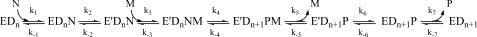Abstract
While matched nucleotide incorporation by DNA polymerase β (Pol β) has been well-studied, a true understanding of polymerase fidelity requires comparison of both matched and mismatched dNTP incorporation pathways. Here we examine the mechanism of misincorporation for wild-type (WT) Pol β and an error-prone I260Q variant using stopped-flow fluorescence assays and steady-state fluorescence spectroscopy. In stopped-flow, a biphasic fluorescence trace is observed for both enzymes during mismatched dNTP incorporation. The fluorescence transitions are in the same direction as that observed for matched dNTP, albeit with lower amplitude. Assignments of the fast and slow fluorescence phases are designated to the same mechanistic steps previously determined for matched dNTP incorporation. For both WT and I260Q mismatched dNTP incorporation, the rate of the fast phase, reflecting subdomain closing, is comparable to that induced by correct dNTP. Pre-steady-state kinetic evaluation reveals that both enzymes display similar correct dNTP insertion profiles, and the lower fidelity intrinsic to the I260Q mutant results from enhanced efficiency of mismatched incorporation. Notably, in comparison to WT, I260Q demonstrates enhanced intensity of fluorescence emission upon mismatched ternary complex formation. Both kinetic and steady-state fluorescence data suggest that relaxed discrimination against incorrect dNTP by I260Q is a consequence of a loss in ability to destabilize the mismatched ternary complex. Overall, our results provide first direct evidence that mismatched and matched dNTP incorporations proceed via analogous kinetic pathways, and support our standing hypothesis that the fidelity of Pol β originates from destabilization of the mismatched closed ternary complex and chemical transition state.
Polymerase fidelity, at its most basic level, is determined by the free energy difference between the highest energy barriers along the correct and incorrect dNTP1 incorporation pathways. Since fidelity requires such a comparison, an ever-increasing gap between the structural and mechanistic information available for matched dNTP incorporation compared with that concerning mismatched dNTP incorporation renders the molecular mechanism of DNA polymerase fidelity unclear (1,2). The thermodynamic instability inherent to polymerase mismatched ternary complexes makes them difficult to crystallize. For this reason, we do not yet have available a structure representing a truly functional prechemistry closed mismatched ternary complex (E′DnN, Scheme 1), although complexes depicting mispairs within the active site (3,4) and mismatch extension (5,6) for various polymerases have been resolved. Likewise, kinetic data for mismatched dNTP incorporations by many DNA polymerases have been reported (7−13), yet commonly used kinetic approaches prevent characterization of individual steps in the mismatched dNTP incorporation pathway.
Scheme 1.
The Ile260 residue of Pol β is located in the hydrophobic hinge region between the palm and the C-terminal fingers subdomain (14,15). Several Pol β variants with alterations in residues lining this hinge region have demonstrated compromised fidelity (16−20). The mutator activity of I260Q was first identified by a genetic screen (21), and subsequent kinetic characterization showed that while this mutant possessed parameters of DNA binding similar to that of wild-type, it interestingly showed loose binding discrimination of mismatched dNTP substrates, and thus a low fidelity (22). These characteristics make the I260Q mutant of specific interest in fidelity studies of Pol β, in order to further understand how one mutation can so drastically alter the mismatch discrimination profile of this enzyme.
Stopped-flow analyses of Pol β, using either 2-aminopurine (2-AP), a fluorescent analogue of adenine and guanine, or Pol βʼs sole Trp325 residue, have been very important in advancing Pol β mechanism studies (23−26). Both probes serve as independent reporters of global enzyme−DNA conformational changes which occur during the catalysis of dNTP incorporation into DNA. While monitoring the fluorescence emission of either 2-AP or Trp325 in stopped-flow, the resultant trace observed during correct dNTP incorporation possesses two phases of fluorescence change. Intricate studies involving a variety of chemical probes have been conducted in order to determine the nature of these two phases. The collective findings of these studies support a model in which the fast fluorescence transition is attributed to dNTP-induced subdomain conformational change (step 2, Scheme 1) to form the ternary closed complex prior to chemistry (step 4, Scheme 1), and the slow fluorescence transition is attributed to subdomain reopening (step 6, Scheme 1) after the rate-limiting chemical step.
This work aims to apply our previously established stopped-flow fluorescence methods, as well as steady-state fluorescence spectroscopy to explore two specific aspects of polymerase fidelity: (i) the mechanism by which WT Pol β discriminates against incorrect dNTP substrate to maintain a moderately high fidelity, and (ii) the deviation of this mechanism in generation of a lower fidelity variant by a single I260Q mutation. Such topics are significant in developing a full understanding of the unique means by which DNA polymerases are able to choose a correct dNTP substrate, whose identity is ever-changing with each round of catalysis, from a pool four nucleotides. Moreover, while it is interesting to comprehend how a polymerase accomplishes this feat to achieve a certain fidelity in DNA replication, it is equally interesting to investigate how a single residue mutation can alter its fidelity. Here, we focus on the kinetic pathway of misincorporation, while another recent paper using small-angle X-ray scattering (SAXS) and molecular modeling lends complementary insights into structural aspects of misincorporation (27). The results of our analyses suggest that Pol β incorporates both correct and mismatched dNTP via analogous kinetic mechanisms differing at the transition state of the chemical step. Furthermore, the divergence in fidelity between WT and I260Q is consequent of differences in ability to stabilize the mismatched ternary complex and chemical transition state.
Materials and Methods
DNA Substrates
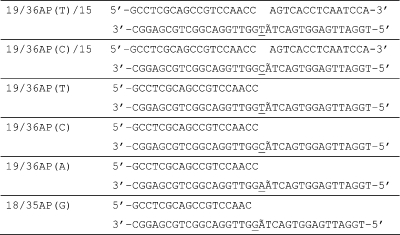
The sequences of primer/template DNA substrates used in the Pol β studies are listed in Table 1. In the text, the notation X:Y indicates [template base]:[incoming dNTP]. Custom synthesized oligomers were purified by denaturing PAGE (15−18% acrylamide, 7 M urea), and desalted using a reverse phase C18 cartridge. Oligomer concentrations were determined by UV absorbance at 260 nm with the following extinction coefficients (M−1 cm−1): 19-mer primer, ε = 166,100; 18-mer primer, ε = 158,900; 36AP template, ε = 335,200; 35AP template, ε = 327,100; and 15-mer downstream oligomer, ε = 144,800. Primer, template, and downstream oligomer (where applicable) were annealed in a 1.1:1:1.2 molar ratio, respectively. Downstream oligomers utilized were 5′-phosphorylated. DNA primers used in the chemical quench experiments were 32P-labeled at the 5′-end using T4 polynucleotide kinase and [γ-32P]ATP (4500 Ci/mol) according to the manufacturer’s protocol. The labeled primers were separated from unreacted ATP using a G-25 microspin column.
Table 1. Pol β DNA Substratesa.
The underlined letter denotes the templating base; Ã denotes 2-aminopurine; dd (in the text) refers to dideoxy-terminated.
Reagents
Ultrapure dNTP’s and G-25 microspin columns were from GE Healthcare (Piscataway, NJ); [γ-32P]ATP from MP Biomedicals (Irvine, CA); T4 polynucleotide kinase from USB (Cleveland, OH); and reverse phase C18 cartridges from Waters (Milford, MA). All synthetic oligonucleotides used in DNA substrate preparation were provided by Integrated DNA Technologies (Coralville, IA). Materials and reagents not listed were of standard molecular biology grade.
