I. Summary
Conceptually, the simplest way to synthesize an organic molecule is to construct it one carbon at a time. The Wood-Ljungdahl pathway of CO2 fixation involves this type of stepwise process. The biochemical events that underlie the condensation of two one-carbon units to form the two-carbon compound, acetate, have intrigued chemists, biochemists, and microbiologists for many decades. We begin this review with a description of the biology of acetogenesis. Then, we provide a short history of the important discoveries that have led to the identification of the key components and steps of this usual mechanism of CO and CO2 fixation. In this historical perspective, we have included reflections that hopefully will sketch the landscape of the controversies, hypotheses, and opinions that led to the key experiments and discoveries. We then describe the properties of the genes and enzymes involved in the pathway and conclude with a section describing some major questions that remain unanswered.
Keywords: Acetogenesis, Nickel, iron-sulfur, cobalamin, methanogenesis, CO2 fixation, carbon cycle, CO dehydrogenase/acetyl-CoA synthase
II. Introduction
In 1945 soon after Martin Kamen discovered how to prepare 14C in the cyclotron, some of the first biochemical experiments using radioactive isotope tracer methods were performed to help elucidate the pathway of microbial acetate formation [1]. Calvin and his coworkers began pulse labeling cells with 14C and using paper chromatography to identify the 14C-labeled intermediates, including the phosphoglycerate that is formed by the combination of CO2 with ribulose diphosphate, in what became known as the Calvin-Benson-Basham pathway. However, though simple in theory, the pathway of CO2 fixation that is used by M. thermoacetica proved to be recalcitrant to this type of pulse-labeling/chromatographic analysis because of the oxygen sensitivity of many of the enzymes and because the key one-carbon intermediates are enzyme bound. Thus, identification of the steps in the Wood-Ljungdahl pathway of acetyl-CoA synthesis has required the use of a number of different biochemical, biophysical, and bioinorganic techniques as well as the development of methods to grow organisms and work with enzymes under strictly oxygen-free conditions.
III. Importance of acetogens
A. Discovery of acetogens
Acetogens are obligately anaerobic bacteria that use the reductive acetyl-CoA or Wood-Ljungdahl pathway as their main mechanism for energy conservation and for synthesis of acetyl-CoA and cell carbon from CO2 [2, 3]. An acetogen is sometimes called a “homoacetogen” (meaning that it produces only acetate as its fermentation product) or a “CO2-reducing acetogen”.
As early as 1932, organisms were discovered that could convert H2 and CO2 into acetic acid (Equation 1) [4]. In 1936, Wieringa reported the first acetogenic bacterium, Clostridium aceticum [5, 6]. M. thermoacetica [7], a clostridium in the Thermoanaerobacteriaceae family, attracted wide interest when it was isolated because of its unusual ability to convert glucose almost stoichiometrically to three moles of acetic acid (Equation 2) [8].
| (1) |
| (2) |
B. Where acetogens are found and their environmental impact
Globally, over 1013 kg (100 billion US tons) of acetic acid is produced annually, with acetogens contributing about 10% of this output [2]. While most acetogens like M. thermoacetica are in the phylum Firmicutes, acetogens include Spirochaetes, δ-proteobacteria like Desulfotignum phosphitoxidans, and acidobacteria like Holophaga foetida. Important in the biology of the soil, lakes, and oceans, acetogens have been isolated from diverse environments, including the GI tracts of animals and termites [9, 10], rice paddy soils [11], hypersaline waters [12], surface soils [13, 14], and deep subsurface sediments [15]. Acetogens also have been found in a methanogenic mixed population from an army ammunition manufacturing plant waste water treatment facility [16] and a dechlorinating community that has been enriched for bioremediation [17].
For organisms that house acetogens in their digestive systems, like humans, termites, and ruminants [18–20], the acetate generated by microbial metabolism is a beneficial nutrient for the host and for other microbes within the community. In these ecosystems, acetogens can compete directly with hydrogenotrophic methanogenic archaea, or interact syntrophically with acetotrophic methanogens that use H2 and CO2 to produce methane [20]. In the termite gut, acetogens are the dominant hydrogen sinks [21], and it has been proposed that acetate is the major energy source for the termite [22]. Like methanogens, acetogens act as a H2 sink, depleting H2 that is generated in anaerobic environments during the natural biodegradation of organic compounds. Since the build-up of H2 inhibits biodegradation by creating an unfavorable thermodynamic equilibrium, acetogens enhance biodegradative capacity by coupling the oxidation of hydrogen gas to the reduction of CO2 to acetate.
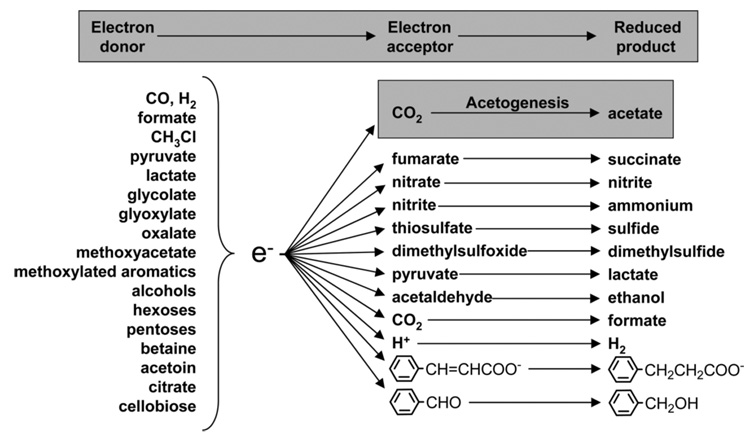
Methanogens are the dominant hydrogenotrophs in many environments since methanogens have a lower threshold for H2 than acetogens [23] and since the energy yield from the conversion of CO2 and H2 to methane is greater than that for conversion to acetate [24, 25]. Under such conditions, acetogens must often resort to other metabolic pathways for growth and, thus, have a highly diverse metabolic menu that comprises the biodegradation products of most natural polymers like cellulose and lignin, including sugars, alcohols, organic acids and aldehydes, aromatic compounds, and inorganic gases like CO, H2, and CO2. They also can use a variety of electron acceptors, e.g., CO2, nitrate, dimethylsulfoxide, fumarate, and protons.
IV. Importance of the Wood-Ljungdahl pathway
A. The Wood-Ljungdahl pathway in diverse metabolic pathways
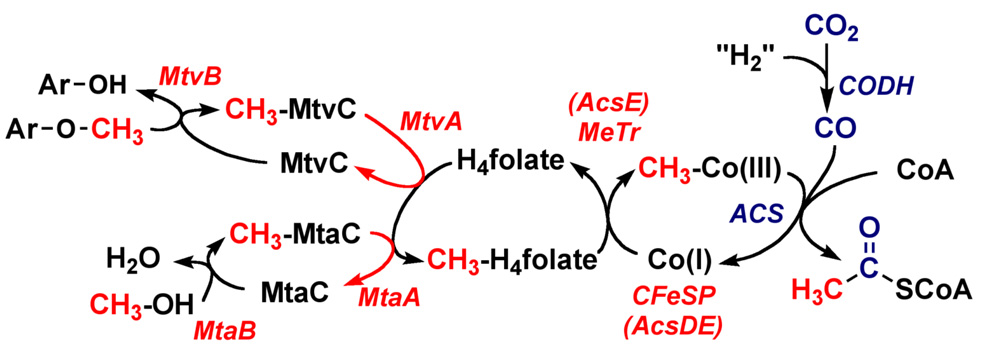
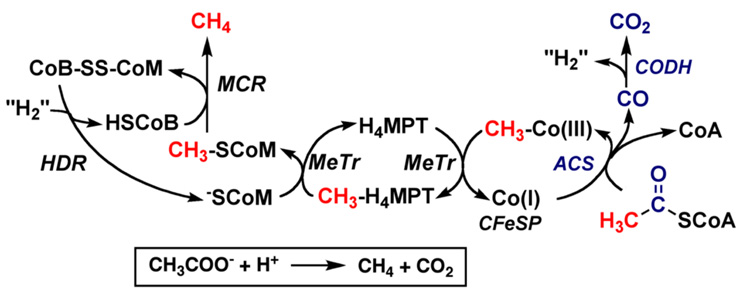
The Wood-Ljungdahl pathway (Figure 1) is found in a broad range of phylogenetic classes, and is used in both the oxidative and reductive directions. The pathway is used in the reductive direction for energy conservation and autotrophic carbon assimilation in acetogens [26–28]. When methanogens grow on H2 + CO2, they use the Wood-Ljungdahl pathway in the reductive direction (like acetogens) for CO2 fixation [29, 30]; however, they conserve energy by the conversion of H2 + CO2 to methane (Equation 2). Given that hydrogenotrophic methanogens assimilate CO2 into acetyl-CoA, it is intriguing that they do not make a mixture of methane and acetate. Presumably this is governed by thermodynamics, since the formation of methane is −36 kJ/mol more favorable (Equation 3) than acetate synthesis. Aceticlastic methanogens [31] exploit this advantageous equilibrium to generate metabolic energy by interfacing the Wood-Ljungdahl pathway to the pathway of methanogenesis (Equation 3). In this reverse direction, the combined actions of acetate kinase [32, 33] and phosphotransacetylase [34] catalyze the conversion of acetate into acetyl-CoA. Sulfate reducing bacteria also run the Wood-Ljungdahl pathway in reverse and generate metabolic energy by coupling the endergonic oxidation of acetate to H2 and CO2 (the reverse of equation 1) to the exergonic reduction of sulfate to sulfide (ΔG0’ = −152 kJ/mol), with the overall process represented by Equation 4 [35–37].
| (2) |
| (3) |
| (4) |
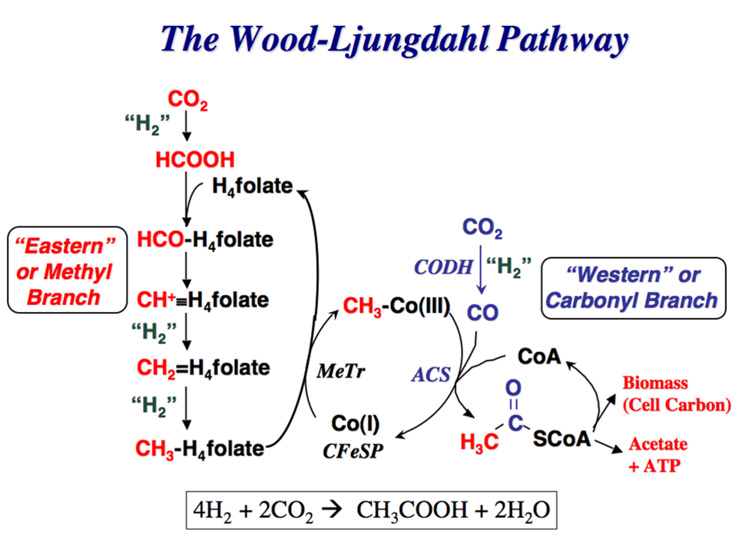
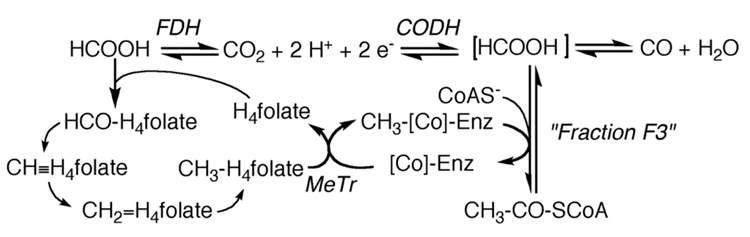
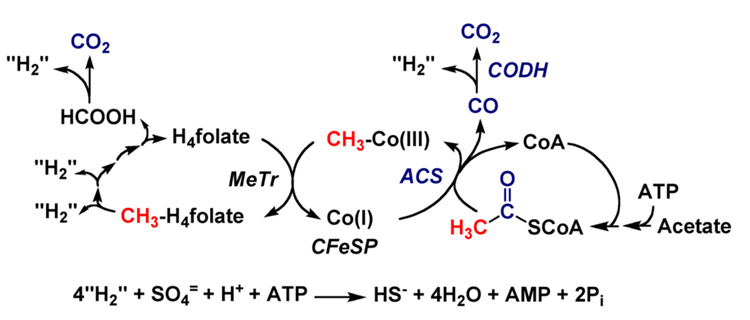
Figure 1.
The Wood-Ljungdahl pathway. “H2” is used in a very general sense to designate the requirement for two electrons and two protons in the reaction.
B. Historical Perspective: Key stages in elucidation of the Wood-Ljungdahl pathway
1. Isolation of Moorella thermoacetica (f. Clostridium thermoaceticum) and demonstration by isotope labeling studies that acetogens fix CO2 by a novel pathway
Chapter 1 of the story of the Wood-Ljungdahl pathway begins with discovery of C. aceticum, the first isolated organism that was shown to grow by converting hydrogen gas and carbon dioxide to acetic acid [5, 6]. Unfortunately, this organism was “lost”, and the baton was passed to Moorella thermoacetica, which was named Clostridium thermoaceticum when it was isolated in 1942 [38] and was so-called until fairly recently when the taxonomy of the genus Clostridium was revised [7]. Thus, M. thermoacetica became the model acetogen, while C. aceticum hid from the scientific community for four decades, until G. Gottschalk, while visiting Barker’s laboratory, found an old test tube containing spores of the original C. aceticum strain and reactivated and reisolated this strain [39, 40]. Although M. thermoacetica did not reveal its potential to grow on H2 + CO2 until 1983 [41], the recognition of a homoacetogenic fermentation of glucose [38], implied that it could use the reducing equivalents from glucose oxidation to convert CO2 to acetate.
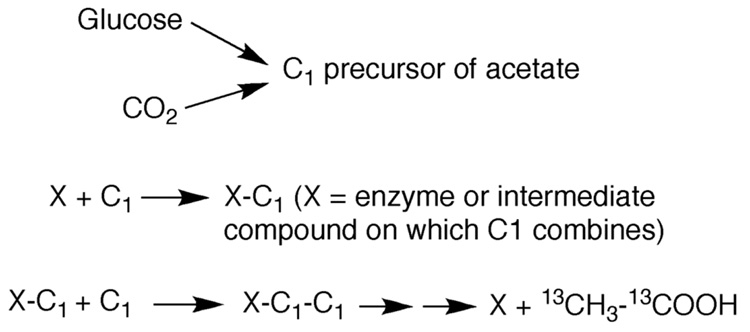
Wood and others embarked on the characterization of this pathway at about the same time that Calvin began to study the CO2 fixation pathway that bears his name [42, 43]. As with the Calvin cycle, isotope labeling studies provided important information regarding the mechanism of CO2 fixation. When 14CO2 was used, approximately an equal concentration of 14C was found in the methyl and carboxyl positions of acetate, leading Barker and Kamen to suggest that glucose fermentation occurred according to Equation 5 and Equation 6, where “*” designates the labeled carbon [1]. Because 14CO2 was used as a tracer in these experiments, one could not distinguish a product that contained an equal mixture of 14CH312COOH/12CH314COOH from one containing doubly labeled 14CH314COOH. Wood used mass spectroscopy to analyze the acetate produced from a fermentation containing a significant percentage (i.e., 25%) of 13CO2 and could conclusively demonstrate that the CO2 is indeed incorporated nearly equally into both carbons of the acetate, confirming equation 6 [44]. It was subsequently shown that carbons 3 and 4 of glucose are the source of the CO2 that is fixed into acetate [45]. The Embden-Meyerhof-Parnas pathway converts glucose to two molecules of pyruvate, which undergo decarboxylation to form two molecules of acetyl-CoA and, thus, two molecules of acetate (Equation 5). The third molecule of acetic acid is generated by utilizing the eight-electrons released during glucose oxidation to reduce the two molecules of CO2 released by pyruvate decarboxylation (Equation 6). Figure 2 describes the level of understanding of the Wood-Ljungdahl pathway in 1951 in a scheme reproduced from a landmark paper by H.G. Wood [44], which was published in the same year that Wood and Utter suggested that the mechanism of acetate synthesis could constitute a previously unrecognized mechanism of autotrophic CO2 fixation [46].
| (5) |
| (6) |
Figure 2.
