Abstract
Viruses often use host machinery in unusual ways to execute different steps during their replication. To identify host factors critical for virus replication, we screened cDNA expression libraries for genes or gene fragments that could interfere with HIV-1 vector transduction. The DNA clone that most potently inhibited HIV-1 expression encoded the N-terminal 91 aa of the eukaryotic initiation factor 3 subunit f (N91-eIF3f). Overexpression of N91-eIF3f or full-length eIF3f drastically restricted HIV-1 replication by reducing nuclear and cytoplasmic viral mRNA levels. N91-eIF3f and eIF3f specifically targeted the 3′ long terminal repeat (3′LTR) region in the viral mRNA. We show that the 3′ end cleavage of HIV-1 mRNA precursors is specifically reduced in N91-eIF3f expressing cells. Our results suggest a role of eIF3f in mRNA maturation and that it can specifically interfere with the 3′ end processing of HIV-1 mRNAs.
Keywords: cDNA genetic screen, HIV-1 restriction, RNA maturation, translation factor
The integrated proviral DNA of HIV-1 functions as a single expression unit with a transcriptional promoter in the 5′ long terminal repeat (LTR) and signals for 3′ end processing in the 3′LTR. The 3′ ends of higher eukaryotic mRNAs are produced by a tightly coupled endonucleolytic cleavage and polyadenylation reaction. Polyadenylation regulates subsequent steps in RNA processing such as stability, nuclear export and translatability (for review, see ref. 1). In mammalian cells 3 elements define the core polyadenylation signal: a highly conserved hexanucleotide AAUAAA found 10–30 nt upstream of the cleavage site; a less well conserved U-rich or GU-rich element located downstream of the cleavage site; and the sequence at the cleavage site itself (CA), which becomes the point of poly(A) addition and is thus referred to as the poly(A) addition site (2, 3). The efficiency of RNA processing correlates with the binding of polyadenylation factors to these elements (2).
HIV-1 polyadenylation at the 3′ end of its viral RNAs, and 3′ processing of both spliced and unspliced RNAs, might be expected to involve a unique manipulation of host cleavage and polyadenylation machineries. The HIV retroviral genome contains a repeat region (R) that forms both the extreme 5′ and 3′ ends. These R regions contain several controlling functional motifs including the poly(A) site, which is part of a highly conserved hairpin structure in various HIV and SIV isolates (4). Several mechanisms have been proposed to regulate utilization of the HIV 5′ and 3′ poly(A) signals. Studies have shown in vitro and in vivo that an upstream stimulatory element (USE), located in the U3 region present only on the 3′ terminus of the viral RNA, is essential for efficient polyadenylation (5, 6).
We performed a genetic screen of a human cDNA library for factors disruptive of fundamental molecular interactions that are critical for HIV replication. Our most potent clone encoded the N-terminal 91 residues of the eukaryotic initiation factor 3 subunit f (N91-eIF3f). eIF3f is a subunit of the multisubunit eIF3 translation factor. Within the cytoplasm, eIF3f functions as a component of the multisubunit eIF3 translation factor that promotes binding of eukaryotic initiation factor 2 (eIF2), GTP, and Met-tRNAi with the 40S ribosome to form a 43S preinitiation complex (7). eIF3 consists of at least 13 nonidentical subunits (8, 9). The active core includes 5 subunits (eIF3a, -b, -c, -g, -i), with the remaining subunits serving to modulate eIF3 activity (10). eIF3f is a member of the Mov34 family, which are involved in regulation of the proteasome, translational initiation, pre-mRNA splicing, and transcription (8, 11, 12). They share a conserved (≈140-aa) domain named MPN (first observed at the N terminus of the yeast Mpr1 and Pad1p, and hence termed “MPN domain”). Given these diverse functions, it is not unexpected that eIF3f might have other roles besides its function in translation initiation, and because it is also found in the nucleus it is plausible that it might have functions in RNA metabolism.
We found that expression of N91-eIF3f or full-length eIF3f severely restricted the replication of HIV-1, by specifically targeting the 3′ end of the viral mRNA. Proviruses were formed normally upon infection but viral mRNA levels thereafter were reduced in the nucleus and cytoplasm. We found that cleavage, rather than polyadenylation, at the 3′ end of HIV mRNAs is reduced in the presence of N91-eIF3f in vitro. Additionally, in the absence of endogenous eIF3f we found that the N91-eIF3f fragment is unable to restrict the virus. Hence, we have identified an unanticipated role for eIF3f in modulating HIV RNA 3′ end processing.
Results
Human cDNA Screen for Genes or Gene Fragments Able to Restrict Genetically Marked Pseudotyped HIV Particles.
