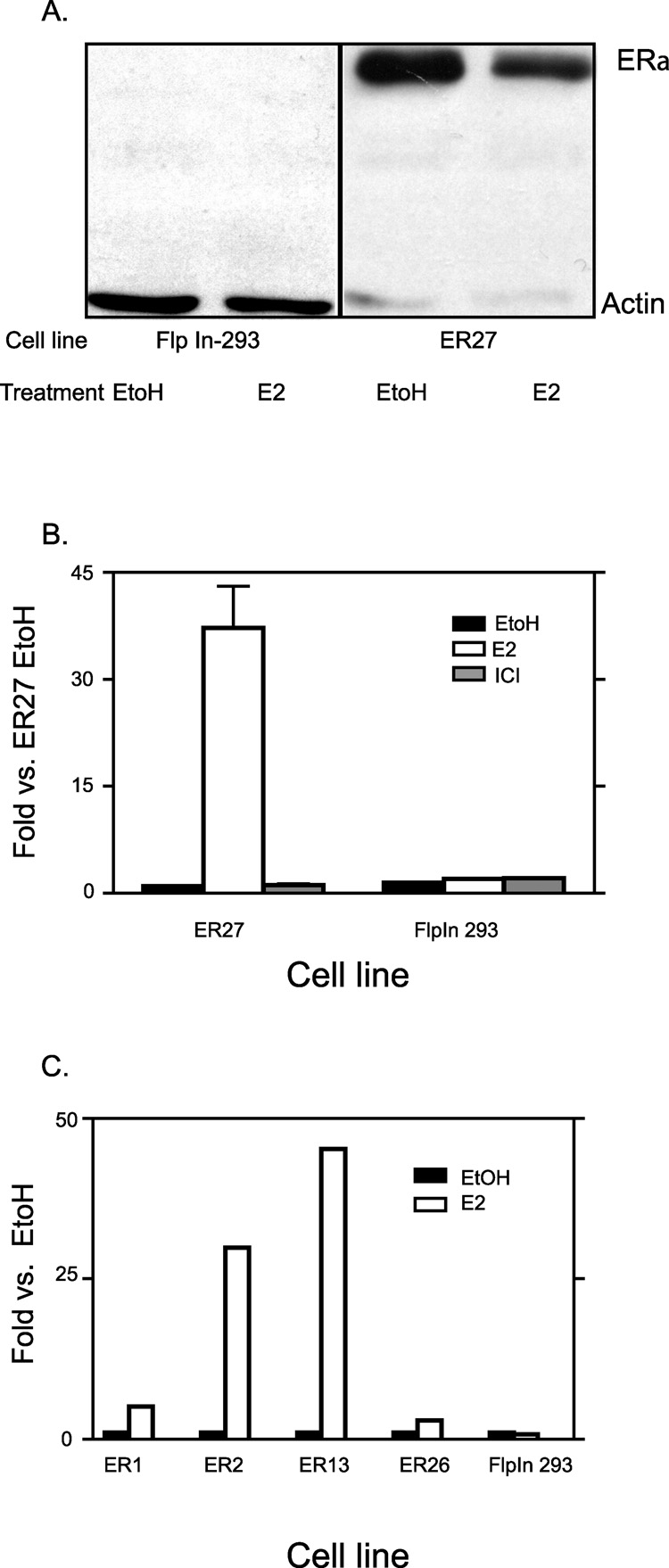
In order to directly compare the structure and activity of pS2 promoters in different chromatin sites, we wanted a cell line that had both the native pS2 gene and a second pS2 promoter stably integrated into the genome. The Flp-In™ system for generating a stable cell line has the advantage of allowing recombination of plasmid DNA (with the aid of flp recombinase) into a single, specific chromatin site as opposed to random and potentially multiple integrations [40]. Other advantages in using this methodolgy include; a site specific integration will enable controls to be performed examining other parameters such as promoter length, whilst keeping the chromatin site constant. Also, integrating a recombinant pS2 in the genome as opposed to transient transfections is more advantageous since in transient transfection, the construct will be poorly chromatinized [40].To this end we used the FlpIn 293 cell line, which has a single FRT site integrated into the genome, to generate stable cell lines. However, FlpIn 293 cells do not express ERα. Therefore, we stably expressed ERα in these cells via random integration of an expression plasmid. Twenty-seven stable lines were assayed for one or all of the following characteristics: ERα expression, E2 inducibility of the native pS2 gene, and for the retention of the FRT site (data not shown). Fig 1A shows one of the clones, ER27 expressing the ERα protein compared to no ERα protein expression in the parental FlpIn 293 cells. As shown in Fig 1B, treating ER27 cells with E2 for 24 hours increased pS2 gene expression by about 35 fold. Treating the ERα negative FlpIn 293 cells with E2 for the same time duration caused no increase in pS2 expression. The EtOH vehicle treatment was the negative control, which showed little or no pS2 expression for both ER27 and FlpIn 293 cells. ICI182780, a pure ERα antagonist that inhibits E2 induction of the pS2 gene was another negative control [41]. The ability of E2 to induce pS2 expression was not unique to ER27 cells alone but was also seen in other clones (Fig 1C). These data demonstrate that stable ERα integration and expression was sufficient and necessary to confer appropriate, estrogen-dependent transcriptional regulation on the native endogenous pS2 gene in these cells. These data are consistent with previously published reports utilizing ERα expressing 293 cells as an estrogen-responsive cell model [42, 43].
Fig. 1. Stable integration of ERα activates native endogenous pS2 gene.
ERα positive cells were generated by stably expressing ERα in FlpIn 293 cells. A, Western blot analysis was done to show ERα protein expression in ER27 cells following 24hr EtOH or E2 treatment after cells were maintained for 4 days in hormone free media. Flpn-293 cells were the negative control and Actin levels were used as the loading control. B, ER27 and FlpIn 293 cells were treated for 24 hrs with EtOH, E2 or ICI after 4 days in hormone free media. Total RNA was isolated and quantitative RT-PCR performed on the pS2 gene. Results were normalized to L19 that was used as loading control. Results were then expressed as fold change over that of EtOH treated ER27 cells. Data represent the mean +/− standard error of three independent experiments of ER27 cells and two independent experiments for FlpIn 293 cells. C, Other ERα positive clones were also treated for 24 hrs with either EtOH or E2 after at least 24 hours in hormone free media. RNA isolation, RT-qPCR and normalization was as in B. Results were expressed as fold change of respective EtOH treated clones. Data represent results from single experiments.
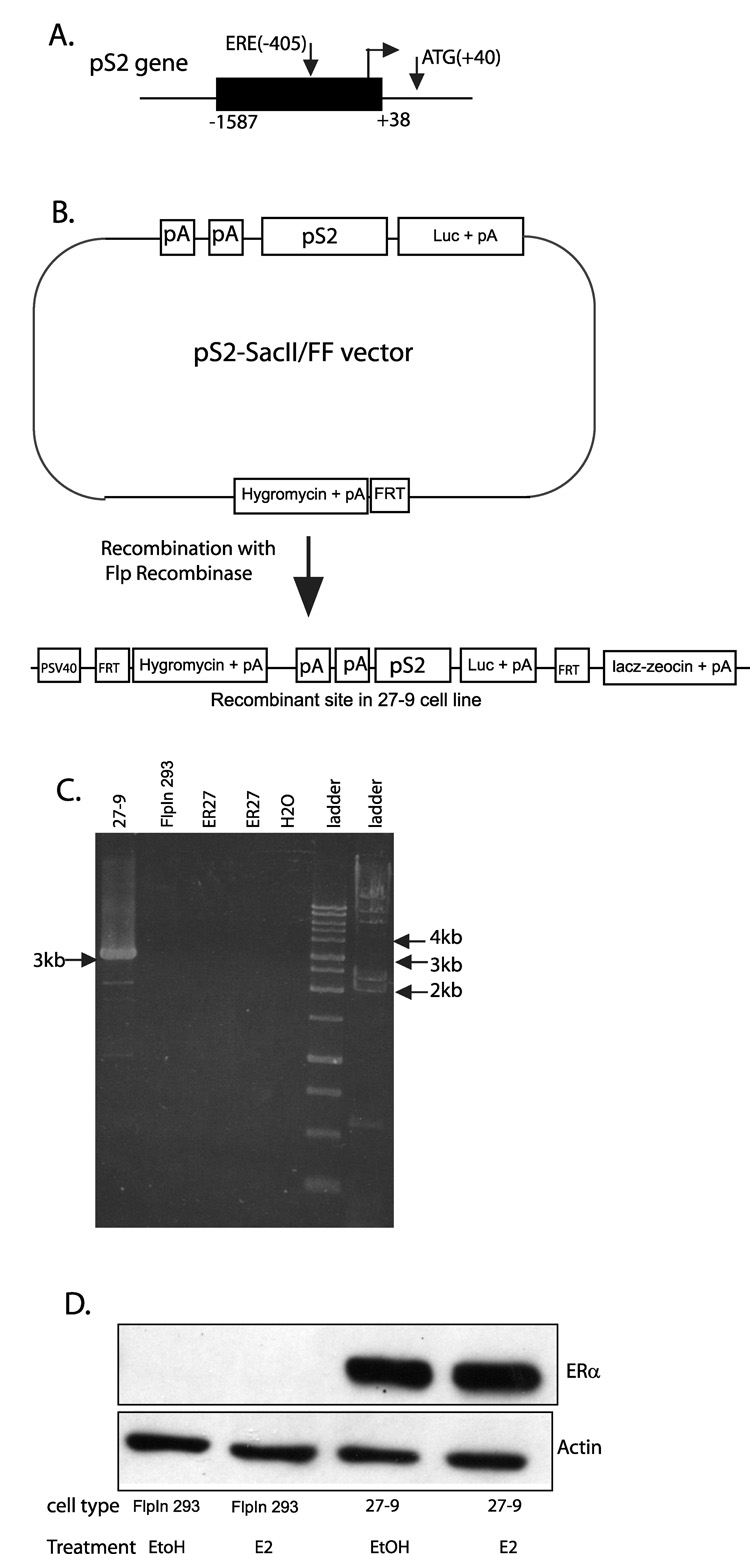
Of the clones that passed screening above, five (namely, ER1, ER2, ER13, ER26 and ER27) were chosen as host cell lines for further work. The pS2 promoter from −1587 to +38, which contains the ERE at −405 bp, was inserted upstream of a firefly luciferase gene in a vector that also contained an FRT site to generate the pS2-SacII/FF construct as diagrammed in Fig 2A and B. This construct was integrated into the FRT site of all five host cell lines by cotransfection with the flp recombinase expression vector (pOG44) to generate 29 total expression cell lines (Fig. 2B). Of the 29 cell lines, 11 were established from the ER1 host cell line, two from ER2, three from ER13, eight from ER26 and five from ER27. All clones were screened for site specific integration using the β–galactosidase assay (data not shown). Seven clones underwent further characterization and the clone named the 27-9 cell line, derived from the ER27 host was chosen for further work. Site specific integration was verified by PCR on genomic DNA to amplify a region spanning a portion of the pS2-SacII/FF construct and a portion of the ER27 genome near the FRT site. Fig 2C shows recovery of the correctly sized PCR product only from the 27-9 cell line and not from either the FlpIn 293 nor the ER27 parental cell lines. The pS2 promoter at the recombinant FRT site will be referred to as pS2-luc to distinguish it from the endogenous pS2 promoter in its native chromatin site. Real time PCR of genomic DNA comparing the pS2-luc to the pS2 gave ratios that fall within the range of a single integration (data not shown). The 27-9 cell line was shown to maintain expression of the ERα protein (Fig. 2D).