Working model of the Wood-Ljungdahl pathway circa 1951.
It is ironic that M. thermoacetica became the model organism for elucidating the Wood-Ljungdahl pathway of autotrophic CO2 fixation, since it was only known as a heterotroph until 1983 [41]. However, as mentioned above, C. aceticum appeared to have been lost, and Acetobacterium woodii (the next acetogen could be cultured on H2 and CO2) was not isolated until 1977 [47]. The anaerobic methods described by Balch et al. [48] have since led to the isolation of many anaerobes, including many acetogens, that can grow autotrophically, and over 100 acetogenic species, representing 22 genera, have so far been isolated [49].
Based on the microbiological, labeling and mass spectrometric results described above and further 14CO2 labeling experiments that ruled out the Calvin cycle and the reductive citric acid cycle [50], it seemed clear that homoaceogenesis involved a novel CO2 fixation pathway. As described in detail below, the “C1” and “X-C1” precursors shown in Figure 2 were found to involve C1 units bound to H4folate, to cobalamin, and to a nickel center in a highly oxygen-sensitive enzyme. Thus, elucidation of the biochemical steps in the Wood-Ljungdahl pathway took many years and required and the use of many biochemical and biophysical methods.
2. Demonstration that corrinoids and tetrahydrofolate are involved in the Wood-Ljungdahl Pathway
Chapter 2 in the history of the Wood-Ljungdahl Pathway is the discovery of the role of corrinoids and tetrahydrofolate (H4folate). Corrinoids contain a tetrapyrrolic corrin ring with a central cobalt atom, which can exist in the 1+, 2+, and 3+ oxidation states. Cobalamin (as vitamin B12) had been identified in the mid-1920’s as the antipernicious anemia factor [51, 52] and had only recently been isolated [53, 54] and crystallized [54] at the time of the early pulse labeling studies of the CO2 fixation pathway. The structure of cobalamin was not determined until 1956 [55]. The initial indication that a corrinoid could be involved in this pathway was based on two key findings: (1) intrinsic factor (known to inhibit vitamin B12-dependent reactions) inhibits incorporation of the methyl group of methyl-B12 into the C-2 of acetate, and (2) in the presence of pyruvate (as an electron and carboxyl group donor), the radioactive methyl group of 14C-methylcobalamin is incorporated into the methyl group of acetate [56] (Equation 7). These results suggested to Wood and coworkers why they had been unable to isolate intermediates in deproteinated solutions by pulse labeling with 14CO2 - the labeled intermediates were bound to cofactors on proteins!
| (7) |
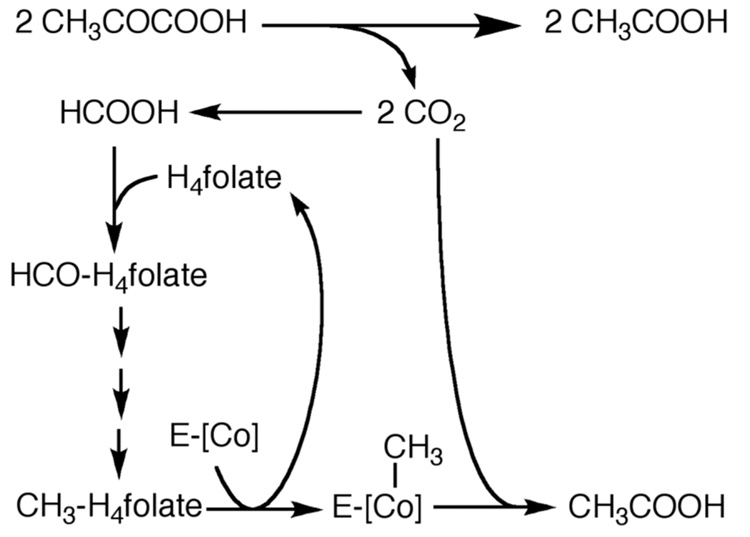
It was then shown that pulse labeling M. thermoacetica cells with 14CO2 led to the formation of 14CH3-labeled corrinoids; furthermore, when the 14CH3-corrinoids were added to cell extracts in the presence of pyruvate and CoA, 14C-labeled acetate was formed [57]. Based on these results, it was proposed that H4folate and cobalamin, shown as [Co] in Figure 3, were involved in this novel CO2 fixation pathway [58]. This proposal was based partly on an analogy with methionine synthase, which requires both H4folate [59] (as methyl-H4folate) and a corrinoid (as methylcobalamin) [60, 61] to convert homocysteine to methionine. Thus, it was postulated [62] that CO2 is converted to HCOOH and then to CH3-H4folate via the same series of H4folate-dependent enzymes (vide infra) that had been studied in Clostridium cylindrosporum and Clostridium acidi-urici by Rabinowitz and coworkers [63]. Subsequently, M. thermoacetica cell extracts were shown to catalyze conversion of the methyl group of 14CH3-H4folate (isolated from whole cells that had been pulse labeled with 14CO2) [64] to 14C-acetate [65]. As described below, Ljungdahl and coworkers isolated and characterized the H4folate-dependent enzymes that catalyze the conversion of formate to methyl-H4folate [66]. Although a role for cobalamin in acetogenesis was described around 1965 [56, 58], the required cobalamin-containing enzyme was isolated nearly two decades later [67] - the first protein identified to contain both cobalamin and an iron-sulfur cluster [68]. The properties of the H4folate- and cobalamin-dependent enzymes involved in the pathway are described below.
Figure 3.
The pathway of CO2 fixation into acetyl-CoA circa 1966, modified from [58].
H4folate and the H4folate-dependent enzymes involved in the acetyl-CoA pathway play key roles in one-carbon transfers for a number of essential cell functions (synthesis of serine, thymidylate, purines, and methionine), as they do in all bacteria and eukaryotes; however, because they are involved in a key catabolic pathway in acetogenic bacteria, they are found at levels 1000-fold higher and with ca. 100-fold higher specific activity than in other organisms.
3. Isolation of CO dehydrogenase/acetyl-CoA synthase and the elusive corrinoid protein and determination of their roles in the Wood-Ljungdahl pathway
Between 1980 and 1985, all components of the Wood-Ljungdahl pathway were purified and their roles were identified. In 1981, Drake, Hu, and Wood isolated five fractions from M. thermoacetica that together could catalyze the conversion of 14CH3-H4folate and pyruvate to acetylphosphate (Equation 8), and purified four of these components to homogeneity. The purified enzymes were pyruvate ferredoxin oxidoreductase, ferredoxin, methyltransferase, and phosphotransacetylase, while the fifth fraction (called Fraction F3) contained several proteins [69]. If CoA was substituted for phosphate, acetyl-CoA was the product (Equation 9) and phosphotransacetylase was not required.
| (8) |
| (9) |
At approximately the same time that Wood’s group was characterizing the five components, Gabi Diekert and Rolf Thauer discovered CO dehydrogenase (CODH) activity in M. thermoacetica [70] and established a growth requirement for nickel [71]. Drake et al. partially purified CODH and provided direct evidence that it contains Ni, using 63Ni [72]. CODH catalyzes the oxidation of CO to CO2 (Equation 10) and transfers the electrons to ferredoxin, or a variety of electron acceptors [73, 74]. Hu et al. then demonstrated that CODH was a component of Fraction F3 and that CO could substitute for pyruvate (Equation 11), and, in this case, only three components were required for acetyl-CoA synthesis: Fraction F3, ferredoxin, and methyltransferase [75]. It was not yet clear whether CODH was a required component or an impurity in Fraction F3, and it was considered that it might act as an electron donor in the reaction. Hu et al. also discovered that Fraction F3 alone could catalyze an exchange reaction between CO and the carbonyl group of acetyl-CoA, as shown in Equation 12 [75]. Discovery of this exchange reaction enabled studies that led to the discovery of the role of CODH in acetyl-CoA synthesis (see below).
| (10) |
| (11) |
| (12) |
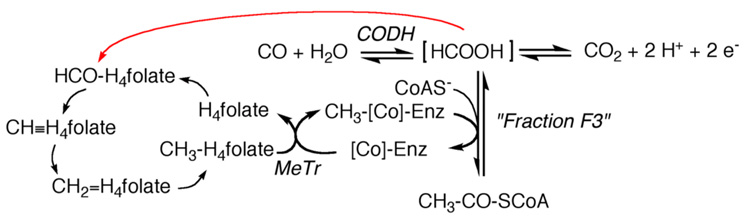
The scheme shown in Figure 4 stirred up the Ljungdahl laboratory, where one of the authors (SWR) was a graduate student. The concept of a bound form of formate was under intense discussion in a number of laboratories. As described in Figure 4, CODH was proposed to generate [HCOOH], which might react with the methylated corrinoid protein and CoA to generate acetyl-CoA [75]. For example, as described below, a carboxymethyl-corrinoid was being discussed as a key intermediate in CO2 fixation. Early in his graduate program at the University of Georgia, SWR had the opportunity to work for a few weeks in the Wood laboratory at Case Western University to learn how to isolate the various fractions and to assay the total synthesis of acetate.
Figure 4.
Scheme circa 1982 proposing roles for the corrinoid protein and CODH in anaerobic autotrophic CO2 fixation, modified from Fig. 2 of Hu et al. [75].
The proposal related to the red arrow shown in Figure 4 became a focus of intense discussion in the Ljungdahl laboratory and a series of studies were initiated to test the hypothesis that the [HCOOH] might directly serve as the source of the formyl group of formyl-H4folate, thus bypassing formate dehydrogenase [75] (Ljungdahl’s favorite enzyme).
Following the aphorism of Efraim Racker ("Don't waste clean thinking on dirty enzymes"), we aimed to purify all the enzymes (including all the H4folate-dependent enzymes, formate dehydrogenase, methyltransferase, and “Fraction F3”) required for the synthesis. Many of these had previously been isolated; however, we wished to also use purified CO dehydrogenase, an enzyme that was known to be extremely oxygen sensitive, and the only reported purification had yielded an impure enzyme that rapidly lost activity [72]. Having visited the Wood laboratory, SWR recognized that they probably had been unsuccessful with the purification because the anaerobic set-up (as described in [69]) was insufficiently rigorous. As students in Ljungdahl’s laboratory, we had experience with the strictly anaerobic methods (including an anaerobic chamber) required for purifying formate dehydrogenase and had purified many of the H4folate-dependent enzymes. So, we set up a small anaerobic chamber in a cold room (maintained at 10 °C) in which the normal light bulb was replaced with a red light in case the reaction proved to be light sensitive. With the anaerobic set-up, the requirement for formate dehydrogenase in the pathway was established [76]; therefore, the red arrow shown in Figure 4 could be erased and replaced with the formate dehydrogenase-and formyl-H4folate synthetasecatalyzed reactions, as shown in Figure 5, and as had been proposed in 1966. Furthermore, we were elated that the first attempt at purifying CODH yielded a homogeneous enzyme with specific activity that was 10-fold higher than had ever been measured [73]. Although CODH was indeed very oxygen sensitive, it was not sensitive to light or heat and, when purified and stored under strictly anaerobic conditions, the enzyme was remarkably stable. The CODHs from M. thermoacetica [73, 77] and Acetobacterium woodii [78] were characterized as two-subunit nickel-iron-sulfur metalloenzymes. However, the role of CODH in the acetyl-CoA pathway remained obscure.
Figure 5.
Scheme circa 1983 after re-establishment of formate dehydrogenase and formyl-H4folate synthetase in the pathway.
Besides CODH, another component of Fraction F3 was the elusive corrinoid enzyme, which Hu purified and showed to undergo methylation to form 14CH3-corrinoid when incubated with the methyltransferase and 14CH3-H4folate [75]. Furthermore, in the presence of Fraction F3, CO, and CoA, the radioactive methyl group of the 14CH3-labeled corrinoid protein was converted to 14C-acetyl-CoA (Equation 13).
| (13) |
Several key questions remained. The roles of CODH and the corrinoid protein in the pathway were unclear. The chemical nature of [HCOOH] was unknown. It also remained to be elucidated how the methyl, carbonyl, and coenzyme A groups were combined in generating the C-C and C-S bonds of acetyl-CoA.
a) Identification of the Corrinoid Protein as Methyl Carrier and CODH as the acetyl-CoA synthase: Bioorganometallic Chemistry
Regarding the roles of CODH and the corrinoid enzyme in acetyl-CoA formation, it was hypothesized (although incorrectly, vide infra) that CO and the methyl group combine on the corrinoid protein to generate a carboxymethyl- or perhaps an acetyl-corrinoid intermediate that would then be cleaved by CoA to generate acetyl-CoA. This hypothesis of the corrinoid protein acting as the site of acetate assembly was partly based on the finding that M. thermoacetica extracts catalyze the formation of acetate from carboxymethylcobalamin and the observation that the conversion of 14CO2 to a 14C-labeled corrinoid, which undergoes photolysis to generate products that had been previously identified as the photolysis products of carboxymethylcobalamin [57]. Furthermore, in the mid-1980’s, it was unclear whether the methyl group reacted as an anion as a cation. Since alkylcobamides had been considered to act as biological Grignard reagents [79] (although we now recognize that the alkyl group is actually transferred as an carbocation, i.e., CH3 + [80]), a mechanism of acetate synthesis involving attack of the methyl carbanion on a carboxy group, forming an acetoxycorrinoid was proposed [81]. This mechanism was based partly on the finding that M. thermoacetica extracts catalyzed the incorporation of approximately 50% of the deuterium from CD3-H4folate or [CD3]-methylcobalamin to trideuteromethyl-acetate. The proposal of a corrinoid-catalyzed methyl radical mechanism also seemed feasible since photolysis of methylcobalamin (in solution) in the presence of a high CO pressure (31 atm.) was shown to generate acetyl-cobalamin via a radical mechanism [82]. Thus, it seemed reasonable that the assembly of acetate occurred on a corrinoid protein though it was unknown whether the synthesis occurred from an acetylcobalt, acetoxycobalt, or a carboxymethylcobalt intermediate. Evidence for an acetyl-intermediate was provided by experiments demonstrating that when the methylated corrinoid protein was incubated with CO, acetate (albeit, low amounts) was formed; however, in the presence of CoA, acetyl-CoA was formed [75].
