Abstract
Background
Past studies in the legume family (Fabaceae) have uncovered several evolutionary trends including differential mutation and diversification rates across varying taxonomic levels. The legume tribe Psoraleeae is shown herein to exemplify these trends at the generic and species levels. This group includes a sizable diversification within North America dated at approximately 6.3 million years ago with skewed species distribution to the most recently derived genus, Pediomelum, suggesting a diversification rate shift. We estimate divergence dates of North American (NAm) Psoraleeae using Bayesian MCMC sampling in BEAST based on eight DNA regions (ITS, waxy, matK, trnD-trnT, trnL-trnF, trnK, trnS-trnG, and rpoB-trnC). We also test the hypothesis of a diversification rate shift within NAm Psoraleeae using topological and temporal methods. We investigate the impact of climate change on diversification in this group by (1) testing the hypothesis that a shift from mesic to xeric habitats acted as a key innovation and (2) investigating diversification rate shifts along geologic time, discussing the impact of Quaternary climate oscillations on diversification.
Results
NAm Psoraleeae represents a recent, rapid radiation with several genera originating during the Pleistocene, 1 to 2 million years ago. A shift in diversification rate is supported by both methods with a 2.67-fold increase suggested around 2 million years ago followed by a 8.73-fold decrease 440,000 years ago. The hypothesis that a climate regime shift from mesic to xeric habitats drove increased diversification in affected taxa was not supported. Timing of the diversification rate increase supports the hypothesis that glaciation-induced climate changes during the Quaternary influenced diversification of the group. Nonrandom spatial diversification also exists, with greater species richness in the American Southwest.
Conclusion
This study outlines NAm Psoraleeae as a model example of a recent, rapid radiation. Diversification rate shifts in NAm Psoraleeae are not due to current climate regimes as represented by habitat, but instead to past global climate change resulting from Quaternary glaciations. NAm Psoraleeae diversification is a good example of how earthly dynamics including global climate change and topography work together to shape biodiversity.
Background
The legume family (Fabaceae) is the third largest plant family on earth with an estimated 19,000 species [1]. A study of diversification across angiosperms found Fabales (that is, Fabaceae, Quillajaceae, Polygalaceae, and Surianaceae) as one of 13 angiosperm orders to exhibit higher than expected diversification rates [2]. Within Fabales, Fabaceae exhibits greater diversification than the other relatively species-poor families. Furthermore, within Fabaceae, the subfamily Papilionoideae contains the bulk of species (~14,000) compared with Mimosoideae and Caesalpinioideae. This uneven distribution of species diversity continues down to lower taxonomic levels in Papilionoideae with Astragalus containing upwards of 2500 species [3,4]. Although Astragalus does not exhibit higher diversification rates relative to other related genera, the Astragalean clade, comprising Astragalus and a subset of the Galegeae tribe, does [3]. Other studies of Fabaceae have examined diversification rates and timing of divergence, focusing on generic or higher levels in the family [5,6]. The collection of research discussed above provides precedent for uneven diversification and shifts in diversification rates within Fabaceae, phenomena relatively unexplored at the species level (but see [7,8]).
Studies based on chloroplast coding genes matK and rbcL found elevated nucleotide substitution rates for the tribe Psoraleeae relative to other legume clades, suggestive of rapid evolution or diversification [5]. In addition, research on transcontinental clades estimated the age of the divergence between two Psoraleeae genera, Cullen of Australia, and Rupertia, endemic to the western United States, at approximately 6.3 million years ago (mya) [5]. This age estimate illustrates the recent diversification of this group, especially as the North American (NAm) clade of Psoraleeae diversified after this transcontinental split. Evidence suggests that Psoraleeae represents a recent, rapid radiation, making this group well suited to a species-level diversification study within Fabaceae.
Current circumscription of the Psoraleeae tribe includes a monophyletic New World group of six genera: Hoita (three spp.), Orbexilum (nine spp.), Pediomelum (29 spp.), Psoralidium (three spp.), and Rupertia (three spp.) from North America and Otholobium (six to eight spp.) in South America [9,10]. Four Old World genera also exist: Cullen, Bituminaria, Psoralea, and Otholobium, the latter of which spans the Old and New Worlds [10]. When considering the NAm clade, the skewed distribution of species richness towards Pediomelum suggests a shift in diversification rate within NAm Psoraleeae.
Diversification is the net speciation and extinction of a group. When coupled with evolutionary and ecological processes, diversification is central to the creation of species richness. Differential species richness across taxonomic and geographic levels has been widely studied, with many hypotheses proposed to explain this variation (reviewed by [11,12]). Most hypotheses concerning species richness focus on the impact of climatic factors on the diversification of species across geographic regions, endeavoring to find and explain global patterns of biodiversity. Hypotheses concerning the cause of diversification rate shifts, or net changes in speciation or extinction rate that bear on relative species diversity, have also been suggested including geographic range fragmentation, ecology, competition, key innovations, and climate change [13-18]. Climatic variables such as water and temperature account for much of the variation in species richness seen globally [19,20]. Indeed, climatic variables have been implicated in studies of species richness and diversification of a wide range of organisms, including angiosperms [21], amphibians [22], reptiles [23], vertebrates [24], insects [25], and birds [26,27], among others [19].
Hypotheses of climate change are particularly relevant to NAm Psoraleeae as climatic variables may affect habitat and range variation past and present. The five NAm genera span a variety of ecological habitats: Hoita is riparian, living strictly in moist soils along streams and springs in California; Rupertia resides on moderately moist slopes in oak-pine communities of California; Orbexilum resides in the semi-moist soils of forest communities of the Southeastern United States; Psoralidium lives in the semi-dry soils of Midwestern and Intermountain grasslands and deserts; and Pediomelum has adapted to well-drained soils in dry, arid ecosystems of the deserts of the Intermountain West and the grasslands and rock outcrops of the Midwestern United States [10,28,29]. Many habitats of endemic Pediomelum species are characterized by arid, hot environments such as the cedar glades of Tennessee, the sandy deserts of the Southwestern United States, or the limestone outcrops of the Edwards Plateau in Texas.
Observation of NAm Psoraleeae in light of evolutionary relationships suggests a shift in climate regimes with Pediomelum, the largest genus, and Psoralidium shifting to more xeric habitats, perhaps leading to increased diversification in these genera relative to other NAm Psoraleeae. This leads to the postulate that adaptation to xeric habitats may have acted as a key innovation, a trait whose origin has spurred diversification within a group [30].
The young age estimate of the tribe suggests that NAm Psoraleeae may have diversified in part in the Quaternary era, a period replete with climate oscillations brought on by glacial cycles. These shifts greatly influenced the range and distribution of species across North America, as ranges were contracted and expanded repeatedly. The onset of climate change required plant species to move, adapt, or go extinct. Upon glacial recession, new environments were open to colonization or expansion, often requiring adaptation of existing flora [31,32]. These cycles could influence speciation either through genetic differentiation spurred by range fragmentation into glacial refugia or through admixture or introgression as previously isolated entities mixed and created new and differing gene pools which contributed to speciation [33]. These events, coupled with selection pressures caused by new environments or competitors, may have spurred genetic and/or morphological change, perhaps influencing the tempo of diversification. The idea of Quaternary climate shifts impacting genetic divergence and diversification is not new [34,35]. While most studies involving the influence of this time period investigate intraspecific and population-level genetic divergence (for example,[36,37]), studies concerning the impact of this era on speciation also exist but are more rare (for example, [31,38]). If NAm Psoraleeae diversified during this time, the tempo of diversification of this group may have been influenced by these climate shifts.
In this study, we present NAm Psoraleeae as a model study system for investigating the impact of climate change on diversification within a recent, rapid radiation. We test the hypothesis of a diversification rate shift within NAm Psoraleeae against the null hypothesis of no shift in diversification rate within the group. We then explore diversification in light of various climate-related hypotheses. In particular, we test the hypothesis that a shift to xeric habitats acted as a key innovation, thus spurring a diversification rate increase in Pediomelum and Psoralidium as opposed to the rest of NAm Psoraleeae against the null hypothesis of no net increase in diversification rate of these genera. We also investigate the impact of Quaternary climate oscillations on diversification of NAm Psoraleeae. To do so, we estimate divergence times of NAm Psoraleeae to determine if the group was diversifying during this period. We then correlate diversification rate shifts to geologic time. The young age of this group provides a glimpse into the emerging evolution of NAm Psoraleeae and the impact of changing environments and climes on its radiation. In addition, the knowledge gained herein may provide insight into other species radiations that have endured similar conditions, and serves as a model for the study of diversification rates and ecological fluctuations in the robust phylogenetic context of a recent, rapid radiation.