Fig. 2. Generation and characterization of 27-9 cell line.
A, Diagram of the pS2 gene. The black rectangle represents the portion of the endogenous promoter cloned into the pS2-SacII/FF vector as described in Materials & Methods. B, Schematic diagram representing the pS2-SacII/FF vector. Integration of the pS2-SacII/FF construct into the FRT site of ER27 cells produced the recombinant DNA region shown in the 27-9 cell line. C, Site specific integration of the pS2-SacII/FF construct into the FRT site verified by PCR on genomic DNA from 27-9 cells. The expected product (3 kb) spans a portion of the construct (unique to 27-9 cells) and a portion of the genome at the FRT site common to ER27 and FlpIn 293 cells (negative controls). D, Western blot showing ERα protein expressed in 27-9 cells, but not in parental FlpIn 293 cells (negative control). Cells were treated for 4hrs with 0.05% EtOH, and for an additional 2hrs with 0.1%EtOH or E2. Actin was used as a loading control. Abbreviations: pA=polyadenylation signal; luc=firefly luciferase coding sequence; FRT=Flp recombination target sequence; PSV40= SV40 early promoter.
ERα recruitment was E2 dependent on both pS2-luc and pS2 promoters
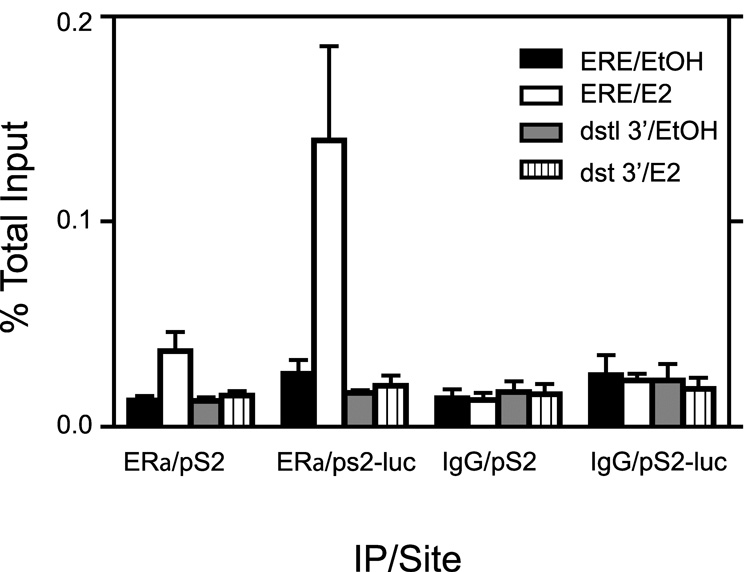
The pS2 promoter has an imperfect estrogen response element that recruits ERα in MCF7 cells following E2 induction [24, 44–46]. We used the chromatin immunoprecipitation (ChIP) assay to determine if ERα bound to both pS2 promoters in our 27-9 cell line and if binding was hormone dependent and specific to the ERE region. Results in Fig 3 show that for EtOH treated cells, ERα detection was similar to IgG controls on both promoters. This implied that in the absence of E2 treatment, there was no recruitment of ERα to either promoter. However, following two hours of E2 treatment, ERα was recruited to both promoters in the ERE region with the pS2-luc promoter showing higher ERα binding signal (5.4 fold over EtOH control) compared to the native pS2 promoter (2.9 fold over EtOH control). Results of E2 treatment for 4 hours showed a similar pattern (data not shown). E2 treatment did not result in ERα binding to the pS2 nor pS2-luc at 3’ distal regions situated 5–6kb downstream of their respective transcriptional start sites. Hence we concluded that ERα was capable of specific binding to both pS2 and pS2-luc promoters around the ERE region in a hormone dependent manner.
Fig. 3. ERα recruitment to the pS2 promoter at both native and recombinant chromatin sites is estrogen dependent and specific to the ERE region.
ChIP analysis with antibody against ERα or an IgG negative control was performed on cells treated for 2 hrs with E2 or 0.1% EtOH vehicle. Analysis by qPCR was performed using primers that amplified around the ERE region and also 5–6 kb downstream of the transcriptional start site (dstl 3’). Data represent the mean +/− standard error for three independent experiments.
Expression from the pS2 gene was E2 inducible whereas pS2-luc was constitutively active
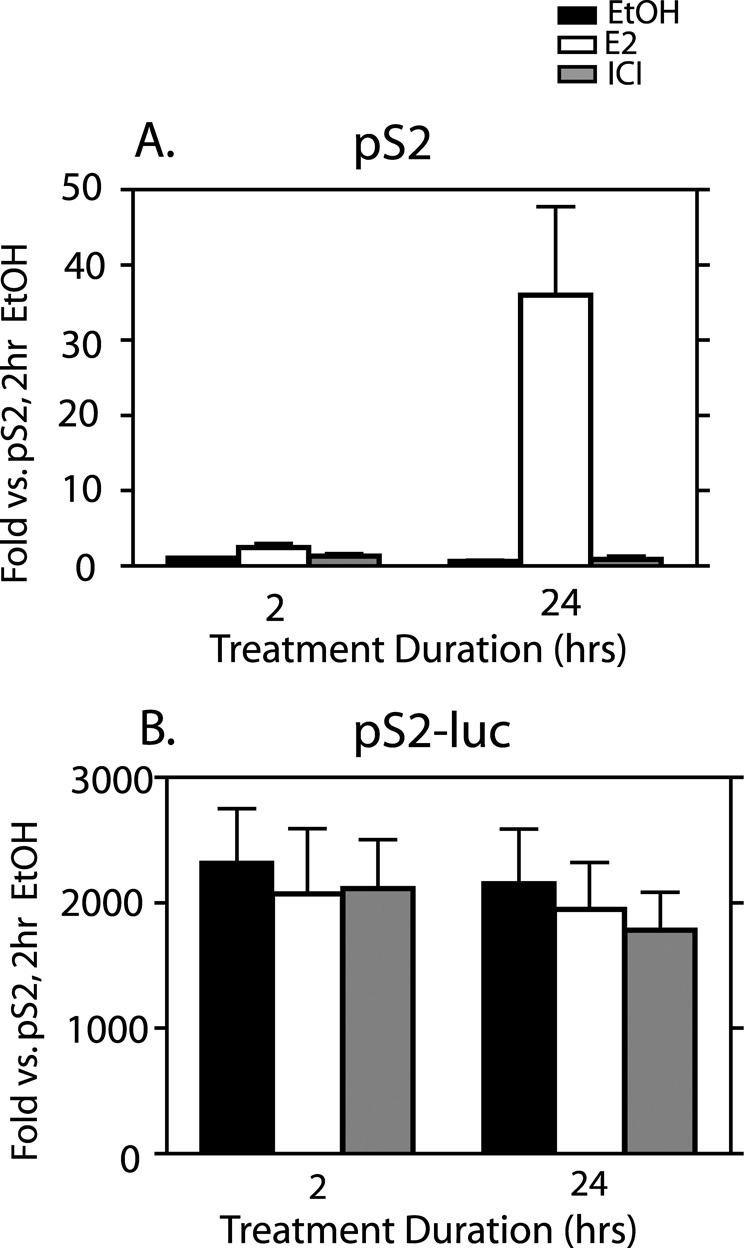
As discussed above, the pS2 gene is estrogen inducible in MCF-7 cells. We assayed for transcription of the pS2 as well as the pS2-luc genes in the 27-9 cells and for E2 inducibility of transcription at both sites by RT-qPCR. The pS2 gene showed low basal expression levels in the EtOH control treated cells and also in the presence of ICI at both 2 and 24 hrs of treatment (Fig. 4A). Treatment with E2 resulted in a 2.5-fold increase in pS2 RNA after 2 hrs of treatment and increased to 36-fold after 24 hrs. Despite the fact that pS2-luc demonstrated higher ERα binding capacity than pS2, this gene showed no E2 inducibility of RNA even after 24 hrs of treatment (Fig. 4B). Unexpectedly, we observed extremely high basal gene expression levels for pS2-luc even after 24 hr of ICI treatment (Fig. 4B). Basal levels of pS2-luc RNA were at least 2000 fold higher than basal levels of pS2 RNA (compare y-axes from Fig. 4A and Fig. 4B). This high basal level of the pS2-luc RNA was not unique to the 27-9 cells alone. Six other pS2-luc expression clones were tested, 1 derived from ER27, 3 from ER13, and 2 from ER1. All showed high, estrogen-independent expression of the pS2-luc RNA (data not shown). The 27-9 clone was the one chosen for full characterization and further work. So, whereas the pS2 in its native site showed E2 dependent transcription, the pS2-luc showed no hormone dependence, but was highly constitutively active.
