Abstract
Tyrosine 411 of human albumin is an established site for covalent attachment of 10-fluoroethoxyphosphinyl-N-biotinamidopentyldecanamide (FP-biotin), diisopropylfluorophosphate, chlorpyrifos oxon, soman, sarin, and dichlorvos. This work investigated the hypothesis that other residues in albumin could be modified by organophosphorus agents (OP). Human plasma was aggressively treated with FP-biotin; plasma proteins were separated into high and low abundant portions using a proteome partitioning antibody kit, and the proteins were digested with trypsin. The FP-biotinylated tryptic peptides were isolated by binding to monomeric avidin beads. The major sites of covalent attachment identified by mass spectrometry were Y138, Y148, Y401, Y411, Y452, S232, and S287 of human albumin. Prolonged treatment of pure human albumin with chlorpyrifos oxon labeled Y138, Y150, Y161, Y401, Y411, and Y452. To identify the most reactive residue, albumin was treated for 2 h with DFP, FP-biotin, chlorpyrifos oxon, or soman, digested with trypsin or pepsin, and analyzed by mass spectrometry. The most reactive residue was always Tyr 411. Diethoxyphosphate-labeled Tyr 411 was stable for months at pH 7.4. These results will be useful in the development of specific antibodies to detect OP exposure and to engineer albumin for use as an OP scavenger.
Introduction
Organophosphorus agents are used in agriculture as pesticides and are stocked by the military as chemical warfare agents. These chemicals are toxic to insects, fish, birds, and mammals. Seizures, respiratory arrest, and death are explained by a cascade of reactions that begins with inhibition of acetylcholinesterase. Although acetylcholinesterase in red blood cells and butyrylcholinesterase in plasma are established biomarkers of organophosphorus ester (OP)1 exposure, additional biomarkers are being sought. Albumin has the potential to serve as a new biomarker of OP exposure (1,2). Albumin has been reported to covalently bind diisopropylfluorophosphate (DFP), sarin, soman, cyclosarin, tabun, 10-fluoroethoxyphosphinyl-N-biotinamido pentyldecanamide (FP-biotin), chlorpyrifos oxon (CPO), and dichlorvos (2−6) and to hydrolyze CPO, paraoxon (7,8), and O-hexyl O-2,5-dichlorophenyl phosphoramidate (9). Mass spectrometry has identified tyrosine 411 of human albumin as the site for covalent attachment of OP nerve agents and OP pesticides (6). A second site for covalent attachment of soman was suggested by experiments that found more fluoride ion released than could be accounted for by one site in albumin (3). Pretreatment of albumin with decanoate, a lipid that binds to the Tyr 411 subdomain, inhibited incorporation of 91% of 3H-DFP, leaving open the possibility that 9% of the 3H-DFP bound to other sites (10). Crystallization trials of CPO-labeled human albumin yielded gelatinous soft amorphous crystals, further suggesting the likelihood that more than one site was labeled and that labeling was not uniform. The goal of this study was to determine if sites in addition to Tyr 411 could make a covalent bond with OP and to identify the labeled residues.
Our starting premise was that OP bound exclusively to Tyr 411 of human albumin. For certain studies, we wanted 100% labeling on Tyr 411. Therefore, we treated albumin with excess CPO. The sample was checked by mass spectrometry to confirm the site of OP labeling, and to our surprise, we found several CPO-labeled peptides.
Our studies with human plasma were initiated with the goal of identifying OP-labeled proteins in human plasma. We had expected to identify several FP-biotin-labeled proteins. However, we found only FP-biotin-labeled albumin. The albumin was covalently modified on five tyrosines and two serines. FP-biotin was used for studies with plasma because biotinylated peptides are readily purified by binding to immobilized avidin beads (1,11).
Experimental Procedures
Materials
FP-biotin (MW 592.32) was custom synthesized in the laboratory of Dr. Charles M. Thompson at the University of Montana (Missoula, MT) (12). FP-biotin was dissolved in methanol and stored at −80 °C. CPO (ChemService Inc. West Chester, PA; MET-674B) was dissolved in ethanol and stored at −80 °C. DFP, a liquid with a concentration of 5.73 M, was from Sigma (D0879). Soman from CEB (Vert-le-Petit, France) was dissolved in isopropanol. A Proteome Partitioning Kit, ProteomeLab IgY-12 High Capacity in Spin Column format contained IgY antibodies directed against the 12 most abundant proteins in human plasma (Beckman Coulter #A24331 S0510903) including albumin, IgG, fibrinogen, transferrin, IgA, IgM, HDL (apo A-I and apo A-II), haptoglobin, α-1-antitrypsin, α-1-acid glycoprotein, and α-2-macroglobulin. Cibacron blue 3GA agarose (Sigma, C1535) bound 10−20 mg of human albumin per mL of gel. Porcine trypsin (Promega, Madison, WI; V5113 sequencing grade modified trypsin) at a concentration of 0.4 μg/μL in 50 mM acetic acid was stored at −80 °C. Pepsin (Sigma, St. Louis, MO; P6887 from porcine gastric mucosa) was dissolved in 10 mM HCl to make a 1 mg/mL solution and stored at −80 °C. Monomeric avidin agarose beads (#20228) were from Pierce Co. NeutrAvidin agarose beads (#29202) were from Thermo Scientific (Rockford, IL). Human plasma (EDTA anticoagulant) was from an adult male, who had fasted overnight before donating blood. Fatty acid free human albumin (Fluka 05418) was from Sigma/Aldrich.
Procedures for FP-Biotin-Labeled Plasma
Separation of Low and High Abundance Proteins in Human Plasma
Two hundred microliters of human plasma were fractionated into low and high abundance proteins by processing 20 μL of plasma at a time on the Beckman Coulter Proteome IgY Spin column depletion kit. The yield of high abundance proteins was 4800 μg in 400 μL. Of this, 123 μL was labeled with FP-biotin, 123 μL was used as a negative control, and the remainder was used for determination of protein concentration.
High Abundance Proteins Labeled with FP-Biotin, Digested with Trypsin, and Purified on Monomeric Avidin
The high abundance fraction of plasma had albumin as its major component. A 123 μL aliquot of the high abundance fraction was treated with 1.25 μL of 20 mM FP-biotin for 48 h at 37 °C at pH 8.0. The final FP-biotin concentration was 200 μM. Excess FP-biotin was removed by dialysis against 2 × 4 L of 10 mM ammonium bicarbonate.
Proteins, in 8 M urea, were reduced with 5 mM dithiothreitol and alkylated with 40 mM iodoacetamide. The samples were diluted to reduce the concentration of urea to 2 M. Proteins were digested with a 1:50 ratio of trypsin to protein at 37 °C overnight. The trypsin was inactivated by heating the sample in a boiling water bath for 10 min. It was necessary to inactivate trypsin because trypsin could have destroyed the avidin protein used in the next step. FP-biotinylated peptides were purified by binding to 0.5 mL of monomeric avidin beads. Nonspecifically bound peptides were washed off with high salt buffers. The column was washed with water to remove salts, and FP-biotinylated peptides were eluted with 10% acetic acid. The eluate was dried in a vacuum centrifuge in preparation for mass spectrometry. The negative control was human plasma treated with everything except FP-biotin.