Results
Divergence dates
A molecular clock was rejected for each data set (p < 0.00001; 52 degrees of freedom), justifying the use of a relaxed molecular clock for divergence dating. The mixed model combined analysis from BEAST yielded a maximum clade credibility tree very similar to previously published phylogenies [9], suggesting a monophyletic NAm Psoraleeae (Figure 1). Individual gene analyses differed somewhat in their divergence date estimates of key nodes across Psoraleeae, illustrating the variation in rates of molecular evolution and phylogenetic information content inherent in each gene region (Table 1). For the most part, date estimates determined using nuclear markers are similar to or younger than dates based on chloroplast markers. Two exceptions to this pattern exist: the node representing the most recent common ancestor (MRCA) of Rupertia has the minimum date (ITS) and the maximum date (waxy) estimated by nuclear markers, while the dates estimated for the MRCA of Glycine and Psoraleeae by nuclear genes exceed those estimated by chloroplast markers.
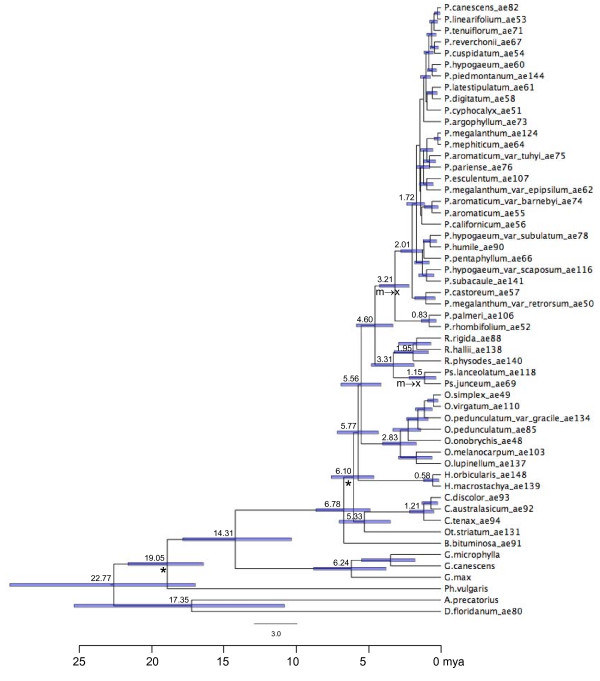
Figure 1.
Chronogram representing the maximum clade credibility tree estimated in BEAST based on total evidence across eight DNA regions using mixed models. Mean divergence dates shown for key nodes. Gray bars represent the 95% high posterior density credibility interval for node age. * nodes used as calibration points. Arrow denotes shift from/to mesic (m) or xeric (x) habitat hypothesized along the lineage shown. Ingroup: P = Pediomelum; Ps. = Psoralidium; R. = Rupertia; O. = Orbexilum; H. = Hoita. Outgroup: C. = Cullen; Ot. = Otholobium; B. = Bituminaria; G. = Glycine; Ph. = Phaseolus; D. = Desmodium. Numbers following taxon names represent accession numbers with reference to previous work (see [9]).
Table 1.
Divergence dates of key nodes estimated using BEAST
| MRCA of: | matK | rpoB- trnC | trnD- trnT | trnL- trnF | trnS- trnG | trnK | waxy | ITS | Ave (Genes) | Total |
| Glycine & Phaseolus* | 18.68 | 18.90 | 18.46 | 18.67 | 18.50 | 18.64 | 18.82 | 18.36 | 18.63 | 19.11 |
| SD | 0.028 | 0.213 | 0.033 | 0.023 | 0.026 | 0.037 | 0.027 | 0.032 | 0.183 | 0.038 |
| 95% HPD lower | 16.01 | 16.16 | 15.77 | 16.06 | 15.61 | 16.02 | 16.20 | 15.56 | 16.54 | |
| 95% HPD upper | 21.42 | 21.51 | 21.17 | 21.43 | 21.15 | 21.51 | 21.56 | 21.03 | 21.80 | |
| Cullen & Rupertia* | 6.62 | 6.31 | 6.82 | 6.56 | 6.74 | 6.59 | 6.36 | 6.90 | 6.61 | 6.08 |
| SD | 0.024 | 0.019 | 0.039 | 0.024 | 0.026 | 0.033 | 0.030 | 0.032 | 0.206 | 0.055 |
| 95% HPD lower | 5.10 | 4.72 | 5.37 | 4.91 | 5.07 | 5.06 | 4.69 | 5.45 | 4.61 | |
| 95% HPD upper | 8.17 | 7.89 | 8.45 | 8.14 | 8.44 | 8.29 | 7.97 | 8.45 | 7.56 | |
| Glycine & Psoraleeae | 13.09 | 14.28 | 14.87 | 13.02 | 16.16 | 12.33 | 18.70 | 16.76 | 14.90 | 14.32 |
| SD | 0.125 | 0.240 | 0.247 | 0.157 | 0.398 | 0.175 | 0.055 | 0.244 | 2.180 | 0.145 |
| 95% HPD lower | 8.27 | 8.89 | 9.17 | 7.75 | 7.14 | 7.61 | 15.58 | 9.80 | 10.39 | |
| 95% HPD upper | 18.64 | 20.83 | 20.54 | 19.05 | 24.42 | 17.46 | 21.97 | 20.77 | 17.95 | |
| Psoraleeae | 6.67 | 9.55 | 7.10 | 6.89 | 9.13 | 8.12 | 7.08 | 7.12 | 7.71 | 6.77 |
| SD | 0.026 | 0.107 | 0.044 | 0.066 | 0.368 | 0.131 | 0.116 | 0.069 | 1.099 | 0.059 |
| 95% HPD lower | 5.01 | 5.85 | 5.13 | 4.62 | 4.73 | 5.24 | 4.19 | 5.27 | 4.91 | |
| 95% HPD upper | 8.32 | 13.82 | 9.34 | 9.58 | 17.36 | 11.63 | 10.57 | 8.84 | 8.69 | |
| Orbexilum | 3.67 | 2.94 | 3.82 | 3.54 | 4.04 | 3.88 | 2.66 | 3.03 | 3.45 | 2.83 |
| SD | 0.064 | 0.049 | 0.034 | 0.055 | 0.132 | 0.065 | 0.056 | 0.107 | 0.506 | 0.038 |
| 95% HPD lower | 1.58 | 1.04 | 2.20 | 1.02 | 0.76 | 1.68 | 0.88 | 1.47 | 1.72 | |
| 95% HPD upper | 6.07 | 5.09 | 5.62 | 6.66 | 8.46 | 6.11 | 4.82 | 5.08 | 4.06 | |
| Leucocraspedon & Psoralidium/Rupertia | 5.07 | 8.60 | 6.05 | 6.08 | 9.16 | 6.51 | 4.66 | 5.26 | 6.42 | 4.60 |
| SD | 0.043 | 0.095 | 0.047 | 0.081 | 0.450 | 0.134 | 0.136 | 0.046 | 1.638 | 0.050 |
| 95% HPD lower | 3.35 | 5.28 | 4.32 | 3.53 | 3.76 | 3.90 | 2.51 | 3.62 | 3.34 | |
| 95% HPD upper | 6.68 | 12.61 | 7.69 | 8.93 | 18.31 | 9.84 | 7.81 | 7.07 | 5.88 | |
| Leucocraspedon & Pediomelum | 4.02 | 6.46 | 4.52 | 4.79 | 6.49 | 4.61 | 3.62 | 3.56 | 4.76 | 3.21 |
| SD | 0.042 | 0.098 | 0.044 | 0.061 | 0.274 | 0.074 | 0.098 | 0.051 | 1.149 | 0.040 |
| 95% HPD lower | 2.51 | 3.62 | 3.04 | 2.89 | 2.83 | 2.52 | 1.85 | 2.11 | 2.21 | |
| 95% HPD upper | 5.49 | 9.73 | 6.08 | 6.93 | 12.14 | 6.57 | 5.74 | 5.12 | 4.27 | |
| Leucocraspedon | 1.46 | 1.96 | 1.51 | 1.10 | 1.88 | 1.70 | 1.29 | 0.12 | 1.38 | 0.83 |
| SD | 0.023 | 0.041 | 0.022 | 0.027 | 0.103 | 0.044 | 0.034 | 0.007 | 0.584 | 0.013 |
| 95% HPD lower | 0.40 | 0.27 | 0.49 | 0.03 | 0.03 | 0.34 | 0.23 | 0.00 | 0.34 | |
| 95% HPD upper | 2.70 | 4.33 | 2.82 | 2.77 | 4.83 | 3.54 | 2.69 | 0.44 | 1.38 | |
| Pediomelum | 3.95 | 3.78 | 2.03 | 2.85 | 6.46 | 2.91 | 2.17 | 2.06 | 3.28 | 2.01 |
| SD | 0.041 | 0.064 | 0.025 | 0.037 | 0.281 | 0.045 | 0.056 | 0.048 | 1.483 | 0.033 |
| 95% HPD lower | 2.49 | 1.76 | 1.13 | 0.92 | 2.62 | 1.27 | 0.88 | 1.08 | 1.25 | |
| 95% HPD upper | 5.53 | 6.15 | 3.09 | 4.69 | 12.18 | 4.55 | 3.70 | 3.23 | 2.87 | |
| Psoralidium & Rupertia | 3.91 | 2.15 | 3.41 | 1.23 | 6.92 | 4.44 | 6.23 | 4.47 | 4.09 | 3.31 |
| SD | 0.035 | 0.031 | 0.035 | 0.022 | 0.401 | 0.082 | 0.147 | 0.053 | 1.901 | 0.051 |
| 95% HPD lower | 2.10 | 0.41 | 1.55 | 0.10 | 0.84 | 2.00 | 3.46 | 2.48 | 1.87 | |