In ref. 13, we described a screen for cDNAs expressing proteins or portions of proteins capable of restricting HIV replication. Briefly, we constructed a cDNA expression library from HeLa cells using an MLV-based vector, pBabe-HAZ (14). The cDNAs of HeLa cells stimulated by IFN were expressed as N-terminal fusion proteins with the zeocin (zeo) resistance marker. The retroviral vector contains a LoxP site in the 3′ LTR, which after reverse transcription, is duplicated onto both LTRs such that the provirus can be excised from the genome by the Cre recombinase. The cDNA library was packaged into virus-like particles (VLPs) by transfection of 293T cells with DNAs encoding the cDNA library, the murine leukemia virus Gag-Pol proteins, and the pantropic envelope of the vesicular stomatitis virus (VSV-G). The recovered virus was used to infect TE671 cells, a human rhabdomyosarcoma cell line highly susceptible to HIV infection. The transduced TE671 cells were selected for zeocin resistance, ensuring expression of at least 1 member of the cDNA library. To isolate rare HIV-resistant clones, pools of transduced TE671 cells were repeatedly infected at low multiplicity with a pseudotyped retrovirus preparation expressing the herpes simplex virus (HSV) thymidine kinase (TK) gene. The virus-susceptible cells, having become HSV-TK+, were then eliminated by exposure to the nucleotide analogue ganciclovir (GCV). The rare surviving HSV-TK-negative clones were recovered as potentially HIV-resistant. A total of 1 × 105 clones were screened and 3 resistant clones were recovered; one expressing the N-terminal region of hnRNPU is reported in ref. 13. The other 2 clones, H2 and H3, were more profoundly resistant and were found to encode the same cDNA. The 3′untranslated region of these 2 clones was found to be different, demonstrating that 2 independent clones had been identified rather than siblings clones. The independent recovery of the same cDNA twice strongly demonstrates the importance of this gene fragment as having antiviral activity. Here, we describe the activity of one of the cDNA clones, dubbed H2, recovered in 1 of these 2 virus-resistant clones.
Characterization of Recovered cDNA and Its Activity.
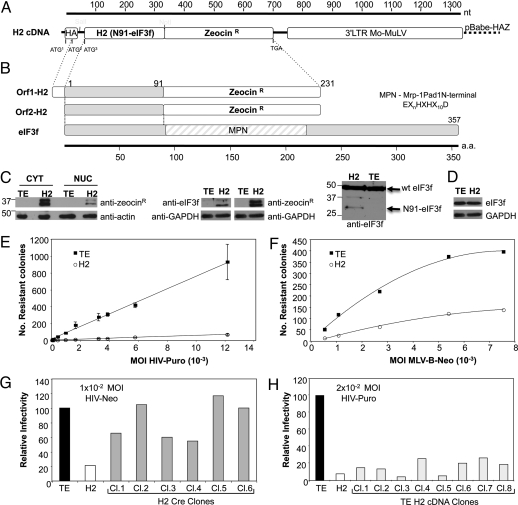
The H2 cDNA was recovered from the H2 resistant cell clone by PCR, and sequence analysis revealed an insert of 330 nt, encoding 91 aa identical to the N-terminal portion of the eukaryotic initiation factor 3 subunit p47 or f (eIF3f) (15) fused to the zeocin resistance gene. The gene product was termed N91-eIF3f (Fig. 1A). An alternative start site was located upstream from the cDNA insert, in frame with the fusion protein, encoding the identical protein but with an additional 16 aa at the N terminus (Fig. 1B). The 2 proteins were expressed at approximately equivalent amounts as determined by Western blot with antiserum detecting the Zeocin resistance protein (Fig. 1C Left) in H2 cells. TE671 cells expressing empty vector control pBabe-HAZ will for simplicity be termed the TE cell line. The protein fragments were observed both in the cytoplasm and in the nucleus of H2 cells. Endogenous eIF3f is reported to reside both in the nucleus and in the cytoplasm of cells (16). Using an antibody specific for eIF3f, only the lower molecular weight form was recognized (Fig. 1C Center), probably because of a different conformation of the longer fusion protein or masking of the recognized epitopes. A slower migrating band on these blots is a nonspecific band in all cell lysates. We have generated an antibody directed against eIF3f in rabbits. This polyclonal antibody more specifically recognizes full-length eIF3f compared with the N91-eIF3f fragment (Fig. 1C Right). It is not possible to correlate the amounts of N91-eIF3f expression to endogenous eIF3f expression by Western blot using this particular antibody. The protein levels of full-length eIF3f, the parental TE cell line, or H2 cells were comparable as judged by Western blot (Fig. 1D), suggesting that the N-terminal fragments did not affect the expression or accumulation of the endogenous protein in the H2 cell line.
Fig. 1.
Characterization of the recovered cDNA. (A) Schematic representation of the H2 cDNA fragment. H2 cDNA inserted into retroviral Mo-MuLV vector, pBabeHAZ. ATG1, start codon at influenza hemaglutinin (HA) tag epitope; ATG2, alternative start codon within HA coding sequence; ATG3 start codon for H2(N91-eIF3f)-Zeo. (B) Homology of H2 cDNA N-terminal portion with eIF3f protein. MPN domain, Mrp-1Pad1N-terminal sequence (EXnHXHX10D). (C) (Left) Western blot analyses (WB) of nuclear and cytoplasmic extracts, from empty vector control TE671 cells (TE) and H2 cellular clone, with an anti-Sh-BLE (zeocin resistance protein). Anti-actin served as a loading control. (Center) detection of both encoded H2 proteins with anti-Sh-BLE or anti-eIF3f antibodies. GAPDH was loading control. (Right) Determination of expression of N91-eIF3f and endogenous eIF3f, using anti-eIF3f antibody in H2 cells; TE cells were used as negative control. (D) Endogenous expression of eIF3f determined using anti-eIF3f antibody. GAPDH was used as a loading control. (E) TE671 cells expressing empty vector control pBabe-HAZ (TE) or H2 cells expressing N91-eIF3f were infected at the indicated multiplicities of infection with VSV-HIV-Puro. Forty-eight hours later, puromycin was added to the medium, and, 5–8 days later, drug-resistant colonies in the plates were counted after Giemsa staining. Results are representative of 3 independent experiments. Error bars represent standard error. (F) Cells tested for susceptibility to a Mo-MuLV vector, VSV-MLV-Neo virus. Transduced drug resistant colonies were scored after Giemsa staining. (G) Cre recombinase was stably introduced into H2 cells by cotransfection with p-Puro. Six puromycin resistant clones were expanded and deletion of H2-Zeo DNA was monitored by PCR. Clones were tested for resistance to VSV-HIV-Neo at a multiplicity of 1 × 10−2. (H) The H2-Zeo fragment was recovered from H2 cells, cloned into pBabe-HAZ, used to generate retroviral supernatants by transfection, and reintroduced into naïve TE671 cells. Zeocin resistant clones were tested for resistance to transduction after infection with VSV-HIV-Puro. Relative infectivity was determined by comparison to the TE cell line. Results shown are typical of those obtained in 3 independent experiments.