Depletion of Albumin on Cibacron Blue, Labeling with FP-Biotin, Digestion with Trypsin, and Purification on NeutrAvidin
An albumin-depleted plasma sample was prepared by binding 0.6 mL of human plasma to 2 mL of Cibacron Blue and collecting the protein that eluted in 10 mL of 10 mM TrisCl, pH 8.0, containing 0.3 M NaCl. About 70% of the albumin was removed from the plasma sample by this procedure. The protein was desalted, concentrated to 0.5 mL, and labeled with 100 μM FP-biotin at 37 °C for 16 h in 10 mM ammonium bicarbonate. The labeled protein was denatured in 8 M urea, reduced with dithiothreitol, carbamidomethylated with iodoacetamide, and desalted on a spin column. The yield was 2000 μg in 500 μL. The entire sample was digested with 40 μg of trypsin (Promega) at 37 °C overnight. The FP-biotinylated tryptic peptides were bound to 0.1 mL of NeutrAvidin beads, washed with high salt buffers and water, and eluted with 45% acetonitrile and 0.1% formic acid.
Mass Spectrometry on QSTAR Elite and QTRAP 2000
Five micrograms of the high abundance FP-biotinylated peptides purified with monomeric avidin beads was analyzed on the QSTAR elite liquid chromatography tandem mass spectrometry (LC/MS/MS) system with ProteinPilot 2.0 software at the Applied Biosystems laboratories (Framingham, MA).
A second 5 μg aliquot from the same protein preparation, a negative control sample, and the NeutrAvidin purified peptides were analyzed by LC/MS/MS on the QTRAP 2000 mass spectrometer (Applied Biosystems) at the University of Nebraska Medical Center with Analyst 1.4.1 software. The digest was dried in a vacuum centrifuge and dissolved in 5% acetonitrile and 0.1% formic acid to make 0.5 μg/μL. A 10 μL aliquot was injected into the HPLC nanocolumn (#218MS3.07515 Vydac C18 polymeric rev-phase, 75 μm i.d. × 150 mm long; P.J. Cobert Assoc, St. Louis, MO). Peptides were separated with a 90 min linear gradient from 0 to 60% acetonitrile at a flow rate of 0.3 μL/min and electrosprayed through a nanospray fused silica emitter (360 μm o.d., 75 μm i.d., 15 μm taper, New Objective) directly into the QTRAP 2000, a hybrid quadrupole linear ion trap mass spectrometer. An ion spray voltage of 1900 V was maintained between the emitter and the mass spectrometer. Information-dependent acquisition was used to collect MS, enhanced MS, and MS/MS spectra for the three most intense peaks in each cycle, having a charge of +1 to +4, a mass between 400 and 1700 m/z, and an intensity >10000 counts per s. All spectra were collected in the enhanced mode, that is, using the trap function. Precursor ions were excluded for 30 s after one MS/MS spectrum had been collected. The collision cell was pressurized to 40 μTorr with pure nitrogen. Collision energies between 20 and 40 eV were determined automatically by the software, based on the mass and charge of the precursor ion. The mass spectrometer was calibrated on selected fragments from the MS/MS spectrum of Glu-Fibrinopeptide B. MS/MS spectra were submitted to Mascot for identification of labeled peptides and amino acids (13). MASCOT identified FP-biotinylated Y*LYEIAR (score 17), HPY*FYAPELLFFAK (score 14), and MPCAEDY*LSVVLNQLCVLHEK (score 15) but none of the other FP-biotinylated peptides. The others were identified by manually searching the MS/MS data files using the Extracted Ion Chromatogram feature of the Analyst software. The scores were low because the software did not recognize the characteristic fragments of FP-biotin at 227, 312, and 329. It also did not recognize the 591 ion of FP-biotin or the FP-biotin-tyrosine immonium ions at 708 and 691 or fragments containing dehydroalanine in place of serine. The ions that Mascot did not recognize were often very intense.
The MASCOT modification file is an open source software called UNIMOD. The FP-biotin modification on serine, threonine, and tyrosine was introduced according to the instructions found on the Web site http://www.unimod.org. Access to the modification is freely available to all MASCOT users in the Variable Modifications menu under the name FP-biotin. Fragments of FP-biotin are not part of the MASCOT modification file. Peptides yielding FP-biotin fragments at 227, 312, and 329 amu were identified using the Extracted Ion Chromatogram feature of ABI’s Analyst software. Neutral loss of fragments of FP-biotin were identified by manual inspection of MS/MS spectra.
Mass Spectrometry by Matrix-Assisted Laser Desorption Tandem Time-of-Flight Mass Spectrometry (MALDI-TOF-TOF) 4800
A 0.5 μL aliquot of essentially salt-free samples was spotted on a MALDI target plate, air-dried, and overlaid with 0.5 μL of 10 mg/mL α-cyano-4-hydroxy cinnamic acid in 50% acetonitrile and 0.1% trifluoroacetic acid. MS spectra were acquired using a MALDI-TOF-TOF 4800 (Applied Biosystems), with a laser power of 3000 V, in positive reflector mode. Each spectrum was the average of 500 laser shots. The mass spectrometer was calibrated against des-Arg-Bradykinin (904.468 Da), angiotensin 1 (1296.685 Da), Glu-Fibrinopeptide B (1570.677 Da), and neurotensin (1672.918 Da) (Cal Mix 1 from Applied Biosystems).
Procedures for Pure Albumin
Percent OP-Labeled Tyr 411 Monitored by MALDI-TOF
A 5 μL aliquot of 10 mg/mL albumin was diluted with 5 μL of 1% trifluoroacetic and digested with 2 μL of 1 mg/mL porcine pepsin for 1−2 h at 37 °C. The digest was diluted with 50% acetonitrile and 0.1% trifluoroacetic acid to give a final protein concentration of about 0.5 mg/mL. A 0.5 μL aliquot was spotted on the MALDI target plate, dried, and overlaid with 0.5 μL of 10 mg/mL α-cyano-4-hydroxy cinnamic acid. MS spectra were acquired with the laser set at 3000 V and were saved to Data Explorer. When the saved spectrum was opened in Data Explorer, the cluster areas appeared in an output window. Percent OP-labeled Tyr 411 was calculated by dividing the cluster area of the labeled peptide by the sum of the cluster areas for the unlabeled and labeled peaks. The unlabeled peptides were 409VRYTKKVPQVSTPTL423 (1717.0 amu) and 408LVRYTKKVPQVSTPTL423 (1830.1 amu). After covalent bond formation with CPO, these masses increased by 136 amu to become 1853.0 and 1966.1 amu. After covalent bond formation with FP-biotin, these masses increased by 572.3 to become 2289.3 and 2402.4 amu.
Prolonged Treatment of Albumin with CPO
At the time that we prepared CPO-labeled human albumin, we knew that Tyr 411 was labeled by CPO and had no reason to suspect that other residues might also be labeled. CPO dissolved in ethanol was added to an albumin solution in 10 mM ammonium bicarbonate, pH 8.3, and 0.01% sodium azide in six additions over a 1 month period.
The labeling efficiency was poor when the albumin concentration was 500 mg/mL, so the albumin was diluted to 35 mg/mL, and then to 5 mg/mL, and finally to 1 mg/mL. The final ratio was 7.7 μmol of albumin to 146 μmol of CPO. During the 1 month labeling time, the decision to add more CPO was based on the percent Tyr 411 labeled. No further additions of CPO were made after 85% of the Tyr 411 had been labeled. The labeled albumin was dialyzed against 10 mM potassium phosphate, pH 7.0, and 0.01% azide and processed for LC/MS/MS analysis in the QTRAP mass spectrometer.