Fig. 4. Differences in transcription are observed at the native pS2 compared to the recombinant pS2-luc gene.
Cells were treated for 2 or 24 hrs with 0.1% EtOH vehicle, E2, or ICI. RT-qPCR for the pS2 (A) and luciferase (B) mRNAs was performed on total RNA. Results were normalized to ribosomal protein L19 and expressed as fold changes relative to the level of pS2 with 2hr EtOH treatment. The data represent the mean +/− standard errors for three independent experiments.
Pol II binding levels higher at the pS2-luc promoter compared to the pS2 promoter in the ERE region
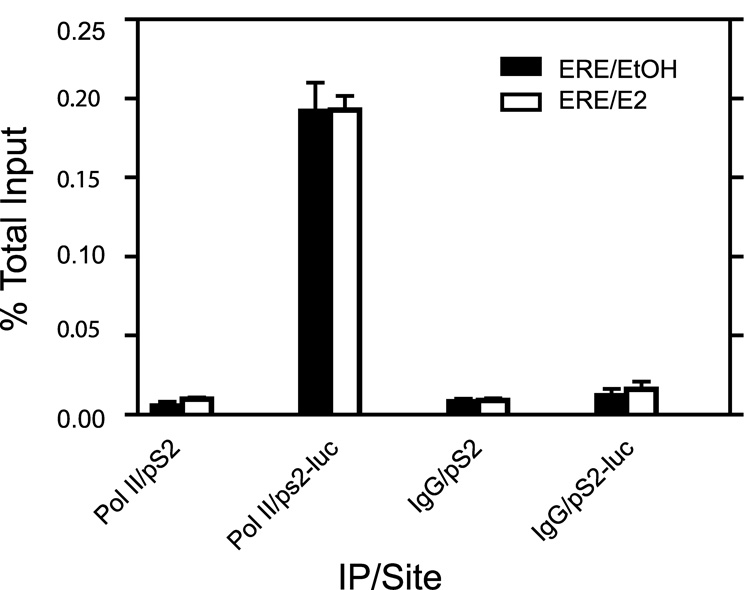
We examined Pol II binding levels at the two promoters in the ERE region to determine if binding levels correlated with the transcriptional activity observed at both sites. We found basal pol II levels to be about 36 fold higher at the pS2-luc promoter compared to the pS2 promoter (Fig 5). Levels at the pS2-luc promoter remained the same following 2 hours of E2 treatment and was about 20 fold higher than the pS2 promoter of E2 treated cells (Fig 5). We also looked at other transcription factors such as Sp1, C-jun and C-Fos, all of which have been implicated in playing various roles in regulating pS2 gene expression. There was no difference seen in levels at the pS2 and pS2-luc promoters (data not shown). Thus, we conclude the Pol II binding levels correlated with transcriptional data observed in Fig 4.
Fig. 5. Differences in Pol II recruitment levels are observed on the pS2 promoter at the recombinant site compared to the native site.
ChIP analysis with antibody against pol II or an IgG negative control was performed on cells treated for 2hrs with E2 or 0.1% EtOH vehicle. Analysis by qPCR was performed using primers that amplified around the ERE region. Data represent the mean +/− standard error for three independent experiments.
Promoter length and presence of ERα played a minor role in high pS2-luc basal transcriptional levels observed
A study by Nunez et al [47] had shown the possibility of higher constitutive activity observed with larger chunks of the pS2 promoter. In order to test the role length of the recombinant pS2 promoter played in contributing to the high basal pS2-luc transcription, we truncated the recombinant pS2 promoter by about 1kb and integrated it into the FRT site of ER27 cells to produce several clones of which Trn-21 and Trn-32 were selected for further study. We examined transcriptional activity of the pS2-luc at the recombinant site of these two clones and compared them to that of the 27-9 cells (Fig 6A). The pS2-luc basal transcriptional levels of both Trn-21 and Trn-32 cell lines were comparable to that of 27-9 cell line. Though pS2-luc expression in the Trn-32 cell line approached two fold less than that of the 27-9 cell line, it was still very high when compared to the 2000 fold increased expression of pS2-luc compared to native pS2 in 27-9 cells (Fig 4). As in the 27-9 cells, in both the Trn-21 and Trn-32 clones, 24 hour E2 treatment did not result in substantial pS2-luc induction (Fig 6A).
Fig. 6. High pS2-luc basal transcriptional levels are neither promoter length dependent nor ERα dependent.
A, Trn-21 and Trn-32 cell lines were made by inserting a 0.6kb pS2 promoter construct into the FRT site of ER27 cells. The two clones together with 27-9 cells (which had a 1.6kb pS2 promoter construct) were then treated for 24 hours with 0.1% EtOH vehicle, E2 or ICI. RT-qPCR for the luciferase mRNA was performed on total RNA for all three cell lines. Results were normalized to ribosomal protein L19 and expressed as fold changes relative to the level in EtOH treated 27-9 cells. The data represent the mean +/− standard errors for three independent experiments. B, Null-ER-99 and Null-ER-91 cell lines which both expressed no ERα but had the 1.6kb pS2 promoter construct inserted in the FRT site were fed, together with the 27-9 cells with hormone free media for 6 days. RT-qPCR for the luciferase mRNA was performed on total RNA for all three cell lines. Results were normalized to ribosomal protein L19 and expressed as fold changes relative to the levels 27-9 cells. The data represent the mean +/− standard errors for three independent experiments for Null-ER-99 cells, and four independent experiments for Null-ER-91 and 27-9 cells. .
Although the recombinant pS2-luc gene was not estrogen dependent for transcription, the possibility existed that the presence of ERα in these cells was important for setting a chromatin structure that was highly active. Despite the absence of added E2, it is possible that ERα was stimulated by other signaling pathways [48]. In order to test the effect of long term ERα activity in remodeling the recombinant site and causing high basal transcription, we generated stable cell lines with the pS2-luc in the same recombinant site using the ERα negative FlpIn 293 cells as the host cell line. We chose two of the clones, Null-ER-99 and Null-ER-91 for further work. Transcriptional activity of pS2-luc in Null-ER-99 was not much different from that of 27-9 (Fig 6B). That of Null-ER-99 was about 3 fold less than the 27-9 cells (Fig 6B) Here too, this difference is still very little compared to the fold difference between basal pS2-luc and native pS2 expression in 27-9 cells (i.e., at least 2000 fold difference). The above results demonstrate a minor role played by ERα expression as well as pS2 promoter length in the high pS2-luc basal transcriptional levels.