The H2 cell line was tested for sensitivity to HIV-based retroviral vectors by counting drug resistant colonies after infection with a reporter virus, VSV-HIV-Puro, and was found to be profoundly resistant to virus transduction. H2 cells yielded ≈30- to 100-fold fewer transductants compared with the TE cell line (Fig. 1E). The H2 cell line was somewhat less resistant to a Moloney murine leukemia virus (MLV) vector; only a 4- to 5-fold restriction was observed (Fig. 1F).
To confirm that the library derived cDNA was responsible for the observed restriction, H2 cells were transfected with a plasmid encoding the Cre recombinase to mediate excision of the cDNA insert via the surrounding LoxP sites in the vector LTRs. Clones in which Cre-induced recombination had deleted the provirus were tested for resistance to infection by VSV-HIV-Neomycin virus. These clones consistently became virus sensitive (Fig. 1G). The correlation between the presence of the insert and resistance to viral infection confirmed that the continuous presence of the cDNA was necessary for the resistance to HIV-1. The cDNA insert present in the H2 clone was recovered by PCR amplification and recloned into the pBabe-HAZ vector. After packaging, the cDNAs were introduced into naïve TE671 cells. The newly transduced cellular clones became highly resistant to infection with VSV-HIV-Puro (Fig. 1H), whereas control cells transduced with pBabe-HAZ vector (TE) alone did not. Expression of the H2 cDNA was thus both necessary and sufficient to confer resistance to HIV-1 infection.
Restriction Ability of N91-eIF3f Not Fused to Zeocin and of Full-Length eIF3f.
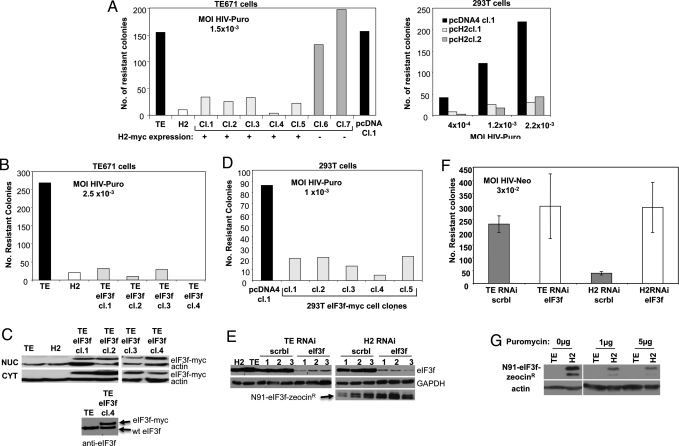
To determine whether the fusion of the N91-eIF3f ORF to the Zeocin ORF was needed to block HIV-1 transduction, TE671 or 293T cells were transfected with an expression vector for N91-eIF3f carrying only a c-myc tag (N91-eIF3f-myc) and clones stably expressing the DNA were selected. The expression of N91-eIF3f-myc in each clone was assessed by RT-PCR and the susceptibility to HIV-1 was determined by scoring for HIV-Puro transduction efficiency (Fig. 2A). The clones expressing N91-eIF3f-myc showed strong resistance to HIV-1 transduction compared with control clones expressing the empty vector or to clones whose expression of the fragment was undetectable. This result indicates that the N91-eIF3f peptide without fusion to Zeocin is sufficient for restriction. Additionally, the phenotype was observed in 2 different cell lines, TE671 and 293T, suggesting that the activity is not cell-type specific.
Fig. 2.