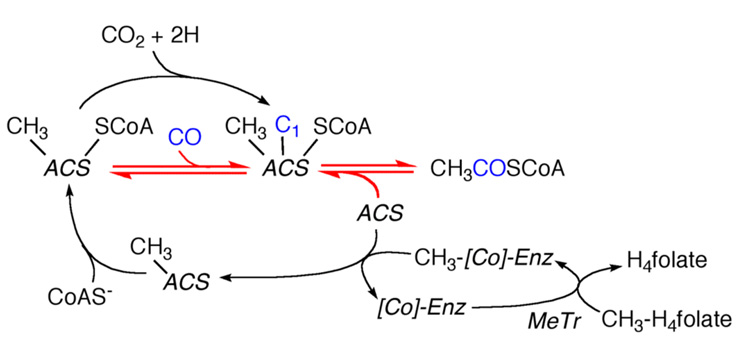
Clarification of the actual roles of the corrinoid protein and CODH exemplifies a remark attributed to Albert von Szent-Györgyi: "Very often, when you look for one thing, you find something else". As noted above, Hu et al. had discovered that Fraction F3 alone could catalyze an exchange reaction between CO and the carbonyl group of acetyl-CoA (Equation 12) [75]. This assay was much less complicated than the typical assay for acetyl-CoA synthesis, which involved 14CH3-H4folate, CO (or pyruvate and pyruvate ferredoxin oxidoreductase), CoA, methyltransferase, the corrinoid protein, plus Fraction F3. Because this assay required the disassembly and reassembly of acetyl-CoA, SWR surmised that purifying the component required in the exchange would uncover the key missing component(s) required for acetyl-CoA synthesis. Since it was thought that the assembly of acetyl-CoA occurred on the corrinoid protein (above), it came as a surprise that the purified corrinoid protein could not catalyze this exchange [83]. This was interpreted to mean that some component of Fraction F3 plus the corrinoid protein was required. However, the shocking results were that CO dehydrogenase was the only required enzyme and that addition of the corrinoid protein had no effect [83]. Thus, as depicted in Figure 6, it was concluded that the role of CODH is to catalyze the assembly of acetyl-CoA from CO, a bound methyl group, and CoA, and that the corrinoid protein serves to accept the methyl group from CH3-H4folate and then transfer it to ACS. As it so happened, what was found was more exciting than what was being looked for. Because CODH played the central role in this new scheme, the enzyme was renamed acetyl-CoA synthase (ACS) [83]. It took several years to recognize that CO oxidation and acetyl-CoA synthesis are catalyzed at separate metal centers [84] and this bifunctional enzyme is now called CODH/ACS [85].
Figure 6.
Scheme circa 1985 showing that CODH is ACS, the central enzyme in the pathway, and that the corrinoid enzyme is a methyl carrier. The red arrows designate the reactions involved in the CO/acetyl-CoA exchange. Modified from [83].
However, the nature of the “C1” shown in Figure 6 (equivalent to [HCOOH] in other schemes) was unknown. Furthermore, it was not yet recognized that CO not only serves as a precursor of the carbonyl group of acetyl-CoA, but it is actually generated from CO2 or pyruvate as an intermediate in the acetyl-CoA synthesis [86]. The nature of “C1” or “[HCOOH]” is described in the next section.
b) Identification of the enzyme-bound precursor of the carbonyl group of acetyl-CoA
Figure 4–Figure 6 portray a “C1” or “HCOOH” intermediate, which designates a bound form of CO that derives from CO2, CO, or the carboxyl group of pyruvate. To attempt to identify this intermediate, various biochemical spectroscopic studies were initiated (see [87–90] for reviews). CODH was incubated with CO and was found to generate an EPR-detectible intermediate [91, 92] that exhibited hyperfine splitting from both 61Ni and 13CO, indicating that an organometallic Ni-CO complex had been formed. This EPR signal eventually was shown to be elicited from a complex between CO and a nickel iron-sulfur cluster and was thus called the “NiFeC species” [93]. The electronic structure of this center is discussed below. However, the identity of this EPR-active species as the active carbonylating agent has been actively debated [87–89, 94], which is also discussed below.
V. General aspects of the Wood-Ljungdahl pathway
A. Coupling of acetogenesis to other pathways allows growth on diverse carbon sources, electron acceptors, electron donors
Acetogens can use a wide variety of carbon sources and electron donors and acceptors. One-carbon compounds that M. thermoacetica and most acetogens can use for growth include H2 + CO2, CO, formate, methanol, and methyl groups from many methoxylated aromatic compounds. In addition, M. thermoacetica can grow on sugars, two-carbon compounds (glyoxylate, glycolate, and oxalate), lactate, pyruvate, and short-chain fatty acids. Besides CO2, electron acceptors include nitrate, nitrite, thiosulfate, and dimethylsulfoxide. Information about the heterotrophic growth characteristics and electron donors/acceptors are described in more detail below.
B. The Wood-Ljungdahl pathway and the emergence and early evolution of life
Based on the patterns of 12C/13C isotopic fractionation, the sedimentary carbon record indicates the emergence of autotrophy soon after the earth became habitable, ca. 3.8 billion years ago [95]. The isotopic fractionation pattern of anaerobic organisms using the Wood-Ljungdahl pathway suggests that they may have been the first autotrophs, using inorganic compounds like CO and H2 as an energy source and CO2 as an electron acceptor approximately 1 billion years before O2 appeared [96].
Acquisition of the core Wood-Ljungdahl genes would allow a wide variety of organisms, including archaea and a broad range of bacterial phyla (Firmicute, Chlorflexi, and Deltaproteobacteria), to fix CO and CO2 by the reductive acetyl-CoA pathway. The simple strategy of the Wood-Ljungdahl pathway of successively joining two one-carbon compounds to make a two-carbon compound has been envisioned to be the earliest form of metabolism [97].
C. Methods used in elucidation of the pathway
As mentioned above, both 14C- and 13C-isotope labeling experiments were key in demonstrating that this pathway is a novel mechanism of CO2 fixation. The 13C-labeling experiments involved mass spectroscopy using a tall column that Wood constructed in the stairwell of the biochemistry department at what was then known as Western Reserve University (“Case-” was appended in 1967). However, these pulse-labeling methods did not elucidate the intermediates in the Wood-Ljungdahl pathway. As one can see in Figure 1, the only products from 14CO2 that would build up in solution are formate and acetate. Various cofactors are used in enzymes in the Wood-Ljungdahl pathway. These include H4folate, cobalamin, [4Fe-4S] clusters, and some unusual metal clusters, including two nickel-iron-sulfur clusters. Eventually, 14CH3-H4folate and 14CH3-corrinoids were identified and shown to be incorporated into the methyl group of acetyl-CoA, but both of these methylated cofactors are bound to enzymes and do not build up in solution.
The hypothesis that H4folate-dependent enzymes were involved was confirmed by isolation of the enzymes and demonstration that the reactions elucidated in other organisms indeed occur in M. thermoacetica. One of the unusual features of this pathway is that a number of intermediates (CH3-Co, CH3-Ni, Ni-CO, acetyl-Ni) are bound to transition metals. Thus, although paper chromatography was not appropriate for isolating intermediates, much more powerful methods that can focus directly on the active site metal centers were enlisted to characterize the properties of the cobalt, nickel, and iron-sulfur clusters in the CFeSP and CODH/ACS. The various oxidation states of these transition metals in the enzyme active sites have been established by electrochemical and spectroscopic methods (EPR, Mossbauer, UV-visible, Resonance Raman, EXAFS, XANES) and the rates of interconversion among the various intermediates have been studied by coupling spectroscopy to rapid mixing methods (stopped flow, chemical quench, rapid freeze quench). Density functional theory is also being used to test various mechanistic hypotheses. The existence of organometallic species has attracted a number of inorganic and bioinorganic chemists, who are developing models of the enzyme active sites to provide a better understanding of the roles of the metals in these intriguing reactions. This combination of approaches has spurred our understanding of the pathway and enriched our understanding of metal-based biochemistry.
D. Insights from the genome sequence
1. What it takes to be an acetogen
The genome of M. thermoacetica was recently sequenced and annotated. This first acetogenic genome to be sequenced is a 2.6 Megabase genome and 70 % of the genes have been assigned tentative functions [98]. Based on the 16S rRNA sequence, M. thermoacetica was classed as a Clostridium within the Thermoanaerobacteriaceae family [7]; after all, it was first known as Clostridium thermoaceticum. Homoacetogenic microbes are widely distributed among a few members of many phyla including Spirochaetes, Firmicutes (e.g., Clostridia), Chloroflexi, and Deltaproteobacteria, clearly indicating that acetogenesis is a metabolic, not a phylogenetic trait. Thus, homoacetogenesis is a rare occurrence within any phylum. For example, two close relatives of M. thermoacetica in the Thermoanaerobacteriaceae family, Thermoanaerobacter tengcongensis [99] and Thermoanaerobacter ethanolicus (draft sequence available at JGI) can grow on various sugars and starch [100], but the other two anaerobes lack acetyl coenzyme A synthase (ACS) and are not homoacetogenic. Among all the genes in the available Firmicutes, Chlorflexi, and Deltaproteobacteria genomic sequences, only acsC and acsD, which encode the two subunits of the CFeSP, co-occur with acetyl-CoA synthase (acsB) gene and are not present in sequences that lack acsB [98]. Surprisingly, acetogens do not seem to require a special set of electron transport-related genes, indicating that acetogens have co-opted electron transfer pathways used in other metabolic cycles.
2. Pathways for heme and cobalamin biosynthesis
In order to grow strictly autotrophically, an organism, of course, should not require any vitamins or cofactors. However, it is likely that in nature, some vitamins could be provided by crossfeeding. M. thermoacetica does require one vitamin, nicotinic acid [101]; however, it contains all the other genes needed to convert nicotinic acid to NAD [98]. This organism also has genes encoding both the de novo and salvage pathways for H4folate synthesis and both the complete anaerobic branch of corrin ring synthesis and the common branch of the adenosyl-cobalamin pathway [98]. M. thermoacetica could be considered to be a cobalamin factory, generating over twenty different types of cobalamin or precursors that total 300 – 700 nmol per gram of cells [102, 103].
VI. Description of the Wood-Ljungdahl Pathway
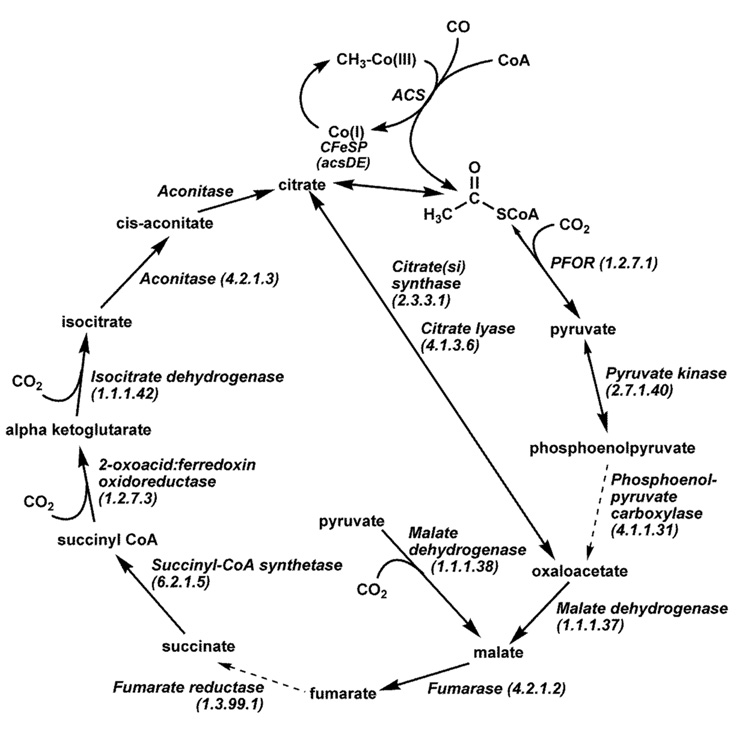
Figure 1 shows the key reactions in the Wood-Ljungdahl pathway of CO2 fixation described to consist of an Eastern (red coloring scheme) and a Western (blue) branch [104]. One molecule of CO2 undergoes reduction by six electrons to a methyl group in the Eastern branch, while the Western branch involves reduction of the other CO2 molecule to carbon monoxide, and condensation of the bound methyl group with CO and coenzyme A (CoA) to make acetyl-CoA. Acetyl-CoA is then either incorporated into cell carbon or converted to acetyl phosphate, whose phosphoryl group is transferred to ADP to generate ATP and acetate, the main growth product of acetogenic bacteria (for reviews see [26, 105, 106]).
A. The Eastern or Methyl Branch of the Wood-Ljungdahl Pathway
The genes encoding enzymes in the Eastern branch of the pathway are scattered around the M. thermoacetica genome. For example, the genes encoding 10-formyl-H4folate synthetase, and the bifunctional 5,10-methenyl-H4folate cyclohydrolase/5,10-methylene-H4folate dehydrogenase are far from each other and are at least 300 genes away from the acs gene cluster.
1. Formate Dehydrogenase (Moth_2312-Moth_2314; EC 1.2.1.43)
The first reaction in the conversion of CO2 to CH3-H4folate is the two-electron reduction of CO2 to formate (Equation 14), which is catalyzed by a tungsten- and selenocysteine-containing formate dehydrogenase [107–109]. CO2, rather than bicarbonate, is the substrate [110–112]. The first enzyme shown to contain tungsten [113], the M. thermoacetica FDH is an α2β2 enzyme (Mr = 340,000) that contains, per dimeric unit, one tungsten, one selenium, and ~ 18 irons and ~ 25 inorganic acid-labile sulfides in the form of iron-sulfur clusters [109]. The α subunit contains the selenocysteine residue, which is encoded by a stop codon in most contexts, and is responsible for binding the molybdopterin cofactor. The acetogenic formate dehydrogenase must catalyze a thermodynamically unfavorable reaction: the reduction of CO2 to formate (Eo’ = −420 [114]) with electrons provided by NADPH, with a half cell potential for the NADP+/NADPH couple of −340 mV. The purified enzyme catalyzes the NADP+-dependent oxidation of formate with a specific activity of 1100 nmol min−1mg −1 (kcat = 6.2 s−1). Another selenocysteine-containing formate dehydrogenase (Moth_2193) was located in the genome that has been proposed to be part of a formate hydrogen lyase system [98].
| (14) |
In the M. thermoacetica FDH, the tungsten is found as a tungstopterin prosthetic group, like the molybdopterin found in the Mo-FDHs and in xanthine oxidase, sulfite oxidase, and nitrate reductase [116, 117]. The M. thermoacetica genome encodes a surprisingly large number of molybdopterin oxidoreductases in the dimethylsulfoxide reductase and xanthine dehydrogenase families [98]. Based on the original [118] and a reinterpreted [119] crystal structure, the mechanism for the Mo-Se-FDH involves formate displacing the Se-Cys residue as it binds to the oxidized Mo(VI) center. Then, two-electron transfer from the substrate to the Mo(VI) generates Mo(IV) and CO2, which is released. Finally, the Se-Cys re-ligates to the Mo and two electrons are transferred through a [4Fe-4S] cluster to an external acceptor, reforming the oxidized enzyme for another round of catalysis. It is likely that the W-Se enzyme follows the same catalytic mechanism.