Identification of the Most Reactive Residues
The conditions reported to label 1 mol of albumin with 1 mol of DFP were used (10). Human albumin (1.8 mg/mL) in 10 mM TrisCl, pH 8.0, was treated with a 20-fold molar excess of DFP for 2 h at room temperature. The reaction was stopped by the addition of solid urea to 8 M and boiling for 10 min in the presence of 10 mM dithiothreitol. Free sulfhydryl groups were alkylated with iodoacetamide. The carbamidomethylated albumin was dialyzed against 2 × 4 L of 10 mM ammonium bicarbonate and digested with trypsin. Tryptic peptides were subjected to LC/MS/MS on the QTRAP 2000.
The experiment was repeated with FP-biotin, soman, and CPO. A 15 μM solution of albumin in 10 mM TrisCl pH 8.0 was treated with 150 μM FP-biotin or 150 μM soman or 150 μM CPO for 2 h at 22 °C. Samples with intact disulfides were digested with pepsin and analyzed by MALDI-TOF. Carbamidomethylated tryptic peptides were analyzed by LC/MS/MS. Tryptic peptides labeled with FP-biotin were also purified on monomeric avidin beads eluted with 50% acetonitrile and 0.1% formic acid and analyzed by MALDI-TOF-TOF.
Stability of CPO-Labeled Tyr 411
The stability of CPO-labeled Tyr 411 in human albumin was tested at pH 1.5, 7.4, and 8.3 after incubation for up to 7 months at 22 and −80 °C. Albumin labeled on 97% of Tyr 411 with diethoxyphosphate was prepared by incubating 1 mg/mL human albumin (15.6 μM) in 10 mM TrisCl, pH 8.0, and 0.01% sodium azide with 118 μM CPO for 2.5 days at 22 °C. Excess CPO was removed by dialysis of the 8.5 mL solution against 2 × 4 L of 10 mM ammonium bicarbonate, pH 8.3, and 0.01% azide.
pH 1.5
The pH of 2.6 mL of the dialyzed CPO-albumin was adjusted to pH 1.5 by adding 2.6 mL of 1% trifluoroacetic acid. Half of the sample was stored at room temperature, and half was divided into 40 μL aliquots and stored at −80 °C.
pH 7.4
The pH was adjusted to pH 7.4 by dialyzing 3.3 mL of the CPO-albumin preparation against 4.5 L of 10 mM potassium phosphate, pH 7.4, and 0.01% azide. To avoid freeze−thaw artifacts, samples intended for storage at −80 °C were divided into 20 μL aliquots so that each tube was thawed only once.
pH 8.3
The pH of 2.6 mL of CPO-albumin was brought to pH 8.3 by dialysis against 10 mM ammonium bicarbonate and 0.01% sodium azide, pH 8.3. Samples to be stored at −80 °C were divided into 65 tubes each containing 20 μL.
After various times, a tube was removed from −80 °C storage, and the entire contents were digested with pepsin. Samples stored at room temperature were also digested with pepsin. The digests were analyzed by MALDI-TOF, and % labeled Tyr 411 was calculated from cluster areas as described above.
The sample stored at −80 °C in pH 7.4 buffer was analyzed by LC/MS/MS to determine whether sites in addition to Tyr 411 were labeled. The CPO-albumin was denatured, reduced, carbamidomethylated, and digested with trypsin in preparation for LC/MS/MS. The diethoxyphosphate group was found on Tyr 411 and Tyr 138.
Results
FP-Biotin Labeled Albumin in Human Plasma
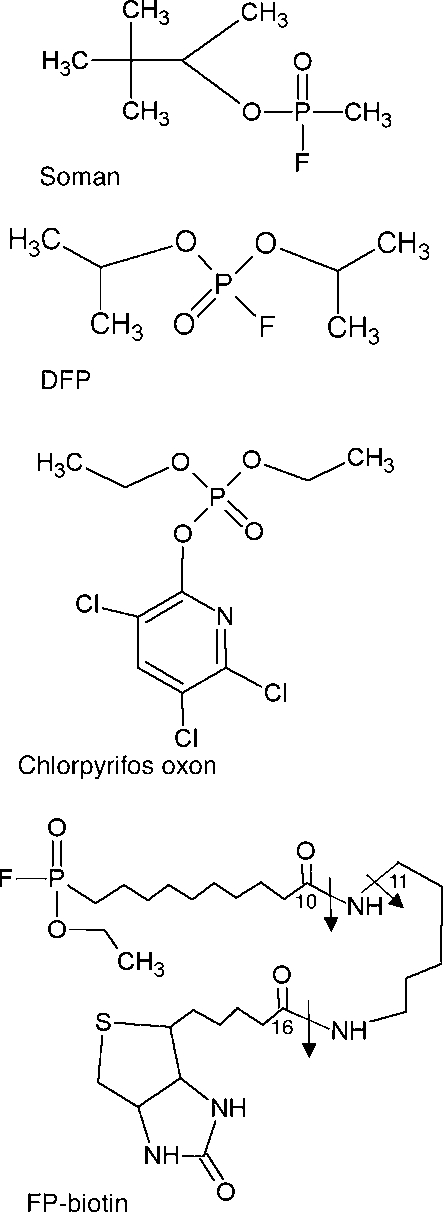
The structures of the organophosphorus agents are shown in Figure 1. Five tyrosines and two serines in human albumin were labeled with FP-biotin including Tyr 138, Tyr 148, Tyr 401, Tyr 411, Tyr 452, Ser 232, and Ser 287 (Table 1).
Figure 1.
Structures of organophosphorus agents. Covalent binding to tyrosine or serine results in loss of the fluoride ion from soman, DFP, and FP-biotin and of the aromatic ring from CPO, so that the added mass is 162.2 for soman, 164.1 for DFP, 136.0 for CPO, and 572.3 for FP-biotin. The arrows in FP-biotin indicate fragmentation sites. A 227 amu ion is produced by cleavage between carbon 16 and the adjacent nitrogen. A 329 amu ion is produced by cleavage between carbon 10 and the adjacent nitrogen. The 312 amu ion is produced by loss of the amine from the 329 ion.
Table 1. FP-Biotinylated Human Albumin Tryptic Peptides Identified by LC/MS/MSa.
| start−end | Mr | sequence | FP-biotinylated |
|---|---|---|---|
| 138−144 | 1499.8 | Y*LYEIAR | Y138 |
| 146−159 | 2315.2 | HPY*FYAPELLFFAK | Y148 |
| 226−233 | 1452.7 | AEFAEVS*K | S232 |
| 287−313 | 3546.6 | S*HCIAEVENDEMPADLPSLAAD-FVESK | S287 |
| 390−402 | 2230.0 | QNCELFEQLGEY*K | Y401 |
| 411−413 | 983.5 | Y*TK | Y411 |
| 446−466 | 3090.5 | MPCAEDY*LSVVLNQLCVLHEK | Y452 |
Residue numbers in accession # gi: 3212456 are for the mature albumin protein and do not include the 24 amino acid signal peptide. The added mass from FP-biotin is 572.3 amu. Cysteines were carbamidomethylated, thus adding a mass of 57 amu.