Expression of N91-eIF3f and eIF3f and resistance to HIV infection. (A) (Left) Susceptibility of TE671 clones expressing N91-eIF3f-myc (H2-myc) to infection as scored by HIV-Puro colony counting. Clones 1, 2, 3, 4, 5 expressed H2-myc; clones 6 and 7 did not. Clone TEpcDNAcl.1 stably expresses empty vector control. The TE and H2 cell lines were included as negative and positive controls, respectively. Cells were infected at an MOI of 1.5 × 10−3 with VSV-HIV-Puro. Forty-eight hours later, puromycin was added to the medium, and, 5–8 days later, resistant colonies in the plates were counted after Giemsa staining. (Right) N91-eIF3f-myc (H2-myc) was transfected into 293T cells, and clones were obtained and tested as for TE671. One empty vector control clone and 2 N91-eIF3f-myc expressing clones were tested for resistance, using the indicated MOIs of VSV-HIV-Puro. (B) Susceptibility of TE671 cells expressing full-length eIF3f-myc. Cellular clones were infected at an MOI of 2.5 × 10−3 with VSV-HIV-Puro, and infectivity assayed as in A. (C) (Upper) Expression of transgene protein was assessed in the nucleus and cytoplasm by WB, using an anti c-myc antibody and anti-actin antibody as a loading control. (Lower) Expression of the transgene was determined in total cellular lysates for comparison with endogenous levels of eIF3f, using anti-eIF3f antibody. (D) Full-length eIF3f-myc was stably expressed in 293T cells; eIF3f-myc expression assayed by WB. Cellular clones were tested for resistance at an MOI of 1 × 10−3 with VSV-HIV-Puro. A, B, and D are representative of as least 3 independent experiments. (E) RNAi-mediated knockdown of eIF3f and restriction to HIV mediated by N91-eIF3f. TE or H2 cells were transduced with pGIPZ-RNAi construct targeting eIF3f or a scrambled negative control. Lysates of individual clones were analyzed by WB for eIF3f, H2-zeo, or GAPDH as a loading control. TE and H2 lysates were loaded as control. (F) Clones knocked down for eIF3f or scrambled control were tested for susceptibility to infection with HIV-Neo at an MOI of 3 × 10−2. The average number of puromycin resistance colonies obtained for each set of 3 clones for 3 independent experiments is presented. Error bars represent standard error. (G) TE and H2 cells were infected with VSV-HIV-Puro and selected for viral expression, using increasing amounts of selective drug (puromycin 1 and 5 μg/mL). Detection of H2 cDNA expression by WB as selective pressure for HIV expression increases, using anti-She-BLE and actin as loading control.
The full-length eIF3f was tested for its ability to block HIV-1. TE671 cellular clones stably expressing the full-length protein fused to the c-myc epitope were created by transfection. The full-length protein was expressed both in the nucleus and in the cytoplasm in TE671 cells (Fig. 2 B and C). The TE671 cellular clones overexpressing eIF3f-myc strongly blocked HIV-Puro infection. These results suggest that overexpression of the full length eIF3f restricts HIV-1 to the same extents as its N-terminal fragment. The overexpression of eIF3f-myc in the 4 independent clones was approximately to the same level as the level of endogenous eIF3f (Fig. 2C Lower). The same results were obtained when eIF3f-myc was overexpressed in 293T cells (Fig. 2D). These results support the notion that the restriction observed is not cell type-dependent. They also show that, remarkably, a 2-fold increase in eIF3f can block virus transduction.
To determine whether N91-eIF3f-mediated restriction of HIV-1 expression acts through an involvement with the endogenous eIF3f, we asked whether RNAi-mediated knockdown of eIF3f in N91-eIF3f-expressing cells would affect viral restriction. We used retroviral vectors encoding appropriate short hairpin RNAs (shRNAs) to knockdown endogenous eIF3f expression in TE or H2 cell lines. A primer targeted to the noncoding sequence of the human eIF3f gene was used and clonal cell lines stably expressing the RNAi construct, and a scrambled vector control, were tested for both eIF3f expression and ability to restrict VSV-HIV-Neo virus in a plate assay (Fig. 2 E and F). In both TE and H2 cells, nearly complete knockdown of eIF3f was achieved (Fig. 2E). Knockdown of endogenous eIF3f had no effect on N91-eIF3f expression in H2 clones, as measured using an anti-zeocin antibody (Fig. 2E), nor did it seem to affect cell viability. The loss of endogenous expression of eIF3f in H2 cells was accompanied by a loss of restrictive HIV-1 activity (Fig. 2F), with a regain of sensitivity to infection to levels equivalent to unaltered TE cells. We noted that the sensitivity to infection of empty vector control TE cells very modestly increased as a result of eIF3f knockdown. Whether this modest increase of infectivity in TE cell is specific to HIV-1 or rather because of an overall increase in translation efficiency needs to be studied further. Nevertheless, our results indicate that N91-eIF3f alone without WT levels of eIF3f is unable to mediate restriction.
We next assessed how N91-eIF3f expressing cells responded to selection for breakthrough expression of an HIV reporter construct. After infection of H2 cells with VSV-HIV-Puro, we forced expression of the vector by culturing the cells in increasing concentrations of puromycin. We observed a loss of N91-eIF3f expression in the cell population with increasing selective pressure for the reporter (Fig. 2G). These results suggest that cells expressing higher amounts of N91-eIF3f were negatively selected in competition with those cells allowing higher HIV-1 gene expression.
Identification of Step in the HIV-1 Life Cycle Blocked by N91-eIF3f.
To determine whether the viral block mediated by N91-eIF3f occurred before or after proviral integration, H2 cells and an empty vector control line were infected with VSV-HIV-Puro virus at several different multiplicities and propagated without puromycin selection. Seven days after infection, total cellular DNA was extracted and the proviral copy number was determined by real time quantitative PCR (qPCR) (Fig. 3A). The proviral copy number in the resistant clone was not significantly different from that found in the TE cells, under the same conditions under which the number of transduced colonies was substantially reduced (Fig. 1E). These results are consistent with a viral block induced by N91-eIF3f at some time after integration of the provirus into the host genome.
Fig. 3.