Cloning and Purification of Full-Length Pol β and I260Q
The plasmid pET17b-Pol β was transformed into Escherichia coli strain BL21(DE3)pLysS (Stratagene) to overexpress and purify rat Pol β (335 amino acids, 39 kDa) as previously described (28). To generate the I260Q mutant, site-directed mutagenesis was carried out with primers: 5′-GGAGAATCGATATCAGGTTGCAGCCCAAAGATCAGTACTACTGTGG-3′ and 5′-CCACAGTAGTACTGATCTTTGGGCTGCAACCTGATATCGATTCTCC-3′ using the QuikChange method (Stratagene). The underlined region represents the site of mutagenesis. Successful mutagenesis was verified by both plasmid sequencing (OSU Plant Microbe Genomics Facility) and electrospray ionization mass spectrometry of the mutant protein (OSU Campus Chemical Instrument Center). The I260Q mutant was overexpressed and purified in the same manner as wild-type (WT) Pol β (28).
Reaction Buffer Composition
Assay buffers consisted of 20 mM BisTrisHCl, 50 mM TrisHCl, 2 mM DTT, 10−35% (w/v) glycerol, with the ionic strength adjusted to 81 mM with KCl, free Mg2+ adjusted to 5 mM with MgCl2, at either pH 7.6 or 8.5 (adjusted at 37 °C). Standard Pol β assay buffer contained 10% glycerol: all deviations from this are noted accordingly in the figure legends. Free Mg2+ concentrations were calculated using the WEBMAXC STANDARD program at http://www.stanford.edu/∼cpatton/webmaxcS.htm, using the appropriate pH, temperature, ionic strength, and nucleotide triphosphate concentration.
Stopped-Flow Fluorescence
Stopped-flow fluorescence assays were performed on an SX 18MV stopped-flow apparatus (Applied Photophysics Ltd., U.K.). The excitation wavelength for 2-aminopurine (2-AP) was 312 nm with a spectral bandpass of 4 nm. Fluorescence emission was monitored using a 360 nm high pass filter (Corion, MA) on a logarithmic time scale for 1 s for correct dNTP incorporation and up to 50 s for mismatched dNTP incorporation (depending on exact conditions). A typical reaction was initiated by rapid mixing of equal volumes of two solutions in degassed Pol β assay buffer: solution A, containing 400 nM DNA substrate preincubated with 1 μM DNA polymerase, and solution B, containing suitable correct or incorrect dNTP. In all cases, both syringes contained equal concentrations of MgCl2, since it is known that this can have a significant effect on observed fluorescence (24). In experiments employing altered buffer viscosity, both stopped-flow solutions A and B contained the indicated concentration of glycerol. Typically, for all reactions a minimum of 7 runs were performed and averaged prior to data analysis.
Rapid chemical quench
Follow-up of stopped-flow fluorescence experiments was performed on a KinTek (State College, PA) rapid chemical quench instrument (29). The rapid chemical quench reactions were initiated exactly as described for stopped-flow, except that the upstream primer was 5′-radiolabeled. Reactions were quenched at various time intervals with 0.6 M EDTA, and the products visualized by denaturing PAGE followed by autoradiography using a STORM 840 PhosphorImager and quantitated using ImageQuant 5.0 software (GE Healthcare, U.K.).
Steady-State Fluorescence Spectroscopy
Steady-state fluorescence experiments were performed on an Aminco-Bowman Series 2 luminescence spectrometer. 2-AP was excited at 312 nm, and the emission spectra recorded from 330 to 500 nm with a step size of 1 nm. In Pol β assay buffer, reactions consisted of 200 nM dideoxy-terminated DNA substrate, 500 nM DNA polymerase, and either correct (300 μM) or incorrect (5 mM) dNTP.
Product and Data Analysis
Rapid chemical quench data were fit using Sigma Plot 9.0 (Jandel Scientific, CA) to a single exponential equation: [DNAn+1] = A(1 − e−kquencht), where A is amplitude and kquench is the observed rate constant of single-nucleotide incorporation. Stopped-flow fluorescence traces (>2 ms) were fit using Applied Photophysics software to a double exponential equation: fluorescence = A1e−kfastt + A2e−kslowt + C, where A1 and A2 represent corresponding amplitudes, C is an offset constant, and kfast and kslow are the observed rate constants for the fast and slow phases of the fluorescence transition, respectively. For fast phase analysis, kfast was plotted as a function of dNTP concentration and the data fit to a hyperbolic equation with a nonzero intercept: kfast = k2[dNTP]/(Kd + [dNTP]) + y0. According to Scheme 1, k2 is the microscopic rate constant for the forward conformational closing step, Kd is the thermodynamic dissociation constant for dNTP (Kd = k−1/k1), and y0 is a function of both microscopic constants k−2 and k4, as well as the binding constant for catalytic Mg2+ as further elaborated in the appendix of ref (30). For slow phase analysis, kslow was plotted as a function of dNTP concentration and the data fit to the hyperbolic equation kslow = kpol[dNTP]/(Kd,app + [dNTP]), where kpol represents the maximum rate of nucleotide incorporation and Kd,app represents the apparent dNTP dissociation constant.
Results
Mismatched dNTP Incorporation by WT Pol β
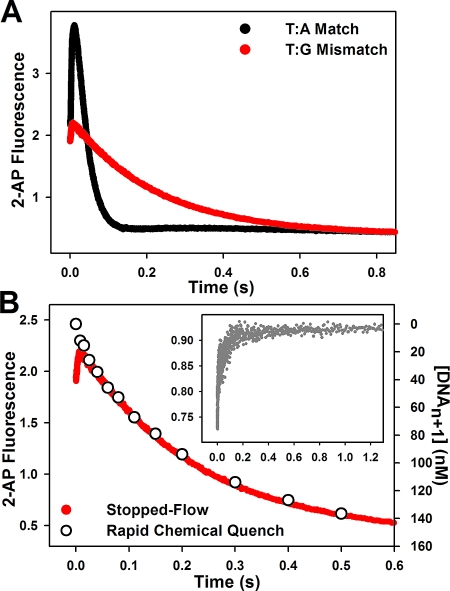
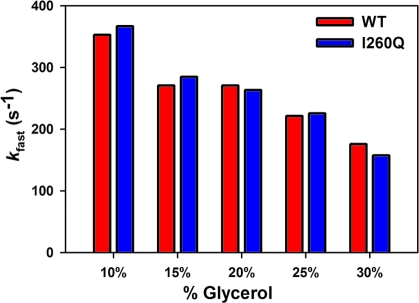
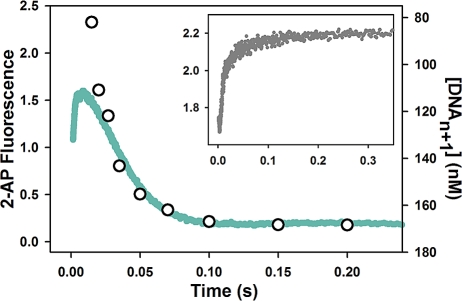
Stopped-flow fluorescence assays and rapid quench experiments have been previously performed in our laboratory under a wide variety of conditions in order to elucidate the mechanism for correct dNTP incorporation by WT Pol β (24−26). Here we report systematic pre-steady-state kinetic analyses of mismatched dNTP incorporation by WT Pol β. As shown in Figure 1, the stopped-flow trace of T:G mismatched incorporation catalyzed by WT follows the same biphasic pattern of fluorescence change observed for correct dNTP incorporation, albeit with lower amplitude. Follow-up of mismatched dNTP incorporation in rapid chemical quench yields rates which correspond precisely to the slow phase rates obtained by stopped-flow (Figure 1B and Table 2). This suggests that the slow phase is attributed to a conformational step limited by the rate of chemistry. Use of dideoxy-terminated primer, which eliminates the chemistry step, abolishes the slow phase completely (Figure 1B inset). This implies that the fast fluorescence transition results from a step before chemistry, while the slow fluorescence transition, analogous to the prior assignment for correct dNTP incorporation (26), likely originates from a step after chemistry. Using the same approaches previously employed for matches (25), we showed that the rate of the fast conformational change is slowed as a function of increasing viscosity for mismatches as well (Figure 2, red bars). Sensitivity to viscosity has been previously interpreted to be indicatory of large conformational movements within the ternary complex during the step responsible for the fast fluorescence transition.