2. 10-Formyl-H4folate synthetase (Moth_0109, EC. 6.3.4.3.)
In the reaction catalyzed by10-formyl-H4folate synthetase, formate undergoes an ATP-dependent condensation with H4folate, forming 10-formyl-H4folate (Equation 15). The formyl-H4folate product is then used in the biosynthesis of fMet-tRNAfMet (which is converted to N-formyl-methionine) and purines, or dehydrated and reduced by the succeeding H4folate-dependent enzymes (cyclohydrolase and dehydrogenase) to generate 5,10-methylene-H4folate, which is used in the biosynthesis of amino acids and pyrimidines [120] or acetate in acetogens.
| (15) |
Like all bacterial formyl-H4folate synthetases that have been studied, the M. thermoacetica enzyme is homotetrameric, with identical substrate binding sites on four 60 kDa subunits [120, 122]. This enzyme has been purified, characterized, and sequenced from a number of sources including M. thermoacetica [123–127], and the structure of the M. thermoacetica enzyme is known [128]. In higher organisms, this enzyme exists as one of the activities of a trifunctional C1-H4folate synthase (also containing a cyclohydrolase and a dehydrogenase) [129, 130], whereas the bacterial formyl-H4folate synthetases are monofunctional and share similar properties [131].
The M. thermoacetica enzyme has been heterologously and actively expressed in E. coli [123]. Based on steady-state kinetic and equilibrium isotope exchange studies, the synthetase appears to follow a random sequential mechanism [125] involving a formyl-phosphate intermediate that suffers nucleophilic attack by the N10 group on H4folate to form the product [132]. Formation of 10-formyl-H4folate by the synthetase from M. thermoacetica occurs with a kcat value of 1.4 s−1 [133]. Monovalent cations activate the enzymes from M. thermoacetica and C. cylindrosporum by decreasing the Km for formate to ~0.1 mM [126, 127].
3. 5,10-methenyl-H4folate cyclohydrolase (EC 3.5.4.9.) and 5,10-methylene-H4folate dehydrogenase (EC 1.5.1.5., NADP; EC 1.5.1.15,NAD) (Moth_1516)
The next two steps in the Ljungdahl-Wood pathway are catalyzed by 5,10-methenyl-H4folate cyclohydrolase (Equation 16) and 5,10-methylene-H4folate dehydrogenase (Equation 17). While the cyclohydrolase and dehydrogenase are part of a bifunctional protein in M. thermoacetica, they are monofunctional proteins in other acetogens, e.g., Clostridium formicoaceticum and Acetobacterium woodii [134–136]. As mentioned above, they are part of the trifunctional C1-synthase in eukaryotes.
| (16) |
| (17) |
The cyclohydrolase reaction strongly favors cyclization, with an equilibrium constant for 5,10-methenyl-H4folate formation of 1.4 × 106 M−1 [137]. The reverse reaction (hydrolysis) occurs at a rate of ~ 250 s−1 (35 °C, pH 7.2) for the C. formicoaceticum enzyme [136]. The NAD(P)H-dependent reduction of methenyl-H4folate to form 5,10-methylene-H4folate is catalyzed by 5,10-methylene-H4folate dehydrogenase (Equation 17). The Eo' for the methenyl-/methylene-H4folate redox couple is −295 mV vs. the standard hydrogen electrode (SHE) [121]; thus, reduction by NAD(P)H is thermodynamically favorable. Generally, microbes contain a bifunctional cyclohydrolase-dehydrogenase. The monofunctional dehydrogenase has so been identified in C. formicoaceticum [136], yeast [138], and A. woodii [135]. One rationale for the bifunctional enzyme may be to protect the highly labile methenyl-H4folate from hydrolysis and channel it [139, 140] to the active site of the dehydrogenase for reduction to the relatively more stable methylene-H4folate. There are both NAD+- and NADP+-dependent forms of this enzyme, yet the NADP-dependent enzyme is most common. A. woodii [135], Ehrlich ascites tumor cells [141], and yeast [142] contain the NAD+-dependent form. Crystal structures have been determined for the monofunctional NAD+-dependent yeast dehydrogenase [142], the bifunctional E. coli dehydrogenase/ cyclohydrolase [143], and the dehydrogenase/cyclohydrolase domains of the human trifunctional C1 synthase [144].
The dehydrogenase reaction has been probed mostly in the direction of 5,10-methenyl-H4folate formation, which is the reverse of the physiological reaction. The A. woodii dehydrogenase catalyzes both the forward and reverse reactions with kcat values of ~1600 s−1 [135]. With the M. thermoacetica dehydrogenase, oxidation of 5,10-methylene-H4folate by NADP+ occurs according to a ternary complex mechanism with a specific activity of 360 s−1 at 37 °C [145] (720 s−1 at 60 °C, D.W. Sherod, W.T. Shoaf, and L.G. Ljungdahl, unpublished).
4. 5,10-methylene-H4folate reductase (Moth_1191, EC 1.1.99.15)
The last step in the eastern branch is catalyzed by 5,10-methylene-H4folate reductase (Equation 18), which in acetogens is an oxygen-sensitive octomeric (αβ)4 enzyme consisting of 35 and 26 kDa subunits [146, 147], while the Peptostreptococcus productus [148] and E. coli [149] enzymes are multimeric consisting of identical 35 kDa subunits. The mammalian enzyme is ~75 kDa, containing a C-terminal extension that binds the allosteric regulator S-adenosyl-L-methionine, which decreases the activity of the reductase by up to 50,000-fold [150]. Studies of the mammalian enzyme are of particular importance because the most common genetic cause of mild homocysteinemia in humans is an A222V polymorphism in the reductase [150, 151]. Among these characterized reductases, all contain FAD but only the acetogenic enzymes contain an iron-sulfur cluster [146, 147, 149]. The acetogenic enzyme uses reduced ferredoxin as an electron donor [146, 147], while the mammalian, E. coli, and P. productus enzymes use NAD(P)H. With the acetogenic proteins, pyridine nucleotides are ineffective electron carriers for the reaction in either direction [146]. The standard reduction potential for the methylene-H4folate/CH3-H4folate couple is −130 mV vs. SHE [152]; thus, this reaction is quite exergonic with either NADH or ferredoxin as electron donor.
| (18) |
The mammalian and E. coli enzymes have been more extensively studied than the acetogenic enzyme [150, 153]. The catalytic domain of all methylene-H4folate reductases consists of an α8β8 TIM barrel that binds FAD in a novel fold and the Ala222 residue, whose polymorphic variant results in hyperhomocysteinemia is located near the bottom of the cavity carved out by the barrel [154].
The acetogenic reductase uses a ping-pong mechanism to catalyze the oxidation of CH3-H4folate with benzyl viologen with a kcat of 300–350 s−1 at its optimal temperature (35 °C and 55 °C for the C. formicoaceticum and M. thermoacetica enzymes, respectively) [146, 147]. The E. coli and pig-liver enzymes also use a ping-pong mechanism and exhibit a significantly lower value of kcat than the acetogenic proteins [149, 155]. For the mammalian enzyme, the first half reaction, the stereospecific reduction of FAD by the pro-S hydrogen of NADPH to form FADH2, is rate limiting during steady-state turnover [153, 155]. In the second half reaction, 5,10-methylene-H4folate undergoes reduction by bound FADH2 to form CH3-H4folate, as hydrogen is stereospecifically transferred to the more sterically accessible face of the pteridine and before undergoing exchange with solvent [153, 156]. It is proposed that during the reduction, the imidazolium ring of 5,10-methylene-H4folate opens to form an iminium cation followed by tautomerization [157–159].
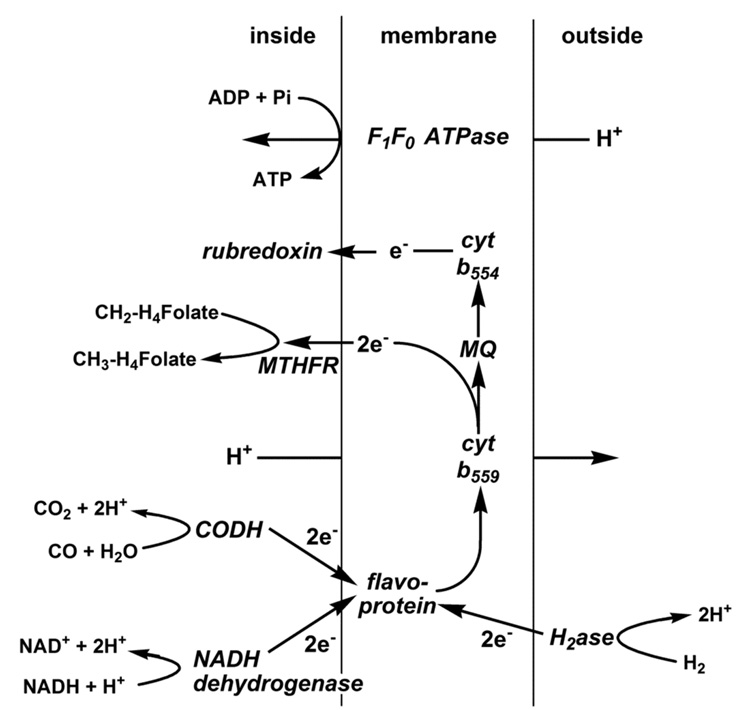
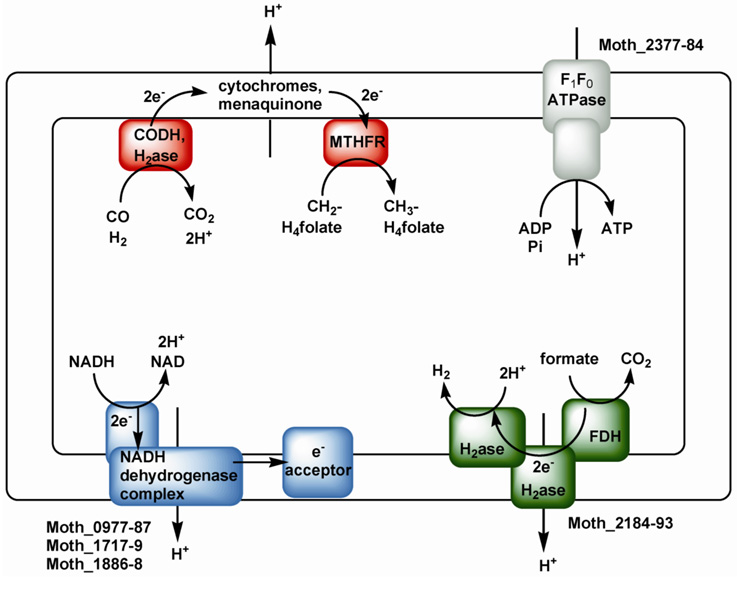
An unsolved question is how acetogens conserve energy by use of the Wood-Ljungdahl pathway. Thauer et al. [160] suggested that energy conservation could occur by linking an electron donor (e.g., H2/hydrogenase, CO/CODH, or NADH dehydrogenase) to a membrane-associated electron transport chain, which would in turn donate electrons to methylene-H4folate and the 5,10-methylene-H4folate reductase. In this scenario, electron transport could result in the generation of a transmembrane proton potential coupled to ATP synthesis. In A. woodii, one of the steps involving methylene-H4folate reductase, the methyltransferase or the CFeSP was shown to generate a sodium motive force across the membrane, which is linked to energy conservation [161]. However, since the reductase seems to be located in the cytoplasm in M. thermoacetica [146] and in other organisms, it appears that this is not the energy-conserving step in the Wood-Ljungdahl pathway [162]. On the other hand, the location of the 5,10-methylene-H4folate reductase gene very near genes encoding a hydrogenase and heterodisulfide reductase (which are directly downstream from the acs gene cluster), suggests a possible mechanism of energy conservation and proton translocation involving the reductase and the membrane-associated hydrogenase and/or heterodisulfide reductase.
B. The Western or Carbonyl Branch of the Wood-Ljungdahl Pathway
1. Genes related to the Western branch of the pathway
While the genes encoding the Eastern branch of the Wood-Ljungdahl pathway are ubiquitous and dispersed on the genome, the Western branch genes are unique to organisms that use the Wood-Ljungdahl pathway and are co-localized in the acs gene cluster (Figure 7) [163]. Interestingly, their functional order in the pathway often matches the gene order: (1) carbon monoxide dehydrogenase (CODH), (2) acetyl-CoA synthase (ACS), (3) and (4) the two subunits of the corrinoid iron-sulfur protein (CFeSP), and (5) methyltransferase (MeTr). The acs gene cluster also includes a gene encoding an iron-sulfur protein of unknown function (orf7), and two genes, cooC and acsF, that are homologous to the Rhodospirillum rubrum cooC, which is required for nickel insertion into carbon monoxide dehydrogenase [164].
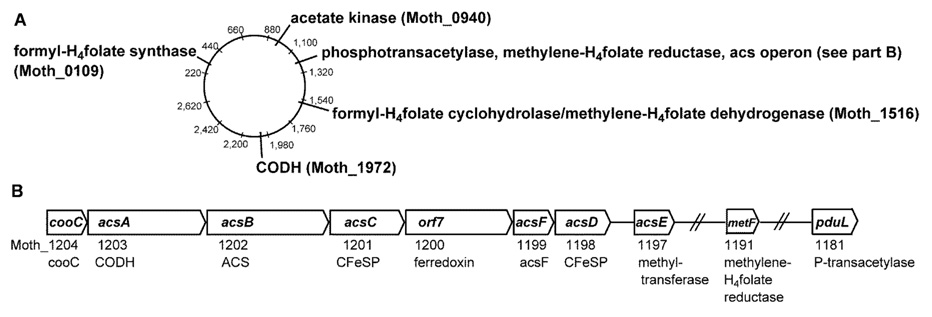
Figure 7.
Arrangement of Wood–Ljungdahl pathway genes in the acs gene cluster and in the chromosome. A. Arrangement of Wood–Ljungdahl pathway genes on the circular chromosome of M. thermoacetica. The numbering shows kilobase pairs from the origin of replication. B. The acs gene cluster that contains core Wood–Ljungdahl pathway genes discussed in the text. From [98].