Analysis of viral block in TE and H2 cell lines. (A) Clones were infected at different multiplicities of VSV-HIV-Puro virus. Total DNA was extracted 7 days after infection and copy number of integrated provirus determined by qPCR. Proviral copy number was normalized per 100 ng of total DNA. Data are representative of 3 independent experiments; error bars represent the standard error. (B) (Left) Analysis of viral mRNA expression. Cytoplasmic mRNA was extracted 7 days after VSV-HIV-Neo infection at the indicated multiplicities. First strand cDNA synthesis and amplification of the target DNA was performed by qPCR, using primers recognizing the neomycin reporter gene. Results were normalized to copies of viral mRNA per copy of GAPDH. (Right) Resistance to infection by HIV-Neo was assessed in parallel. Neomycin was added to the medium 48 h after infection, and, 5–8 days later, resistant colonies were counted after Giemsa staining. Results shown are typical of those obtained in 3 independent experiments. (C) (Left) Nuclear and cytoplasmic RNA was extracted 7 days after infection with 10-fold decreasing dilutions of VSV-HIV-TK. After first strand cDNA synthesis, TK and GAPDH cDNA was amplified by PCR. (Right) TE and H2 cells were infected at an MOI of 3 × 10−2 with HIV-Puro. Seven days after infection; nuclear RNA was extracted and quantified by qRT-PCR, using primers recognizing the puromycin reporter gene. Results are relative to values of GAPDH and the ratio found in TE cells was taken as reference. All data are representative of 3 independent experiments. Errors are standard error of the mean. HI, heat-inactivated.
To define the postintegration block, the levels of viral mRNAs present in the cytoplasm of the resistant clones were assessed after infection of TE and H2 cells with VSV-HIV-Neo. Assays of viral mRNA levels were determined by qRT-PCR, using primers directed to the neomycin resistance gene (Fig. 3B Left). The number of neomycin resistant colonies was determined in parallel (Fig. 3B Right). We observed a >10-fold reduction in the mRNA levels in the cytoplasm of H2 cells compared with cells expressing the empty vector control, corresponding well with the reduction in neomycin resistant colonies. We also evaluated the expression of viral mRNA in the nucleus of H2 and TE cells upon infection with VSV-HIV-TK. RT-PCR (Fig. 3C Left) and qRT-PCR (Fig. 3C Right) assays of nuclear RNA revealed that the levels of nuclear viral RNA in H2 cells were drastically reduced: The levels were reduced >85% in the H2 cell line compared with control. These results suggest that H2 cDNA expression prevents the accumulation of viral mRNAs in the nucleus and in the cytoplasm. The reduction of viral mRNAs was likely to be posttranscriptional, because different vectors with different promoters all exhibited the same reduction of cytoplasmic and nuclear viral RNA.
Target of N91-eIF3f Restriction.
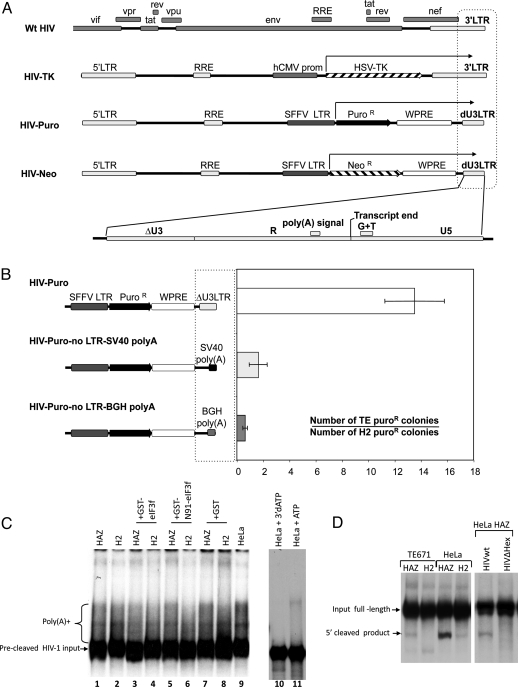
Inspection of the different retroviral vectors used as reporters of susceptibility to transduction of H2 cells identified the 3′LTR as the only common feature of the transcripts subject to negative selection (Fig. 4A). This region contains many regulatory sequences including the putative AAUAAA poly(A) signal, a G+U rich region, and the USE element, all required for efficient cleavage and polyadenylation of nascent HIV-1 transcripts (17). To test the role of the 3′LTR as a target of N91-eIF3f restriction, we replaced the 3′LTR region of the HIV-Puro vector with the simian virus 40 (SV40) or the bovine growth hormone (BGH) 3′ regions. TE and H2 cells were transfected with these DNAs, or the control expression constructs, and then scored for their ability to produce puromycin resistant colonies (Fig. 4B). An ≈14-fold specific restriction of the HIV-puro reporter was observed in H2 compared with TE cells. In contrast, parallel constructs carrying SV40 or BGH poly(A) sequences showed no restriction, and comparable reporter gene expression in both cell lines. This result shows that the HIV-1 3′LTR is the target for N91-eIF3f mediated restriction.
Fig. 4.