Figure 1.
Pol β mismatched dNTP incorporation in stopped-flow. Reactions were initiated by rapid mixing of equal volumes of solution A (containing 1 μM Pol β and 400 nM DNA substrate) and solution B (containing either 110 μM dATP or 4 mM dGTP). Reactions were performed at 37 °C in Pol β assay buffer at pH 8.5 containing 10% glycerol and 5 mM free Mg2+. (A) Stopped-flow traces of WT T:A matched (black) and T:G mismatched incorporation (red) into 19/36AP(T)/15 DNA, yielding slow phase rates of 37.0 ± 0.2 s−1 and 4.4 ± 0.01 s−1, respectively. (B) Stopped-flow trace of T:G mismatch (red) incorporation with rapid quench overlay (open circles) yielding a rate of 4.8 ± 0.3 s−1. The inset shows the corresponding T:G mismatch reaction using dideoxy-terminated primer (gray trace).
Table 2. Kinetic Rate and Binding Constants for WT versus I260Q Nucleotide Incorporation into 19/36AP(T)/15 and 19/36AP(C)/15 DNA Substrates at pH 8.5a.
| T:A (matched) |
T:G (mismatched) |
|||
|---|---|---|---|---|
| wild-type | I260Q | wild-type | I260Q | |
| k2 | 116 ± 5 s−1 | 108 ± 7 s−1 | 255 ± 9 s−1 | 485 ± 25 s−1 |
| Kd | 30 ± 4 μM | 6.6 ± 2.1 μM | 433 ± 72 μM | 232 ± 37 μM |
| kpol | 42.9 ± 0.6 s−1 | 43.6 ± 1.4 s−1 | 5.7 ± 0.1 s−1 | 13.6 ± 0.2 s−1 |
| Kd,app | 6.8 ± 0.5 μM | 7.1 ± 1.2 μM | 489 ± 26 μM | 49 ± 3 μM |
| kquench | 45.7 ± 2.8 s−1 | 47.6 ± 2.5 s−1 | 4.8 ± 0.3 s−1 | 14.8 ± 0.5 s−1 |
| kpol/Kd,appb | 6.3 | 6.2 | 0.012 | 0.28 |
| fidelityc | 541 | 23 | ||
| C:G (matched) |
C:A (mismatched) |
|||
|---|---|---|---|---|
| wild-type | I260Q | wild-type | I260Q | |
| kpol | 20.8 ± 0.5 s−1 | 18.3 ± 0.4 s−1 | 1.9 ± 0.1 s−1 | 9.4 ± 0.2 s−1 |
| Kd,app | 2.2 ± 0.3 μM | 1.2 ± 0.1 μM | 227 ± 22 μM | 45 ± 3 μM |
| kpol/Kd,appb | 9.7 | 15 | 0.0082 | 0.21 |
| fidelityc | 1180 | 74 | ||
The k2, Kd, kpol and Kd,app values were obtained from hyperbolic fit of the dNTP concentration dependence of the observed rates of the fast and slow fluorescence phases (kfast and kslow) as described in . The rate of dNTP incorporation (kquench) was obtained from single exponential fit of rapid chemical quench data at a saturating dNTP concentration. The k2 value represents the rate constant of forward conformational closing, while Kd reflects the stability of the ternary complex before closing. Therefore, WT and I260Q show little difference during the initial dNTP binding before conformational change. The kpol value represents the maximum rate of dNTP incorporation, while the Kd,app value possesses a contribution from all steps up to the rate-limiting step and can be thought as the dissociation constant of the closed ternary complex.
Catalytic efficiency as measured in units of μM−1 s−1.
Fidelity defined as [(kpol/Kd,app)cor + (kpol/Kd,app)inc]/(kpol/Kd,app)inc, where the subscripts “cor” and “inc” indicate correct (matched) and incorrect (mismatched) nucleotide incorporation, respectively.
Figure 2.
Viscosity dependence of kfast in stopped-flow during mismatched dNTP incorporation by WT Pol β and I260Q. Reactions were initiated by rapid mixing of equal volumes of solution A (containing 1 μM Pol β and 400 nM 19/36AP(T)/15) and solution B (containing 4 mM dGTP). Reactions were performed at 37 °C in Pol β assay buffer at pH 8.5 containing 5 mM free Mg2+ and differential glycerol in both syringes.
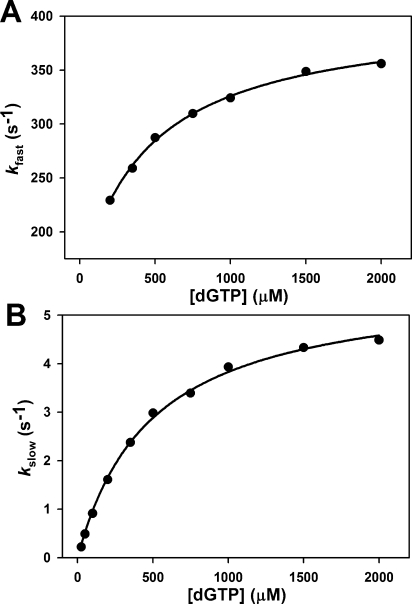
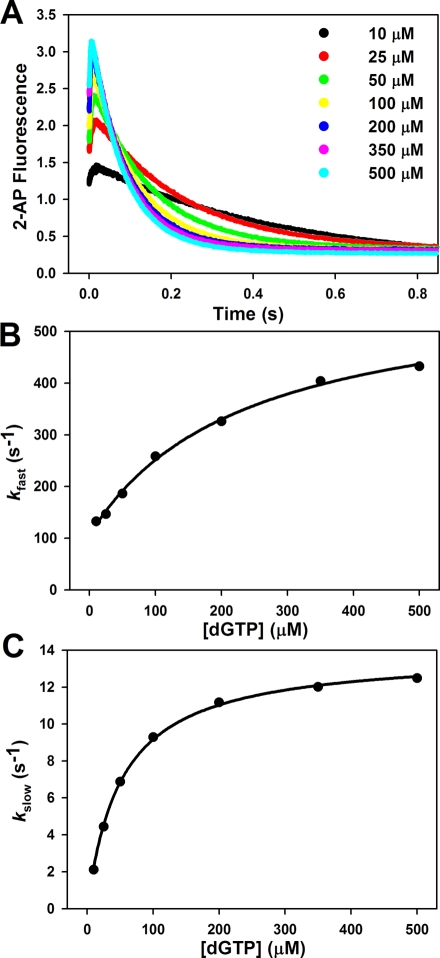
dNTP Concentration Dependence of the Fast and Slow Fluorescence Transitions during Mismatch Incorporation
Previous stopped-flow fluorescence assays investigating matched dNTP incorporation showed that both the fast and slow fluorescence transitions demonstrated a hyperbolic dependence on dNTP concentration (24,30). Here, we analyze the dNTP dependence of both the fast and slow fluorescence phases during mismatched dNTP incorporation in stopped-flow. The observed rate constants for the fast and slow phases (kfast and kslow), obtained from a double exponential fit of each stopped-flow trace (see ), individually plotted as a function of dNTP concentration, revealed that both phases demonstrate a hyperbolic dependence on dNTP concentration (Figure 3). In the case of the fast phase, a plot of the observed rate constant as a function of dNTP was fit to a hyperbola with a nonzero intercept to obtain values for k2 (see Scheme 1), the rate constant of forward conformational closing, and Kd, the thermodynamic dNTP dissociation constant (see ). If the fast phase were resultant from the bimolecular binding of mismatched dNTP, one would expect a linear dependence on dNTP concentration. However, the observed hyperbolic dependence of the fast phase on mismatched dNTP largely indicates that this phase originates from a conformational change induced by mismatched dNTP binding. Similarly, for slow phase analysis, the observed rate constant was plotted against dNTP concentration and fit to a hyperbola to obtain values for kpol, the maximum rate of nucleotide incorporation, and Kd,app, the apparent dNTP dissociation constant (see ). These parameters correspond to the pre-steady-state values of kpol and Kd,app traditionally obtained by rapid chemical quench. The parameters for k2, Kd, kpol, and Kd,app obtained for T:A match and T:G mismatch incorporation by WT Pol β are recorded in Table 2.