Chromatin accessibility at the pS2 promoter and the pS2-luc promoter, as assayed by micrococcal nuclease (Mnase) sensitivity, were similar
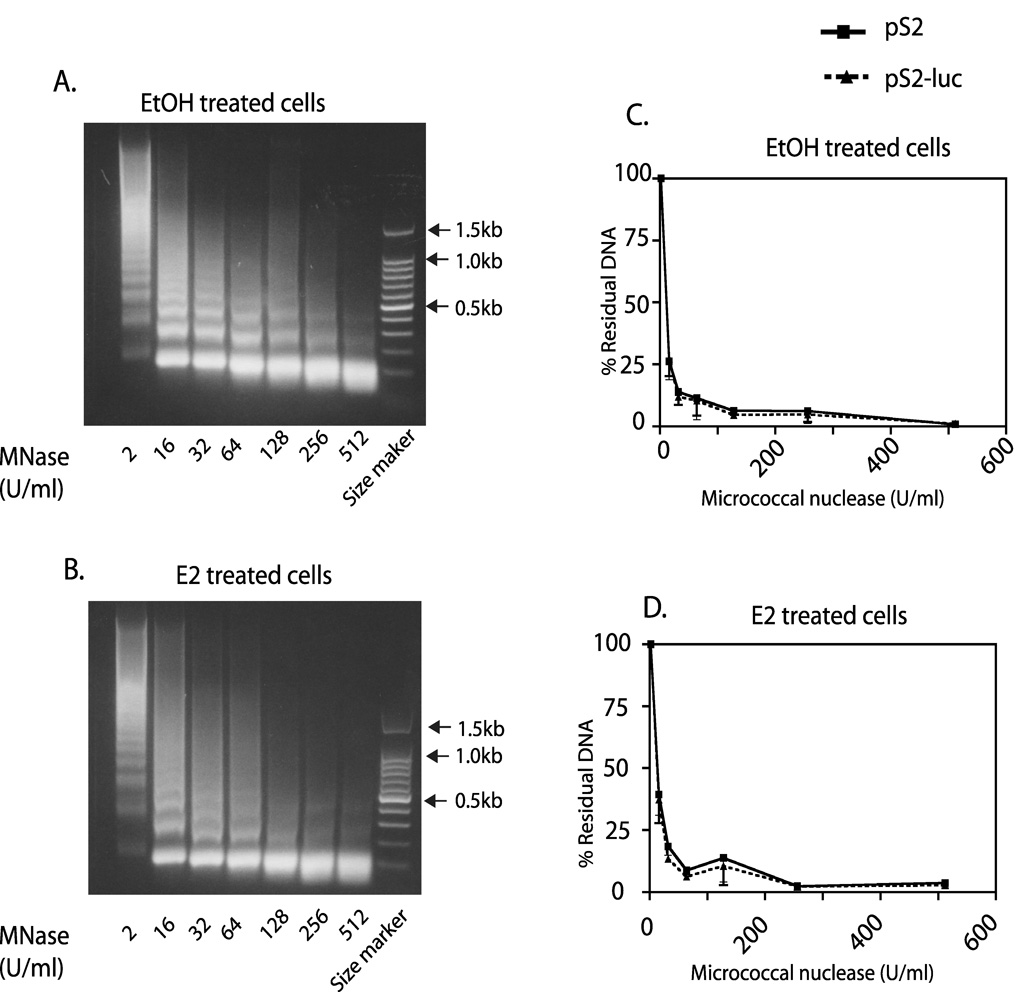
Overall chromatin structure can act as a predictor of gene expression status and reflects multiple elements including nucleosome positioning, patterns of histone modifications, the distribution of transcription factors, and even tertiary structure. The differences in transcriptional activity observed between pS2 and pS2-luc made us predict that there would be differences in the chromatin structures of the two promoters near the ERE. We tested this possibility by examining how sensitive the DNA in the two promoters was to Mnase digestion, the idea being that the pS2-luc will be more sensitive to Mnase digestion. We used a ChART PCR (chromatin accessibility by real time PCR) assay for this study [37, 38]. This assay is capable of distinguishing between chromatin regions with differential Mnase and other nuclease accessibility [37, 38]. Mnase digestion of chromatin packaged DNA in the 27-9 cell nuclei yielded DNA ladders with about 150 bp space between each band (Fig. 7A and B). Increasing Mnase concentrations led to the gradual disappearance of the larger sized DNA fragments (Fig. 7A and B). PCR primers were designed that distinguished the two promoters and encompassed the transcriptional start site close to the ERE for a region of ~424 bp for pS2 and ~416 bp for pS2-luc. Realtime PCR analysis showed that the relative degree of DNA degradation was the same for the pS2 and pS2-luc promoters in both EtOH vehicle and E2 treated cells (Fig 7C and D). This suggested a similar Mnase accessibility at both sites despite very different gene expression levels. This result demonstrated that both promoters were in an accessible chromatin structure without any major differences in nucleosome positioning, packaging, or tertiary structure. However, these results did not rule out the possibility that the levels of active histone modifications might differ.
Fig. 7. No difference is observed in MNase sensitivity of the native pS2 compared to the recombinant pS2-luc promoter.
Cells were treated for 2hrs with E2 or 0.1% EtOH vehicle. Nuclei were subjected to digestion with increasing concentrations of MNase and DNA purified as described in Experimental Procedures. 1% agarose gels showing ladders typical of total genomic DNA digested with MNase from EtOH (A) and E2 (B) treated 27-9 cells. C & D are graphs of residual native pS2 or recombinant pS2-luc promoter DNA as detected by qPCR from EtOH (C) and E2 (D) treated cells. The data are expressed as a percentage of residual DNA from the 2 U/ml digested samples. Data points are the mean +/− standard error for three independent experiments.
Differences in histone modifications in the ERE regions of the pS2 and pS2-luc promoters correlated with the gene expression levels
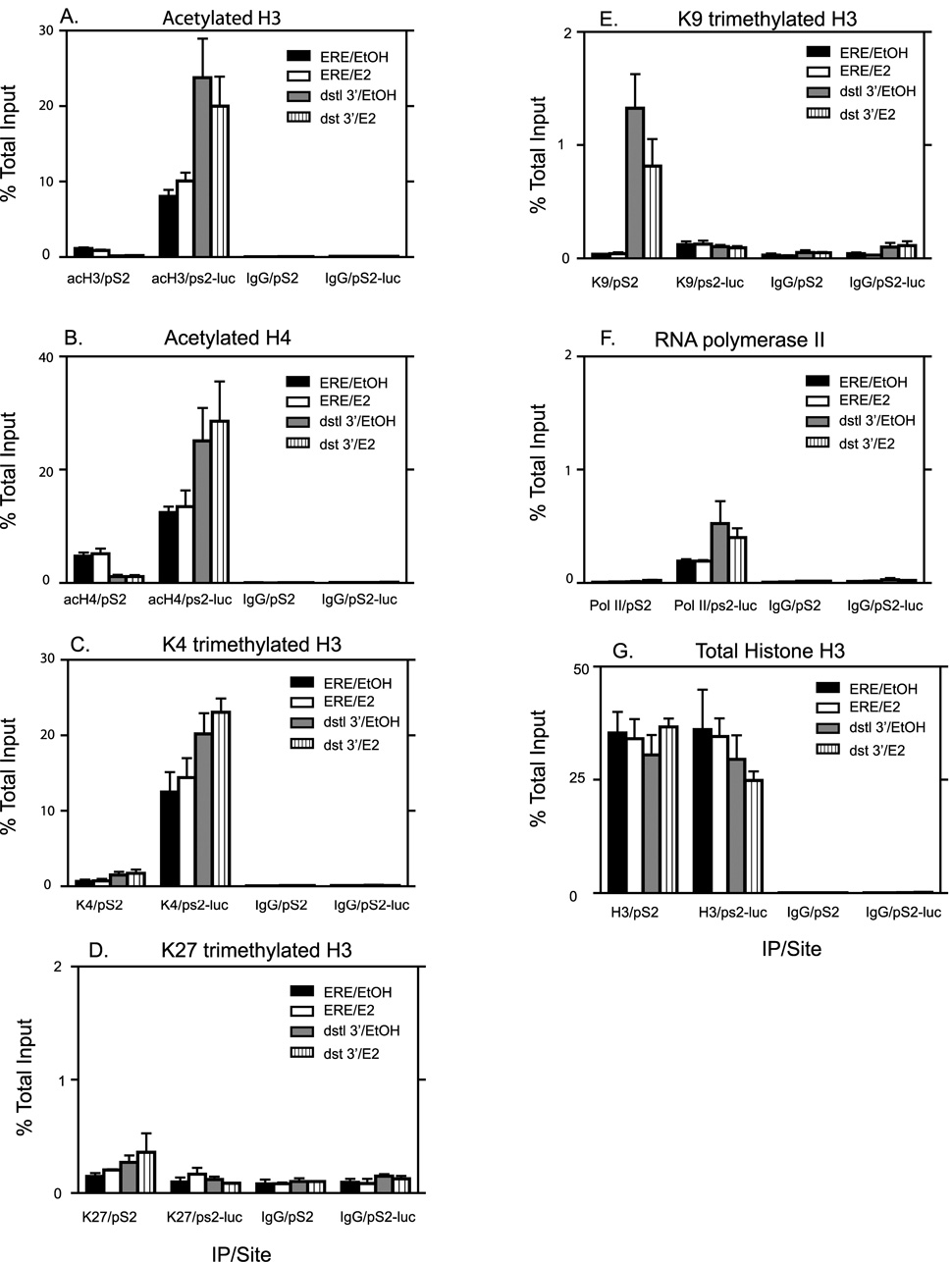
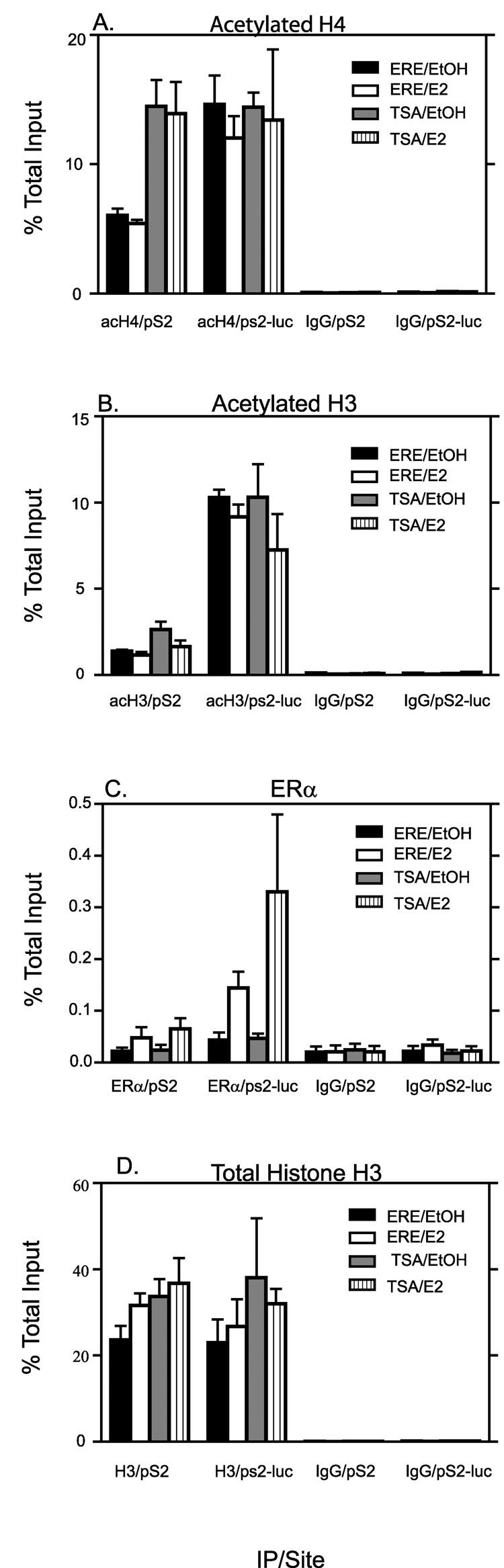
Since we detected dramatic differences in transcription from pS2 and pS2-luc, we hypothesized that one possible reason could be differences in histone modifications between the two promoters in the ERE region that could account for the differential regulation of gene expression. In order to determine if differences existed at the two chromatin sites, we used ChIP to analyze three histone modifications associated with activation. Acetylated histones H3 and H4 as well as trimethylation of lysine 4 on H3 (H3K4me3) have been implicated in transcriptional activation [11, 49, 50]. Basal levels of acH3 were much higher at the pS2-luc promoter (average of 7 fold higher) than at the pS2 promoter as seen in Fig. 8A. E2 treatment had little or no effect on acH3 levels at either the pS2 or the pS2-luc promoter. Basal acH4 levels (Fig. 8B) were also higher at the pS2-luc (average of 2.6 fold higher) compared to the native pS2 promoter though the difference was not as great as that seen for acH3. Similar to acH3, E2 treatment did not increase acH4 levels at either promoter (Fig. 8B). Similar patterns were observed for both acH3 and acH4, at both 45 minutes and 4 hours of E2 treatment as that seen for 2 hr (data not shown). The greatest difference in the level of a histone modification between the two chromatin sites was observed for H3K4me3. The pS2-luc promoter exhibited a 19 fold higher signal for H3K4me3 compared to that observed at the pS2 promoter (Fig. 8C). Here too, E2 treatment had no effect on either promoter (Fig. 8C). Repressive histone modifications such as trimethylation of lysine 27 on H3 (H3K27me3) (Fig 8D), dimethylation of lysine 9 on H3 (H3K9me2) (data not shown) as well as trimethylation of lysine 9 on H3 (H3K9me3) (Fig 8E) were also studied. Both the pS2-luc and the pS2 promoters exhibited very low levels of these three repressive histone marks comparable to IgG levels and E2 had no effect on their levels. All the above observations were not due to differences in histone density of the two sites since ChIP analysis of total Histone H3 showed similar results between pS2 and pS2-luc promoters (Fig 8G). This is consistent with the Mnase accessibility data, which also showed no difference in accessibility between the two sites (Fig 7).