Viral target for H2 mediated resistance. (A) Comparison between different WT HIV and retroviral vectors gene products (arrow) restricted by H2 cells. Amplified dashed box represents the common features: the U3 deleted 3′LTR (dU3LTR), containing the poly(A) signal and a GT rich region, both defining the transcript end. RRE, Rev-responsive element; SFFV, spleen focus forming virus; WPRE, Woodchuck posttranscriptional regulatory element. (B) HIV-puro and a modified HIV-puro with SV40 poly(A) or a BGH poly(A) sequence replacing dU3LTR were transiently transfected into TE or H2 cell lines. Two days after transfection, puromycin was added to the medium, and, 5–8 days later, resistant colonies were counted after Giemsa staining. Results are shown as the ratio between the numbers of TE versus H2 resistant colonies and are representative of 3 independent experiments. Error bars represent standard error. (C) In vitro polyadenylation reaction. Poly(A) addition. The 32P-labeled precleaved RNA substrate was incubated in nuclear extracts from HeLa-CD4-pBabe-HAZ, HeLa-CD4-H2, or HeLa cells and the recombinant eIF3f-GST, N91-eIF3f-GST, and GST proteins, with ATP for 30 min at 30 °C. The RNA products were isolated and resolved on a denaturing 10% polyacrylamide gel. Lanes 10 and 11 are controls of polyadenylation. Precleaved HIV-1 input and HeLa nuclear extracts with 3′dATP or ATP respectively. (D) Poly(A) site cleavage. The 32P-labeled uncleaved 3′LTR HIV RNA substrate was incubated with nuclear extracts of TE671 or HeLa cells expressing empty vector control (HAZ) or N91-eIF3f (H2) for 30 min at 30 °C. The RNA products were isolated and resolved on a denaturing 10% polyacrylamide gel. The 3′LTR RNA substrate (HIVwt) or a substrate with a core poly(A) hexamer deletion (HIVΔHex) were incubated with HeLa HAZ extracts for 5′cleavage product control.
The drastic reduction of viral nuclear RNA in H2 cells and the involvement of the 3′LTR in the restriction hinted at the possibility that RNA precursor cleavage and/or polyadenylation might be affected by N91-eIF3f. To test whether the HIV-1 polyadenylation signal could be properly recognized in the presence of N91-eIF3f, a 32P-labeled fragment of the HIV-1 RNA precursor was synthesized with the correct 3′end for polyadenylation (Fig. S1). Nuclear extracts from HeLa cells expressing the H2 cDNA or pBabe-HAZ or control HeLa cells were prepared and incubated with the 32P-labeled precleaved RNA and its polyadenylation was monitored by gel electrophoresis (Fig. 4C). We did not observe any significant decrease in polyadenylated RNA species when nuclear extracts from cells expressing N91-eIF3f were used compared with empty vector control, suggesting that polyadenylation of a precleaved RNA from the HIV 3′LTR is not affected by N91-eIF3f.
We next assessed the specific activity of nuclear extracts from TE671 or HeLa cells expressing empty vector control or N91-eIF3f for cleavage of RNA at the site of polyadenylation. A longer (338-nt) uncleaved precursor 32P-labeled RNA was incubated with extracts in vitro, and cleavage was assessed by gel electrophoresis (Fig. 4D). When proper cleavage occurs, a 237-nt 5′ cleaved product is detected. Extracts from TE671 or HeLa cells expressing N91-eIF3f consistently produced significantly less cleavage product compared with extracts of empty vector control cells. The RNA precursor HIV 3′LTR (HIVwt) or HIV 3′LTR with a deletion in the core poly(A) hexamer (HIVΔHex) were incubated with HeLa HAZ extracts and included as a 5′ cleavage control. These results clearly show that the expression of N91-eIF3f specifically reduces the cleavage at the 3′LTR of HIV-1 preventing maturation of viral RNAs, which consequently renders them more prone to degradation, explaining the drastic reduction of HIV-1 nuclear mRNA in N91-eIF3f expressing cells.
Discussion
The results presented here show that a fragment of eukaryotic initiation factor 3 subunit f (eIF3f), or the inappropriate overexpression of the WT protein, can modulate the host cleavage and polyadenylation machinery to specifically decrease 3′end processing of HIV-1 mRNA transcripts. A cDNA encoding the N-terminal portion of eIF3f, N91-eIF3f, was recovered from a library screen in 2 independent virus-resistant clones, and was found to be necessary and sufficient to confer a severe block to HIV replication. Less efficient was the restriction observed to MLV-VLPs (4- to 5-fold). Overexpressed full-length eIF3f was also able to mediate the inhibition of HIV expression. Expression of N91-eIF3f did not alter the expression of endogenous eIF3f. However, if expression of endogenous eIF3f was knocked down by RNAi, the overexpression of N91-eIF3f was no longer sufficient to mediate restriction to the virus. This result strongly suggests that the activity of the fragment involves the contribution of the WT protein. The knockdown of eIF3f in WT cells was associated with a slight increase (≈30%) in HIV-1 expression, although it is not clear whether this is specific for HIV or rather is due to an overall increase in translation efficiency, on the concept that eIF3f might be a negative regulator of translation (18).
These results provide evidence for an effect of eIF3f on HIV gene expression. Overexpression of the N91-eIF3f fragment or the WT protein has a dominant effect to reduce HIV-1 3′ end processing. This effect seems to require the presence of endogenous amounts of eIF3f. Whether the endogenous levels of eIF3f have already a potential restrictive activity needs to be further studied. The fragment N91-eIF3f is not a natural occurring proteolytic product of eIF3f, and it does not occur naturally in vivo. Its activity was identified from a screen of an overexpression cDNA library that includes many gene fragments. However, its independent recovery twice as a restrictive element of HIV gene highlights the importance of eIF3f in 3′end processing and raises the expectation that additional components of these pathways will also be important in viral mRNA formation.