| 95% HPD upper | 5.77 | 4.20 | 5.55 | 2.94 | 15.51 | 7.17 | 9.92 | 6.39 | 4.84 | |
| Psoralidium | 2.76 | 0.41 | 0.55 | 0.33 | 2.41 | 1.22 | 1.87 | 1.50 | 1.38 | 1.15 |
| SD | 0.033 | 0.010 | 0.011 | 0.077 | 0.192 | 0.030 | 0.040 | 0.040 | 0.925 | 0.021 |
| 95% HPD lower | 0.97 | 0.00 | 0.00 | 0.00 | 0.01 | 0.03 | 0.23 | 0.34 | 0.36 | |
| 95% HPD upper | 4.60 | 1.30 | 1.54 | 1.09 | 8.68 | 3.05 | 3.91 | 2.95 | 2.23 | |
| Rupertia | 1.99 | 2.28 | 1.96 | 3.13 | 5.14 | 2.10 | 5.92 | 1.97 | 3.06 | 1.95 |
| SD | 0.027 | 0.032 | 0.032 | 0.035 | 0.162 | 0.042 | 0.092 | 0.048 | 1.583 | 0.048 |
| 95% HPD lower | 0.55 | 0.54 | 0.59 | 0.84 | 0.74 | 0.57 | 3.14 | 0.55 | 0.87 | |
| 95% HPD upper | 3.79 | 4.36 | 3.50 | 5.65 | 10.49 | 4.05 | 8.81 | 3.61 | 3.24 | |
| Hoita | 0.92 | 1.51 | 0.44 | 1.39 | 1.58 | 2.87 | 1.22 | 0.75 | 1.34 | 0.58 |
| SD | 0.018 | 0.026 | 0.010 | 0.028 | 0.062 | 0.120 | 0.035 | 0.032 | 0.735 | 0.011 |
| 95% HPD lower | 0.02 | 0.07 | 0.01 | 0.02 | 0.05 | 0.08 | 0.03 | 0.05 | 0.15 | |
| 95% HPD upper | 2.42 | 3.59 | 1.25 | 3.55 | 4.13 | 7.24 | 3.56 | 1.80 | 1.21 | |
| Cullen | 1.15 | 2.12 | 1.46 | 1.45 | 1.57 | 0.99 | 1.76 | 1.52 | 1.50 | 1.21 |
| SD | 0.018 | 0.034 | 0.028 | 0.022 | 0.030 | 0.028 | 0.045 | 0.037 | 0.347 | 0.023 |
| 95% HPD lower | 0.08 | 0.32 | 0.22 | 0.08 | 0.01 | 0.02 | 0.45 | 0.40 | 0.50 | |
| 95% HPD upper | 2.65 | 4.20 | 3.04 | 3.52 | 4.08 | 2.56 | 3.28 | 3.13 | 2.19 | |
| Glycine | 4.50 | 5.78 | 6.47 | 6.26 | 5.45 | 5.77 | 8.25 | 4.99 | 5.93 | 6.24 |
| SD | 0.090 | 0.143 | 0.135 | 0.106 | 0.167 | 0.107 | 0.272 | 0.136 | 1.132 | 0.115 |
| 95% HPD lower | 1.11 | 1.31 | 2.50 | 2.30 | 0.67 | 1.85 | 3.89 | 1.82 | 3.82 | |
| 95% HPD upper | 8.43 | 10.84 | 10.75 | 11.05 | 11.54 | 10.02 | 14.07 | 9.31 | 8.87 |
MRCA is most recent common ancestor. SD is standard deviation. 95% upper and lower HPD is high posterior density credibility interval. Ave is the average of the dates across individual gene estimates with accompanying standard deviation below. Total is the date based on the combined dataset using mixed models.
Considering the gene regions as a whole, the combined mixed model analysis estimated the divergence date of calibrated nodes closer to the date expected than an analysis that averaged date estimates across all gene regions. For example, the calibrated node of the MRCA of Glycine and Phaseolus should have had a mean of 19.2 mya; the combined analysis estimated this divergence date at 19.1 mya while the average across the individual genes for this node was 18.6 mya (Table 1). With the exception of Glycine and the MRCA of Glycine and Phaseolus, the mean divergence date summarized across individual genes is older than the mean divergence date estimated under the combined mixed model analysis.
The minimum and maximum divergence date estimates across the individual genes could be taken as a range of uncertainty surrounding the divergence date of respective nodes, much the way we utilize the credibility intervals from Bayesian analyses. The ranges suggested across individual gene regions differ somewhat from the 95% high posterior density (HPD) credibility interval determined in the combined mixed model analysis. With the exception of the MRCAs of Glycine/Psoraleeae, Orbexilum, Cullen, and Glycine, the range obtained from the minimum and maximum estimates across individual gene regions is more conservative (wider) than the Bayesian credibility interval from the combined analysis (see Table 1).
While divergence estimates from the combined mixed model analysis differed from the average across individual gene estimates, the difference is slight. Furthermore, the divergence ranges suggested by utilizing the minimum and maximum estimates across individual genes or the range suggested by the 95% confidence interval (+/- 2 standard deviations) inferred using the mean across individual genes both exhibit considerable overlap with the range suggested by the 95% HPD credibility interval. We therefore confine our discussion to the dates and ranges observed under the combined mixed model framework.
As estimated in the combined analysis, Psoraleeae began diversifying in North America around 5.8 mya with the split of the genus Hoita from the remaining taxa (Figure 1). Orbexilum split from the ancestor of Pediomelum, Psoralidium, and Rupertia shortly thereafter, with the genus beginning to diversify around 2.8 mya (see Table 1 for credibility intervals). Around 4.6 mya, the Pediomelum lineage split from the lineage leading to Psoralidium and Rupertia, two genera which would later split around 3.3 mya. Rupertia began diversifying around 1.95 mya while Psoralidium speciated around 1.15 mya. Pediomelum, the largest genus, began diversifying around 3.2 mya.
Differential diversification rates
The results of the seven whole-tree statistics computed in SymmeTREE using the 12 maximum parsimony tree topologies suggest a significant shift in diversification rates within NAm Psoraleeae with strong support (all statistics' p-value < 0.00001). The delta statistics, those that help to localize the diversification rate shift to a certain area of the phylogeny, did not find statistical support for a single shift point of diversification rate. However, the test does support that a shift took place, either gradually over the whole phylogeny or in multiple places over time.