Fig. 8. Comparing levels of activating and repressive histone modifications on the pS2 and pS2-luc sites in the ERE region and the distal 3’ regions of pS2 and pS2-luc promoters.
ChIP analysis for acetylated histone H3 (A), acetylated histone H4 (B), H3K4me3 (C), H3K27me3 (D), H3K9me3 (E), pol II (F) and total Histone H3 (G) with IgG as a negative control was performed on cells treated for 2 hr with E2 or 0.1% EtOH vehicle. The data represent the mean +/− standard errors for four independent experiments for A–C, two independent experiments for D, three independent experiments for E and F, and five independent experiments for G.
Differences in histone modifications in the 3’ distal regions of the pS2 and pS2-luc promoters correlated with the gene expression levels
In order to determine if the differences in histone modifications were unique to the ERE region, we looked at 3’ distal sites 5–6kb downstream for both the pS2 region and the pS2-luc region. For the pS2-luc, this region was in the lacZ-zeocin gene, which now lacked a promoter, or an ATG for initiation of protein synthesis. Hence the lacZ-Zeocin gene in pS2-luc cells did not produce protein as detailed in Materials and Methods. Whereas levels of acH3 were diminished at the 3’ distal region compared to the ERE region in the pS2 site, levels were actually elevated at the 3’distal region of the pS2-luc site compared to its ERE region (Fig 8A). Results similar to that observed for acH3 was seen also for acH4 (Fig 8B). H3k4me3 levels for the distal 3’ regions were higher at the pS2-luc compared to the pS2 although levels were similar within the same gene site. H3K27me3 levels (Fig 8D) and H3K9me2 levels (data not shown) were comparable to IgG levels. Levels of H3K9me3 were a bit elevated in the 3’distal region of the pS2 site whereas levels at the pS2-luc site were comparable to IgG levels. We also examined Pol II binding in the distal 3’ regions and found levels elevated in the pS2-luc site compared to the pS2 site (Fig 8F). Fig 8F includes PolII binding in the ERE region as shown in Fig 5 as a comparison to binding in the 3’ distal region. Total histone H3 levels were similar between gene sites and regions implying uniform histone density (Fig 8G). These results showed that activating histone modifications were not limited to the ERE region alone but spread to a wide region at the pS2-luc site, whereas these modifications were a bit more limited to the ERE region of the pS2 site. Whereas other histone repressive marks, were also undetectable in the 3’distal region of both gene sites, H3K9me3 was elevated further downstream in the pS2 site but absent in the pS2-luc site (Fig 8E).
LacZ-zeocin gene highly transcribed despite absence of a promoter, and is polycistronic to the pS2-luc
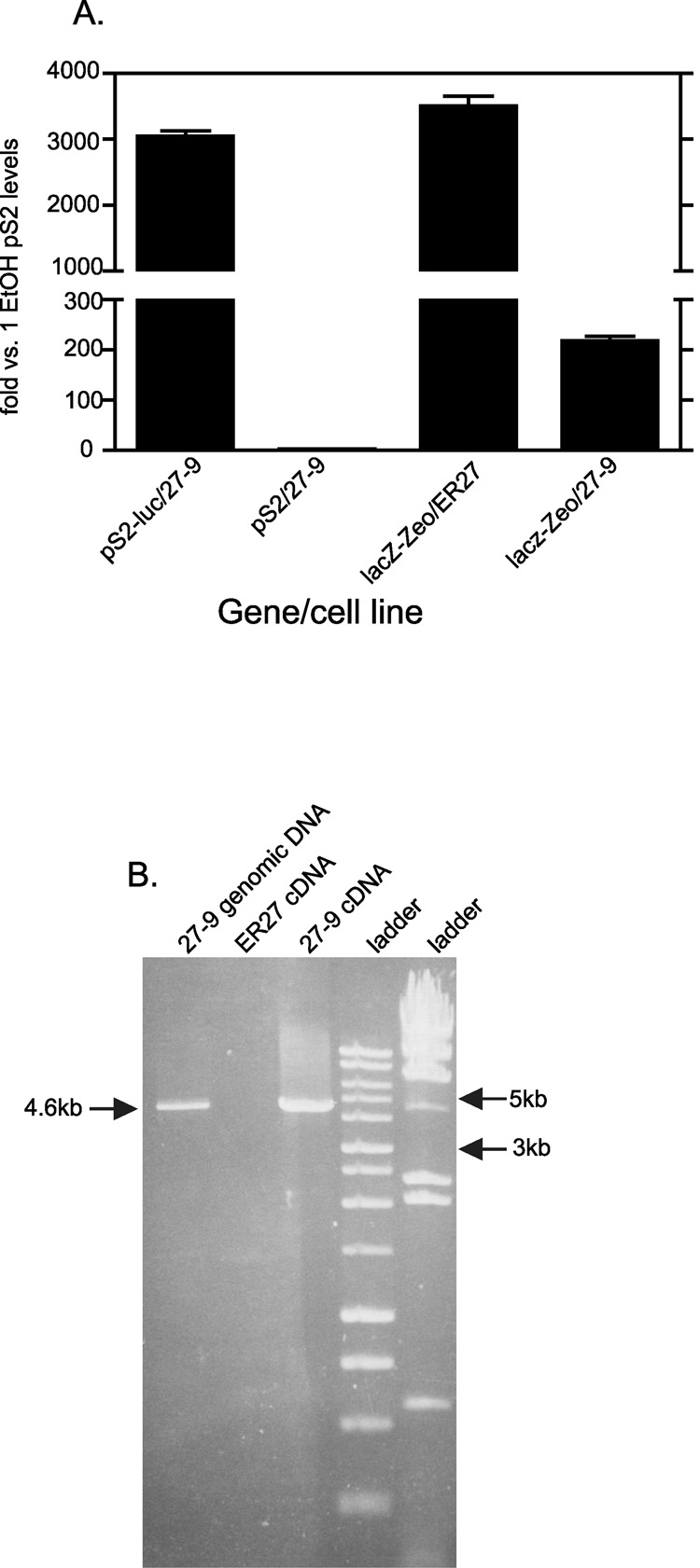
As shown in Fig 8, the distal 3’ region of pS2-luc, which encodes the promoterless lacZ-zeocin gene showed elevated levels of activating histone modifications as well as Pol II. This was the case even though it had no promoter regulating it and there was a polyadenylation signal between the closest promoter, the recombinant pS2-luc, and it. In the Flpn system, the parental cell lines, FlpIn 293 and ER27, have the lacZ-zeocin gene driven by an SV40 promoter. LacZ-zeocin transcription levels are therefore very high, and comparable to that of the pS2-luc gene in the 27-9 cells (Fig 9A). Insertion of the recombinant pS2 constructs displaces the LacZ-zeocin gene from the SV40 promoter and the ATG codon for initiation of protein synthesis. Fig 9A shows that, even though LacZ-zeocin transcription levels are diminished by 16 fold in 27-9 cells compared to ER27 cells, levels are still over 200 fold higher than the native pS2 gene in EtOH treated 27-9 cells (Fig 9A). To determine if this lacZ-zeocin gene expression is polycistronic, we used primers spanning the luciferase gene in the pS2-luc construct and that of the lacZ-zeocin gene to perform a conventional RT-PCR reaction on EtOH treated 27-9 cells. We observed a product of an expected 4.6kb size in these cells, which was absent in the negative control ER27 cells (Fig 9B). These data demonstrated that at the pS2-luc recombinant site, pol II achieved processive transcription initiating from the pS2 promoter and continuing through the firefly luciferase gene, the polyadenylation signal, and the lacZ-zeocin gene to produce the observed polycistronic message.