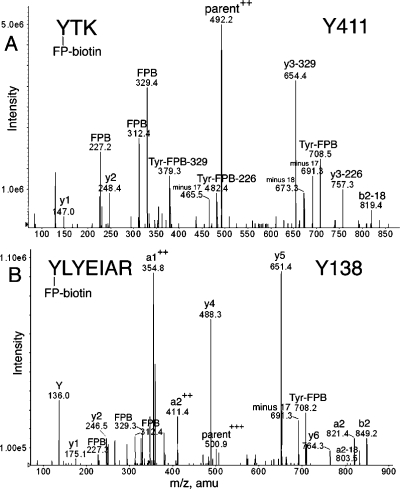
Supporting MS/MS spectra for these assignments are in Figures 2−6. A peptide labeled with FP-biotin had ions at 227, 312, and 329 atomic mass units (amu) resulting from fragmentation of FP-biotin (12). Two ions characteristic of covalent binding of FP-biotin to the hydroxyl group of tyrosine are the immonium ion of tyrosine-FP-biotin at 708 amu and its partner ion at 691 amu, produced by loss of NH3. The 708 and 691 amu masses are prominent in Figure 2A,B but barely visible in Figure 3A,B. An additional complexity in Figure 2A is the presence of ions that had lost a 329 or 226 amu fragment from FP-biotin.
Figure 2.
MS/MS spectra of albumin peptides labeled with FP-biotin on Tyrosine. (A) Tyr 411 in peptide YTK and (B) Tyr 138 in peptide YLYEIAR are covalently modified by FP-biotin. The characteristic fragments of FP-biotin at 227.2, 312.4, and 329.3 amu are present. Ions characteristic of FP-biotin modification on tyrosine are the immonium ion at 708 amu and the immonium ion minus 17 at 691 amu. Support for modification of the first tyrosine rather than the second in YLYEIAR is the mass of ions a2, b2, and a2+2. The doubly charged parent ion in panel A had a mass of 492.2 amu. The triply charged parent ion in panel B had a mass of 500.9 m/z.
Figure 6.
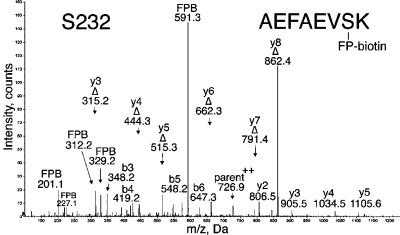
MS/MS spectrum of albumin peptide labeled with FP-biotin on Ser 232. This spectrum was acquired on the QSTAR elite mass spectrometer. The doubly charged parent ion is at 726.9 amu. The peak at 591.3 is FP-biotin released from serine, carrying a hydroxyl group in place of fluorine. Four y-ions (y2, y3, y4, and y5) carry FP-biotin on serine, whereas six y-ions (Δy3−Δy8) have lost FP-biotin as well as a molecule of water, thus converting serine to dehydroalanine.
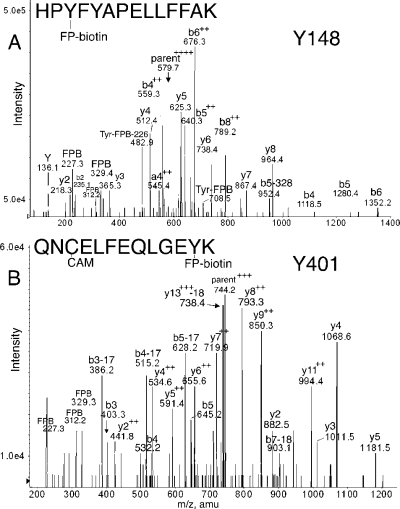
Figure 3.
MS/MS spectra of albumin peptides labeled with FP-biotin on Tyrosine. (A) Tyr 148 in peptide HPYFYAPELLFFAK is labeled with FP-biotin. The quadruply charged parent ion has a mass of 579.7 m/z. The FP-biotin tyrosine immonium ion is at 708.5; after neutral loss of 226, its mass is 482.9. (B) Tyr 401 in peptide QNCELFEQLGEYK is covalently modified by FP-biotin. The triply charged parent ion in B has a mass of 744.2 m/z.
The masses in Figure 2A are consistent with the sequence YTK where the added mass of 572 amu from FP-biotin is on Tyr. The complete y-ion series is present (y1, 147.0 amu, Lys; y2, 248.4 amu, LysThr; and the doubly charged, FP-biotinylated parent ion). Peaks at 227.2, 312.4, 329.4, 691.3, and 708.5 amu are indicative of the presence of FP-biotinylated tyrosine. The remaining major peaks are consistent with various FP-biotinylated tyrosine fragments missing pieces of the FP-biotin moiety.
Peptide YLYEIAR has two tyrosines. A y-ion series (y1−y6) indicates that the FP-biotin label is on the N-terminal Tyr. Additional evidence for labeling on Tyr 138 rather than on Tyr 140 was the presence of the a2 ion at 821.4 amu, the b2 ion at 849.2 amu, the a1+2 ion at 354.8, and the a2+2 ion at 411.4 m/z (Figure 2B). If the FP-biotin had been attached to Tyr 140, the masses would have been a2 = 249, b2 = 277, a1+2 = 68, and a2+2 = 125 amu. Peaks at 226.3, 312.4, and 329.3 are fragments of FP-biotin. Masses at 708.2 and 691.3 amu for FP-biotinylated tyrosine confirm the presence of FP-biotinylated tyrosine in the peptide. Analysis of the missed cleavage peptide KYLYEIAR supports labeling on Tyr 138 (data not shown).
Peptide HPYFYAPELLFFAK in Figure 3A also has two tyrosines. Evidence for labeling on Tyr 148 rather than on Tyr 150 is the presence of the b4 ion at 1118.5 amu, the a4+2 ion at 545.4 amu, and the b4+2 ion at 559.3 m/z. The total mass of the b4 fragment (1117.6 amu) is equal to the fragment HPYF (545 amu) plus the added mass from FP-biotin (572 amu). Of the four residues in the b4 fragment, Tyr 148 is the most reasonable candidate for labeling. Fragment masses for b5 and b6 also support labeling of Tyr 148 rather than Tyr 150. An extensive y-ion series (y2−y8) supports the assignment of this peptide. Masses at 227.3, 312.2, and 329.4 indicate the presence of FP-biotin. Masses at 691.2 and 708.5 amu indicate the presence of FP-biotinylated tyrosine. A similar analysis was made for peptides RHPYFYAPELLFFAK and HPYFYAPELLFFAKR, which differ from HPYFYAPELLFFAK by virtue of missed cleavages (data not shown).
Peptide QNCELFEQLGEYK in Figure 3B is FP-biotinylated on Tyr 401 as demonstrated by the y2 ion at 882.5 amu, the y3 ion at 1011.5 amu, the y4 ion at 1068.5 amu, and the y5 ion at 1181.8 amu. The y2 mass is equal to the sum of Lys (147 amu), Tyr (163 amu), and the added mass of FP-biotin (572 amu). A variety of larger, multiply charged y-ion fragments support the labeling assignment. Prominent b-ion fragments confirm the identity of the peptide. Fragments at 227.2, 312.2, and 329.3 amu indicate the presence of FP-biotin in the peptide.