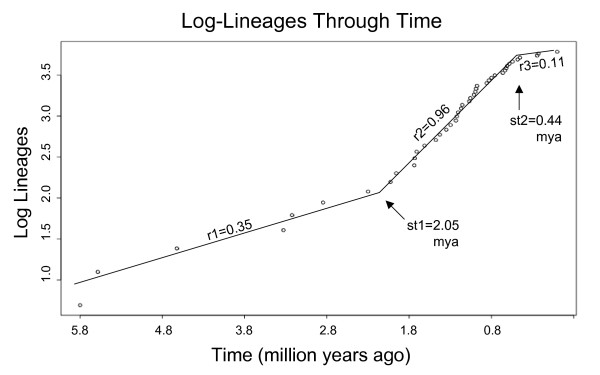
The birth-death likelihood (BDL) analysis based on the maximum clade credibility tree from BEAST determined the maximum likelihood estimate of the speciation rate under the pureBirth model to be 0.58 speciation events per million years for the combined data. BDL chose pureBirth as the best rate-constant model (Table 2). It was therefore used as the null to calculate ΔAICrc for the rate variable models. The critical value for ΔAICrc, as compared with the best rate-constant model, to maintain a significance level of α = 0.05 is 7.96. BDL found yule3rate as the best rate-variable model (ΔAICrc = 11.732). We therefore reject the null hypothesis of rate-constancy because the observed ΔAICrc > 7.96. According to the scenario suggested by the yule3rate model, NAm Psoraleeae began diversifying with a net diversification rate of 0.35 speciation events per million years. A shift in net diversification rate took place 2.05 mya in which the rate shifts dramatically to 0.96 speciation events per million years. The net diversification rate shifted again around 0.44 mya, decreasing to 0.11 speciation events per million years (Table 2). A lineage through time plot illustrates the definitive 3-rate model as outlined in the BDL analysis (Figure 2). Combined evidence from the BDL and divergence date analyses places the diversification rate shift at the origin of the main Pediomelum radiation.
Table 2.
Results of fitting diversification models to North American Psoraleeae using Birth-Death Likelihood
| pureBirth | BD | DDL | DDX | yule2rate | yule3rate | |
| Parameters | r 1 = 0.58 | r 1 = 0.58 | r 1 = 0.93 | r 1 = 0.74 | r 1 = 0.76 | r 1 = 0.35 |
| a = 0 | k = 68.18 | x = 0.081 | r 2 = 0.11 | r 2 = 0.96 | ||
| st = 0.45 | r 3 = 0.11 | |||||
| st 1 = 2.05 | ||||||
| st 2 = 0.44 | ||||||
| Ln(L) | 54.723 | 54.723 | 56.242 | 54.829 | 61.654 | 64.588 |
| AIC | -107.445 | -105.445 | -108.484 | -105.658 | -117.308 | -119.177 |
| ΔAIC | 0 | 2 | -1.039 | 1.787 | 9.863 | 11.732 |
The maximum clade credibility chronogram was calibrated such that the divergence of Hoita and Pediomelum was scaled to 5.8 mya. r = net diversification rate (speciation events per million years); a = extinction fraction; st = time of rate shift (mya); k = carrying capacity parameter; x = rate change parameter; Ln(L) = Log-likelihood; AIC = Akaike information criterion; ΔAIC = change in AIC relative to pureBirth.
Figure 2.
Lineage through time plot for diversification in North American Psoraleeae based on the total evidence maximum clade credibility chronogram (see Figure 1). All outgroups were culled from the chronogram until only North American Psoraleeae taxa remained, leaving Hoita, Rupertia, Psoralidium, Orbexilum, and Pediomelum. st = time of diversification rate shifts from yule3rate model estimates. r = diversification rate. Mya = million years ago.
Testing of macroevolutionary hypotheses
The hypothesis of xeric habitats acting as a key innovation in the diversification of NAm Psoraleeae based on the 1000 posterior distribution trees from the BEAST analysis was rejected (p-value = 0.80) using Ree's key innovation test. The hypothesis that Quaternary climate shifts influenced diversification of NAm Psoraleeae is supported by the acceptance of the yule3rate model having a diversification rate shift, st, between 2.5 and 0.8 mya, the time span equating to glaciation-induced climate oscillations during the Quaternary [39]. This translates to a 2.67-fold increase in net diversification rate around 2 mya (Table 2; Figures and 2).
Discussion
Recent, rapid radiations
The legume family is somewhat remarkable in that age estimates of this incredibly diverse group are relatively young, with transcontinental crown clade ages ranging from 8 to 16 mya and intracontinental crown clade ages ranging from 4 to 6 mya, providing a mostly Quaternary to Neogene age distribution [5,6]. While Astragalus is the legume poster child for rapid radiations, other recent, rapid radiations exist within Fabaceae [40,41]. Such radiations are often characterized by short branch lengths, little sequence variation, and diverse morphology and ecological habitats [42]. NAm Psoraleeae, especially Pediomelum, exhibits all of these characteristics [9,10]. This, coupled with evidence of increased rates of molecular evolution and recent age estimates of transcontinental clades, suggests that tribe Psoraleeae is a recent, rapid radiation [5].
Divergence dates
Our divergence date estimates place the origin of NAm Psoraleeae between 4.2 and 7.5 mya with a mean estimate of 5.8 mya, concomitant with many other legume genera [6]. Of particular interest in terms of divergence dates is the largest genus, Pediomelum. Grimes' circumscription segregates Pediomelum into three subgenera based on branching pattern and dehiscence of inflorescences: subgenus Leucocraspedon includes P. rhombifolium and P. palmeri with remaining taxa split between subgenera Disarticulatum and Pediomelum [10]. The diversification of this genus began around 3 mya (Figure 1, Table 1) with the split of subgenus Leucocraspedon from the remainder of Pediomelum. The two species in subgenus Leucocraspedon differ greatly from the rest of the genus both morphologically and genetically [9,10], this evidenced in the divergence of the two main Pediomelum lineages. The remainder of Pediomelum began diversifying around 2 mya, constituting the main body of species richness of NAm Psoraleeae. To place this in perspective with other rapid radiations, the Hawaiian silverswords, the model for adaptive radiations, diverged around 5 mya [43]. Within Fabaceae, Astragalus was estimated to have diverged from Oxytropis around 12 to 16 mya with Neo-Astragalus radiating approximately 4.4 mya [44], while the Andean radiation of Lupinus is dated around 1.5 mya [7]. These dates place the diversification of Pediomelum on level ground with other well-known examples of recent radiations.
Diversification rates
A rapid, family-wide diversification is strongly supported in the legume family [5]. Psoraleeae is no exception. Estimates of diversification rates vary depending on the model of diversification used in BDL on the maximum clade credibility tree from BEAST (Table 2). However, all our estimates of NAm Psoraleeae diversification are as fast or faster than rates determined for other notable rapid radiations: under the pureBirth model, a diversification rate of 0.58 for NAm Psoraleeae is similar to other rapid radiations including the Hawaiian Silverswords [43], mosses [45], squamate reptiles [46], and salamanders [47], all of which range between 0.4 and 0.8 speciation events per million years.
Skewed species richness to Pediomelum suggests a shift in net diversification rates within NAm Psoraleeae. Our analyses, including both topological and temporal methods, confirmed this hypothesis. Whole-tree statistics based on topology suggested a shift in diversification rate mid-depth in the phylogeny of NAm Psoraleeae. While a shift was strongly supported, the topological method was unable to decipher between the presence of multiple shifts in diversification rates along the phylogeny or a gradual change in diversification rate throughout the phylogeny as a whole. In addition, the topological method could not determine whether the shift or shifts were net increases or decreases in diversification rate. This lack of information is due to limitations of the topological method stemming from the fact that it relies solely on the order of branching events as opposed to the timing of branching events. The greater power of BDL, a temporal method, found two shifts in net diversification rate along the phylogeny. According to diversification rates suggested by the yule3rate model and BDL, NAm Psoraleeae diversification rates shifted dramatically with a net increase from 0.35 to 0.96 speciation events per million years approximately 2 mya. The increased rate is one of the fastest diversification rates published to date, rivaling those of ice plants in South Africa [48] and the Andean radiation of Lupinus [7]. The increase in net diversification rate did indeed occur at mid-depth in the phylogeny and represents a large increase in the diversification of the tribe within North America. The concordance of dates between the divergence date and BDL analyses confirms that the diversification rate shift took place as the main radiation of Pediomelum (subgenera Disarticulatum and Pediomelum), began diversifying around 2 mya (Figure 1, Table 2).