Fig. 9. LacZ-Zeocin gene is still highly expressed in 27-9 cells and the mRNA is polycistronic to pS2-luc.
A, 27-9 cells and ER27 cells were all treated with EtOH for 24 hours after being in hormone free media for 4 days. Total RNA was isolated and quantitative RT-PCR performed on the pS2-luc, pS2 and lacZ-Zeocin genes for 27-9 cells and lacZ-Zeocin gene for ER27 cells. Results were normalized to L19 that was used as loading control. Results were then expressed as fold change over one of the pS2 expression results of 27-9 cells. Data represent the means +/− standard errors of three experiments. B, RNA was isolated from 24 hour EtOH treated ER27 and 27-9 cells and converted to cDNA. Conventional PCR was performed with primers spanning the pS2-luc gene and the lacZ-Zeocin gene. The expected product length of recombinant region in 27-9 cells was polycistronic was ~4.6kb. 27-9 genomic DNA was used as a positive control and ER27 cDNA was used as a negative control
TSA treatment selectively increased acH4 at the pS2 promoter, but had minimal effect on transcription
As shown in Fig 8, the pS2-luc promoter had higher levels of acH3 and acH4 than the pS2 promoter did. We hypothesized that these high acetylation levels played a role in the high pS2-luc transcription even in the absence of E2. Therefore, we expected that increasing the acH3 and acH4 levels at the pS2 promoter in its native site would also result in higher constitutive transcription levels without E2. To test this hypothesis, we examined the effect of histone deacetylase (HDAC) inhibition on the pS2 promoter and gene expression. Trichostatin A (TSA) is an HDAC inhibitor and has been shown to increase global and gene specific acH3 and acH4 [51–53]. The 27-9 cells were treated with either TSA + EtOH vehicle or TSA + E2 and the ERE region of both sites were examined. Both treatments resulted in an increase in acH4 levels of the pS2 promoter comparable to that of the pS2-luc promoter (Fig. 10A). TSA did not increase the acH4 at the pS2-luc promoter (Fig. 10A). AcH3 levels showed a slight increase at the pS2 promoter for both TSA+EtOH and TSA+E2 treated cells (Fig. 10B). However, the levels were still much lower than at the pS2-luc promoter. AcH3 levels did not increase at the pS2-luc promoter with TSA treatment (Fig. 10B), neither was there a statistically significant decrease. ERα binding levels showed no statistically significant increase at the pS2 promoter following the TSA+E2 treatment compared to the E2 only treated cells (Fig. 10C). Total ERα binding was still lower than at the pS2-luc promoter in E2 treated cells. TSA+E2 seemed to result in increased ERα binding at the pS2-luc promoter when compared to the E2 only treated cells. This was, however, not statistically significant due to huge experiment to experiment variation observed (E2 treatment gave an average percent input value of 0.144 +/− 0.031, whereas TSA +E2 treatment gave an average percent input value of 0.330 +/− 0.150). For both the pS2 and the pS2-luc promoters, TSA treatment without addition of E2 (TSA+EtOH) did not show an increase in ERα binding (Fig. 10C). TSA treatment had little effect on histone density of either promoter (Fig 10D).
Fig. 10. Effect of TSA treatment on histone modifications and ERα recruitment to the native and recombinant pS2 promoters around the ERE region.
Cells were treated with TSA or EtOH vehicle for a total of 6hrs and E2 or EtOH vehicle was added to appropriate plates during the last 2hrs. The total EtOH (vehicle) concentration in all plates was 0.05% for first 4hrs, then 0.1% for final 2hrs. ChIP analysis for acetylated histone H4 (A), acetylated histone H3 (B), ERα (C) and total histone H3 (D), with IgG as the negative control, at the native pS2 or recombinant pS2-luc site was performed. All data represent means +/− standard errors of three independent experiments.
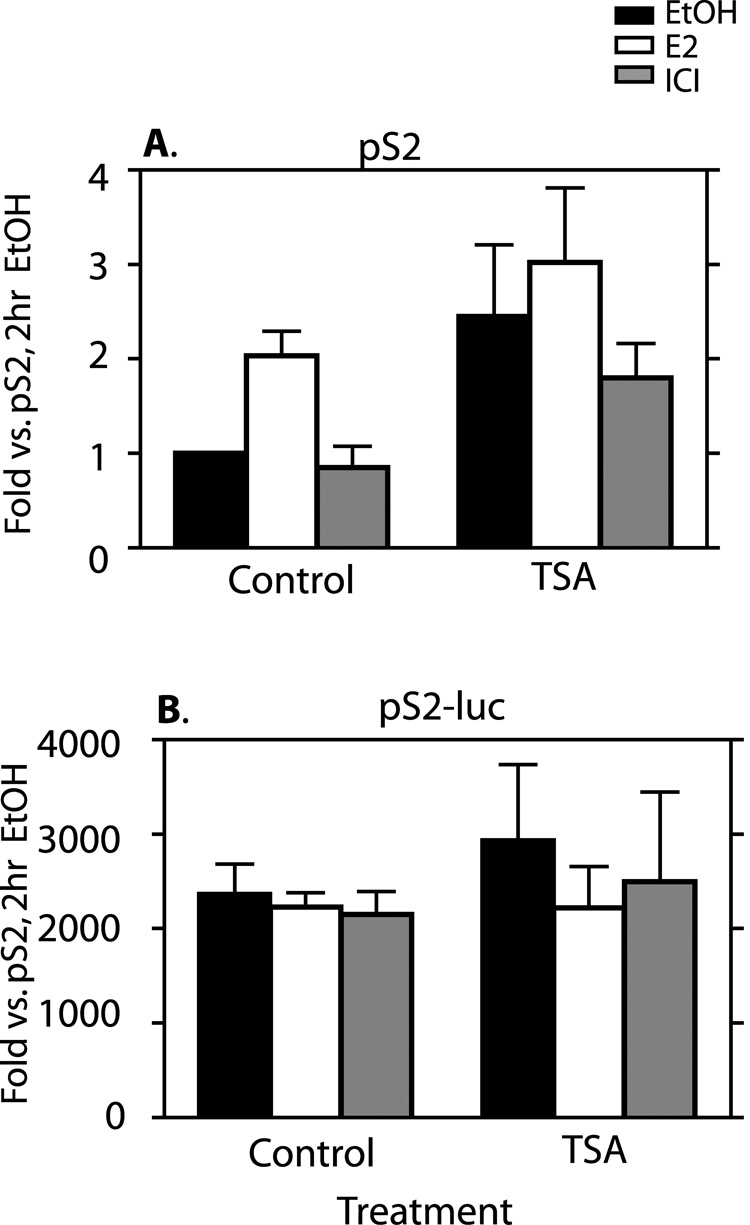
The effect of TSA on gene expression from the two promoters is shown in Fig 11. RNA accumulation from the pS2 gene showed a slight increase following TSA treatment in all cases when compared to their non-TSA treated counterparts (Fig. 11A). TSA+EtOH showed a 2.5 fold increase above EtOH only. TSA+E2 showed a 1.5 fold increase above E2 only (which was not statistically significant), and TSA+ICI showed a 2 fold increase above ICI only. Note that increased basal expression of pS2 with TSA treatment translated to a loss of E2 regulation despite generally higher overall expression (Fig 11A). The TSA treated cells still showed much lower gene expression from pS2 compared to levels observed at the pS2-luc (at least 2000 fold, Fig. 11B). No effect of TSA treatment on gene expression from the pS2-luc gene was observed (Fig. 11B).
Fig. 11. TSA treatment does not substantially alter transcription from either chromatin location.
Cells were treated as in Fig. 5. Transcriptional analysis was performed as described in Fig. 3 for mRNA from the native pS2 (A) and the recombinant pS2-luc genes (B). All data represent means +/− standard errors of three independent experiments.
DISCUSSION
Chromatin context can affect the transcriptional status of a gene [30, 54–56]. Clearly, heterochromatin is one extreme in which the chromatin structure renders the DNA binding sites for transcription factors inaccessible despite their presence in the cell. Euchromatin can show a varying level of permissiveness for transcription dependent on the level of activating histone modifications. Work with constitutive, viral promoters suggest that the strength of the promoter, which may reflect basal transcription factor binding, can overcome less permissive euchromatin and remain transcriptionally active [30]. The question of the ability of an inducible promoter to be regulated by its appropriate transcription factor in a highly permissive chromatin site is addressed in this study. We examined the relative importance of chromatin structure versus the binding of a transcription factor (ERα) at an inducible promoter on regulation of transcriptional activity in euchromatin. Our results on the E2 inducibility, transcriptional activity, and chromatin structure of the pS2 promoter in its native site compared to a recombinant site (pS2-luc) showed that chromatin marked by very high levels of activating histone modifications can override the role of transcription factor binding at an inducible promoter. We found that even though ERα was recruited to both promoters in a ligand dependent manner, transcription was E2 inducible only at the native chromatin site and essentially constitutive at the recombinant pS2-luc site. We measured activating as well as repressive histone modifications at the pS2 and pS2-luc promoters. The level of repressive marks (H3K27me3, H3K9me2, H3K9me3) was very low at both sites. Activation marks (acH3, acH4, and H3K4me3) however, were present at both sites, with basal levels of all three marks being substantially elevated at the pS2-luc promoter.