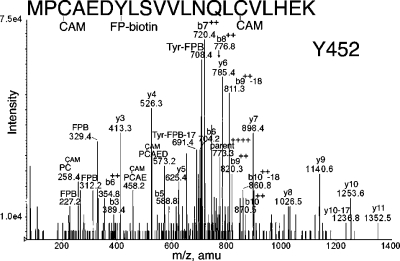
Peptide MPCAEDYLSVVLNQLCVLHEK in Figure 4 is FP-biotinylated on Tyr 452. The best evidence in support of this interpretation is the doubly charged mass at 720.4 m/z, which is consistent with the b7 ion plus the added mass from FP-biotin. The b7 ion consists of MPCAEDY. Of these residues, only Tyr 452 is a reasonable candidate for FP-biotinylation. The b8+2, b9+2, and b10+2 ions support this interpretation. The y-series (y3−y11) supports identification of this peptide. Masses at 227.2, 312.2, and 329.4 indicate the presence of FP-biotin. Masses at 691.4 and 708.4 amu indicate the presence of FP-biotinylated tyrosine in this peptide. A missed cleavage form of this peptide, RMPCAEDYLSVVLNQLCVLHEK, was also analyzed, and the results support labeling of Tyr 452 (data not shown).
Figure 4.
MS/MS spectrum of albumin peptide labeled with FP-biotin on tyrosine 452. The quadruply charged parent ion has a mass of 773.3 m/z. The carbamidomethylated cysteine is indicated as CAM. Internal fragmentation at proline yielded the 458.2 ion for PC(CAM)AE and the 573.2 ion for PC(CAM)AED. The characteristic fragments of FP-biotin at 227.2, 312.2, and 329.4 are present. The FP-biotin-tyrosine immonium ion is at 708.4 amu. After loss of an amine, the FP-biotin tyrosine immonium ion has a mass of 691.4 amu.
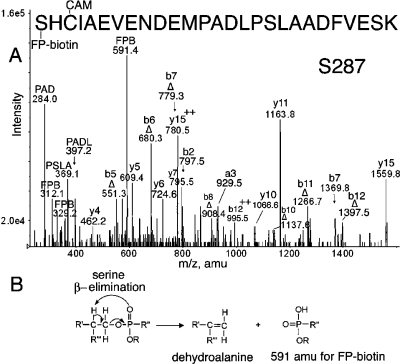
Peptide SHCIAEVENDEMPADLPSLAADFVESK in Figure 5A is FP-biotinylated on Ser 287. Existence of an FP-biotinylated serine is indicated by the major peak at 591.4 amu. This is a characteristic mass from FP-biotin that appears as the result of β-type elimination of FP-biotin from a serine adduct (Figure 5B), during collision-induced dissociation in the mass spectrometer (12). The complementary peptide fragment arising from this fragmentation contains a dehydroalanine in place of serine. The masses of a b-series (Δb5−Δb12) containing a dehydroalanine residue support the elimination of FP-biotin from serine. Of the residues in the b5 fragment (SHCIA), serine at position 287 is a candidate for FP-biotinylation. The cysteine might have been considered a target for labeling, but the overall mass of the fragment is consistent with carbamidomethylation on the cysteine. A y-ion series (y4−y15) supports the identification of the peptide. Additional support for the presence of FP-biotin in the peptide comes from characteristic masses at 312.1 and 329.2 amu. The absence of the characteristic mass at 227 amu is common for FP-biotinylated serine.
Figure 5.
(A) MS/MS spectrum of albumin peptide labeled with FP-biotin on Ser 287. The triply charged parent ion has a mass of 1183.8 m/z. The carbamidomethylated (CAM) peptide carried the FP-biotin on Ser 287. The evidence for modification on serine is the presence of a b-ion series for the dehydroalanine form of the peptide, designated Δ. The 591.4 amu ion is FP-biotin released from serine where the fluoride ion has been replaced by a hydroxyl group. Release of the entire OP accompanied by formation of dehydroalanine is a characteristic of OP bound to serine. Internal fragmentation at proline yielded masses at 284.0 for PAD, 369.1 for PSLA, and 397.2 for PADL. (B) Scheme for β-elimination of the OP label from serine. Fragmentation in the mass spectrometer eliminates the OP from serine and simultaneously converts serine to dehydroalanine.
The MS/MS spectrum for peptide AEFAEVSK labeled by FP-biotin on Ser 232 is in Figure 6. The b- and y-ion masses support the assigned sequence. Peaks not assigned by Protein Pilot included six dehydroalanine fragments as well as the 591 amu ion of FP-biotin and the 227, 312, and 329 amu fragments of FP-biotin. These additional peaks strongly support the conclusion that Ser 232 of albumin was labeled by FP-biotin. This labeled peptide was detected by the sensitive QSTAR elite mass spectrometer but not by the QTRAP 2000 mass spectrometer. No FP-biotinylated peptides were found in the control plasma that had not been treated with FP-biotin.
Search for Other FP-Biotin-Labeled Proteins in Human Plasma
The present method identified 7 FP-biotin-labeled albumin peptides but no FP-biotin-labeled peptides from any other protein. A Western blot hybridized with Streptavidin Alexafluor-680 showed many FP-biotinylated bands in human plasma treated with FP-biotin under our conditions (data not shown). One such protein is FP-biotinylated plasma butyrylcholinesterase (1,14). However, the FP-biotinylated butyrylcholinesterase peptide was not found with the present methods. FP-biotinylated peptides from proteins other than albumin are difficult to detect in the presence of the overwhelmingly high concentration of albumin. Even after depletion of albumin with Cibacron Blue, the concentration of albumin was still too high to allow detection of other FP-biotinylated peptides. Human plasma contains 5 mg of butyrylcholinesterase and 50000 mg albumin per L. In experiments not described in this report, we found OP-labeled butyrylcholinesterase in human plasma only after the butyrylcholinesterase had been purified by binding to procainamide affinity gel, thus eliminating more than 95% of the albumin.
Albumin Residues Labeled by CPO
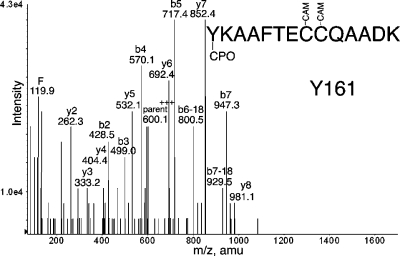
Prolonged labeling of pure human albumin with CPO resulted in labeling of six tyrosines: Y138, Y150, Y161, Y401, Y411, and Y452 (Table 2). Four of these sites were also labeled by FP-biotin (Y138, Y401, Y411, and Y452). The HPYFYAPELLFFAK peptide was labeled on Tyr 150 by CPO, whereas it was labeled on Tyr 148 by FP-biotin. A new peptide YKAAFTECCQAADK was labeled by CPO (Figure 7) and not by FP-biotin. Labeling on tyrosine is supported by the b ion series. The identity of the peptide is supported by the y2−y8 ions. Additional MS/MS spectra for CPO-labeled peptides are in the Supporting Information (Figures S1−S5).