Surprisingly, the M. thermoacetica genome sequence reveals an additional carbon monoxide dehydrogenase (Moth_1972), which is similar to an uncharacterized carbon monoxide dehydrogenase from Clostridium cellulolyticum and to CODH IV from Carboxydothermus hydrogenoformans [165], and is unlinked to the acs gene cluster.
2. Enzymology and bioinorganic chemistry of the Western branch of the pathway
a) Methyl-H4folate:CFeSP Methyltransferase (MeTr) (Moth_1197,EC 2.1.1.X)
MeTr catalyzes the transfer of the methyl group of methyl-H4folate to the cobalt center of the corrinoid iron-sulfur protein (CFeSP) (Equation 19). This reaction forms the first in a series of enzyme-bound bioorganometallic intermediates in the Wood-Ljungdahl pathway (methyl-Co, methyl-Ni, Ni-CO, acetyl-Ni), a strategy that is a novel feature of the Wood-Ljungdahl pathway. Equation 19 uses the term cobamide, instead of cobalamin, because the CFeSP contains methoxybenzimidazolylcobamide, not dimethylbenzimidazolylcobamide (cobalamin), although the enzyme is fully active with cobalamin [166]. This reaction is similar to the first step in the reaction mechanism of cobalamin-dependent methionine synthase [80, 167]. As noted in Equation 19, cobalt undergoes redox changes during the reaction and, as in other cobalamin-dependent methyltransferases [168], Co(I) is the only state that can catalyze methyl transfer, forming an alkyl-Co(III) product. The redox chemistry involving the CFeSP will be covered in the next section. Here we will focus on the structure and function of MeTr.
| (19) |
While MeTr catalyzes methyl transfer from methyl-H4folate, various cobalamin-dependent methyltransferases are found in biology, which react with a wide range of natural methyl donors, including methanol, methylamines, methyl thiols, and aromatic methyl ethers [168]. Structural and kinetic studies have been performed to attempt to understand how the methyltransferases catalyze the methyl transfer reaction. A key issue is that the methyl group requires activation, since the bond strengths of the C-O, C-N, or C- bonds in methanol, methylamines, and methyl thiols are quite large (356, 305, and 272 kJ/mol, respectively). As discussed in a recent review [168], there are at least three ways that MeTr enzymes activate the methyl donor: general acid catalyzed protonation of the N5 of pterins in methyl-H4folate (and probably in methyltetrahydromethanopterin) through a H-bonding network, Lewis acid catalysis using a Zn active site near the cobalamin, and covalent catalysis using a novel amino acid (pyrrolysine). Protonation of the N-5 group of N5-methyl-H4folate would lead to electrophilic activation of the methyl group. Thus, one of the goals of structural studies described below was to identify how substrates bind and undergo activation in the MeTr active site.
All of these methyltransferases bind the methyl donor within an α/β TIM barrel structure, as observed in the crystal structures of the methanol binding protein MtaB [169], the methyl-H4folate binding domain of methionine synthase [170], the methylamine binding protein MtmB [171], and the methyl-H4folate:CFeSP methyltransferase, shown in Figure 8 [172, 173]. In all of these proteins, the methyl donor binds within the cavity formed by the TIM barrel, as shown in (Figure 8B). The red surface indicates the negative charge in the cavity, which complements positive charges on methyl-H4folate, which is tightly bound by hydrogen bonds involving residues D75, N96, and D160 that are conserved between MeTr and methionine synthase and have been referred to as the “pterin hook” [172]. Another conserved feature among the various MeTr structures is the lack of an obvious proton donor near the N-5 group. How does the methyl group then undergo activation?
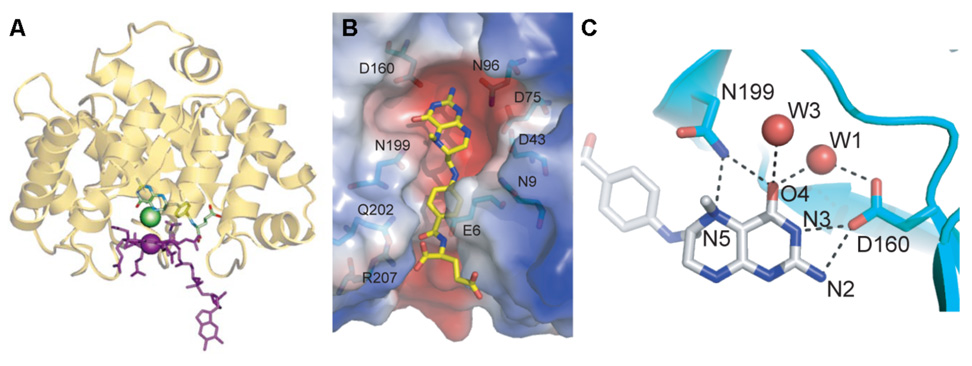
Figure 8.
A. Structure of MeTr. Methyl-H4folate and base-off cobalamin were modeled into the active site of MeTr, with the methyl group of methyl-H4folate shown in green. From Fig. 5, [172]. B. The active site cavity of MeTr, based on the structure of the MeTr-methyl-H4folate binding site. From Fig. 5, [173]. C. Hydrogen bonding network near the N-5 of methyl-H4folate in the crystal structure of the binary complex of MeTr with methyl-H4folate [173].
All of the cobalamin-dependent methyltransferases appear to use a similar principle to activate the methyl group that will be transferred: to donate positive charge to the heteroatom (O, N, or S) attached to the methyl group. This mechanism of electrophilic activation of the methyl group is supported by studies of the transfer of methyl groups in solution in inorganic models. Quaternary amines, with a full positive charge on nitrogen, do not require activation, as shown by studies of methyl transfer from a variety of quaternary ammonium salts to Co(I)-cobaloxime [174], trimethylphenylammonium cation to cob(I)alamin [175], and dimethylaniline at low pH to Co(I)-cobyrinate [176]. Furthermore, Lewis acids, such as Zn(II), can catalyze methyl transfer [177].
The mechanism of protonation of the N5 group of methyl-H4folate, which leads to electrophilic activation of the methyl group, has been extensively studied in methionine synthase and the M. thermoacetica MeTr by transient kinetic and NMR measurements of proton uptake [178, 179], by measurements of the pH dependencies of the steady-state and transient reaction kinetics [167, 180], and by kinetic studies of variants that are compromised in acid-base catalysis [173]. These studies cumulatively show that general acid catalysis is involved in the methyl transfer reaction, but whether this occurs in the binary or ternary complex, or perhaps in the transition state for the methyl transfer reaction remains under discussion.
If protonation of the N5 group of methyl-H4folate is key to the mechanism of methyl transfer, one would expect that the crystal structure would uncover the residue that acts as the general acid catalyst. However, in the crystal structures of the binary complexes of CH3-H4folate bound to methionine synthase [170] and the methyl-H4folate:CFeSP methyltransferase [173], no obvious proton donor is within H-bonding distance of the N5 position of CH3-H4folate, and the only amino acid located near enough to N5 to participate in H-bonding is the side chain of Asn199, which is a conserved residue among all methyltransferases (Figure 8) [173]. Although this is an unlikely proton donor, upon binding CH3-H4folate, Asn199 swings by ~ 7 Å from a istant position into the H-bonded location shown in Figure 8C. Furthermore, the N199A variant has a mildly lower (~20-fold) affinity for CH3-H4folate, but a marked (20,000–40,000) effect on catalysis, suggesting that Asn199 plays an important role in stabilizing a transition state or high-energy intermediate for methyl transfer [173]. These experiments are consistent with the involvement of an extended H-bonding network in proton transfer to N5 of the folate that includes Asn199, a conserved Asp (Asp160), and a water molecule. Thus, although Asn199 is certainly not a chemically suitable proton donor, it becomes part of an extended H-bonding network that includes several water molecules that are also conserved in the methionine synthase structure.
The lack of a discernable proton donating residue is seen in a number of enzymes, including methionine synthase [170], dihydrofolate reductase [181], and purine nucleoside phosphorylase [182]. An extended H-bond network that includes water molecules, a Glu residue, and an Asn residue has been proposed to protonate the purine N7 in the transition state of the reaction catalyzed by purine nucleoside phosphorylase [183]. Thus, it has been speculated that the Asn residue in MeTr plays a key role in the transition state for methyl transfer and would slightly shift its position to allow both the carboxamide oxygen and nitrogen atoms to engage in H-bonding interactions with the pterin [173]. This dual H-bonding function could rationalize the placement of Asn at this key position in the methyltransferases, i.e., a typical general acid would be less apt in this bifurcated H-bonding stabilization of the transition state.
Most of the information related to the MeTr in this section dealt with binding and activation of its smaller substrate, the methyl group donor. We will now address the methyl acceptor, which is a cobamide that is tightly bound to an 88 kDa heterodimeric protein, the CFeSP.
b) Corrinoid iron-sulfur protein (CFeSP) (Moth_1198, Moth_1201,EC 2.1.1.X)
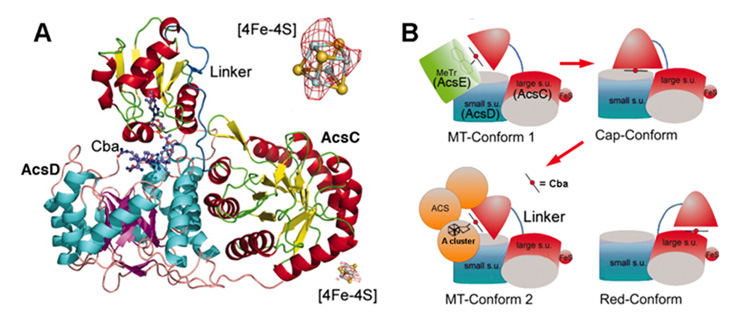
Twenty years after the discovery (described above) that cobalamin is involved in the pathway of anaerobic CO2 fixation, Hu et al. partially purified a corrinoid protein that accepts the methyl group of CH3-H4folate to form a methylcorrrinoid species that, when incubated with CO, CoA, and a protein fraction containing CODH activity, is incorporated into the methyl group of acetyl-CoA [67] (Equation 20). When this 88 kDa protein was purified to homogeneity and characterized by various biophysical methods, it was found to contain an iron-sulfur cluster in addition to the corrinoid; therefore, it was named the corrinoid iron-sulfur protein (CFeSP) [68]. Besides this CFeSP protein involved in acetogenesis and its homolog in methanogens [184], the only other corrinoid proteins so far known to contain an iron-sulfur cluster are the dehalogenases, which catalyze the reductive removal of the halogen group in the process linked to growth of microbes on halogenated organics [185, 186]. The structure of the CFeSP (Figure 9A) is divided into three domains: the N-terminal domain that binds the [4Fe-4S] cluster, the middle TIM-barrel domain, and the C-terminal domain (also adopts a TIM barrel fold), which interacts with the small subunit to bind cobalamin [187]. The two TIM barrel domains of the CFeSP appear to be related to the MeTr and it has been proposed that the C-terminal domain undergoes conformational changes to alternatively bind MeTr/methyl-H4folate and ACS (Figure 9B).
| (20) |
| (21) |
.
Figure 9.
Structure of the CFeSP. (A) From Figure 1 of [187], AcsC and AcsD are the large and small subunits, respectively, and Cba is the corrinoid cofactor. This figure shows the middle and C-terminal domains of the large subunit, while the N-terminal domain that contains the [4Fe-4S] cluster was disordered and could not be modeled. An expanded view of the [4Fe4S] cluster is shown in the upper right hand corner. (B) Modified from Fig. 3 of [187], proposed conformational changes of the CFeSP during methyl transfer from methyl-H4folate/MeTr to the corrinoid (MT-Conform 1) to a resting state (Cap-Conform shown in Fig. A), and then from methyl-corrinoid to the A cluster on ACS (MT-Conform 2). The reductive activation conformation is also depicted (Red-Conform).
Cobalt in cobalamin can access the (I), (II) and (III) redox states. As shown in Equation 20, the Co center cycles between the Co(I) and methyl-Co(III) states during the reaction, with the Co(I) state being required to initiate catalysis, as established by stopped-flow studies with the CFeSP [180] and with methionine synthase [188]. In the Co(I) state, cobalt has a d8 configuration. In B12 and related corrinoids, Co(I) is a supernucleophile [189, 190] and is weakly basic, with a pKa below 1 for the Co(I)-H complex [191]. Protein-bound Co(I) is also highly reducing with a standard reduction potential for the Co(II)/(I) couple below −500 mV [192–194]. This high level of reactivity comes with a price - Co(I) can easily undergo oxidative inactivation to the Co(II) state; for example, in MeTr and in methionine synthase, the Co(I) center succumbs to the 2+ state once in every 100–2,000 turnovers [195–197].
Resuscitation of inactive MeTr requires reductive activation, which occurs by the transfer of one electron from the [4Fe-4S] cluster within the large subunit (product of the acsC gene) of the CFeSP [197]. This cluster has a reduction potential of −523 mV, which is slightly more negative than that of the Co(II)/Co(I) couple of the cobamide (−504 mV) in the CFeSP, making reductive activation a thermodynamically favorable electron-transfer reaction [193]. The [4Fe-4S] cluster can accept electrons from a low-potential ferredoxin as well as directly from CO/CODH, H2/hydrogenase, or pyruvate/pyruvate ferredoxin oxidoreductase [197]. It has been demonstrated that the cluster is involved in reductive activation, but does not participate directly in the methyl transfer from methyl-H4folate or to the Ni center in ACS [198, 199]. Unlike the CFeSP, in methionine synthase, the electron donors (methionine synthase reductase in mammals [200], flavodoxin in E. coli [188]) are flavins, with a higher redox potential that that of the Co(II)/(I) couple; therefore, a reductive methylation system involving S-adenosyl-L-methionine is used to drive the uphill reactivation [192].
Although the redox potential of −504 mV for the Co(II)/Co(I) couple of the cobamide in the CFeSP is fairly low, it is well above the standard potential for the same couple in cobalamin in solution (~-610 mV) [201]. It appears that control of the coordination state of the Co(II) center plays a key role in increasing the redox potential into a range that would make the reduction feasible for biologically relevant electron donors. Instead of having a strong donor ligand like imidazole or benzimidazole, which is found in most other corrinoid proteins, the Co center in the CFeSP is base-off and the only axial ligand is a weakly coordinated water molecule [187, 202]. Interestingly, removing the dimethylbenzimidazole ligand is an intermediate step in the reductive activation of the cobalt center in methionine synthase [203] and in the electrochemical reduction of Co(II) to the Co(I) state of B12 in solution [201]. Thus, the CFeSP appears to have evolved a mechanism to facilitate reductive activation that can be explained by some well-studied electrochemical principles.