Figure 3.
Pol β T:G mismatch titration at pH 8.5. Reactions were initiated by rapid mixing of equal volumes of solution A (containing 1 μM Pol β and 400 nM 19/36AP(T)/15) and solution B (containing varied dGTP concentration). Reactions were performed at 37 °C in Pol β assay buffer containing 5 mM free Mg2+. (A) A hyperbolic fit of the fast phase rate (kfast) versus dGTP concentration (after mixing) yields a k2 of 255 ± 9 s−1, a Kd of 433 ± 72 μM, and a y0 of 148 ± 13 s−1. (B) A hyperbolic fit of the slow phase rate (kslow) versus dGTP concentration (after mixing) yields a Kd,app of 489 ± 26 μM and a kpol of 5.7 ± 0.1 s−1.
Overall, all the aforementioned results support that, analogous to our prior assignments for correct dNTP incorporation, the fast and slow fluorescence changes observed for mismatched incorporation (Figure 1) can be assigned to the dNTP-induced subdomain-closing conformational change and the chemical step (which likely limits the reopening step), respectively. An important observation is that the forward rate of conformational closing (k2, Scheme 1) for mismatched dNTP incorporation is comparable with that for correct dNTP incorporation (though with significant increase in Kd), while the maximum rate of nucleotide incorporation (kpol) is substantially slower (also with significant increase in Kd,app) (Table 2). This suggests that overall mismatched incorporation follows a similar pathway, though both the Kd and Kd,app values are higher and the rate of the chemical step is slower.
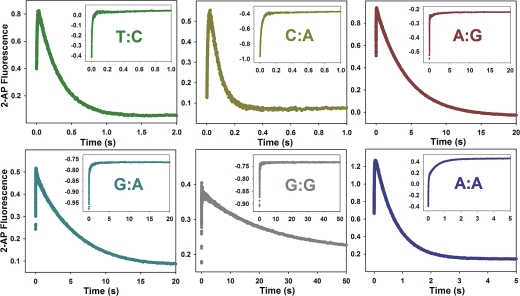
A Biphasic Trace Is Observed for Multiple Mismatches
To further substantiate the conclusions drawn from the investigation of T:G mismatch incorporation in stopped-flow, several additional mismatches were investigated (T:C, C:A, A:A, A:G, G:A, G:G). A biphasic trace was observed for all mismatches examined (Figure 4), and only the fast phase was present when dideoxy-terminated primer was employed (Figure 4, insets). These results lend credence to the notion that while Pol β incorporates mismatches via the same overall mechanism, involving fast subdomain closure upon dNTP binding, each mismatch is unique in its approach to the transition state, as exemplified by the variance in slow phase rates among the mismatches examined.
Figure 4.
Biphasic fluorescence trace observed for multiple mismatched dNTP incorporation by WT Pol β. Reactions were initiated by rapid mixing of equal volumes of solution A (containing 1 μM Pol β and 400 nM 19/36AP(T,C,A) or 18/35AP(G)), and solution B (containing 6 mM mismatch dNTP). Reactions were performed at 37 °C in Pol β assay buffer at pH 8.5 containing 5 mM free Mg2+ and 35% glycerol. The outside traces demonstrate the biphasic trace observed for regular primer, while the insets depict the corresponding reaction with dideoxy-terminated primer. Fast and slow phase misincorporation rates are as follows: T:C mismatch (dark green), 122 ± 1 s−1 and 3.2 ± 0.02 s−1; C:A mismatch (dark yellow), 114 ± 1 s−1 and 14.0 ± 0.1 s−1; A:G mismatch (dark red), 113 ± 2 s−1 and 0.23 ± 0.001 s−1; G:A mismatch (dark cyan), 115 ± 2 s−1 and 0.19 ± 0.001 s−1; G:G mismatch (dark gray), 97 ± 3 s−1 and 0.040 ± 0.001 s−1; A:A mismatch (dark blue), 92 ± 1 s−1 and 1.3 ± 0.01 s−1.
Kinetic Properties of I260Q and WT Are Similar for Correct dNTP Incorporation
As an extension of the WT studies delineated above, further studies on the I260Q mutant were conducted in the interest of kinetic comparison. Stopped-flow analysis of correct dNTP incorporation (Figure 5 and Table 2) indicates that I260Q behaves similarly to WT in that (a) it possesses a biphasic trace with similar rate constants compared to WT, (b) use of dideoxy-terminated primer abolishes the slow phase, (c) the rapid chemical quench rate matches that of the slow phase, and (d) the viscosity dependence pattern of the fast and slow phases is identical to that of WT (Figure 6). Furthermore, dNTP concentration dependence analysis revealed that for T:A correct incorporation both enzymes have similar k2 values (116 ± 5 s−1 and 108 ± 7 s−1 for WT and I260Q, respectively), kpol values (42.9 ± 0.6 s−1 and 43.6 ± 1.4 s−1 for WT and I260Q, respectively) and Kd,app values (6.8 ± 0.5 μM and 7.1 ± 1.2 μM for WT and I260Q, respectively), while demonstrating a 4.5-fold difference in Kd (30 ± 4 μM and 6.6 ± 2.1 μM for WT and I260Q, respectively) (Table 2). Likewise, analysis of C:G matched incorporation yielded similar values of kpol and Kd,app between the two enzymes (Table 2).
Figure 5.
I260Q correct dNTP incorporation in stopped-flow. I260Q T:A matched incorporation into 19/36AP(T)/15 (teal trace). Reactions were initiated by rapid mixing of equal volumes of solution A (containing 1 μM Pol β and 400 nM DNA substrate) and solution B (600 μM dATP) at pH 8.5. The slow phase fits to a rate of 48.5 ± 0.4 s−1, while rapid quench overlay (open circles) fits to a rate of 47.6 ± 2.5 s−1. The inset shows the corresponding T:A match reaction using dideoxy-terminated primer (gray trace).
Figure 6.
Viscosity dependence of kfast and kslow in stopped-flow during correct dNTP incorporation by WT Pol β versus I260Q. Reactions were initiated by rapid mixing of equal volumes of solution A (containing 1 μM Pol β or I260Q and 400 nM 19/36AP(T)/15) and solution B (containing 600 μM dATP). Reactions were performed at 37 °C in Pol β assay buffer at pH 7.6 containing 5 mM free Mg2+ and differential glycerol in both syringes.