Table 2. CPO-Labeled Human Albumin Peptidesa.
| start−end | Mr | sequence | CPO-labeled |
|---|---|---|---|
| 139−144 | 1234.6 | K(CONH2)Y*LYEIAR | Y138 |
| 146−159 | 1877.9 | HPYFY*APELLFFAK | Y150 |
| 161−174 | 1797.8 | Y*KAAFTECCQAADK | Y161 |
| 390−402 | 1791.8 | QNCELFEQLGEY*K | Y401 |
| 411−413 | 547.2 | Y*TK | Y411 |
| 446−466 | 2653.2 | MPCAEDY*LSVVLNQLCVLHEK | Y452 |
Peptide KYLYEIAR was carbamylated on the N-terminal Lys by degradation products in urea, adding a mass of 43. Chorpyrifos oxon adds a mass of 136 to the labeled tyrosine.
Figure 7.
MS/MS spectrum of albumin peptide labeled with CPO on Tyr 161. The b2 and b3 ions support labeling on tyrosine.
Tyr 411 Reacts Most Readily with OP
The finding that seven tyrosines and two serines make a covalent bond with OP led to the question of which amino acid reacts most readily with OP. To answer this question, we duplicated the conditions reported to label one molar equivalent of human albumin with DFP (10). MALDI-TOF analysis of pepsin-digested, DFP-treated human albumin suggested that 80% of Tyr 411 was labeled with DFP. MS/MS analysis of a tryptic digest of carbamidomethylated DFP-treated albumin confirmed that Tyr 411 in peptide Y*TK was labeled. In addition, less than 10% of peptide EFNAETFTFHADICT*LS*EK was labeled (on residues T515 and S517).
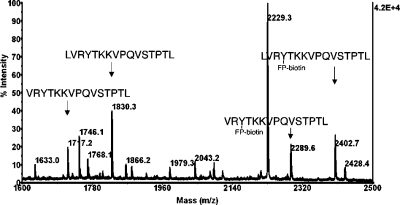
Albumin treated with FP-biotin for 2 h and digested with pepsin had 52% of its Tyr 411 labeled in peptide VRY*TKKVPQVSTPTL as calculated by MALDI-TOF mass spectrometry (Figure 8). The carbamidomethylated, trypsin-digested preparation analyzed by LC/MS/MS confirmed that Tyr 411 in peptide Y*TK was labeled with FP-biotin. Peptide HPY*FYAPELLFFAK was labeled on Tyr 148 with FP-biotin but to less than 10%. A third method to identify FP-biotinylated peptides was purification on monomeric avidin beads followed by MALDI-TOF-TOF analysis. This method yielded only one FP-biotinylated peptide, the Y*TK peptide labeled on Tyr 411.
Figure 8.
MALDI-TOF spectrum of pepsin-digested albumin to show labeling of Tyr 411 by FP-biotin. Pepsin digestion of albumin yields two unlabeled peptides at 1717.2 (VRYTKKVPQVSTPTL) and 1830.3 (LVRYTKKVPQVSTPTL) amu, both containing Tyr 411. Both peptides have a mass shift of 572.3 after reaction with FP-biotin, yielding the peaks at 2289.6 and 2402.7 amu. The FP-biotin is covalently bound to Tyr 411. About 50% of the Tyr 411 is labeled as calculated from cluster areas of the labeled and unlabeled peaks.
Soman-treated albumin (150 μM soman for 2 h) analyzed by MALDI-TOF and LC/MS/MS yielded only one labeled peptide. The soman was on Tyr 411.
Albumin treated with CPO for 2 h before digestion with pepsin or trypsin and analyzed by MALDI-TOF and LC/MS/MS was labeled on Tyr 411. Approximately 30% of the Tyr 411 sites were labeled in peptides VRY*TKKVPQVSTPTL and LVRY*TKKVPQVSTPTL. In addition, less than 5% of Thr 566 in peptide ET*CFAEEGKK and less than 5% of Thr 236 and Thr 239 in peptide LVT*DLT*KVHTECCHGDLLECADDR were labeled. We conclude that Tyr 411 is the most OP reactive residue in human albumin.
Support for the conclusion that Tyr 411 is the most OP reactive residue in albumin comes from ref (2). Williams incubated the albumin fraction of human plasma with radiolabeled sarin, digested with trypsin, purified the radiolabeled peptides by HPLC, and analyzed by LC/MS/MS. A single radiolabeled peptide was isolated. Its sequence was YTK with the isopropyl methylphosphonyl group on tyrosine.
Unstable OP-Ser and OP-Thr but Stable OP-Tyr
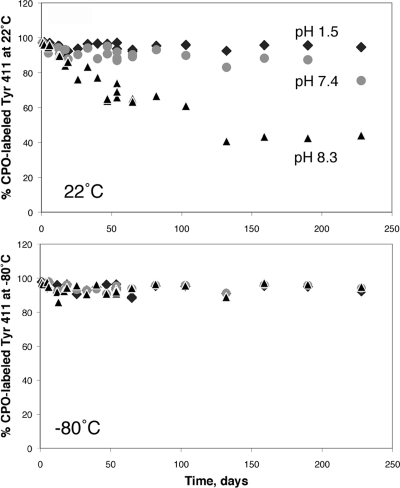
It was noted that serine and threonine residues were labeled in addition to tyrosine when samples had been incubated at pH 8.0−8.3 for 2−48 h but were not found in samples incubated at pH 8.3 for a month. In contrast, OP-labeled tyrosines were found even after 1 month of incubation at pH 8.3. Our stability study of CPO-labeled albumin confirmed that the Tyr 411 adduct was stable (Figure 9). Incubation at pH 7.4 and 22 °C resulted in almost no loss of the CPO label on Tyr 411 after 7 months. In contrast, about half of the label was lost after 3.6 months at pH 8.3 and 22 °C. The CPO-labeled Tyr 411 was stable at pH 1.5 and 22 °C and was stable at all pH values when the labeled albumin was stored at −80 °C. These results suggest that OP-labeled serine and threonine adducts are unstable as compared to OP-labeled tyrosine.
Figure 9.
Stability of the diethoxyphosphate adduct of human albumin on Tyr 411. Albumin was treated with CPO to achieve 97% labeling of Tyr 411. Excess CPO was removed by dialysis. The pH of the dialyzed albumin was adjusted to 1.5, 7.4, and 8.3. CPO-albumin samples were stored at 22 and −80 °C. After various times of storage, samples were digested with pepsin and % labeling of Tyr 411 was calculated from cluster areas of labeled and unlabeled peptides in the MALDI-TOF mass spectrometer. CPO-labeled Tyr 411 was stable at pH 1.5 and 7.4 when CPO-albumin was stored at 22 °C (top panel), and it was stable at all pH values when CPO-albumin was stored at −80 °C (bottom panel). Storage of CPO-albumin at pH 8.3 at 22 °C resulted in release of half of the diethoxyphosphate group from Tyr 411 after 3.6 months.
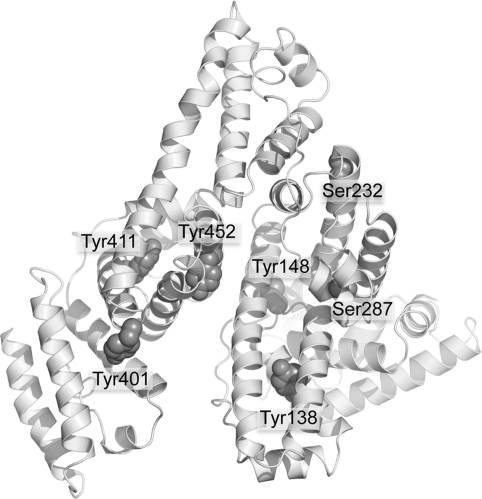
Surface Location of OP Reactive Residues
The crystal structure in Figure 10 shows the five tyrosines and two serines that become labeled by FP-biotin. These residues are located on the surface of the albumin molecule where they are available for reaction with OP.