We raised histone acetylation levels by treating cells with TSA in order to assess their role. Our results suggest that levels of acH4 play a minor role at best in the different basal transcriptional activity of the two promoters since elevating acH4 at pS2 to levels similar to the pS2-luc promoter using TSA treatment had a slight effect on transcription. These results are consistent with a study reported by Yan and Boyd [30]. They showed that transgenes have higher transcriptional activity at chromatin sites expressing elevated levels of acH3 and H3K4me3. However, they saw little difference in acH4 levels between highly transcriptionally active and less active sites. However, the slight increases seen in basal transcription following TSA treatment and apparent loss of E2’s effect on transcription following 2 hours of E2 treatment suggests that elevated acH4 may not be totally eliminated as responsible for these results though elevated acH3 and H3K4me3 might be needed for very obvious effects. In both our system and that of Yan and Boyd’s [30], histone repressive marks were also less important, although, in our system, H3K9me3 was a bit elevated 5–6kb downstream of the transcriptional start site of the native pS2. In our system, the transcriptional activity was not only higher at the site with elevated levels of acH3 and H3K4me3; it was also independent of the transcription factor that is required at the native site. TSA treatment did not raise the level of acH3 at the native pS2 to the levels measured at the pS2-luc promoter, limiting the conclusions we can draw about this modification. The individual or combined high levels of acH3 and H3K4me3 may play a role in the mechanism for regulating high and constitutive transcription from pS2-luc.
E2 treatment did not induce changes in histone modifications at either the native or recombinant pS2 promoter. This is in contrast to the models that suggest nuclear receptor binding changes the histone acetylation status of a hormone responsive promoter which will, in turn, result in transcriptional induction [45, 46, 57]. Rather in the 27-9 cells, existing levels of histone acetylation and methylation appear to set the amount of ERα that can be recruited to each of the pS2 promoters upon E2 treatment. Surprisingly, increased capacity for ERα recruitment to the pS2-luc promoter does not lead to ligand dependent regulation. We did not observe induction by E2 nor repression by ICI, an ERα antagonist, at the pS2-luc site. We conclude that the chromatin context can override the effect of a bound transcription factor, in this case ERα, on transcriptional regulation. In addition, we observed a robust production of a polycistronic message from the pS2-luc promoter that spanned the luciferase gene and the lacZ-Zeocin fusion gene. This transcript transversed a polyadenylation signal between the two genes that is generally thought to promote RNA synthesis termination. The high level of activating histone modifications seen in the pS2-luc promoter were also observed in the downstream lacZ-Zeocin region. These data indicate a high level of processive transcription that overrides both transcription factor binding and polyadenylation signal elements. It has been shown that histone acetylation can facilitate binding of basal transcription factors such as TBP, TFIID or RNA pol II to a promoter [58–60]. The level of pol II binding at the pS2-luc gene and in the lacZ-Zeocin gene was elevated constitutively, consistent with the high histone acetylation and transcription observed. We speculate that the high levels of activating histone modifications at the pS2-luc promoter may allow increased binding of other, non-ligand regulated or basal transcription factors leading to high, constitutive transcription.
We observed a trend towards increase in ligand-dependent ERα binding to the pS2-luc promoter upon TSA treatment of the cells, despite no increase in histone acetylation at this site. Although the means of the individual experiments all showed an increase in ligand dependent ERα binding to the pS2-luc promoter with TSA treatment, there were huge differences in the mean values from experiment to experiment, which made a paired comparison t-test performed on these values render the overall observation as statistically insignificant. A number of studies have shown that nuclear receptors undergo acetylation [61–63]. Kim, et al in their study also showed that acetylation of ERα results in increased DNA binding [64]. We considered the possibility that the increased ERα binding seen on pS2-luc was due to increased ERα acetylation following TSA treatment. We performed western blot analysis on immunoprecipitated ERα with antibody against ERα acetylated on lysines 266/268. However, though we could detect increased acetylation of ERα from TSA (2µM) + nicotinamide (5mM) treated cells, no increase was seen with TSA (200nM) alone (data not shown). It remains possible that acetylation of ERα on another site [61] or acetylation of another protein that stabilizes ERα DNA binding is responsible for the increased ER binding seen with TSA. However, it still does not account for the differences in transcription observed between the two sites.
Several studies show that chromatin remodeling is a feature seen in regions of transcribed genes. Differences in nucleosomal mobilization for instance can sometimes be observed in an induced versus an uninduced gene [65–68]. Differences in chromatin structure can be detected by measuring sensitivity to nuclease activity. We hypothesized that one possible reason for the differences observed in transcription at the two different pS2 promoter chromatin sites was due to altered chromatin structure close to the ERE region. We reasoned that since the pS2-luc was constitutively active whereas the pS2 in its native site was essentially silent without E2 induction, pS2-luc would be more sensitive to Mnase. However, there is no difference in MNase sensitivity between the pS2 promoters at both chromatin sites in either E2 induced or uninduced cells. This suggests that altered nucleosome mobilization is not responsible for the differences in transcription seen at the two chromatin sites.
Our overall conclusion from these studies is that a highly active chromatin structure can override the requirement for ERα transcription factor on the inducible pS2 promoter leading to high levels of uncontrolled transcription. A similar dysregulation of chromatin structure at ERα regulated genes could be a pathway for the development of estrogen independence seen in breast tumors. Breast tumors acquiring hormone independence is a problem since hormonal therapy becomes ineffective. Various mechanisms have been proposed to explain the acquisition of endocrine resistance [69, 70]. Some of these include ERα phosphorylation, gene mutations, hypoxia and loss of ERα [70]. Understanding all proposed mechanisms is important in developing various new therapeutic alternatives. We hope to further elucidate the mechanisms underlying the differences seen between the two pS2 promoters in our cell line in future studies.
ACKNOWLEDGEMENTS
We thank Flavien Leclere, Ruben Alexanian, Brittany Conrad and Rebecca McDermid for technical support; and Ali Wright and Flavien Leclere for reading the manuscript and their suggestions. We also thank Dr Elaine Alarid (UW Madison) for supplying us with the pCLBabepuro/ERα plasmid and Dr. W.L. Krause (Cornell University) for generously providing us with antibody against ERα acetylated on lysines 266/268. This work was supported in part by DOD grant W81XWH-05-1-0474 to FEM, NIH DK64243 and UW Graduate School grants to MKF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Knezetic JA, Luse DS. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 2.Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 3.O'Neill TE, Roberge M, Bradbury EM. Nucleosome arrays inhibit both initiation and elongation of transcripts by bacteriophage T7 RNA polymerase. J Mol Biol. 1992;223:67–78. doi: 10.1016/0022-2836(92)90716-w. [DOI] [PubMed] [Google Scholar]
- 4.Laybourn PJ, Kadonaga JT. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 5.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 6.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nightingale KP, Wellinger RE, Sogo JM, Becker PB. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. Embo J. 1998;17:2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 13.Wolffe AP. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 14.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 15.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 16.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg AP. The epigenetics of cancer etiology. Semin Cancer Biol. 2004;14:427–432. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 19.Rice KL, Hormaeche I, Licht JD. Epigenetic regulation of normal and malignant hematopoiesis. Oncogene. 2007;26:6697–6714. doi: 10.1038/sj.onc.1210755. [DOI] [PubMed] [Google Scholar]
- 20.Weidman JR, Dolinoy DC, Murphy SK, Jirtle RL. Cancer susceptibility: epigenetic manifestation of environmental exposures. Cancer J. 2007;13:9–16. doi: 10.1097/PPO.0b013e31803c71f2. [DOI] [PubMed] [Google Scholar]
- 21.Bruserud O, Stapnes C, Tronstad KJ, Ryningen A, Anensen N, Gjertsen BT. Protein lysine acetylation in normal and leukaemic haematopoiesis: HDACs as possible therapeutic targets in adult AML. Expert Opin Ther Targets. 2006;10:51–68. doi: 10.1517/14728222.10.1.51. [DOI] [PubMed] [Google Scholar]