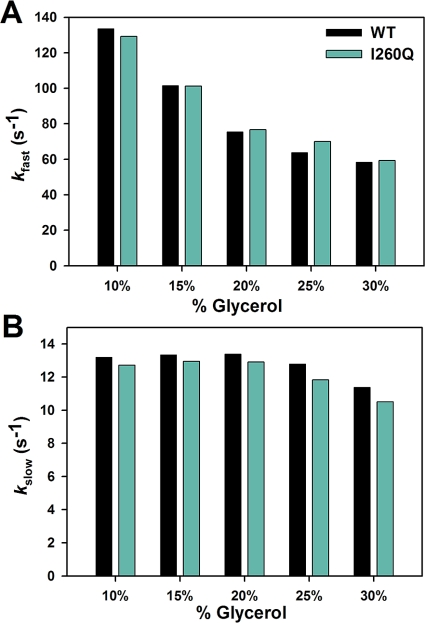
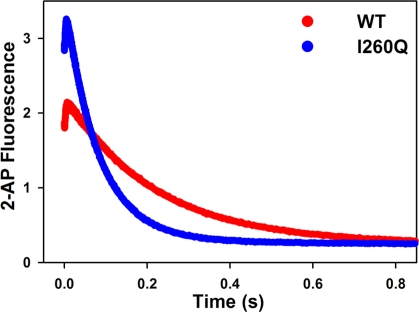
Kinetic Properties of I260Q and WT Are Different for Mismatched dNTP Incorporation
There are clear kinetic differences between I260Q and WT during mismatched incorporations. The similarities and differences are summarized here: (a) A comparison in Figure 7 demonstrates that the fluorescence change of I260Q mismatch incorporation follows the same pattern as that of WT, though the slow phase is faster by a factor of 2−3 (Table 2). The assignments of the two phases have again been verified by viscosity dependence (Figure 2) and rapid quench (Table 2). (b) Note that the rate of the conformational change, k2, is not slowed down for I260Q relative to WT (Figure 8A,B and Table 2), suggesting that the dNTP-induced conformational closing is not disrupted during mismatched dNTP incorporation by I260Q. This is in contrast to what has been suggested previously (22). (c) The main difference between I260Q and WT lies in the ability of I260Q to more efficiently stabilize the mismatched ternary complex, as suggested by the 10-fold decrease in Kd,app of I260Q (49 ± 3 μM, see Figure 8C) relative to WT (489 ± 26 μM) for T:G mismatch incorporation and the 5-fold decrease in Kd,app of I260Q (45 ± 3 μM) relative to WT (227 ± 22 μM) for C:A mismatch incorporation (Table 2). This is in agreement with the tight mismatch dNTP binding of this mutant formerly reported (22). The combination of the lower Kd,app and faster kpol values observed for I260Q mismatched incorporation yields catalytic efficiencies of 0.28 μM−1 s−1 for T:G mismatch and 0.21 μM−1 s−1 for C:A mismatch, which are significantly larger than those recorded for WT (Table 2). Note that the catalytic efficiencies for both enzymes during matched incorporation show little difference. Selective enhancement of mismatch incorporation by I260Q translates into a fidelity 24-fold lower for T:G mismatch incorporation and 16-fold lower for C:A mismatch.
Figure 7.
Comparison of WT Pol β and I260Q T:G mismatched incorporation at pH 8.5. Reactions were initiated by rapid mixing of equal volumes of solution A (containing 1 μM Pol β or I260Q and 400 nM 19/36AP(T)/15) and solution B (containing 4 mM dGTP) and performed at 37 °C in Pol β assay buffer at pH 8.5 containing 5 mM free Mg2+ and 10% glycerol. Fast and slow phase misincorporation rates were 323 ± 6 s−1 and 4.5 ± 0.01 s−1 correspondingly for WT (red trace), and 383 ± 6 s−1 and 12.2 ± 0.03 s−1 correspondingly for I260Q (blue trace).
Figure 8.
I260Q T:G mismatch titration at pH 8.5. Reactions were initiated by rapid mixing of equal volumes of solution A (containing 1 μM I260Q and 400 nM 19/36AP(T)/15) and solution B (containing varied dGTP concentration). Reactions were performed at 37 °C in Pol β assay buffer containing 5 mM free Mg2+. (A) Stopped-flow traces as a function of increasing dGTP concentration (after mixing) are pictured. (B) A hyperbolic fit of the fast phase rate (kfast) versus dGTP concentration (after mixing) yields a k2 of 485 ± 25 s−1, a Kd of 232 ± 37 μM, and a y0 of 106 ± 8 s−1. (C) A hyperbolic fit of the slow phase rate (kslow) versus dGTP concentration (after mixing) yields a Kd,app of 49 ± 3 μM and a kpol of 13.6 ± 0.2 s−1.
The Fast Fluorescence Transition Is Also Present at Physiological pH
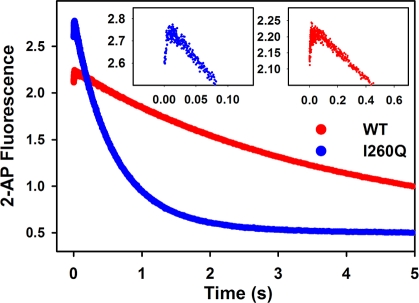
To ensure that the conclusions based on the results obtained at pH 8.5 are also applicable under physiological conditions, we repeated analysis of mismatched dNTP incorporation at pH 7.6. At this pH, a biphasic trace of fluorescence change is still observed in stopped-flow for both WT and I260Q during T:G misincorporation (Figure 9). Similar to that observed for pH 8.5, the observed rate of the dNTP induced conformational change is not slowed down for I260Q relative to WT at pH 7.6, again pointing out that the dNTP-induced conformational closing of I260Q is not disrupted during matched or mismatched dNTP incorporation. A rigorous dNTP titration analysis of the fast phase was not possible for mismatches at this pH due to a combination of the low amplitude of this phase at dNTP concentrations below 2 mM and a noted pattern of inhibition at high dNTP concentration. Slow phase analyses for WT versus I260Q nucleotide incorporation at pH 7.6 are reported in Table 3. Again, for matched incorporation, little difference is observed for kpol and Kd,app between WT and I260Q at this pH, leading to similar catalytic efficiencies (Table 3). However, a 7.4-fold decrease in the Kd,app of I260Q (88 ± 6 μM) relative to WT (654 ± 42 μM) for T:G mismatch incorporation, and a 4.3-fold decrease in the Kd,app of I260Q (69 ± 3 μM) relative to WT (298 ± 23 μM) for C:A mismatch incorporation at pH 7.6 further supports that the main difference between I260Q and WT lies in the enhanced ability of I260Q to stabilize the mismatched ternary complex. Again, the combination of the lower Kd,app and faster kpol values observed for I260Q mismatched incorporation at this pH translates into a higher catalytic efficiency for mismatch incorporation by I260Q compared to WT, and thus a lower fidelity (Table 3).
Figure 9.
Comparison of WT Pol β and I260Q T:G mismatched incorporation at pH 7.6. Reactions were initiated by rapid mixing of equal volumes of solution A (containing 1 μM Pol β or I260Q and 400 nM 19/36AP(T)/15) and solution B (containing 4 mM dGTP) and performed at 37 °C in Pol β assay buffer at pH 7.6 containing 5 mM free Mg2+ and 10% glycerol. Fast and slow phase misincorporation rates were 70 ± 5 s−1 and 0.25 ± 0.0004 s−1 respectively for WT, and 314 ± 22 s−1 and 1.7 ± 0.003 s−1 respectively for I260Q. The insets show a close-up of the fast phases for WT (red trace) and I260Q (blue trace).