In addition to a net increase, analyses also provide evidence for a net decrease in diversification rate from 0.96 to 0.11 speciation events per million years around 440,000 years ago. This rate is far lower than the previous rates, suggesting a decrease in the amount of speciation or an increase in the amount of extinction or both. The low speciation rate seen at this recent time point could represent the speciation/population boundary. Diversification from this point forward to the present could be seen as existing on a population-level scale, as genetic differentiation between populations not sufficiently diverged to be distinguished as species. Furthermore, this decrease in diversification rate may be indirectly influenced by recent events, including those of anthropogenic origin. Indeed, we know for certain that human influence has been the cause of extinction within NAm Psoraleeae: Orbexilum stipulatum, once endemic to Rock Island, Kentucky, at Falls of the Ohio River, is confirmed extinct after the island was destroyed for construction of a dam; Orbexilum macrophyllum was collected from a single site in North Carolina and is presumed extinct [49]. Still other species are severely threatened, such as Pediomelum pentaphyllum and P. humile, due to the elimination of extant populations through urban development, leaving only one or two known populations alive. Estimates of more recent diversification shifts are especially dependent on the number of extant species. If the number of species is depleted through recent events such as human influence or through the lack of species recognition, it will more likely be reflected in diversification rate shifts taking place at more recent times, this due to the fact that we are estimating the diversification rate based on the number of species, instead of the number of clades.
Diversification shifts – causes?
Uneven diversification is evident in both taxonomic and geographical realms, exemplified in the ideas of species richness and the latitudinal diversity gradient. A great number of hypotheses have been proposed as explanations for uneven diversification with many centering on key innovations or climate. While the pattern of the latitudinal diversity gradient, meaning the existence of greater species numbers centered around lower latitudes, is well documented, the mechanism(s) behind the phenomenon is unclear, leading to a plethora of hypotheses [11]. It is possible and even probable that no single hypothesis can explain such patterns [50]. Furthermore, these and other patterns could be the results of purely stochastic processes. However, evidence has shown that climatic factors are especially influential on the evolutionary history of many organismal groups (see below). As NAm Psoraleeae is within temporal and geographical range of Quaternary climate oscillations and exhibits a wide variety of climate-based habitats, we chose to address hypotheses involving climate change as possible effectors of diversification rate shifts.
Habitat shifts
Past climate change has impacted current climate regimes, creating a spectrum of climate-based habitats across the earth. Movement of ancestral species from one habitat to another may also have spurred diversification, relegating climate-based habitat as a key innovation. Climate regime shifts have been found to have impacted diversification across various organismal groups, including plants [51] and lizards [52,53]. Hoita, Orbexilum, and Rupertia all reside in more mesic environments characterized by warm, moist, humid climates whereas Psoralidium and Pediomelum reside in more xeric habitats characterized by hot, dry environments such as the deserts of the Intermountain and Southwest United States (see Figure 1). In testing habitat shifts from mesic to xeric habitats, our analyses suggest that past climate change affected diversification rates in NAm Psoraleeae more than shifts in present climate regimes as defined by current habitats. The shift to xeric habitats was not considered a key innovation in terms of increasing diversification rates within the group. Instead, current habitats could be seen as relicts from the effects of past climate change on its influence on the diversification of North American Psoraleeae.
Quaternary climate oscillations
The recent origin of the main radiation of Pediomelum around 2 mya places its diversification in the middle of the Pleistocene epoch of the Quaternary era, a time period replete with dramatic glaciation-induced climate oscillations. These glacial fluxes include Milankovitch cycles which consist of repeated formation and recession of glacial ice sheets having periodicities of 20,000 to 100,000 years, resulting in wide-ranging climate fluxes [33]. It has been well documented that much of the Earth's biodiversity and many species' evolutionary trajectories were drastically influenced by the climate fluxes brought on by Quaternary glaciations (for example, [36,37,39,54]). In North America, the sheer presence of ice sheets shifted climates and strongly influenced the range and distribution of species [55]. Ice formation may have influenced speciation via genetic differentiation through range fragmentations such as glacial refugia. Upon glacial recession, new environments were formed and subsequently colonized through species expansion and adaptation [31,32]. Given the geographical and temporal distribution of NAm Psoraleeae, these cycles may have influenced its evolutionary history. Indeed, the shift point for the increase in diversification along the phylogeny at 2 mya and the lineage through time plot from our analyses support the hypothesis that Quaternary climate fluxes, occurring between 2.5 and 0.8 mya, influenced the diversification of NAm Psoraleeae.
Geography
The effects of these oscillations are not homogenous, however. The Quaternary ice ages and climate oscillations affected geographic regions across North America differently. The Southeast provided several refugia in which species could survive [31] while the montane regions of the West were characterized by more varied environments such as ice, tundra, pluvial lakes, and deserts [56]. In addition, elevation played an important role in species distributions as elevational ranges of plants were 600 to 1200+ meters below modern ranges, an effect felt more readily in the topographically diverse regions of the West [57]. Upon glacial recession, the Intermountain West offered a greater variety of habitats for colonization and adaptation as a result of topography. This is in line with Cracraft's lithospheric complexity hypothesis, stating that geographic separation spurred by topographical changes promotes speciation [58]. In addition to orogeny, glacial cycles contributed to topographical change, and thus are linked with the lithospheric hypothesis.
Geographical trends are seen in the diversity of Psoraleeae. NAm Psoraleeae ranges throughout the United States: Orbexilum is mostly confined to the southeast United States, with one species reaching into Mexico, Pediomelum is mostly found in central and Intermountain states, Hoita is endemic to California, Rupertia ranges along the West Coast, and Psoralidium is spread over the great plains with one species endemic to Utah. Of interest is the seeming correlation of lineage age with southerly distribution. Orbexilum is one of the oldest NAm Psoraleeae genera, diversifying earlier than most other lineages. This could be demonstrative of long-established ranges of Orbexilum through Pleistocene refugia in the Southeast United States. Of import also is the age of Pediomelum as a whole, beginning its diversification around 3 mya with the break off of subgenus Leucocraspedon. The two Leucocraspedon species exhibit the most southern distribution of all Pediomelum species with P. rhombifolium being distributed throughout Texas and into Mexico while P. palmeri is almost exclusively found in Mesoamerica. This southern distribution perhaps allowed this lineage to persist through the Quaternary climate oscillations as the effects of glacial climate change were moderated toward the equator. The main radiation of Pediomelum exhibits the greatest number of species with the lowest intraspecific variation, findings in accord with the lithospheric complexity hypothesis, geographic separation, ecological diversification due to habitat diversity, and range fragmentation brought on by Quaternary climate shifts. The pattern of less diversity in the Southeast (as evidenced by Orbexilum) and greater diversity in the Southwest (as evidenced by Pediomelum) is also found in Cicindela beetles [59] and Agelenopsis spiders [60], among others.
Conclusion
NAm Psoraleeae has proved useful for studying evolutionary divergence and the impact of climate change on diversification within a recent, rapid radiation. This work effectively demonstrates differential rates of diversification at the species and generic levels, trends manifested across taxonomic levels within Fabaceae. An estimation of divergence dates within Psoraleeae found that the main radiation of Pediomelum, comprising the bulk of species richness in NAm Psoraleeae, originated 1 to 2 mya during the Quaternary era, concurrent with the Milankovitch climate oscillations brought on by glacial cycles. The hypothesis of a diversification rate shift in NAm Psoraleeae was confirmed by both topological and temporal methods. We demonstrated a large increase in diversification rate near the beginning of the Pleistocene epoch. Our findings suggest that the diversification rate increase in NAm Psoraleeae is not due to current climate regimes as represented by habitat, but rather imply that Quaternary climate oscillations impacted the diversification of the group. We show nonrandom spatial diversification as well, with higher species diversity in the American Southwest.
NAm Psoraleeae diversification is a good example of how earthly dynamics including global climate change and topography work together to shape biodiversity. In a time when the issue of global climate change pervades society and science, this study aptly illustrates the control that global climate change can have over the evolutionary trajectories and fates of lineages and species.
Methods
Taxon sampling and DNA sequences
This study includes a thorough sampling of NAm Psoraleeae, with only four of 47 recognized NAm Psoraleeae taxa unsampled. Of these, Hoita strobilina is critically endangered, Orbexilum macrophyllum and O. stipulatum are presumed extinct, and Pediomelum latestipulatum var. latestipulatum was not collected. In addition to North American taxa, representatives of Otholobium, Cullen, and Bituminaria are included for estimating the dates of divergence within the tribe. Six outgroup taxa are incorporated for calibration: Abrus precatorius, Phaseolus vulgaris, Desmodium floridanum, Glycine microphylla, G. canescens, and G. max cv. Essex. In total, 54 taxa are included as described in Egan and Crandall [9].
All phylogenetic analyses were based on eight DNA regions: the Internal Transcribed Spacer (ITS) and a single-copy gene, Granule Bound Starch Synthase I (Waxy) from the nuclear genome and the trnL-trnF, trnD-trnT, trnS-trnG and rpoB-trnC intergenic spacers, the trnK intron regions, and the matK protein-coding gene from the chloroplast genome. DNA isolations, primer specifications, PCR amplification, DNA sequencing, voucher specimen information, GenBank numbers, and maximum parsimony phylogenetic analyses are described in Egan and Crandall [9].