Formation of the first organometallic intermediate (methyl-Co) in the Wood-Ljungdahl pathway precedes methyl group transfer from the methylated CFeSP to a NiFeS cluster in acetyl-CoA synthase (ACS) (Equation 21). This reaction and the associated carbonylation and methyl migration to form an acetyl-ACS intermediate and then thiolysis to form acetyl-CoA are the subject of the next section.
c) CO dehydrogenase/acetyl-CoA synthase (CODH/ACS)
The stepwise formation of a series of organometallic intermediates (methyl-Co, Ni-CO, methyl-Ni, acetyl-Ni) is one of the novel features of the Wood-Ljungdahl pathway. The key enzyme in this pathway, CODH/ACS, has been recently reviewed [204–206]. Figure 1 shows that when acetogens grow on H2 + CO2, one molecule of CO2 is reduced to CO by CODH, which becomes the carbonyl group of acetyl-CoA, and another CO2 is reduced to formate, which serves as the precursor of the methyl group of acetyl-CoA. Under heterotrophic growth conditions, e.g., sugars, CO2 and electrons are generated from the decarboxylation of pyruvate by PFOR (see below). When CO is the growth substrate, one molecule of CO must be converted to CO2, which is then reduced to formate for conversion to the methyl group of acetyl-CoA, while another molecule of CO can be incorporated directly into the carbonyl group.
Both CO and CO2 are unreactive without a catalyst, but the enzyme-catalyzed reactions involving these one-carbon substrates are fast, with turnover numbers as high as 40,000 s−1 at 70 °C reported for CO oxidation by the Ni-CODH from Carboxydothermus hydrogenoformans [207]. Ni-CODHs are classified into two groups: the monofunctional enzyme that functions physiologically in catalyzing CO oxidation (Equation 10), allowing microbes to take up and oxidize CO at the low levels found in the environment, while CODH in the bifunctional protein functions to generate CO by the reverse of Eq. 10 and to couple to ACS to generate acetyl-CoA (Equation 11).
(1) CO dehydrogenase structure and function
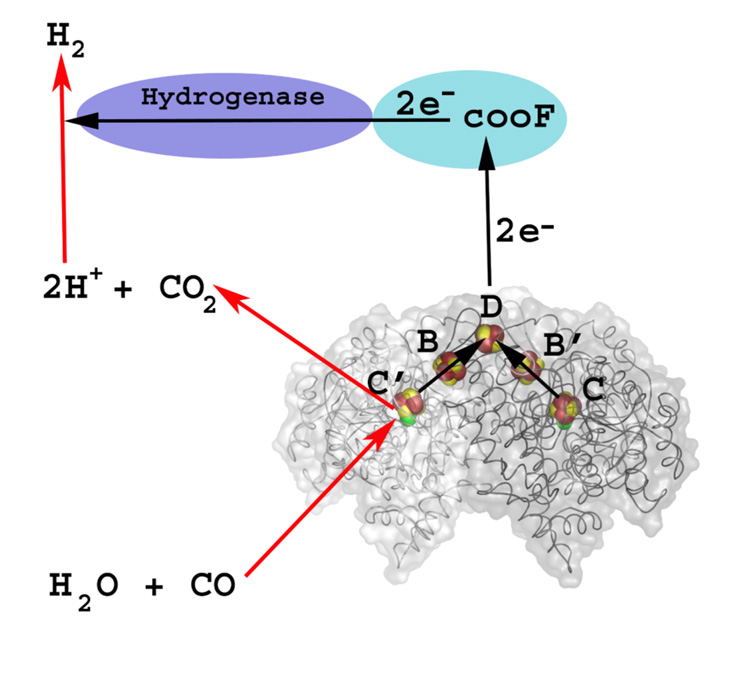
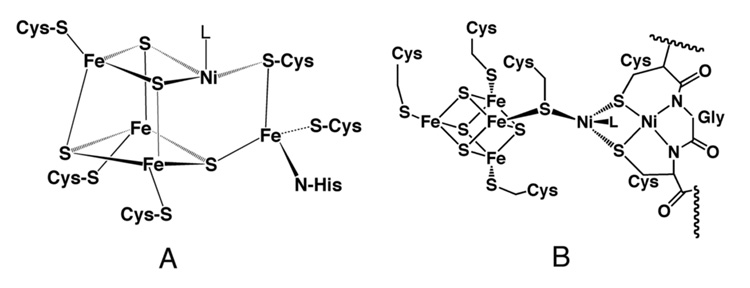
Regardless of whether the Ni-CODH is monofunctional or part of the CODH/ACS bifunctional CODH/ACS complex, the CODH components are homodimeric enzymes that contain five metal clusters, including two C-clusters, 2 B-clusters, and a bridging D-cluster as diagrammed in Figure 10. The B-cluster is a typical [4Fe-4S]2+/1+ cluster, while the D-cluster is a [4Fe-4S]2+/1+ cluster that bridges the two identical subunits, similar to the [4Fe-4S]2+/1+ cluster in the iron protein of nitrogenase. Spectroscopic studies and metal analyses identified four of the five metal clusters present in CODH [91, 92, 208–210], but did not identify the D-cluster and were unable to predict the correct arrangement of metals at the catalytic site for CO oxidation, the C-cluster. The entire complement of metal clusters and their locations were provided by the crystal structures of the monofunctional Ni-CODHs [211, 212]. A nearly superimposable structure including identical metal clusters were found for the CODH component of the bifunctional CODH/ACS [213, 214]. Buried 18 Å below the protein surface, as shown in Figure 11, the C-cluster can be described as a [3Fe-4S] cluster that is bridged to a binuclear NiFe site, in which the iron in the binuclear cluster is bridged to a His ligand. Because of its redox state, this iron has been called Ferrous Component II (FCII). The C. hydrogenoformans C-cluster may have a sulfide bridge between Ni and FCII [215, 216]; however, recent studies may have ruled out the catalytic relevance of this bridge [217, 218]. Furthermore, there is evidence for a catalytically important persulfide at the C-cluster [219].
Figure 10.
From Figure 1 of [87].The two subunits are shown with light and dark wire tracings. Electrons generated by the oxidation of CO at the C and C’ clusters are transferred to the internal redox chain in CODH, consisting of the B (and B’) and D clusters. The D cluster, located at the interface between the two subunits, is proposed to transfer electrons to the electron transfer protein (CooF), which is coupled to hydrogenase.
Figure 11.
A. The C-cluster of CODH and (B) the A-cluster of ACS. From [204].
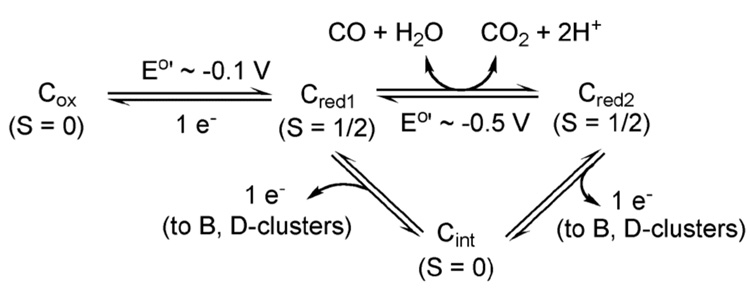
As shown in Figure 12, the C-cluster can exist in four redox states (Cox, Cred1, Cint, and Cred2), which differ by one electron. Cox is an inactive state and can undergo reductive activation to the Cred1 form [220, 221]. Reaction of the active Cred1 form of CODH with CO, generates the Cred2 state [84], which is considered to be two electrons more reduced than Cred1.
Figure 12.
Redox states of the CODH catalytic center, the C-cluster. From Figure 2 of [335].
The CODH reaction mechanism has been reviewed [204]. Following a ping-pong mechanism, CODH is reduced by CO in the “Ping” step and the reduced enzyme transfers electrons to an external redox mediator like ferredoxin (or a specialized ferredoxin-like protein, CooF, in R. rubrum) in the “Pong” step. CooF then couples to a membrane hydrogenase as shown in Fig. 10, or to other energy requiring cellular processes. Recent direct electrochemical studies of CODH linked to a pyrolytic graphite electrode show complete reversibility of CO oxidation and CO2 reduction without any overpotential; in fact, at low pH values, the rate of CO2 reduction exceeds that of CO oxidation [221].
The CODH mechanism described in Figure 13 was derived from NMR studies that will be described below [222] and from crystallographic, kinetic (steady-state and transient), and spectroscopic studies (reviewed in [87]). The reaction steps are analogous to those of the water-gas shift reaction in that the mechanism includes a metal-bound carbonyl, a metal bound hydroxide ion, and a metal-carboxylate, which is formed by attack of the M-OH on M-CO. Elimination of CO2 either leaves a metal-hydride or a two-electron-reduced metal center and a proton. CODHs have a weak CO-dependent hydrogen evolution activity that might suggest a metal-hydride intermediate [223–225]. However, a key difference between the water-gas shift and the enzymatic reaction is that H2 is the product of the nonenzymatic reaction, whereas protons and electrons are the product of the CODH reaction. This indicates that in the enzyme, electron transfer is very rapid relative to H2 evolution, perhaps because of the placement of the B and D clusters as electron acceptors, like a wire, between the C-cluster and the site at which external electron carriers bind. Furthermore, the enzyme uses distinct pathways for delivery/egress of protons and electrons.
Figure 13.
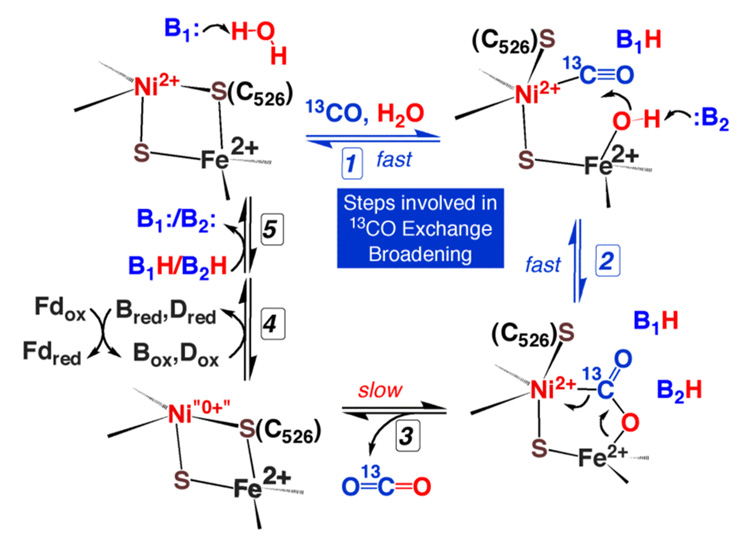
Proposed mechanism of CO oxidation at the C-cluster of CODH, from Figure 4 of [222]. B1 and B2 designate active site bases. Recent information about early stages in CO oxidation was gleaned through NMR studies of a rapid reaction involving the interconversion between CO and bound CO2 [222]. See the text for a detailed explanation of each step.
The sequential steps in the CODH mechanism have been discussed in a recent review [74], so here we will only summarize earlier studies and focus on work published since 2004. Not shown in Figure 13 are two proposed channels that connect the C-cluster to substrates in the solvent: a hydrophobic channel for CO and a hydrophilic water channel [212]. NMR experiments indicate that movement of gas molecules through the hydrophobic channel is very rapid, i.e., ~33,500 s−1 at 20 °C [222], which would extrapolate to 106 s−1, if the rate doubles with every 10 °C rise in temperature. In fact, migration of CO to the C-cluster occurs at diffusion-controlled rates even at 20 °C (3.3 × 109 M-1 s−1 at the published Kd of 10 µM).
As shown in Step 1, CO appears to bind to Ni, based on crystallographic [211, 212], FTIR [226], and X-ray absorption studies of cyanide (a competitive inhibitor) binding to CODH [227]. However, the M-CO has not yet been observed in any crystal structure, and there is also evidence that CN− binds to iron [228]. It is likely that water (or hydroxide) binds to the special iron (ferrous component II, above), based on ENDOR studies [229], which would lower the pKa for water and facilitate formation of an active hydroxide, as described for other enzymes [230]. A recent crystal structure shows a bridging OH− in a sample prepared at mild redox potentials [227], so perhaps the OH− can equilibrate between bridging and Fe-bound states. The nonbridging hydroxide would be expected to be the most active nucleophilic state for attack on the Ni-CO.
Following substrate binding in this proposed bimetallic mechanism, the bound hydroxide would be poised to rapidly attack the Ni-CO to generate a bound carboxyl group, which is shown in Figure 13 to be bridged between Ni and Fe, based on the recent structure of bound CO2 in the C. hydrogenoformans CODH [218]. The intermediacy of a bound CO2 intermediate is consistent with NMR experiments described in the next paragraph and with FTIR studies in which IR bands assigned to M-CO disappear as bands for metal-carboxylates (1724 and 1741 cm−1) and CO2 (2278 cm−1 in the 13CO sample) appear [226].
The first two steps in the CODH reaction were followed by NMR experiments with the monofunctional C. hydrogenoformans enzyme [222]. When CODH is incubated with 13CO, the13CO linewidth markedly increases. This linewidth broadening was concluded to arise from a chemical exchange between solution 13CO and a bound form of 13CO2 (presumably the bridged form shown in Figure 13). Thus, the 13CO exchange broadening mechanism involves steps 1 and 2 of Figure 13, and, although it is in the slow exchange regime of the NMR experiment, occurs with a rate constant (1080 s−1 at 20 °C) that is slightly faster than the rate of CO oxidation at 20 °C. The 13CO exchange broadening is pH independent, unlike the overall CO oxidation reaction, which has a well defined pKa value of 6.7, suggesting the presence of an internal proton reservoir that equilibrates with solvent more slowly than the rate of CO exchange with bound CO2 [222]. A Lys and several His residues were earlier suggested as acid-base catalysts [211, 219], and might account for the internal proton reservoir, depicted as B1 and B2 in Figure 13.
Since a ping-pong mechanism requires that electron acceptors bind only after CO2 is released, presumably the protein is in a two-electron reduced state when CO2 is bound. Then in slightly slower reactions, CO2 is proposed to dissociate (step 3) and protons and electrons are transferred to solvent (steps 4, 5).
(2) The final steps in acetyl-CoA synthesis catalyzed by CODH/ACS
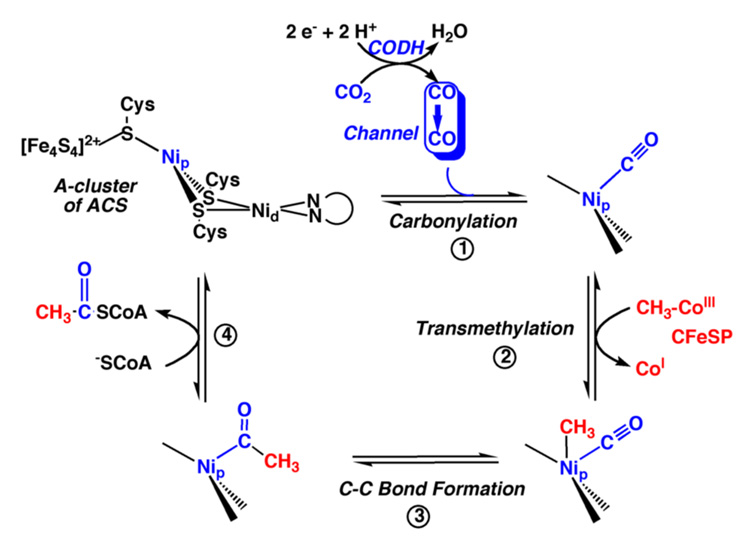
By the principle of microreversibility, CO2 reduction should occur by a direct reverse of the steps shown in Figure 13. This reverse reaction generates CO as a key one-carbon intermediate in the Wood-Ljungdahl pathway [86]. Thus, as shown in Figure 1, the bifunctional CODH/ACS, encoded by the acsA/acsB genes, is a machine that converts CO2, CoA, and the methyl group of the methylated CFeSP to acetyl-CoA, which is a precursor of cellular material (protein, DNA, etc.) and a source of energy.