Table 3. Kinetic Rate and Binding Constants for WT versus I260Q Nucleotide Incorporation into 19/36AP(T)/15 and 19/36AP(C)/15 DNA Substrates at pH 7.6.
| T:A (matched) |
T:G (mismatched) |
|||
|---|---|---|---|---|
| wild-type | I260Q | wild-type | I260Q | |
| kpol | 13.4 ± 0.5 s−1 | 13.4 ± 0.4 s−1 | 0.26 ± 0.01 s−1 | 1.8 ± 0.03 s−1 |
| Kd,app | 2.2 ± 0.3 μM | 1.1 ± 0.2 μM | 654 ± 42 μM | 88 ± 6 μM |
| kpol/Kd,appa | 6.1 | 12 | 0.00040 | 0.021 |
| fidelityb | 15100 | 590 | ||
| C:G (matched) |
C:A (mismatched) |
|||
|---|---|---|---|---|
| wild-type | I260Q | wild-type | I260Q | |
| kpol | 7.2 ± 0.1 s−1 | 6.6 ± 0.2 s−1 | 0.19 ± 0.01 s−1 | 1.1 ± 0.01 s−1 |
| Kd,app | 1.0 ± 0.1 μM | 0.46 ± 0.07 μM | 298 ± 23 μM | 69 ± 3 μM |
| kpol/Kd,appa | 7.0 | 14 | 0.00064 | 0.016 |
| fidelityb | 10900 | 891 | ||
Catalytic efficiency as measured in units of μM−1 s−1.
Fidelity defined as [(kpol/Kd,app)cor + (kpol/Kd,app)inc]/(kpol/Kd,app)inc, where the subscripts “cor” and “inc” indicate correct (matched) and incorrect (mismatched) nucleotide incorporation, respectively.
Steady-State Fluorescence Studies
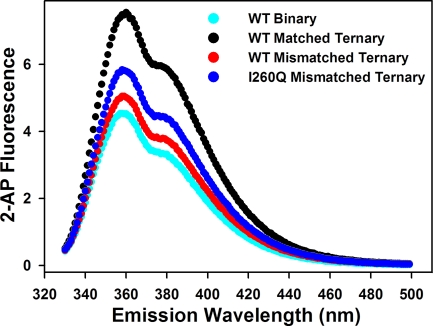
Steady-state fluorescence emission spectra of WT versus I260Q demonstrate identical binary complex traces. For both enzymes there is an equivalent increase in 2-AP fluorescence intensity at the emission maximum (360 nm) upon addition of saturated concentrations of correct dNTP (Figure 10). Addition of mismatched dNTP also results in a fluorescence change in the same direction, though with lower amplitude. Importantly, I260Q demonstrates higher fluorescence intensity (Figure 10) and lower saturating mismatched dNTP concentration compared to WT (data not shown). The data correlate well to the observed amplitude differences in the stopped-flow fluorescence assays (Figure 1 and Figure 7). Overall, these results are consistent with the proposal that both matches and mismatches are incorporated via a similar mechanism, as both correct and mismatched dNTPs elicit the same direction of fluorescence change. In combination with conclusions from recent SAXS structural studies (27), they also qualitatively support the hypothesis that the enhanced ability of I260Q to incorporate mismatched dNTP may arise from a more “partially closed” mismatched ternary complex structure than WT (see ).
Figure 10.
Steady-state fluorescence spectra of matched and mismatched ternary complexes for WT Pol β and I260Q. Reactions contained 200 nM 19dd/36AP(T)/15 DNA, 500 nM enzyme in the presence of either no dNTP, 300 μM matched dNTP, or 5 mM mismatched dNTP in Pol β assay buffer at pH 8.5. Emission spectra shown include WT binary complex (cyan), WT T:A matched ternary complex (black), WT T:G mismatched ternary complex (red), I260Q T:G mismatched ternary complex (blue). Omitted for simplification are the I260Q binary and T:A matched ternary complexes, which overlay accurately with the corresponding WT traces.
Discussion
As an extension of previous methodology used to delineate Pol βʼs correct dNTP incorporation mechanism (24,25), similar techniques were employed to examine the mechanism of mismatched dNTP incorporation by Pol β and its I260Q “mutator” mutant. Detailed studies of these two enzymes led to the following important conclusions: (i) Stopped-flow fluorescence analyses indicate that there is, in fact, a conformational closing event that occurs for mismatched dNTP incorporation, as evidenced by the existence of a fast fluorescence phase preceding chemistry. Furthermore, the rate of conformational change induced by mismatched dNTP is comparable to that of subdomain closing induced by correct dNTP. (ii) Comparison studies of WT and I260Q show that I260Q possesses fast conformational closing steps during both matched and mismatched dNTP incorporation as well. This argues against a hindered dNTP closing for this enzyme previously hypothesized to account for its altered fidelity (22). (iii) The main difference between the two enzymes lies in more effective stabilization of the mismatched ternary complex by I260Q. (iv) Steady-state fluorescence studies demonstrate that while both matched and mismatched dNTP elicit the same direction of fluorescence change, the I260Q mismatched ternary complex fluorescence emission spectrum bears more resemblance to that of the matched ternary complex. Recent SAXS and molecular modeling studies from our laboratory suggest that for both enzymes the mismatched ternary complex lies between the open and the closed forms (27). The combination of these conclusions with the steady-state fluorescence studies presented here provides grounds for speculation that, on the global conformational closing pathway ranging from fully open (binary complex) to fully closed (matched ternary complex), mismatches may induce a less “partially closed” (more open) conformation for WT than for I260Q.
Although our observation of an incorrect dNTP-induced conformational change by Pol β may appear to contradict the findings of two FRET-base studies monitoring the conformational motions of Klentaq (31) and Klenow (32), the differences could be reconciled. Both studies report an increase in FRET signal, upon addition of correct dNTP, yet do not observe any significant change upon addition of incorrect dNTP. In contrast to moderate and low fidelity polymerases, including Pol β, since Klentaq and Klenow are higher fidelity enzymes, it is likely that they more effectively destabilize the mismatched ternary complex to the extent that no fluorescence change is observable for mismatch binding. Our observation of differences in the amplitude of stopped-flow fluorescence traces (Figure 7) and steady-state fluorescence spectra (Figure 10) between WT Pol β and the lower fidelity I260Q variant further illustrates an extension of the correlation between fidelity and mismatch destabilization.
Our observation that the conformational change occurs with a similar rate for both correct and mismatched incorporation by Pol β differs from the conclusion of the recent single molecule kinetic analysis of T7 DNA polymerase (33), which reported a significantly reduced rate of conformational closing induced by mismatched dNTP binding. It remains to be established whether such a discrepancy reflects differences in the mismatched discrimination mechanism employed by two polymerases, or whether it results from different experimental systems and conditions. However, another study on T7 reported little difference in the forward rates of conformational closing between matches and mismatches, while noting a large difference in the reverse rates of conformational closing (34).
On the other hand, our results are in full agreement with recent reports on the linear free energy relationship (LFER) of pyrophosphate leaving group elimination for both correct and mismatched dGTP incorporations utilizing dNTP analogues in which the β,γ-bridging oxygen is substituted with various halomethylene moieties (35,36). Based on observation that the Brønsted correlation between log kpol and the leaving group pKa is very similar between correct and mismatched incorporations, the authors conclude that their results support our earlier prediction that the rate-limiting step is the chemical step, not the conformational change (1). Our observation of a fast conformational change for mismatched dNTP incorporations supports the interpretation of this work, and additionally argues against the notion that conformational closing is a major contributor to fidelity.