Figure 10.
Crystal structure of human albumin showing surface location of Tyr 138, Tyr 148, Tyr 401, Tyr 411, Tyr 452, Ser 232, and Ser 287. The residues are shown as space-filled structures. The picture was made with PyMol software using the structure in PDB code 1bm0(28).
Human albumin has 18 tyrosines and 24 serines but only five tyrosines and two serines made a covalent bond with FP-biotin. Their special reactivity may be explained by a nearby arginine or lysine that stabilizes the ionized hydroxyl of tyrosine or serine.
Discussion
Many OP-Reactive Residues in Human Albumin
Although Tyr 411 is the most OP reactive residue in human albumin, an additional eight amino acids were labeled when the OP concentration was high and the reaction time was prolonged. The reaction with FP-biotin at pH 8.0 resulted in the labeling of five tyrosines and two serines in albumin. CPO labeled six tyrosines (two of which were different from those labeled by FP-biotin) and no serines. We agree with Means and Wu (10) that about 90% of the label is on Tyr 411 and 10% is on other residues.
The pKa of the tyrosine hydroxyl group is 10.1 and of the serine hydroxyl group is approximately 16, based on comparison to ethanol (15). In the absence of special activation, less than 1% of the tyrosines and less than 0.000001% of the serines would be expected to be ionized at pH 8.0. Ionized forms react preferentially with OP, so the reactivity of tyrosine and especially of serine with OP would be expected to be poor at pH 8. The special reactivity of Tyr 411 suggests that the pKa of this particular tyrosine has been lowered. Means and Wu identified an OP reactive residue in albumin that had a pKa of 8.3 (10). It is likely that Tyr 411 corresponds to that residue.
Albumin as an OP Scavenger
Our results show that albumin is an OP scavenger, undergoing a covalent reaction with OP. As such, albumin contributes to detoxication of OP. A significant amount of OP can be bound by albumin because the concentration of albumin in serum is high (≈0.6 mM), even though the rate of reaction with OP is slow (3).
Tyrosines with an abnormally low pKa are involved directly or indirectly in the catalytic activity of numerous enzymes including glutathione S-transferase (16), asparaginase (17), β-lactamases (18), and old yellow enzyme (19). Lowering the pKa of tyrosines in albumin by modifying their environment, either by mutagenesis or by chemical modification of vicinal residues, would increase the reactivity of albumin with OP. Specific nitration of tyrosine by tetranitromethane was found to lower the pKa of tyrosine to 6.8 (20). Another nitration reagent of tyrosine, peroxynitrite, was found to increase the catalytic activity of a few enzymes (21). Thus, specific nitration of tyrosine residues in albumin could also lead to a gain in reactivity of this protein, increasing its scavenging properties.
No Aging of OP-Tyrosine Adducts
When soman or DFP are bound to acetylcholinesterase or butyrylcholinesterase, the OP loses an alkyl group in a process called aging (22−25). An aged soman-labeled peptide would have an added mass of 78 rather than 162; an aged DFP-labeled peptide would have an added mass of 122 rather than 164; an aged CPO-labeled peptide would have an added mass of 108 rather than 136. Masses corresponding to aged OP-labeled peptides were not found in MS scans. We conclude that albumin OP adducts on tyrosine do not age.
Support for this conclusion comes from the work of others (2,26). Human albumin covalently labeled with soman or sarin and treated with sodium fluoride to release the OP yielded intact soman and sarin. Soman-tyrosine adducts isolated from nerve agent-treated guinea pigs contained the pinacolyl group of soman.
The absence of aging is a special advantage for OP-albumin as a biomarker because it allows for a more precise identification of the OP. In contrast, soman and sarin exposure cannot be distinguished when the biomarker is cholinesterase, where aging of OP adducts occurs rapidly.
OP Labeling of Albumin in Living Animals
Guinea pigs treated with the nerve agents soman, sarin, cyclosarin, or tabun have nerve agent-labeled albumin in their blood (2). The OPs are bound to tyrosine. The tabun-tyrosine and soman-tyrosine adducts were detected in blood 7 days postexposure, indicating that the adducts are stable. The adducts had not undergone aging and had not been released from tyrosine by treatment of the guinea pigs with oxime. These are characteristic features of OP adducts on albumin. Mice treated with a nontoxic dose of FP-biotin by intraperitoneal injection had FP-biotinylated albumin in blood and muscle (1). These examples show that OP binds covalently to albumin under physiological conditions and that OP-albumin adducts could therefore be useful as biomarkers of OP exposure (27). Low OP doses make a covalent bond with Tyr 411 of albumin.
Significance
Our results suggest that OP exposure could be monitored by mass spectrometry of OP-albumin adducts or with antibodies against OP-albumin adducts. The surface location of the OP-binding sites in albumin suggests that these epitopes may be available for reaction with antibodies. This is in distinct contrast with acetylcholinesterase and butyrylcholinesterase where the OP binding site is buried deep within the molecule, making it unavailable to antibodies. Antibodies to OP-albumin would be primarily against OP-labeled Tyr 411 because Tyr 411 is the most reactive residue at low OP concentrations. The common OP pesticides yield either diethoxyphosphate or dimethoxyphosphate adducts. Therefore, only two antibodies would be needed for detection of exposure to common OP pesticides. The studies described here support investigation into whether albumin could be engineered to become a more efficient OP scavenger.
Acknowledgments
Mass spectra were obtained with the support of the Mass Spectrometry & Proteomics core facility at the University of Nebraska Medical Center. This work was supported by U.S. Army Medical Research and Materiel Command W81XWH-07-2-0034 (to O.L.), W81XWH-06-1-0102 (to S.H.H.), NIH CounterACT U01 NS058056-02 (to O.L.), NIH Eppley Cancer Center Grant P30CA36727, NIH Grant U01 ES016102, and NIH CounterACT U44 NS058229 (to C.M.T.), DGA/PEA 08co501 (to F.N.), and DGA Grant 03co010-05/PEA01 08 7 (to P.M.).
Supporting Information Available
MS/MS spectra for CPO-labeled peptides (Figures S1−S5). This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: CPO, chlorpyrifos oxon; DFP, diisopropylfluorophosphate; FP-biotin, 10-fluoroethoxyphosphinyl-N-biotinamido pentyldecanamide; OP, organophosphorus ester; LC/MS/MS, liquid chromatography tandem mass spectrometry; MALDI-TOF-TOF, matrix-assisted laser desorption tandem time-of-flight mass spectrometry.