Acetyl-CoA synthesis is catalyzed by ACS at the A-cluster, which consists of a [4Fe-4S] cluster that is bridged via a cysteine residue (Cys509 in M. thermoacetica) to a Ni site (called the proximal Ni, Nip) that is linked to the distal Ni ion (Nid) in a thiolato- and carboxamido-type N2S2 coordination environment [213, 214, 231], as shown in Figure 11B.
One of the proposed mechanisms of acetyl-CoA synthesis is shown in Figure 14. Two competing mechanisms for acetyl-CoA synthesis have been proposed, which differ mainly in the electronic structure of the intermediates: one proposes a paramagnetic Ni(I)-CO species as a central intermediate [74] and the other (the “diamagnetic mechanism”) proposes a Ni(0) or, as recently proposed, a spin-coupled [4Fe-4S]1+-Nip1+ intermediate [88, 232]. In this review, a generic mechanism is described that emphasizes the organometallic nature of this reaction sequence.
Figure 14.
(A) Proposed mechanism of acetyl-CoA synthesis at the A-cluster of ACS, from Figure 3 of [206]. See the text for details.
Before the first step in the ACS mechanism, CO migrates from its site of synthesis at the C-cluster of CODH to the A-cluster of ACS through a 70 Å channel [213, 233–235]. By determining the structure of CODH crystals incubated with high pressures of Xe, which has a molecular size that approximates that of CO, a series of hydrophobic gas binding pockets have been located within this tunnel [235]. The residues that make up these cavities are conserved among the CODH/ACSs in their hydrophobic character, but not in sequence identity. One of the Xe binding sites is within 4 Å of the binickel center and likely represents a portal near the A-luster for CO, just prior to its ligation to the metal center, shown in Figure 14, reminiscent of the “inland lake”, which has been suggested as the entry point for O2 into the active site of the copper-containing amine oxidase [236, 237].
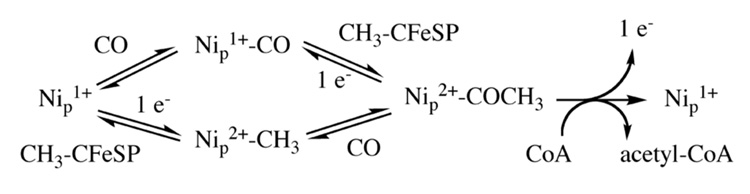
Although Figure 14 simplistically shows an ordered mechanism, recent pulse-chase studies demonstrate that binding of CO and the methyl group in Steps 1 and 2 occurs randomly as shown in Figure 15 [238]. As mentioned above, after CO travels through the intersubunit channel, it binds to the Nip site in the A-cluster to form an organometallic complex, which according to the paramagnetic mechanism, is the so-called “NiFeC species” that has been characterized by a number of spectroscopic and kinetic approaches [74]. The electronic structure of the NiFeC species is described as a [4Fe-4S]2+ cluster linked to a Ni1+ center at the Nip site, while Nid apparently remains redox-inert in the Ni2+ state [89]. Another view is that the NiFeC species is not a true catalytic intermediate in acetyl-CoA synthesis, but is an inhibited state, and that a Ni(0) state is the catalytically relevant one [88, 232], as recently discussed [204]. Gencic and Grahame recently suggested a spin-coupled [4Fe-4S]1+ cluster linked to a Ni1+ site as the active intermediate [239], although no spectroscopic studies were reported in the paper describing this proposal. Evidence for a such a spin-coupled state was recently provided by Mossbauer spectroscopy, and Tan et al. suggest that the methyl group of the methylated CFeSP could be transferred as a methyl cation (see the CFeSP section above) to the spin-coupled Nip 1+-[4Fe-4S]1+ center to form a CH3-Nip2+-[4Fe-4S]2+ state [240]. One possibility is that the paramagnetic state and the spin-coupled state are in equilibrium and that the paramagnetic state is favored with CO as the first substrate, while the spin-coupled state is favored when the methyl group binds first. If so, a redox shuttle is required since the spin-coupled cluster is one-electron more reduced than the NiFeC species.
Figure 15.
Random mechanism of acetyl-CoA synthesis, from Figure 3 of [238]. See the text for details.
Following transfer of the methyl group of the methylated CFeSP apparently to the Nip site of the A-cluster [241–244], Step 3 involves carbon-carbon bond formation by condensation of the methyl and carbonyl groups to form an acetyl-metal species. Experimental evidence for an acetyl-enzyme intermediate was proposed because an exchange reaction between CoA and acetyl-CoA, which involves C-S bond cleavage and resynthesis, was found to occur significantly faster than another exchange reaction between CO and the carbonyl group of acetyl-CoA, which involves cleavage and resynthesis of both C-C and C-S bonds [245–247]. Thus, many cycles of cleavage and re-synthesis of the C-S bond of acetyl-CoA can occur for each cycle of C-C bond synthesis/cleavage. Further evidence for an acetyl-ACS intermediate was recently provided by burst kinetic studies [239].
In the last step, CoA binds to ACS triggering thiolysis of the acetyl-metal bond to form the C-S bond of acetyl-CoA, completing the reactions of the Western branch of the Wood-Ljungdahl pathway. The CoA binding site appears to be within the second domain of ACS, which includes residues 313–478, based on evidence that Trp418 undergoes fluorescence quenching on binding CoA [248]. Furthermore, this domain contains six Arg residues near Trp418, and CoA binding is inhibited by phenylglyoxal, which is a rather specific Arg modification reagent [83].
C. Phosphotransacetylase (E.C. 2.3.1.8) and acetate kinase (E.C.2.7.2.1.)
Phosphotransacetylase catalyzes the conversion of acetyl-CoA to acetyl-phosphate. Although the M. thermoacetica phosphotransacetylase has been purified and characterized enzymatically [249], the gene encoding this protein could not be easily annotated because the protein sequence has not been determined and, when the genome sequence was first determined, no candidates were found that were homologous to known phosphotransacetylase genes. Fortuitously, a novel phosphotransacetylase (PduL) involved in 1,2-propanediol degradation was recently identified in Salmonella enterica [250] that is homologous to two M. thermoacetica genes (Moth_1181 and Moth_0864). Thus, one of these genes is likely to encode the M. thermoacetica phosphotransacetylase that is involved in the Wood-Ljungdahl pathway. Located only ten genes downstream from the gene encoding methylene-H4folate reductase, Moth_1181 is a likely candidate. However, the phosphotransacetylase associated with the Wood-Ljungdahl pathway needs to be unambiguously identified by sequencing the active protein, because phosphotransacetylase is a member of a rather large family of CoA transferases.
Conversion of of acetyl-phosphate to acetate is catalyzed by acetate kinase, which is encoded by the Moth_0940 gene. This gene is distant from, and thus is not linked to Moth_1181, Moth_0864, or the acs gene cluster.
D. The Secret to Metabolic Diversity in Acetogens: Coupling of the Wood-Ljungdahl to Other Pathways
1. Pyruvate ferredoxin oxidoreductase (PFOR) (EC1.2.7.1,Moth_0064)
Pyruvate:ferredoxin oxidoreductase (PFOR) catalyzes the oxidative cleavage of pyruvate and attachment of the two-carbon fragment (carbons 2 and 3) to CoA, forming acetyl-CoA, reducing ferredoxin, and eliminating CO2, (Equation 22) [251–256]. A fairly recent review on PFOR is available [257]. There are five enzymatic activities that can oxidize pyruvate. While PFOR transfers its electrons to a low-potential reductant (ferredoxin or flavodoxin), pyruvate dehydrogenase reduces NAD to NADH (also generating acetyl-CoA), and pyruvate oxidase transfers its electrons to oxygen to make hydrogen peroxide and acetyl-phosphate. Pyruvate decarboxylase, which is a key enzyme in ethanol fermentation, retains its electrons in the substrate to generate acetaldehyde and pyruvate formate lyase transfers the electrons to the carboxyl group to generate formate and acetyl-CoA. Among these five pyruvate metabolizing enzymes, only pyruvate formate lyase does not use thiamine pyrophosphate (TPP).
PFORs have been isolated from several bacteria and the enzymes from M. thermoacetica [69, 86], D. africanus [258–260] have been best characterized. In addition, the enzymes from Pyrococcus [261–263] and from the methanogenic archaea, Methanosarcina barkeri [264] and Methanobacterium thermoautotrophicum [265] have been studied. PFOR plays an important role in the oxidation of acetogens since it catalyzes the first step in the metabolism of pyruvate, supplied as a growth substrate. PFOR is also essential in sugar metabolism, since it couples the Embden-Meyerhof-Parnas pathway to the Wood-Ljungdahl pathway. Furthermore, for anaerobes like methanogens and acetogens that fix CO2 by the Wood-Ljungdahl pathway, PFOR is also a pyruvate synthase that catalyzes the conversion of acetyl-CoA to pyruvate, the first step in the incomplete reductive tricarboxylic acid cycle (TCA, see below) [30, 266].
PFOR is an ancient molecule that apparently predated the divergence of the three kingdoms of life (prokarya, archaea, and eukarya). All archaea appear to contain PFOR and it is widely distributed among bacteria, and even some anaerobic protozoa like Giardia [267]. A heterotetrameric enzyme, like the archaeal PFORs, has been proposed to be a common ancestor [254, 268] that underwent gene rearrangement and fusion to account for the hetero- and homodimeric enzymes. These are represented by COG0674, COG1013, and COG1014, which are found as separate alpha, beta, and gamma subunits in some organisms [98]. The ancestral δ subunit is an 8 Fe ferredoxin-like domain (COG1014) that binds two [4Fe-4S] clusters, while the beta subunit (COG1013) binds the essential cofactor TPP and an iron-sulfur cluster. The PFOR that has been purified from M. thermoacetica is a homodimer (a fusion of all four COGs), with a subunit mass of 120 kDa [269]. Five other sets of genes belonging to the same COGs as PFOR could encode authentic pyruvate:ferredoxin oxidoreductases, but they could also encode other proteins in the oxoglutamate oxidoreductase family, like 2-ketoisovalerate oxidoreductase, indolepyruvate oxidoreductase, and 2-ketoglutarate oxidoreductase.
| (22) |
The PFOR mechanism is summarized in Figure 16. As with all TPP-dependent enzymes, PFOR forms an "active acetaldehyde" intermediate, as first proposed by Breslow in 1957 [270], which involves the formation of a hydroxyethyl adduct between the substrate and the C-2 of the thiazolium of TPP. This is followed by one-electron transfer to one of the three [4Fe-4S] clusters to generate a radical intermediate. The radical intermediate was first identified in the early 1980’s, and was considered to be a pi radical with spin density delocalized over the aromatic thiazolium ring as shown in Figure 16 [252, 271]. However, based on the crystal structure, this intermediate was proposed to be a novel sigma-type acetyl radical [272]. However, spectroscopic studies recently provided unambiguous evidence that negated the sigma radical formulation and provided strong support for the pi radical model [273, 274]. The [4Fe-4S] cluster to which the HE-TPP radical is coupled also was identified by recent spectroscopic studies [273, 274]. Rapid freeze quench EPR and stopped flow studies demonstrated that this radical forms and decays significantly faster than the kcat value of the steady-state reaction (5 s−1 at 10°C for the M. thermoacetica enzyme), demonstrating its catalytic competence as an intermediate in the PFOR reaction mechanism [275, 276]. One of the key remaining questions in the PFOR mechanism is how CoA stimulates decay of the HE-TPP radical intermediate by at least 100,000-fold [276]. Several hypotheses have been proposed [257, 276].
Figure 16.
PFOR Mechanism showing the ylide nucleophile and the HE-TPP anionic and radical intermediates.
Perhaps one reason that PFOR, instead of pyruvate dehydrogenase, is used in acetogens and other anaerobes is that PFOR uses low-potential electron-transfer proteins like ferredoxin and flavodoxin, which is likely to make the pyruvate synthase reaction feasible as the first step for converting acetyl-CoA into cell material. The requisite electron donor for the PDH complex, NADH, is much too weak an electron source to reduce acetyl-CoA to pyruvate (the NAD/NADH half reaction is 200 mV more positive than the acetyl-CoA/pyruvate couple). However, one should consider the possibility that even the ferredoxin-coupled PFOR reaction requires some type of additional driving force for the energetically uphill synthesis of pyruvate, as suggested for some other systems. In methanogens, the pyruvate synthase reaction appears to be linked to H2 oxidation by the membrane-associated Ech hydrogenase and to require reverse electron transfer [277]. The M. barkeri enzyme was shown to catalyze the oxidative decarboxylation of pyruvate to acetyl-CoA and the reductive carboxylation of acetyl-CoA with ferredoxin as an electron carrier [278]. Yoon et al. have also studied the pyruvate synthase reaction of PFOR from Chlorobium tepidum [279], which, like the methanogens, links to the incomplete TCA cycle to convert oxaloacetate (derived from pyruvate by the action of pyruvate carboxylase or the linked activities of phosphoenolpyruvate (PEP) synthetase and PEP carboxylase) into malate, fumarate, succinate, succinyl-CoA, and alpha-ketoglutarate [266].
2. Incomplete TCA cycle
The tricarboxylic acid (TCA) cycle is used oxidatively by aerobic organisms to generate reducing equivalents that eventually couple to the respiratory chain to generate ATP. Many autotrophic anaerobes use the TCA cycle in the reductive direction to incorporate the acetyl group of acetyl-CoA into cell carbon and to generate metabolic intermediates. The reductive TCA cycle is also used for autotrophic growth by some green sulfur bacteria [280] and Epsilonproteobacteria [281]. The reductive TCA cycle has three enzymes that are distinct from the oxidative pathway enzymes: 2-oxoglutarate:ferredoxin oxidoreductase, fumarate reductase, and ATP citrate lyase [280, 282]. In some anaerobes, like acetogens, the reverse TCA cycle is incomplete (Figure 17). The incomplete TCA cycle is used in both the oxidative and reductive directions to generate metabolic intermediates [283, 284], like α-ketoglutarate and oxaloacetate for amino acid synthesis.
Figure 17.
Incomplete TCA cycle allowing conversion of acetyl-CoA to cellular intermediates. Dashed arrows represent enzymes that not identified in the M. thermoacetica genome. From [98].