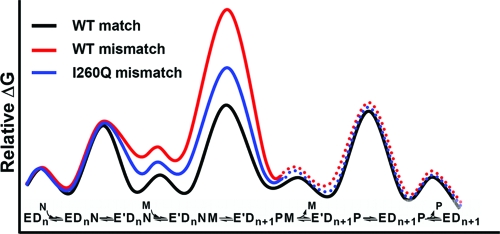
Figure 11 illustrates the current knowledge of the kinetic scheme of Pol β and its free energy diagram for matched dNTP incorporation (black profile) (26). What remain to be established are the free energy diagram (i.e., kinetic mechanism) and the intermediate structures (i.e., structural mechanism) for mismatched incorporations. Multiple and recent proposals contemplate a perturbed conformation of the ternary complex and/or a different reaction pathway may be engaged by a mismatched dNTP incorporation in DNA polymerases (3,34,37−40). Our steady-state and stopped-flow fluorescence studies strongly support that both correct and mismatches are incorporated via a similar mechanism, and that the fidelity of Pol β is controlled, at least partly, by destabilization of the mismatched ternary complex (higher Kd,app values) and the chemical transition state (smaller kpol and kquench values) in the same reaction pathway (Figure 11, red profile).
Figure 11.
Qualitative free energy profile of matched and mismatched dNTP incorporation by Pol β versus I260Q. E = DNA polymerase in open conformation; E′ = closed conformation; Dn = DNA; N = Mg·dNTP; M = catalytic Mg2+; P = Mg·PPi.
The loss of fidelity of I260Q has previously been explained by a hindrance of dNTP-induced subdomain closure (21,22). It was suggested that upon the substitution of Ile with Gln in I260Q, rotamers are generated which cause a reduction in an important cavity space near residue 260, thus perturbing subdomain motion. However, our results indicate the contrary: that the kinetic properties of I260Q, including the rate of the dNTP-induced subdomain closure, are largely unperturbed from WT Pol β for correct dNTP incorporations. Thus, for correct dNTP incorporation its free energy profile is assumed to be the same as that of WT for the purpose of discussion here. The main difference leading to the low fidelity of I260Q appears to be a loss in ability to destabilize the mismatched ternary complex (Figure 11, blue profile). This conclusion is based on the observation that the low fidelity of I260Q mutant is primarily determined by enhanced efficiency of mismatched incorporation, whereas for most low fidelity DNA polymerase variants, reduced fidelity correlates with low efficiency of correct dNTP incorporation (41). Based upon the two mismatches examined, I260Q incorporates mismatches 23- to 53-fold more efficiently than WT (see Tables 2 and 3). The observed change in kpol/Kd,app for mismatched incorporations by I260Q suggests that the “overstabilization” of the mismatched ternary complex by I260Q is further enhanced in the transition state. Molecular modeling of the I260Q mismatched ternary complex predicts the presence of additional hydrogen bonds, involving Gln260, Tyr296, Glu295, and Arg258, which may be a plausible source of this stabilization (27).
Overall, kinetic behavioral comparisons between WT Pol β and I260Q allow us to conclude that infidelity of I260Q originates from enhanced stabilization of the mismatched ternary complex and the chemical transition state. Regarding the mechanism of mismatched discrimination, our laboratory previously suggested that DNA polymerase fidelity originates primarily from differential substrate binding at the highest energy transition state and suggested a most likely model (Figure 1D in ref (1)), which is highly consistent with the results and conclusions of this work. Of future interest to enhance our knowledge of the mechanism of polymerase fidelity would be crystallization of the I260Q mismatched ternary complex. This would provide insight into the extent of this variant’s conformational closing profile and delineate a structural basis for understanding which specific residues might contribute to this variant’s unique capability to support mismatched dNTP incorporation more efficiently.
The insights into the mechanism of misincorporation presented here add to the growing body of knowledge ultimately desired to consist of the microscopic rates of all steps in Pol βʼs matched and mismatched incorporation mechanism, in order to fully and confidently assess which step is the most important contributor to this enzyme’s fidelity. Recent illumination of the importance of the reverse rate of the dNTP-induced conformational closing step and its effect on enzyme specificity further stresses the necessity for complete evaluation of all microscopic rate constants (34). Ongoing studies in our laboratory are examining this step for Pol β during both matched and mismatched incorporation. On a broader scale, through the use of Pol β we continue to develop a more precise understanding of the basis of DNA polymerase fidelity by establishing our techniques as a systematic framework of investigation that can be applied to multiple polymerases and additional site-specific mutants (42) during both matched and mismatched nucleotide incorporations.
Acknowledgments
We thank Dr. Ross Dalbey for the generous use of his FPLC and fluorimeter.
Funding Statement
National Institutes of Health, United States
Footnotes
Abbrevations: 2-AP, 2’-deoxy-2-aminopurine; dNTP, 2′-deoxynucleoside 5′-triphosphate; Pol β, Rattus norvegicus DNA polymerase β; WT, wild-type.
References
- Showalter A. K.; Tsai M. D. (2002) A reexamination of the nucleotide incorporation fidelity of DNA polymerases. Biochemistry 41, 10571–10576. [DOI] [PubMed] [Google Scholar]
- Showalter A. K.; Lamarche B. J.; Bakhtina M.; Su M. I.; Tang K. H.; Tsai M. D. (2006) Mechanistic comparison of high-fidelity and error-prone DNA polymerases and ligases involved in DNA repair. Chem. Rev. 106, 340–360. [DOI] [PubMed] [Google Scholar]
- Krahn J. M.; Beard W. A.; Wilson S. H. (2004) Structural insights into DNA polymerase beta deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure 12, 1823–1832. [DOI] [PubMed] [Google Scholar]
- Johnson S. J.; Beese L. S. (2004) Structures of mismatch replication errors observed in a DNA polymerase. Cell 116, 803–816. [DOI] [PubMed] [Google Scholar]
- Trincao J.; Johnson R. E.; Wolfle W. T.; Escalante C. R.; Prakash S.; Prakash L.; Aggarwal A. K. (2004) Dpo4 is hindered in extending a G.T mismatch by a reverse wobble. Nat. Struct. Mol. Biol. 11, 457–462. [DOI] [PubMed] [Google Scholar]
- Batra V. K.; Beard W. A.; Shock D. D.; Pedersen L. C.; Wilson S. H. (2005) Nucleotide-induced DNA polymerase active site motions accommodating a mutagenic DNA intermediate. Structure 13, 1225–1233. [DOI] [PubMed] [Google Scholar]
- Wong I.; Patel S. S.; Johnson K. A. (1991) An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry 30, 526–537. [DOI] [PubMed] [Google Scholar]
- Showalter A. K.; Tsai M. D. (2001) A DNA polymerase with specificity for five base pairs. J. Am. Chem. Soc. 123, 1776–1777. [DOI] [PubMed] [Google Scholar]
- Roettger M. P.; Fiala K. A.; Sompalli S.; Dong Y.; Suo Z. (2004) Pre-steady-state kinetic studies of the fidelity of human DNA polymerase mu. Biochemistry 43, 13827–13838. [DOI] [PubMed] [Google Scholar]
- Kati W. M.; Johnson K. A.; Jerva L. F.; Anderson K. S. (1992) Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267, 25988–25997. [PubMed] [Google Scholar]