Supplementary Material
References
- Peeples E. S.; Schopfer L. M.; Duysen E. G.; Spaulding R.; Voelker T.; Thompson C. M.; Lockridge O. (2005) Albumin, a new biomarker of organophosphorus toxicant exposure, identified by mass spectrometry. Toxicol. Sci. 83, 303–312. [DOI] [PubMed] [Google Scholar]
- Williams N. H.; Harrison J. M.; Read R. W.; Black R. M. (2007) Phosphylated tyrosine in albumin as a biomarker of exposure to organophosphorus nerve agents. Arch. Toxicol. 81, 627–639. [DOI] [PubMed] [Google Scholar]
- Li B.; Nachon F.; Froment M. T.; Verdier L.; Debouzy J. C.; Brasme B.; Gillon E.; Schopfer L. M.; Lockridge O.; Masson P. (2008) Binding and hydrolysis of soman by human serum albumin. Chem. Res. Toxicol. 21, 421–431. [DOI] [PubMed] [Google Scholar]
- Schwartz M. (1982) A serine protease activity of human serum albumin towards 4-methylumbelliferyl-guanidinobenzoate (MUGB) and diisopropyl fluorophosphate (DEP): Implications for the use of MUGB reactivity in amniotic fluid in prenatal diagnosis of cystic fibrosis. Clin. Chim. Acta 124, 213–223. [DOI] [PubMed] [Google Scholar]
- Hagag N.; Birnbaum E. R.; Darnall D. W. (1983) Resonance energy transfer between cysteine-34, tryptophan-214, and tyrosine-411 of human serum albumin. Biochemistry 22, 2420–2427. [DOI] [PubMed] [Google Scholar]
- Li B.; Schopfer L. M.; Hinrichs S. H.; Masson P.; Lockridge O. (2007) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry assay for organophosphorus toxicants bound to human albumin at Tyr411. Anal. Biochem. 361, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultatos L. G.; Basker K. M.; Shao M.; Murphy S. D. (1984) The interaction of the phosphorothioate insecticides chlorpyrifos and parathion and their oxygen analogues with bovine serum albumin. Mol. Pharmacol. 26, 99–104. [PubMed] [Google Scholar]
- Ortigoza-Ferado J.; Richter R. J.; Hornung S. K.; Motulsky A. G.; Furlong C. E. (1984) Paraoxon hydrolysis in human serum mediated by a genetically variable arylesterase and albumin. Am. J. Hum. Genet. 36, 295–305. [PMC free article] [PubMed] [Google Scholar]
- Sogorb M. A.; Vilanova E. (2002) Enzymes involved in the detoxification of organophosphorus, carbamate and pyrethroid insecticides through hydrolysis. Toxicol. Lett. 128, 215–228. [DOI] [PubMed] [Google Scholar]
- Means G. E.; Wu H. L. (1979) The reactive tyrosine residue of human serum albumin: Characterization of its reaction with diisopropylfluorophosphate. Arch. Biochem. Biophys. 194, 526–530. [DOI] [PubMed] [Google Scholar]
- Kidd D.; Liu Y.; Cravatt B. F. (2001) Profiling serine hydrolase activities in complex proteomes. Biochemistry 40, 4005–4015. [DOI] [PubMed] [Google Scholar]
- Schopfer L. M.; Champion M. M.; Tamblyn N.; Thompson C. M.; Lockridge O. (2005) Characteristic mass spectral fragments of the organophosphorus agent FP-biotin and FP-biotinylated peptides from trypsin and bovine albumin (Tyr410). Anal. Biochem. 345, 122–132. [DOI] [PubMed] [Google Scholar]
- Perkins D. N.; Pappin D. J.; Creasy D. M.; Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567. [DOI] [PubMed] [Google Scholar]
- Schopfer L. M.; Voelker T.; Bartels C. F.; Thompson C. M.; Lockridge O. (2005) Reaction kinetics of biotinylated organophosphorus toxicant, FP-biotin, with human acetylcholinesterase and human butyrylcholinesterase. Chem. Res. Toxicol. 18, 747–754. [DOI] [PubMed] [Google Scholar]
- Ballinger P.; Long F. A. (1960) Acid ionization constants of alcohols. II. Acidities of some substituted methanols and related compounds. J. Am. Chem. Soc. 82, 795–798. [Google Scholar]
- Atkins W. M.; Wang R. W.; Bird A. W.; Newton D. J.; Lu A. Y. (1993) The catalytic mechanism of glutathione S-transferase (GST). Spectroscopic determination of the pKa of Tyr-9 in rat alpha 1-1 GST. J. Biol. Chem. 268, 19188–19191. [PubMed] [Google Scholar]
- Derst C.; Wehner A.; Specht V.; Rohm K. H. (1994) States and functions of tyrosine residues in Escherichia coli asparaginase II. Eur. J. Biochem. 224, 533–540. [DOI] [PubMed] [Google Scholar]
- Lamotte-Brasseur J.; Dubus A.; Wade R. C. (2000) pK(a) calculations for class C beta-lactamases: The role of Tyr-150. Proteins 40, 23–28. [DOI] [PubMed] [Google Scholar]
- Kohli R. M.; Massey V. (1998) The oxidative half-reaction of Old Yellow Enzyme. The role of tyrosine 196. J. Biol. Chem. 273, 32763–32770. [DOI] [PubMed] [Google Scholar]
- Cuatrecasas P.; Fuchs S.; Anfinsen C. B. (1968) The tyrosyl residues at the active site of staphylococcal nuclease. Modifications by tetranitromethane. J. Biol. Chem. 243, 4787–4798. [PubMed] [Google Scholar]
- Ji Y.; Neverova I.; Van Eyk J. E.; Bennett B. M. (2006) Nitration of tyrosine 92 mediates the activation of rat microsomal glutathione s-transferase by peroxynitrite. J. Biol. Chem. 281, 1986–1991. [DOI] [PubMed] [Google Scholar]
- Michel H. O.; Hackley B. E. Jr.; Berkowitz L.; List G.; Hackley E. B.; Gillilan W.; Pankau M. (1967) Ageing and dealkylation of soman (pinacolylmethylphosphonofluoridate)-inactivated eel cholinesterase. Arch. Biochem. Biophys. 121, 29–34. [DOI] [PubMed] [Google Scholar]
- Millard C. B.; Kryger G.; Ordentlich A.; Greenblatt H. M.; Harel M.; Raves M. L.; Segall Y.; Barak D.; Shafferman A.; Silman I.; Sussman J. L. (1999) Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level. Biochemistry 38, 7032–7039. [DOI] [PubMed] [Google Scholar]
- Nachon F.; Asojo O. A.; Borgstahl G. E.; Masson P.; Lockridge O. (2005) Role of water in aging of human butyrylcholinesterase inhibited by echothiophate: The crystal structure suggests two alternative mechanisms of aging. Biochemistry 44, 1154–1162. [DOI] [PubMed] [Google Scholar]
- Jansz H. S.; Brons D.; Warringa M. G. (1959) Chemical nature of the DFP-binding site of pseudocholinesterase. Biochim. Biophys. Acta 34, 573–575. [DOI] [PubMed] [Google Scholar]
- Adams T. K.; Capacio B. R.; Smith J. R.; Whalley C. E.; Korte W. D. (2004) The application of the fluoride reactivation process to the detection of sarin and soman nerve agent exposures in biological samples. Drug Chem. Toxicol. 27, 77–91. [DOI] [PubMed] [Google Scholar]
- Carter W. G.; Tarhoni M.; Rathbone A. J.; Ray D. E. (2007) Differential protein adduction by seven organophosphorus pesticides in both brain and thymus. Hum. Exp. Toxicol. 26, 347–353. [DOI] [PubMed] [Google Scholar]
- Sugio S.; Kashima A.; Mochizuki S.; Noda M.; Kobayashi K. (1999) Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 12, 439–446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.