Even though we used a cDNA library of cells stimulated by the IFN to increase our chance of finding an antiviral gene, it is worth noting here that eIF3f expression is not stimulated by IFN expression as shown by WB. Thus, both constitutively expressed and induced genes can be recovered from these cell-based screens.
Experiments to probe the step of viral replication blocked by N91-eIF3f revealed a drastic reduction of retroviral transcripts in the nucleus and cytoplasm of infected cells. The loss of viral nuclear mRNA was not apparently caused by reduced efficiency of transcription initiation, because different vectors with different promoters driving the reporter gene were all affected. Moreover, expression of N91-eIF3f did not cause loss of nuclear mRNA expression of a nonretroviral construct carrying the same promoters and reporter genes. We determined that the target for N91-eIF3f-mediated restriction was the 3′LTR of the HIV pre-mRNA: replacement of this region with the BGH or SV40 poly(A) sites abolished the restriction. This result also supports the notion that a reduced level of viral mRNA is not due to reduced transcription initiation.
We studied both cleavage and polyadenylation activities of nuclear extracts from empty vector or N91-eIF3f-expressing cells. The specific cleavage activity of the HIV-1 poly(A) site by the latter extracts was significantly reduced compared with empty vector control extract. The presence of N91-eIF3f resulted in the reduction of premRNA 3′ processing. A potential insight into the mechanism of N91-eIF3f inhibition of HIV mRNA 3′ processing is provided by the observation that eIF3f is known to interact directly, via its MPN domain, with the cyclin-dependent kinase CDK11 (16); CDK11 interacts with the SR protein 9G8, and in turn, 9G8 interacts with the mammalian poly(A) site recognition factor CFIm (19). We suggest that CDK11 might serve to link eIF3f and mRNA 3′ processing. The binding of 9G8 to sequences upstream of the poly(A) site of the avian retrovirus RSV has been shown to promote mRNA 3′ processing both in vivo and in vitro (20). Intriguingly, multiple sequences related to the consensus 9G8 binding sites are present in the HIV-1 3′LTR. The interrelated functions of these various gene products in poly(A) site cleavage remain to be explored.
It is of note that the 2 cDNAs discovered as a result of this screen, N86-hnRNPU (13) and N91-eIF3f, both target the 3′LTR region of the HIV-1 mRNA. We can hypothesize why this happened: One can argue that the pressure exerted during the selection process for an antiviral gene was harsh and that we have abrogated all of the preintegration blocks. Alternatively, we can postulate that the 3′LTR region of the viral mRNA is a uniquely sensitive target for interference by host proteins. HIV-1 replication requires the 3′ processing and export of multiple alternatively spliced transcripts, and unspliced mRNA. The use of a single poly(A) site to produce both spliced and unspliced mRNAs might involve a unique manipulation of the host 3′ processing and export machinery.
Our data are the first indication that the cleavage at the 3′ end of HIV-1 mRNA is sensitive to specific manipulation by host factors with a large impact on viral production. We offer the suggestion that eIF3f is a new player in 3′end cleavage in some cellular mRNAs, and provide evidence that a protein fragment can interfere with this unanticipated function to block HIV-1 mRNA processing. Further analysis of these interactions might result in a clearer understanding of the regulation and function of eIF3f in the 3′end processing of HIV-1 pre mRNA, and manipulating these factors and their interactions may ultimately offer novel means to suppress viral replication.
Experimental Procedures
HeLa IFN cDNA Library in pBabe-HAZ.
HeLa cells were stimulated by IFN-γ (1 μg/mL) overnight and with IFN-α (1,000 units/mL) for 3 h. Construction of the library was described in ref. 13.
Production of VSV-HIV-1 Reporter Viruses.
To generate VSV HIV-HSV-TK virus, 2 μg of the retrovirus vector TRIP-TK [a generous gift from G. Gao (Chinese Academy of Sciences, Beijing)] was transiently transfected with fugene (Roche) into 293T cells, along with 1 μg of HIV-Gag-pol DNA (p8.91) and 1 μg of pMDG. The culture supernatant was collected at 48, 72, and 96 h, filtered, aliquoted and titrated on TE671 cells. To generate VSV-HIV-Neomycin and VSV-HIV-Puromycin viruses, the retroviral vectors pSCNW and pSCPW, a kind gift from G. Towers (University College of London, London), were used, respectively.
Titrations Using VSV-HIV-1 Reporter Viruses.
A total of 105 cells to be tested were seeded in 10-cm plates and the next day cells were infected with serial dilutions of VSV-pseudotyped viruses in 8 mL (total volume) of complete medium. Forty-eight hours after infection, the selective drug was added to the culture media. TE671 cells were cultured with 5 μg/mL Puromycin, 1.5 mg/mL Neomycin or 50 μg/mL Zeocin according to the viral vector used. Culture media was changed every 2 days until visible colonies were detected. Colonies were stained with Giemsa and counted.
Isolation of H2 cDNA from an H2 Cell Line.
The recovery of the cDNA insert was as described in ref. 14.
Fractionation of Nuclear and Cytoplasmic Protein Extracts.
This was performed according to Dignam protocol (21).
Western Blot Analyses.