Divergence dates
Departure from a molecular clock was determined using likelihood ratio tests with scores estimated using rate-constant and rate-variable models [61] in PAUP* 4.0 [62]. Models were estimated as described in Egan and Crandall [9] using ModelTest 3.7 [63]; matK, trnK, trnD-trnT, rpoB-trnC, ITS, and the combined data set used the general time reversible (GTR) model plus proportion of invariable sites (I) and the gamma distribution (G) to account for rate heterogeneity, trnL-trnF used GTR+I, trnS-trnG used GTR+G, and waxy used HKY+G.
Divergence dates were estimated for individual genes and a combined total evidence dataset via Bayesian MCMC sampling conducted in BEAST v1.4.7 [64] under a relaxed clock model with log-normally distributed, uncorrelated rates of substitution between branches [65]. No topological constraints were employed, allowing topological uncertainty to be taken into account. We analyzed each of the eight DNA regions individually and the combined total evidence dataset, allowing the investigation of how rates of molecular evolution vary among DNA regions.
Two calibration points were used for estimating divergence dates in BEAST: the most recent common ancestor of Glycine and Phaseolus has a mean estimate of 19.2 mya accompanied by a standard deviation (SD) of 1.4 mya. The minimum and maximum age estimates for this node are 15.5 to 22.4 mya. The most recent common ancestor of Cullen and Rupertia was estimated to range between 4.1 to 8.8 mya with a mean date of 6.3 mya (0.9 SD). These dates were estimated from a family-wide divergence date analysis using penalized likelihood in r8s using Bayesian-inferred phylogenies for matK and based on 13 fossil calibration points [5]. We chose to use their matK date estimates over rbcL because the matK dataset incorporated more fossil calibration points (13 as opposed to 9) and the estimate of dates was more stable in the matK dataset as shown by a lower SD. We incorporated these dates as calibration points into our BEAST analyses by the use of a normal prior using their respective mean and SD estimates. This strategy allows uncertainty surrounding these dates as calibration points to be accounted for in our divergence date analyses.
Each gene or data set used the model of evolution determined by ModelTest mentioned above. The combined dataset incorporated mixed models into the analysis, allowing each gene to evolve under the model and parameters best suited to the gene region. The tree prior was modeled under the Yule speciation process. With the exception of the normally-distributed priors on calibration points, all other priors were set as default values in the program BEAUti v1.4.7 which was used to help create the XML files for input to BEAST [64]. Final analyses consisted of two separate MCMC runs of 5 to 10 million generations sampled every 1000 generations for the individual gene analyses, while the combined dataset was run for 15 million generations. Tracer v1.4 [66] was used to confirm likelihood stationary and adequate mixing of the MCMC chains and whether the two separate runs had converged to the same results. If runs had converged, results of both runs were combined using LogCombiner v1.4.7 [64]. Tracer v1.4 was then used to determine adequate burn-in (10%), confirm mixing and stationarity of the combined runs, and summarize the posterior distribution for divergence date estimates across key nodes. The maximum clade credibility tree was computed for the combined dataset using TreeAnnotator v.1.4.7, part of the BEAST software package.
Differential diversification rates
Two methodological strategies exist for estimating differential diversification rates across phylogenies: comparisons of topological species distribution to a random distribution of diversity; and comparisons of temporal distribution of divergence events to a randomly created distribution [67]. Topological methods are generally employed as a first test to determine if diversification rate shifts exist across lineages, while temporal methods are more powerful at addressing specific questions of when, how much, and where shifts in diversification took place along a phylogeny. Here, we employ both strategies to test both general and specific hypotheses concerning diversification rates.
The topological program SymmeTREE [68] was used to test the general hypothesis of variation in diversification rates across NAm Psoraleeae. This tests for deviations of observed topological distribution of species diversity to that expected under the Yule model [69] of an equal-rates Markov random-branching process. Seven whole-tree tests of differential diversification rates, each sensitive to different scenarios and nodal depths, are employed. MR tests for diversification at deeper nodes, or at the root of the tree, by determining if the observed trees are more or less asymmetric than expected [70]. IC is similar but is a calculated measure instead of a probability [70]. MΠ and MΣ consider the asymmetry of internal nodes, with MΠ more sensitive to deeper nodes through multiplication of individual node probabilities and MΣ more sensitive to nodes closer to the tips of the tree through addition of node probabilities. MΠ* and MΣ* vary from the above by having each node probability weighted by its size [71]. Unlike the above tests, B1 measures tree balance as opposed to imbalance. This statistic is sensitive to changes at the tips of the tree as it excludes the root [72]. As a topological method, SymmeTREE does not make use of branch length data, but uses order of branching events only. For the SymmeTREE analysis, we therefore used the previously estimated set of 12 fully resolved, most parsimonious trees from the total evidence maximum parsimony analysis that included gaps [9]. This allowed us the benefit of completely resolved trees and incorporation of a measure of phylogenetic uncertainty while avoiding the duplication of exact topologies that may be evident in the set of trees from the Bayesian posterior distribution. Bonferroni corrections were used to adjust significance thresholds across multiple tests.
The temporal method BDL was used to test specific hypotheses of diversification rate shifts [39,53]. This method calculates maximum likelihood estimates of speciation rate parameters and a likelihood score per tree. The null model of rate-constancy is tested against rate-variable models using the Akaike Information Criterion [73]. BDL has been found to perform as well or better than the popular γ-statistic, which only identifies temporal decreases in diversification [74], and has greater power to detect shifts in diversification rates in the presence of extinction [53]. By comparing the sum of the likelihood of internode distances across the tree under varying models, BDL can decipher between increased diversification rate vs. rate-constancy with background extinction [53]. Several diversification models were tested using LASER [75]. Rate-constant models included pureBirth, a constant speciation model (Yule) with zero extinction [69] and birth-death (BD), a constant speciation-constant extinction model [76]. Rate-variable models include density-dependent speciation models with exponential (DDX) and logistic (DDL) variants [77,78], yule2rate and yule3rate models. The yule2rate and yule3rate models are multi-rate variants of the Yule model, allowing two and three shifts in speciation rates respectively at times st across a tree. Branching times only were allowed as possible rate shift points. We used the maximum clade credibility tree from the combined dataset estimated in BEAST (chronogram shown in Figure 1) to test for diversification shifts using BDL. This chronogram was chosen in part due to the need for complete resolution and also to make use of divergence date estimates based on the full dataset. Outgroups were excised and the root, the ancestor of Hoita and Pediomelum, scaled to 5.8 mya – the divergence date estimated in BEAST based on the combined dataset (see Figure 1). Significance of the change in Akaike Information Criterion (AIC) scores was determined by creating a distribution of AIC scores. This was done by simulating 1000 trees using yuleSim in LASER having the same number of taxa and the same speciation rate as that estimated under the pureBirth model.
Testing of macroevolutionary hypotheses
The factors that we tested as influencing the tempo of diversification in NAm Psoraleeae include Quaternary climate changes and shifts in climate regime from moist, warm climes (mesic) to dry, hot climes (xeric). The adaptation to or colonization of xeric environments was hypothesized as a key innovation, a trait or character change whose origin may have spurred diversification. Pediomelum and Psoralidium live in xeric environments while other NAm Psoraleeae are in more mesic habitats. Xeric environment was tested to see if its origin was associated with increased diversification in NAm Psoraleeae against the null hypothesis of no increase in diversification rates across the tribe. We used the key innovation test [79] which employs Bayesian techniques to assess correlations between the timing of branching events (representative of speciation) and the evolutionary history of the character of interest. If acting as a key innovation, lineages with the new regime will have shorter waiting times before branching, as represented by internode length, than those without it. Tree length is calibrated to equal the total amount of change in the character over the whole phylogeny, such that waiting times represent the relative time spent in one character state as opposed to the other. Significance is determined by comparison to a null distribution of random waiting times between branching events. This goes beyond clade size methods by incorporating the time of branching events and allowing the trait to be both acquired and lost. Other tests assume acquisition only, not allowing loss of the trait, thereby excluding potential information from the analysis [79]. We used a set comprising the last 1000 trees from the Bayesian (BEAST) posterior distribution based on the combined dataset to test the hypothesis of xeric environment acting as a key innovation. This treeset was optimal for this analysis because the trees were already ultrametric and allowed incorporation of phylogenetic uncertainty. Trees were scaled so that the total tree length was equal to total character change from mesic to xeric habitats. In this case the tree length was 2, illustrating two changes from mesic to xeric environment at the base of the lineages leading to Psoralidium and Pediomelum (see Figure 1).