- Fiala K. A.; Abdel-Gawad W.; Suo Z. (2004) Pre-steady-state kinetic studies of the fidelity and mechanism of polymerization catalyzed by truncated human DNA polymerase lambda. Biochemistry 43, 6751–6762. [DOI] [PubMed] [Google Scholar]
- Eger B. T.; Benkovic S. J. (1992) Minimal kinetic mechanism for misincorporation by DNA polymerase I (Klenow fragment). Biochemistry 31, 9227–9236. [DOI] [PubMed] [Google Scholar]
- Ahn J.; Werneburg B. G.; Tsai M. D. (1997) DNA polymerase beta: structure-fidelity relationship from pre-steady-state kinetic analyses of all possible correct and incorrect base pairs for wild type and R283A mutant. Biochemistry 36, 1100–1107. [DOI] [PubMed] [Google Scholar]
- Pelletier H.; Sawaya M. R.; Wolfle W.; Wilson S. H.; Kraut J. (1996) Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry 35, 12742–12761. [DOI] [PubMed] [Google Scholar]
- Sawaya M. R.; Pelletier H.; Kumar A.; Wilson S. H.; Kraut J. (1994) Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science 264, 1930–1935. [DOI] [PubMed] [Google Scholar]
- Shah A. M.; Li S. X.; Anderson K. S.; Sweasy J. B. (2001) Y265H mutator mutant of DNA polymerase beta. Proper teometric alignment is critical for fidelity. J. Biol. Chem. 276, 10824–10831. [DOI] [PubMed] [Google Scholar]
- Opresko P. L.; Sweasy J. B.; Eckert K. A. (1998) The mutator form of polymerase beta with amino acid substitution at tyrosine 265 in the hinge region displays an increase in both base substitution and frame shift errors. Biochemistry 37, 2111–2119. [DOI] [PubMed] [Google Scholar]
- Clairmont C. A.; Narayanan L.; Sun K. W.; Glazer P. M.; Sweasy J. B. (1999) The Tyr-265-to-Cys mutator mutant of DNA polymerase beta induces a mutator phenotype in mouse LN12 cells. Proc. Natl. Acad. Sci. U.S.A. 96, 9580–9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. X.; Vaccaro J. A.; Sweasy J. B. (1999) Involvement of phenylalanine 272 of DNA polymerase beta in discriminating between correct and incorrect deoxynucleoside triphosphates. Biochemistry 38, 4800–4808. [DOI] [PubMed] [Google Scholar]
- Shah A. M.; Maitra M.; Sweasy J. B. (2003) Variants of DNA polymerase Beta extend mispaired DNA due to increased affinity for nucleotide substrate. Biochemistry 42, 10709–10717. [DOI] [PubMed] [Google Scholar]
- Starcevic D.; Dalal S.; Sweasy J. (2005) Hinge residue Ile260 of DNA polymerase beta is important for enzyme activity and fidelity. Biochemistry 44, 3775–3784. [DOI] [PubMed] [Google Scholar]
- Starcevic D.; Dalal S.; Jaeger J.; Sweasy J. B. (2005) The hydrophobic hinge region of rat DNA polymerase beta is critical for substrate binding pocket geometry. J. Biol. Chem. 280, 28388–28393. [DOI] [PubMed] [Google Scholar]
- Arndt J. W.; Gong W.; Zhong X.; Showalter A. K.; Liu J.; Dunlap C. A.; Lin Z.; Paxson C.; Tsai M.-D.; Chan M. K. (2001) Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes. Biochemistry 40, 5368–5375. [DOI] [PubMed] [Google Scholar]
- Dunlap C. A.; Tsai M. D. (2002) Use of 2-aminopurine and tryptophan fluorescence as probes in kinetic analyses of DNA polymerase beta. Biochemistry 41, 11226–11235. [DOI] [PubMed] [Google Scholar]
- Bakhtina M.; Lee S.; Wang Y.; Dunlap C.; Lamarche B.; Tsai M. D. (2005) Use of viscogens, dNTPalphaS, and rhodium(III) as probes in stopped-flow experiments to obtain new evidence for the mechanism of catalysis by DNA polymerase beta. Biochemistry 44, 5177–5187. [DOI] [PubMed] [Google Scholar]
- Bakhtina M.; Roettger M. P.; Kumar S.; Tsai M. D. (2007) A unified kinetic mechanism applicable to multiple DNA polymerases. Biochemistry 46, 5463–5472. [DOI] [PubMed] [Google Scholar]
- Tang K. H.; Niebuhr M.; Tung C. S.; Chan H. C.; Chou C. C.; Tsai M. D. (2008) Mismatched dNTP incorporation by DNA polymerase beta does not proceed via globally different conformational pathways. Nucleic Acids Res. 36, 2948–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werneburg B. G.; Ahn J.; Zhong X.; Hondal R. J.; Kraynov V. S.; Tsai M. D. (1996) DNA polymerase beta: pre-steady-state kinetic analysis and roles of arginine-283 in catalysis and fidelity. Biochemistry 35, 7041–7050. [DOI] [PubMed] [Google Scholar]
- Johnson K. A. (1986) Rapid kinetic analysis of mechanochemical adenosinetriphosphatases. Methods Enzymol. 134, 677–705. [DOI] [PubMed] [Google Scholar]
- Zhong X.; Patel S. S.; Werneburg B. G.; Tsai M. D. (1997) DNA polymerase beta: multiple conformational changes in the mechanism of catalysis. Biochemistry 36, 11891–11900. [DOI] [PubMed] [Google Scholar]
- Rothwell P. J.; Mitaksov V.; Waksman G. (2005) Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol. Cell 19, 345–355. [DOI] [PubMed] [Google Scholar]
- Stengel G.; Gill J. P.; Sandin P.; Wilhelmsson L. M.; Albinsson B.; Norden B.; Millar D. (2007) Conformational dynamics of DNA polymerase probed with a novel fluorescent DNA base analogue. Biochemistry 46, 12289–12297. [DOI] [PubMed] [Google Scholar]
- Luo G.; Wang M.; Konigsberg W. H.; Xie X. S. (2007) Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 104, 12610–12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. C.; Johnson K. A. (2006) A new paradigm for DNA polymerase specificity. Biochemistry 45, 9675–9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucato C. A.; Upton T. G.; Kashemirov B. A.; Osuna J.; Oertell K.; Beard W. A.; Wilson S. H.; Florian J.; Warshel A.; McKenna C. E.; Goodman M. F. (2008) DNA polymerase beta fidelity: halomethylene-modified leaving groups in pre-steady-state kinetic analysis reveal differences at the chemical transition state. Biochemistry 47, 870–879. [DOI] [PubMed] [Google Scholar]
- Sucato C. A.; Upton T. G.; Kashemirov B. A.; Batra V. K.; Martinek V.; Xiang Y.; Beard W. A.; Pedersen L. C.; Wilson S. H.; McKenna C. E.; Florian J.; Warshel A.; Goodman M. F. (2007) Modifying the beta,gamma leaving-group bridging oxygen alters nucleotide incorporation efficiency, fidelity, and the catalytic mechanism of DNA polymerase beta. Biochemistry 46, 461–471. [DOI] [PubMed] [Google Scholar]
- Joyce C. M.; Benkovic S. J. (2004) DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry 43, 14317–14324. [DOI] [PubMed] [Google Scholar]
- Purohit V.; Grindley N. D.; Joyce C. M. (2003) Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I (Klenow fragment). Biochemistry 42, 10200–10211. [DOI] [PubMed] [Google Scholar]
- Florian J.; Goodman M. F.; Warshel A. (2005) Computer simulations of protein functions: searching for the molecular origin of the replication fidelity of DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 102, 6819–6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y.; Oelschlaeger P.; Florian J.; Goodman M. F.; Warshel A. (2006) Simulating the effect of DNA polymerase mutations on transition-state energetics and fidelity: evaluating amino acid group contribution and allosteric coupling for ionized residues in human pol beta. Biochemistry 45, 7036–7048. [DOI] [PubMed] [Google Scholar]
- Beard W. A.; Shock D. D.; Vande Berg B. J.; Wilson S. H. (2002) Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J. Biol. Chem. 277, 47393–47398. [DOI] [PubMed] [Google Scholar]
- Kraynov V. S.; Showalter A. K.; Liu J.; Zhong X.; Tsai M.-D. (2000) DNA polymerase β: contributions of template-positioning and dNTP triphosphate-binding residues to catalysis and fidelity. Biochemistry 39, 16008–16015. [DOI] [PubMed] [Google Scholar]