Immunodetection of H2-zeo protein was performed by resolving proteins on a 15% SDS-polyacrylamide gel and electroblotting onto a 0.2-μm nitrocellulose membrane. A 1:3,000 dilution of an anti-Sh ble (Zeocin resistance protein) rabbit antiserum (Cayla) was incubated with the membrane followed by a secondary incubation with a 1:10,000 dilution of sheep anti-rabbit antibody conjugated to horseradish peroxidase (HRP) (Amersham). The detection of H2-myc or eIF3f-myc was performed using mouse anti-c-myc antibody (Santa Cruz Biotechnologies) at a 1:10,000 dilution followed by a secondary incubation with sheep anti-mouse HRP. Detection of endogenous eIF3f was made using an antiserum raised in rabbit, immunized with recombinant N91-eIF3f-GST. Loading controls were performed using antibodies against actin or GAPDH (Sigma).
Preparation of RNA for Analysis of Viral Block.
Cells of each cell line were infected with HIV-Puromycin or HIV-Neomycin for at least 4 h. Five to 7 days after infection, cells were trypsinized and collected for RNA preparation. The cytoplasmic and nuclear RNA fractions were separated using the RNA extraction kit (Qiagen) following the manufacturer's instruction. The nuclear pellet was washed twice with the cytoplasmic lysis buffer and the nuclear RNA was extracted as for total RNA.
Real-Time PCR.
PCRs were as described in ref. 13.
Virology.
Titration of WT HIV was done as described in ref. 13.
Protein Expression.
Full-length eIF3f or N91-eIF3f were cloned into pGEX-5X-1 (Amersham), expressed in Escherichia coli as a C-terminal fusion with GST (GST) and purified from bacterial lysates, using glutathione sepharose.
Cleavage and Polyadenylation Assays.
Preparation of RNA extracts, nuclear protein extractions, and 3′ processing reactions were performed as described in refs. 22 and 23.
Supplementary Material
Acknowledgments.
We thank Guangxia Gao (Chinese Academy of Science, Beijing), Greg Towers (University College of London, London), and Uriel Hazan (Institut Cochin de Génétique Moléculaire, Paris), for reagents and helpful discussion. This work was supported by the Howard Hughes Medical Institute (to S.T.V.), the American Foundation for AIDS Research, the Portuguese Ministry of Education, Praxis XXI. S.P.G. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900557106/DCSupplemental.
References
- 1.Wahle E, Keller W. The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 2.Weiss EA, Gilmartin GM, Nevins JR. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 1991;10:215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gil A, Proudfoot NJ. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′ end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout B, Klaver B, Das AT. A conserved hairpin structure predicted for the poly(A) signal of human and simian immunodeficiency viruses. Virology. 1995;207:276–281. doi: 10.1006/viro.1995.1077. [DOI] [PubMed] [Google Scholar]
- 5.Gilmartin GM, Fleming ES, Oetjen J. Activation of HIV-1 pre-mRNA 3′ processing in vitro requires both an upstream element and TAR. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valsamakis A, Schek N, Alwine JC. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol Cell Biol. 1992;12:3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinnebusch AG. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Hershey JW, et al. Conservation and diversity in the structure of translation initiation factor EIF3 from humans and yeast. Biochimie. 1996;78:903–907. doi: 10.1016/s0300-9084(97)86711-9. [DOI] [PubMed] [Google Scholar]
- 9.Browning KS, et al. Unified nomenclature for the subunits of eukaryotic initiation factor 3. Trends Biochem Sci. 2001;26:284. doi: 10.1016/s0968-0004(01)01825-4. [DOI] [PubMed] [Google Scholar]
- 10.Asano K, Kinzy TG, Merrick WC, Hershey JW. Conservation and diversity of eukaryotic translation initiation factor eIF3. J Biol Chem. 1997;272:1101–1109. doi: 10.1074/jbc.272.2.1101. [DOI] [PubMed] [Google Scholar]
- 11.Aravind L, Ponting CP. Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci. 1998;7:1250–1254. doi: 10.1002/pro.5560070521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grainger RJ, Beggs JD. Prp8 protein: At the heart of the spliceosome. Rna. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valente S, Goff SP. Inhibition of HIV-1 gene expression by a fragment of hnRNP U. Mol Cell. 2006;23:597–605. doi: 10.1016/j.molcel.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 14.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 15.Asano K, et al. Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J Biol Chem. 1997;272:27042–27052. doi: 10.1074/jbc.272.43.27042. [DOI] [PubMed] [Google Scholar]
- 16.Shi J, et al. The p34cdc2-related cyclin-dependent kinase 11 interacts with the p47 subunit of eukaryotic initiation factor 3 during apoptosis. J Biol Chem. 2003;278:5062–5071. doi: 10.1074/jbc.M206427200. [DOI] [PubMed] [Google Scholar]
- 17.Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- 18.Doldan A, et al. Loss of the eukaryotic initiation factor 3f in pancreatic cancer. Molecular carcinogenesis. 2008;47:235–244. doi: 10.1002/mc.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein–protein interactions, and subcellular localization. J Biol Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 20.Maciolek NL, McNally MT. Serine/arginine-rich proteins contribute to negative regulator of splicing element-stimulated polyadenylation in rous sarcoma virus. J Virol. 2007;81:11208–11217. doi: 10.1128/JVI.00919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilmartin G. M. In: mRNA formation and function. Richter JD, editor. New York.: Academic; 1997. pp. 79–98. [Google Scholar]
- 23.Gilmartin GM, Fleming ES, Oetjen J, Graveley BR. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: Multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.