The impacts of Quaternary climate shifts were tested using lineage-through-time plots [39] and BDL using LASER. These used the maximum clade credibility chronogram from the combined dataset estimated in BEAST. A shift in diversification rates taken between 2.5 and 0.8 mya, the time of Quaternary climate oscillations, is taken as support for their influence on diversification in this group.
Authors' contributions
ANE gathered data, ran analyses, and wrote the paper. KAC suggested programs for analyses, gave feedback and guidance on hypotheses, and provided comments on the manuscript. Both authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank B Moore and R Ree for help with programs and L Johnson and anonymous reviewers for comments on the manuscript. This research was funded in part by fellowships and grants to ANE from Brigham Young University Graduate Studies, the American Society of Plant Taxonomists and the Society for Systematic Biologists.
Contributor Information
Ashley N Egan, Email: ane23@cornell.edu.
Keith A Crandall, Email: keith_crandall@byu.edu.
References
- Lewis G, Schrire BP, MacKinder B, Lock M. Legumes of the World. Kew: Royal Botanic Gardens; 2005. [Google Scholar]
- Magallon S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, Wojciechowski MF. Diversification rates in a temperate legume clade: Are there 'so many species' of Astragalus (Fabaceae)? Am J Bot. 1996;83:1488–1502. doi: 10.2307/2446103. [DOI] [Google Scholar]
- Wojciechowski M, Sanderson MJ, Baldwin BG, Donoghue MJ. Monophyly of aneuploid Astragalus (Fabaceae): evidence from nuclear ribosomal DNA-ITS sequences. Am J Bot. 1993;80:711–722. doi: 10.2307/2445441. [DOI] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Lavin M, Schrire BP, Lewis G, Pennington RT, Delgado-Salinas A, Thulin M, Hughes CE, Matos AB, Wojciechowski MF. Metacommunity process rather than continental tectonic history better explains geographically structured phylogenies in legumes. Phil Trans R Soc Lond B. 2004;359:1509–1522. doi: 10.1098/rstb.2004.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C, Eastwood R. Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. PNAS. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM. Rapid diversification of a species-rich genus of neotropical rain forest trees. Science. 2001;293:2242–2245. doi: 10.1126/science.1061421. [DOI] [PubMed] [Google Scholar]
- Egan AN, Crandall KA. Incorporating gaps as phylogenetic characters across eight DNA regions: ramifications for North American Psoraleeae (Leguminosae) Mol Phylogenet Evol. 2008;46:532–546. doi: 10.1016/j.ympev.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Grimes JW. A revision of the New World species of Psoraleeae (Leguminosae: Papilionoideae) Mem New York Bot Gard. 1990;61:1–113. [Google Scholar]
- Willig MR, Kaufmann DM, Stevens RD. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Ann Rev Ecol Syst. 2003;34:273–309. doi: 10.1146/annurev.ecolsys.34.012103.144032. [DOI] [Google Scholar]
- Rosenzweig ML. Species Diversity in Time and Space. Cambridge, UK: Cambridge University Press; 1995. [Google Scholar]
- Dodd ME, Silvertown J, Chase MW. Phylogenetic analysis of trait evolution and species diversity variation among angiosperm families. Evolution. 1999;53:732–744. doi: 10.2307/2640713. [DOI] [PubMed] [Google Scholar]
- Farrell BD. 'Inordinate fondness' explained: Why are there so many beetles? Science. 1998;281:555–559. doi: 10.1126/science.281.5376.555. [DOI] [PubMed] [Google Scholar]
- Hodges SA, Arnold ML. Spurring plant diversification: Are floral nectar spurs a key innovation? Proc R Soc Lond B. 1995;262:343–348. doi: 10.1098/rspb.1995.0215. [DOI] [Google Scholar]
- Niklas KJ, Tiffney BH, Knoll AH. Patterns in vascular land plant diversification. Nature. 1983;303:614–616. doi: 10.1038/303614a0. [DOI] [Google Scholar]
- Owens IPF, Bennett PM. Ecological basis of extinction risk in birds: Habitat loss versus human persecution and introduced predators. PNAS. 2000;97:12144–12148. doi: 10.1073/pnas.200223397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JE, Weitz FM, Fay MF, Cronk QCB, Linder HP, Reeves G, Chase MW. Rapid and recent origin of species richness in the Cape Flora of South Africa. Nature. 2001;412:181–183. doi: 10.1038/35084067. [DOI] [PubMed] [Google Scholar]
- Hawkins BA, Field R, Cornell HV, Currie DJ, Guegan JF, Kaufmann DM, Kerr JT, Mittelbach GG, Oberdorff T, O'Brien EM, Porter EE, Turner JRG. Energy, water, and broad-scale geographic patterns of species richness. Ecology. 2003;84:3105–3117. doi: 10.1890/03-8006. [DOI] [Google Scholar]
- Wright DH, Currie DJ, Maurer BA. Energy supply and patterns of species richness on local and regional scales. In: Ricklefs RE, Schluter D, editor. Species Diversity in Ecological Communities: Historical and Geographical Perspectives. Chicago, IL: University of Chicago Press; 1993. pp. 66–74. [Google Scholar]
- Francis AP, Currie DJ. A globally consistent richness-climate relationship for angiosperms. Am Nat. 2003;161:523–536. doi: 10.1086/368223. [DOI] [PubMed] [Google Scholar]
- Wiens J. Global patterns of diversification and species richness in amphibians. Am Nat. 2007;170:S86–S106. doi: 10.1086/519396. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Donnellan SC, Talaba AL, Lovette IJ. Exceptional among-lineage variation in diversification rates during the radiation of Australia's most diverse vertebrate clade. Proc R Soc Lond B. 2007;274:2915–2923. doi: 10.1098/rspb.2007.0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PR, McKay JK. Latitudinal variation in genetic divergence of populations and the potential for future speciation. Evolution. 2004;58:938–945. doi: 10.1111/j.0014-3820.2004.tb00428.x. [DOI] [PubMed] [Google Scholar]
- Sanders NJ, Lessard J-P, Fitzpatrick MC, Dunn RR. Temperature, but not productivity or geometry, predicts elevational diversity gradients in ants across spatial grains. Glob Ecol Biogeogr. 2007;16:640–649. doi: 10.1111/j.1466-8238.2007.00316.x. [DOI] [Google Scholar]
- Cardillo M, Orme CDL, Owens IPF. Testing for latitudinal bias in diversification rates: an example using new world birds. Ecology. 2005;86:2278–2287. doi: 10.1890/05-0112. [DOI] [Google Scholar]
- Weir J, Schluter D. Ice sheets promote speciation in boreal birds. Proc R Soc Lond B. 2004;271:1881–1887. doi: 10.1098/rspb.2004.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isely D. Native and Naturalized Leguminosae (Fabaceae) of the United States (exclusive of Alaska and Hawaii) Provo: MLBM Press; 1998. [Google Scholar]
- Ockendon DJ. A taxonomic study of Psoralea subgenus Pediomelum (Leguminosae) Southwest Nat. 1965;10:81–124. doi: 10.2307/3669160. [DOI] [Google Scholar]
- Maynard Smith J, Szathmary E. The major transitions in evolution. Oxford, UK: Freeman; 1995. [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Lond B Biol Sci. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett KD. Milankovitch cycles and their effects on species in ecological and evolutionary time. Paleobiology. 1990;16:11–21. [Google Scholar]
- Axelrod DI. Quaternary extinctions of large mammals. Univ Calif Publ Geol Sci. 1967;74:1–42. [Google Scholar]
- Kershaw AP. Quaternary vegetation and environments. In: Keast A, editor. Ecological Biogeography in Australia. Amsterdam: Dr. W. Junk; 1981. [Google Scholar]
- Ayoub NA, Riechert SE. Molecular evidence for Pleistocene glacial cycles driving diversification of a North American desert spider, Agelenopsis aperta. Mol Ecol. 2004;13:3453–3465. doi: 10.1111/j.1365-294X.2004.02335.x. [DOI] [PubMed] [Google Scholar]