- 22.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manni A, Arafah B, Pearson OH. Estrogen and progesterone receptors in the prediction of response of breast cancer to endocrine therapy. Cancer. 1980;46:2838–2841. doi: 10.1002/1097-0142(19801215)46:12+<2838::aid-cncr2820461421>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 24.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 25.Prest SJ, May FE, Westley BR. The estrogen-regulated protein, TFF1, stimulates migration of human breast cancer cells. Faseb J. 2002;16:592–594. doi: 10.1096/fj.01-0498fje. [DOI] [PubMed] [Google Scholar]
- 26.Abba MC, Hu Y, Sun H, Drake JA, Gaddis S, Baggerly K, Sahin A, Aldaz CM. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6:37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Olson JA, Jr, Marks JR, Nevins JR. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tozlu S, Girault I, Vacher S, Vendrell J, Andrieu C, Spyratos F, Cohen P, Lidereau R, Bieche I. Identification of novel genes that co-cluster with estrogen receptor alpha in breast tumor biopsy specimens, using a large-scale real-time reverse transcription-PCR approach. Endocr Relat Cancer. 2006;13:1109–1120. doi: 10.1677/erc.1.01120. [DOI] [PubMed] [Google Scholar]
- 29.Lacroix M. Significance, detection and markers of disseminated breast cancer cells. Endocr Relat Cancer. 2006;13:1033–1067. doi: 10.1677/ERC-06-0001. [DOI] [PubMed] [Google Scholar]
- 30.Yan C, Boyd DD. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol Cell Biol. 2006;26:6357–6371. doi: 10.1128/MCB.00311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beato M, Klug J. Steroid hormone receptors: an update. Hum Reprod Update. 2000;6:225–236. doi: 10.1093/humupd/6.3.225. [DOI] [PubMed] [Google Scholar]
- 32.Robyr D, Wolffe P. Hormone action and chromatin remodelling. Cell Mol Life Sci. 1998;54:113–124. doi: 10.1007/s000180050130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kishimoto M, Fujiki R, Takezawa S, Sasaki Y, Nakamura T, Yamaoka K, Kitagawa H, Kato S. Nuclear receptor mediated gene regulation through chromatin remodeling and histone modifications. Endocr J. 2006;53:157–172. doi: 10.1507/endocrj.53.157. [DOI] [PubMed] [Google Scholar]
- 34.Lee ER, McCool KW, Murdoch FE, Fritsch MK. Dynamic changes in histone H3 phosphoacetylation during early embryonic stem cell differentiation are directly mediated by mitogen- and stress-activated protein kinase 1 via activation of MAPK pathways. J Biol Chem. 2006;281:21162–21172. doi: 10.1074/jbc.M602734200. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 37.Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J Immunol. 2001;167:4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- 38.Francis J, Babu DA, Deering TG, Chakrabarti SK, Garmey JC, Evans-Molina C, Taylor DG, Mirmira RG. Role of chromatin accessibility in the occupancy and transcription of the insulin gene by the pancreatic and duodenal homeobox factor 1. Mol Endocrinol. 2006;20:3133–3145. doi: 10.1210/me.2006-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okino ST, Whitlock JP., Jr Dioxin induces localized, graded changes in chromatin structure: implications for Cyp1A1 gene transcription. Mol Cell Biol. 1995;15:3714–3721. doi: 10.1128/mcb.15.7.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan C, Wang H, Aggarwal B, Boyd DD. A novel homologous recombination system to study 92 kDa type IV collagenase transcription demonstrates that the NF-kappaB motif drives the transition from a repressed to an activated state of gene expression. Faseb J. 2004;18:540–541. doi: 10.1096/fj.03-0960fje. [DOI] [PubMed] [Google Scholar]
- 41.Wiseman LR, Wakeling AE, May FE, Westley BR. Effects of the antioestrogen, ICI 164,384, on oestrogen induced RNAs in MCF-7 cells. J Steroid Biochem. 1989;33:1–6. doi: 10.1016/0022-4731(89)90349-x. [DOI] [PubMed] [Google Scholar]
- 42.Valley CC, Metivier R, Solodin NM, Fowler AM, Mashek MT, Hill L, Alarid ET. Differential regulation of estrogen-inducible proteolysis and transcription by the estrogen receptor alpha N terminus. Mol Cell Biol. 2005;25:5417–5428. doi: 10.1128/MCB.25.13.5417-5428.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preisler-Mashek MT, Solodin N, Stark BL, Tyriver MK, Alarid ET. Ligand-specific regulation of proteasome-mediated proteolysis of estrogen receptor-alpha. Am J Physiol Endocrinol Metab. 2002;282:E891–E898. doi: 10.1152/ajpendo.00353.2001. [DOI] [PubMed] [Google Scholar]
- 44.Berry M, Nunez AM, Chambon P. Estrogen-responsive element of the human pS2 gene is an imperfectly palindromic sequence. Proc Natl Acad Sci U S A. 1989;86:1218–1222. doi: 10.1073/pnas.86.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 46.Wagner S, Weber S, Kleinschmidt MA, Nagata K, Bauer UM. SET-mediated promoter hypoacetylation is a prerequisite for coactivation of the estrogen-responsive pS2 gene by PRMT1. J Biol Chem. 2006;281:27242–27250. doi: 10.1074/jbc.M605172200. [DOI] [PubMed] [Google Scholar]
- 47.Nunez AM, Berry M, Imler JL, Chambon P. The 5' flanking region of the pS2 gene contains a complex enhancer region responsive to oestrogens, epidermal growth factor, a tumour promoter (TPA), the c-Ha-ras oncoprotein and the c-jun protein. Embo J. 1989;8:823–829. doi: 10.1002/j.1460-2075.1989.tb03443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coleman KM, Smith CL. Intracellular signaling pathways: nongenomic actions of estrogens and ligand-independent activation of estrogen receptors. Front Biosci. 2001;6:D1379–D1391. doi: 10.2741/coleman. [DOI] [PubMed] [Google Scholar]
- 49.Wolffe AP, Guschin D. Review: chromatin structural features and targets that regulate transcription. J Struct Biol. 2000;129:102–122. doi: 10.1006/jsbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- 50.Norton VG, Imai BS, Yau P, Bradbury EM. Histone acetylation reduces nucleosome core particle linking number change. Cell. 1989;57:449–457. doi: 10.1016/0092-8674(89)90920-3. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 52.Gregory RI, O'Neill LP, Randall TE, Fournier C, Khosla S, Turner BM, Feil R. Inhibition of histone deacetylases alters allelic chromatin conformation at the imprinted U2af1-rs1 locus in mouse embryonic stem cells. J Biol Chem. 2002;277:11728–11734. doi: 10.1074/jbc.M105775200. [DOI] [PubMed] [Google Scholar]
- 53.McCool KW, Xu X, Singer DB, Murdoch FE, Fritsch MK. The role of histone acetylation in regulating early gene expression patterns during early embryonic stem cell differentiation. J Biol Chem. 2007;282:6696–6706. doi: 10.1074/jbc.M609519200. [DOI] [PubMed] [Google Scholar]
- 54.Wakimoto BT. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- 55.Guy LG, Kothary R, DeRepentigny Y, Delvoye N, Ellis J, Wall L. The beta-globin locus control region enhances transcription of but does not confer position-independent expression onto the lacZ gene in transgenic mice. Embo J. 1996;15:3713–3721. [PMC free article] [PubMed] [Google Scholar]
- 56.Lewis EB. The phenomenon of position effect. Adv Genet. 1950;3:73–115. doi: 10.1016/s0065-2660(08)60083-8. [DOI] [PubMed] [Google Scholar]
- 57.Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 58.Lu H, Pise-Masison CA, Fletcher TM, Schiltz RL, Nagaich AK, Radonovich M, Hager G, Cole PA, Brady JN. Acetylation of nucleosomal histones by p300 facilitates transcription from tax-responsive human T-cell leukemia virus type 1 chromatin template. Mol Cell Biol. 2002;22:4450–4462. doi: 10.1128/MCB.22.13.4450-4462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biswas D, Imbalzano AN, Eriksson P, Yu Y, Stillman DJ. Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex. Mol Cell Biol. 2004;24:8312–8321. doi: 10.1128/MCB.24.18.8312-8321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sewack GF, Ellis TW, Hansen U. Binding of TATA binding protein to a naturally positioned nucleosome is facilitated by histone acetylation. Mol Cell Biol. 2001;21:1404–1415. doi: 10.1128/MCB.21.4.1404-1415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem. 2001;276:18375–18383. doi: 10.1074/jbc.M100800200. [DOI] [PubMed] [Google Scholar]
- 62.Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275:20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- 63.Popov VM, Wang C, Shirley LA, Rosenberg A, Li S, Nevalainen M, Fu M, Pestell RG. The functional significance of nuclear receptor acetylation. Steroids. 2007;72:221–230. doi: 10.1016/j.steroids.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim MY, Woo EM, Chong YT, Homenko DR, Kraus WL. Acetylation of estrogen receptor alpha by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol Endocrinol. 2006;20:1479–1493. doi: 10.1210/me.2005-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Almer A, Rudolph H, Hinnen A, Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. Embo J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Belikov S, Gelius B, Almouzni G, Wrange O. Hormone activation induces nucleosome positioning in vivo. Embo J. 2000;19:1023–1033. doi: 10.1093/emboj/19.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reik A, Schutz G, Stewart AF. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: induction leads to a reversible disruption of nucleosomes over an enhancer. Embo J. 1991;10:2569–2576. doi: 10.1002/j.1460-2075.1991.tb07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Angelov D, Charra M, Seve M, Cote J, Khochbin S, Dimitrov S. Differential remodeling of the HIV-1 nucleosome upon transcription activators and SWI/SNF complex binding. J Mol Biol. 2000;302:315–326. doi: 10.1006/jmbi.2000.4069. [DOI] [PubMed] [Google Scholar]
- 69.Massarweh S, Schiff R. Unraveling the mechanisms of endocrine resistance in breast cancer: new therapeutic opportunities. Clin Cancer Res. 2007;13:1950–1954. doi: 10.1158/1078-0432.CCR-06-2540. [DOI] [PubMed] [Google Scholar]
- 70.Kurebayashi J. Endocrine-resistant breast cancer: underlying mechanisms and strategies for overcoming resistance. Breast Cancer. 2003;10:112–119. doi: 10.1007/BF02967635. [DOI] [PubMed] [Google Scholar]