- Knowles LL. Did the Pleistocene glaciations promote divergence? Tests of explicit refugial models in montane grasshoppers. Mol Ecol. 2001;10:691–701. doi: 10.1046/j.1365-294x.2001.01206.x. [DOI] [PubMed] [Google Scholar]
- Zhang LB, Comes HP, Kadereit JW. The temporal course of Quaternary diversification in the European high mountain endemic Primula sect. Auricula (Primulaceae) Int J of Plant Sci. 2004;165:191–207. doi: 10.1086/380747. [DOI] [Google Scholar]
- Barraclough TG, Vogler AP. Recent diversification rates in North American tiger beetles estimated from a dated mtDNA phylogenetic tree. Mol Biol Evol. 2002;19:1706–1716. doi: 10.1093/oxfordjournals.molbev.a003993. [DOI] [PubMed] [Google Scholar]
- Wagstaff SJ, Heenan PB, Sanderson MJ. Classification, origins, and patterns of diversification in New Zealand Carmichaelinae (Fabaceae) Am J Bot. 1999;86:1346–1356. doi: 10.2307/2656781. [DOI] [PubMed] [Google Scholar]
- Mitchell AD, Heenan PB. Sophora sect. Edwardsia (Fabaceae): further evidence from nrDNA sequence data of a recent and rapid radiation around the Southern Oceans. Bot J Linn Soc. 2002;140:435–441. doi: 10.1046/j.1095-8339.2002.00101.x. [DOI] [Google Scholar]
- Wagstaff SJ, Breitwieser I. Phylogeny and classification of Brachyglottis (Senecioneae, Asteraceae): An example of a rapid species radiation in New Zealand. Syst Bot. 2004;29:1003–1010. doi: 10.1600/0363644042450991. [DOI] [Google Scholar]
- Baldwin BG, Sanderson MJ. Age and rate of diversification of the Hawaiian silversword alliance (Compositae) Proc Nat Acad Sci USA. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski MF. Astragalus (Fabaceae): A molecular phylogenetic perspective. Brittonia. 2005;57:382–396. doi: 10.1663/0007-196X(2005)057[0382:AFAMPP]2.0.CO;2. [DOI] [Google Scholar]
- Wall DP. Origin and rapid diversification of a tropical moss. Evolution. 2005;59:1413–1424. [PubMed] [Google Scholar]
- Ricklefs RE, Losos JB, Townsend TM. Evolutionary diversification of clades of squamate reptiles. J Evol Biol. 2007;20:1751–1762. doi: 10.1111/j.1420-9101.2007.01388.x. [DOI] [PubMed] [Google Scholar]
- Kozak KH, Weisrock DW, Larson A. Rapid lineage accumulation in a non-adaptive radiation: phylogenetic analysis of diversification rates in eastern North American woodland salamanders (Plethodontidae: Plethodon) Proc R Soc Lond B Biol Sci. 2006;273:539–546. doi: 10.1098/rspb.2005.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klak C, Reeves G, Hedderson T. Unmatched tempo of evolution in Southern African semi-desert ice plants. Nature. 2004;427:63–65. doi: 10.1038/nature02243. [DOI] [PubMed] [Google Scholar]
- NatureServe Explorer: An online encyclopedia of life [web application]. Version 6.2 http://www.natureserve.org/explorer
- Clarke A, Gaston KJ. Climate, energy and diversity. Proc R Soc Lond B Biol Sci. 2006;273:2257–2266. doi: 10.1098/rspb.2006.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Cook L, Steane D. Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities? Phil Trans R Soc Lond B. 2004;359:1551–1571. doi: 10.1098/rstb.2004.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon LJ, Schulte JA, Larson A, Losos JB. Tempo and mode of evolutionary radiation in iguanian lizards. Science. 2003;301:961–964. doi: 10.1126/science.1084786. [DOI] [PubMed] [Google Scholar]
- Rabosky DL. Likelihood methods for detecting temporal shifts in diversification rates. Evolution. 2006;60:1152–1164. [PubMed] [Google Scholar]
- Smith CI, Farrell BD. Range expansions in the flightless longhorn cactus beetles, Moneilema gigas and Moneilema armatum, in response to Pleistocene climate changes. Mol Ecol. 2005;14:1025–1044. doi: 10.1111/j.1365-294X.2005.02472.x. [DOI] [PubMed] [Google Scholar]
- Clark PU, MacAyeal DR, Andrews JT, Barltein PJ. Ice sheets play important role in climate change. Eos. 1995;76:265–270. doi: 10.1029/95EO00157. http://www.agu.org/sci_soc/clark.html [DOI] [Google Scholar]
- Owen LA, Finkel RC, Minnich RA, Perez AE. Extreme southwestern margin of late Quaternary glaciation in North America: Timing and controls. Geology. 2003;31:729–732. doi: 10.1130/G19561.1. [DOI] [Google Scholar]
- Thompson RS, Whitlock C, Bartlein PJ, Harrison SP, Spaulding WG. Climatic changes in the western United States since 18,000 year bp. In: Wright HEJ, Kutzback JE, Webb III T, Ruddman WE, Street-Perrott FA, Barltein PJ, editor. Global Climates Since the Last Glacial Maximum. Minneapolis, MN: University of Minnesota Press; 1993. [Google Scholar]
- Cracraft J. Biological diversification and its causes. Ann Missouri Bot Gard. 1985;72:794–822. doi: 10.2307/2399222. [DOI] [Google Scholar]
- Vogler AP, Welsh A, Barraclough TG. Molecular phylogeny of the Cicindela maritima (Coleoptera: Cicindelidae) group indicates fast radiation in western North America. Ann Entom Soc Am. 1998;91:185–194. [Google Scholar]
- Ayoub NA, Riechert SE, Small RL. Speciation history of the North American funnel web spiders, Agelenopsis (Areneae: Agelenidae): Phylogenetic inferences at the population-species interface. Mol Phylogenet Evol. 2005;36:42–57. doi: 10.1016/j.ympev.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Version 40b10. Sunderland, MA: Sinauer Associates; 2002. PAUP*. Phylogenetic analysis using parsimony (* and other methods) [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. Tracer v1.4. 2007. http://beast.bio.ed.ac.uk/Tracer
- Sanderson MJ, Donoghue MJ. Reconstructing shifts in diversification rates on phylogenetic trees. Tree. 1996;11:15–20. doi: 10.1016/0169-5347(96)81059-7. [DOI] [PubMed] [Google Scholar]
- Chan KMA, Moore BR. SymmeTREE: whole-tree analysis of differential diversification rates. Bioinformatics. 2005;21:1709–1710. doi: 10.1093/bioinformatics/bti175. [DOI] [PubMed] [Google Scholar]
- Yule G. A mathematical theory of evolution, based on the conclusions of Dr JC Willis. Phil Trans R Soc Lond B. 1924;213:21–87. doi: 10.1098/rstb.1925.0002. [DOI] [Google Scholar]
- Page RDM. On islands of trees and the efficacy of different methods of branch swapping in finding most-parsimonious trees. Syst Biol. 1993;42:200–210. doi: 10.2307/2992542. [DOI] [Google Scholar]
- Chan KMA, Moore BR. Whole-tree methods for detecting differential diversification rates. Syst Biol. 2002;51:855–865. doi: 10.1080/10635150290102555. [DOI] [PubMed] [Google Scholar]
- Shao K-T, Sokal RR. Tree Balance. Syst Zool. 1990;39:266–276. doi: 10.2307/2992186. [DOI] [Google Scholar]
- Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- Pybus OG, Harvey PH. Testing macroevolutionary models using incomplete molecular phylogenies. Proc R Soc Lond B. 2000;267:2267–2272. doi: 10.1098/rspb.2000.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL. LASER: A maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol Bioinform Online. 2006;2:247–250. http://www.la-press.com/laser-a-maximum-likelihood-toolkit-for-detecting-temporal-shifts-in-di-a135 [PMC free article] [PubMed] [Google Scholar]
- Kendall DG. On the generalized 'birth-and-death' process. Ann Math Stat. 1948;19:1–15. doi: 10.1214/aoms/1177730285. [DOI] [Google Scholar]
- Nee S, Mooers AÃ, Harvey PH. Tempo and mode of evolution revealed from molecular phylogenies. PNAS. 1992;89:8322–8326. doi: 10.1073/pnas.89.17.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nee S, May RM, Harvey PH. The reconstructed evolutionary process. Phil Trans R Soc Lond B. 1994;344:305–311. doi: 10.1098/rstb.1994.0068. [DOI] [PubMed] [Google Scholar]
- Ree RH. Detecting the historical signature of key innovations using stochastic models of character evolution and cladogenesis. Evolution. 2005;59:257–265. [PubMed] [Google Scholar]