As shown in Figure 17, pyruvate:ferredoxin oxidoreductase can catalyze the formation of pyruvate from acetyl-CoA and CO2 (see secton IV D1). Generally in the reductive TCA cycle, pyruvate is converted to phosphoenolpyruvate, which then undergoes carboxylation to generate oxaloacetate, which is reduced to fumarate. However, since homologs of phosphoenolpyruvate carboxylase are not found in the M. thermoacetica genome, M. thermoacetica may be able to convert pyruvate to malate, using a malate dehydrogenase homologous to the NADP-dependent malic enzyme of E. coli (encoded by maeB). Although this enzyme generally functions in the malate decarboxylation direction, the pyruvate carboxylation reaction is thermodynamically favorable [25], and expression of the other E. coli malate dehydrogenase (encoded by maeA) has been shown to allow malate formation from pyruvate in a pyruvate-accumulating E. coli strain [285]. The genome of M. thermoacetica appears to lack any homolog of known fumarate reductases; therefore, either M. thermoacetica lacks this enzyme, or it is encoded by a novel gene. On the other hand, the closely related acetogens Clostridium formicoaceticum and Clostridium aceticum can reduce fumarate to succinate [286].
The pathway from succinate to citrate appears to be present in M. thermoacetica. The presence of several sets of genes predicted to encode 2-oxoacid:ferredoxin oxidoreductases [98] and of a likely citrate lyase gene open the possibility that the complete reductive TCA cycle can be used by M. thermoacetica, or that M. thermoacetica has lost this ability during its evolution, with loss of fumarate reductase. M. thermoacetica also encodes a putative citrate synthase, which could allow generation of compounds from citrate to succinate in the oxidative direction.
3. Various cobalamin-dependent methyltransferases allow growth on diverse methyl group donors
Acetogenic bacteria are able to grow on a variety of methyl donors (methanol, aromatic O-methyl ethers, and aromatic O-methyl esters) by coupling different methyltransferase systems to the Wood-Ljungdahl pathway, as shown in Figure 18. The cobalamin dependent methyltranferases have recently been reviewed [80, 168]. Each of the acetogenic methyltransferase systems conforms to a three-module arrangement for catalysis plus a separate module for reductive activation. The three catalytic components comprise a binding site for the methyl donor, a corrinoid binding module, and a module for binding the methyl acceptor. The methyl group is transferred from the donor to the corrinoid, generating an intermediate methyl-Co species, from which the methyl group is transferred to methyl acceptor. The reductive activation module comes into play when the Co center undergoes oxidation. Because the Co2+/1+ couple has such a low redox potential, reductive activation often requires energy input in the form of ATP or adenosylmethionine (as in methionine synthase). In the Wood-Ljungdahl pathway, the methyl donor module is the methyltransferase that binds methyl-H4folate, the corrinoid binding module is the CFeSP, and the A-cluster in ACS is the methyl group acceptor module. The reductive activation module for the CFeSP, as described above, does not require extra energy input (i.e., ATP, etc.) because the one-electron reductant is a low-potential [4Fe-4S cluster] within the large subunit of the CFeSP. It is likely that the modular structural arrangement of methyl donors and acceptors allows for diverse metabolism of methyl groups, with different metabolic modules interfacing at the level of methyl-H4folate.
Figure 18.
Coupling of various methyl donors to the Wood-Ljungdahl pathway. The methyltransferase systems involved in transferring the methyl groups from aromatic methyl ethers (Mtv) or methanol (Mta) to methyl-H4folate have been identified by genomic and enzymatic studies. See the text for details. Ar-O-CH3 designates the aromatic methyl ether that serves as the methyl donor, and Ar-OH signifies the alcohol, which is the demethylation product.
Many of the aromatic compounds that are utilized are products of the degradation of lignin, which is a polymer that constitutes about 25% of the earth’s biomass and is composed of hydroxylated and methoxylated phenylpropanoid units. Acetogenic bacteria have a special propensity for metabolism of methoxylated aromatics and this property has been exploited to selectively isolate acetogens [287]. M. thermoacetica has been shown to utilize at least 20 methoxylated aromatics [288].
Metabolism of the various methyl donors involves three successive two-electron oxidations of one of the methyl groups, generating six electrons that are used by CODH to convert three mol of CO2 to CO, each of which then reacts at the ACS site with a methyl group and CoA to make acetyl-CoA. The methyltransferase systems that catalyze transfer of the methyl groups of methanol and methoxylated aromatic compounds to tetrahydrofolate have been identified by genomic and biochemical studies. A three-component system in M. thermoacetica that initiates the metabolism of methoxylated aromatic compounds is encoded by the mtvABC gene cluster (identified in the genomic sequence as Moth_0385-7 and Moth_1316-8) [98]. This system has been purified and identified biochemically by enzymatic function and protein sequence analysis [289]. A similar system has been identified in Acetobacterium dehalogenans, which also is linked to an ATP-dependent activating protein that can accept electrons from a hydrogenase [290, 291]. The Mtv system is coupled to the Wood-Ljungdahl pathway as shown in Figure 18. M. thermoacetica also encodes several homologs of MtvA, MtvB and MtvC that may used for growth on methyl donors other than vanillate and methanol [98].
Many acetogens can use methanol as a methyl donor by expressing the methanol methyltransferase system (MtaABC), encoded by Moth_1208-9 and Moth_2346 in the M. thermoacetica genome [98]). The M. thermoacetica methanol:methyl-H4folate system was recently characterized and the crystal structure of the MtaC component was determined [292].
4. Heterotrophic Growth
In environments where both acetogens and methanogens are found, methanogens are usually the dominant hydrogenotrophs (see section IB). Acetogenic bacteria can grow in these environments because they have the ability to use a great variety of carbon sources and electron donors and acceptors. A long-recognized characteristic of acetogens is their ability to convert sugars stoichiometrically to acetate [38, 293]. A typical acetogen can use most of the substrates shown in Figure 19. M. thermoacetica, for example, can grow heterotrophically on fructose, glucose, xylose, ethanol, n-propanol, n-butanol [294], oxalate and glyoxylate [295], glycolate [296], pyruvate [293], and lactate [297], and acetate yields from growth on these substrates are consistent with oxidation to CO2, and formation of acetate from all available electrons.
Figure 19.
Redox couples that can be used by acetogens. Figure taken from [336]. CO2 reduction to acetate is one of many possible electron-accepting processes.
Genes encoding all enzymes of the Embden-Meyerhof-Parnas pathway pathway and the oxidative branch of the pentose phosphate pathway [98] are found in the M. thermoacetica genome. Thus, one mole of xylose is converted to 2.5 moles of acetate. It seems likely that fructose is metabolized by the same pathway as glucose, since growth yields on the two substrates are very similar [298]. Ethanol, propanol, and butanol are oxidized to acetate, propionate, and butyrate, respectively, [294] and genes encoding alcohol dehydrogenases and aldehyde:ferredoxin oxidoreductases have been located on the M. thermoacetica genome. When M. thermoacetica is grown on lactate, Moth_1826 appears to be the lactate dehydrogenase that catalyzes the conversion of lactate to pyruvate. Pyruvate is then metabolized using pyruvate:ferredoxin oxidoreductase (PFOR) and the Wood-Ljungdahl pathway enzymes, as described above.
The two-carbon substrates oxalate, glyoxylate and glycolate can be oxidized by two, four and six electrons, respectively. Thus, during growth on these substrates and CO2, for each mole of acetate produced, approximately two moles of glyoxylate or four moles of oxalate are oxidized, and four moles of glycolate are oxidized to make three moles of acetate [295]. It is not clear from the genome sequence what pathway is used by M. thermoacetica for growth on the two-carbon substrates oxalate, glyoxylate, and glycolate, because only partial pathways known to be involved in glycolate metabolism in other organisms are present in M. thermoacetica [98]. Thus, post-genomic studies will be required to elucidate the pathways for growth on these compounds.
The Wood-Ljungdahl pathway serves two functions in acetogenic bacteria: as an electron-accepting, energy-conserving pathway, and as a pathway for carbon assimilation. Yet, as shown in Figure 19, reduction of CO2 to acetate by the Wood-Ljungdahl pathway is only one of many electron-accepting pathways that can be used by acetogens. Respiration with higher redox potential acceptors than CO2, such as nitrate, could provide more energy to the cell, thus it is not surprising that nitrate is preferred over CO2 as electron acceptor for M. thermoacetica [299]. Accordingly, in undefined medium (with yeast extract to provide precursors for cell carbon), the growth of M. thermoacetica on a variety of electron donors (H2, CO, formate, vanillate, ethanol, and n-propanol) produces more biomass with nitrate as electron acceptor than with CO2 [300].
The presence of nitrate blocks carbon assimilation by the Wood-Ljungdahl pathway, by transcriptional regulation of genes encoding Wood-Ljungdahl enzymes, at least under some culture conditions, and by decreasing levels of cytochrome b in membranes [300, 301]. A similar down-regulation of acetogenesis has been seen in nitrite-supplemented cultures. Because of this repression, some substrates (e.g., methanol, vanillate, formate, CO, and H2 + CO2) cannot be used by M. thermoacetica for growth in nitrate media that lacks yeast extract (provides precursors for cell carbon). Glyoxylate is the only electron donor that has been shown to support growth with nitrite. In these cultures, ammonia was the reduced end-product, and CO2 reduction to acetate only occurred in stationary phase, and cytochrome b was absent during the exponential growth phase [302].
Oxalate is a unique substrate in that M. thermoacetica preferentially reduces CO2 over nitrate in the presence of oxalate, and produces acetate during exponential growth on oxalate, CO2 and nitrate during exponential growth, [303]. Some substrates, including sugars, pyruvate, oxalate and glyoxylate support growth in basal medium with nitrate. M. thermoacetica can also grow in basal medium with nitrate if vanillate, which provides pre-formed methyl groups, and CO are supplied together.
To our knowledge, the ability of M. thermoacetica to make acetate in the presence of electron acceptors besides nitrate and nitrite has not been tested. In contrast to the tight regulation of acetogenesis by these electron acceptors in M. thermoacetica, other organisms have been shown to use other electron acceptors simultaneously with CO2. With either fructose or H2 as the electron source, A. woodii can reduce caffeate simultaneously with reduction of CO2 to acetate, even though the reduction of caffeate is more favorable (ΔEo’ = +32 mV for caffeate reduction vs. SHE, by analogy to the fumarate/succinate couple). However, methanol or betaine, when provided as the electron donors, inhibit caffeate reduction during acetogenesis [304]. Peptostreptococcus productus and Clostridium formicoaceticum both reduce fumarate to succinate simultaneously with CO2 reduction to acetate [305, 306], although in P. productus, both activities are stimulated by increasing the levels of CO2 in the medium [305]. Exogenous CO2 levels can also affect utilization of other substrates; for example, C. formicoaceticum does not convert fructose to acetate in the absence of added CO2 [307]. P. productus directs electron flow away from the Wood-Ljungdahl pathway when the CO2 concentration is low, and although some electron donors were used more efficiently at high CO2 concentrations, increasing the CO2 concentration during growth on xylose decreased the cell yield, indicating that acetogenesis may not be the most energetically favorable metabolic pathway for this organism [305]. Clearly, acetogens have well-developed strategies for adjusting their growth to environments that compromise their ability to use the Wood-Ljungdahl pathway.
E. Reverse acetogenesis: Acetate catabolism by methanogens and sulfate reducers
As mentioned in the introduction, several classes of anaerobes other than acetogens, including methanogens and anammox bacteria, use the Wood-Ljungdahl pathway of CO2 reduction to allow them to grow autotrophically by generating cell carbon from H2 + CO2. Furthermore, reverse acetogenesis enables the use of acetate as a carbon and electron source; however as indicated by Equation 1, acetate oxidation to H2 + CO2 is thermodynamically unfavorable. Therefore, the reaction has to be coupled to a thermodynamically favorable process, like methanogenesis or sulfate reduction.
The pathway of acetate catabolism by methanogens is shown in Figure 20 (color-coded as in Figure 1) (for review see [31]). Acetoclastic methanogens convert acetate in the growth medium to acetyl-CoA by the actions of acetate kinase and phosphotransacetylase. Then, the C-C and C-S bonds of acetyl-CoA are cleaved to release CoA and CO, which is converted to CO2 by CODH, leaving a methyl-ACS intermediate on the A-cluster. Continuing the reverse of the acetogenesis pathway, the methyl group is transferred to the corrinoid iron-sulfur protein component of the methanogenic CODH/ACS (or acetyl-CoA decarbonylase synthase) complex. Two consecutive methyltransferase reactions then are involved in transfer of the methyl group from the methyl-[Co] intermediate to CoM to generate methyl-SCoM, which, in the presence of Coenzyme B, is converted to methane and the heterodisulfide product (CoB-SS-CoM) by the nickel metalloenzyme, methyl-SCoM reductase. In the overall process, the methyl and carboxyl groups of acetate are converted to methane and CO2, respectively, while the two electrons released in the oxidation of CO to CO2 are used by heterodisulfide reductase to reduce the heterodisulfide product of the MCR reaction back to the free thiolate state.
Figure 20.
Acetoclastic methanogenesis: coupling methanogenesis to the Wood-Ljungdahl pathway (reverse acetogenesis). MCR, methyl-SCoM reductase; HDR, heterodisulfide reductase
An interesting example of metabolic coupling of acetogenic and methanogenic metabolism is observed in co-cultures of acetogens (growing on sugars) and methanogens. The reducing equivalents produced during glycolysis and pyruvate oxidation are transferred to the methanogen, perhaps as H2, to reduce CO2 to methane [308]. Thus, per mol of glucose, two mol of acetate and one mol each of CO2 and CH4 are produced. Furthermore, when acetogens are co-cultured with acetate utilizing methanogenic strains, the three mol of acetate produced from glycolysis are converted to three mol each of CO2 and CH4 [309]. This metabolic interdependence was described for a co-culture of the mesophiles, A. woodii and Methanosarcina barkeri, and for a thermophilic co-culture [310, 311]. The physiology, biochemistry, and bioenergetics related to the syntrophic growth of acetogens with other organisms has been recently reviewed [312]. General principles include the requirement for a syntrophic association when free energy for the metabolic process is very low; for example, coupling anaerobic methane oxidation to sulfate reduction (with a free energy associated with this process of only -18 kJ/mol). On the other hand, various sulfate reducers are capable of coupling acetate oxidation to H2S production as described in Figure 21 [35–37], a process for which the free energy is much more significant (Equation 4).
Figure 21.
Coupling reverse acetogenesis to sulfate reduction.
VII. Energy metabolism associated with acetogenesis
It has been long recognized that autotrophic growth by the Wood-Ljungdahl pathway must be linked to an energy-generating anaerobic respiratory process, since during autotrophic growth, there is no net ATP synthesis by substrate-level phosphorylation. However, the chemiosmotic pathway(s) that is connected to the Wood-Ljungdahl pathway has not been identified. Evidence for chemiosmotic ATP synthesis has been found in studies with A. woodii, M. thermoacetica, and Moorella thermoautotrophica (formerly, Clostridium thermoautotrophicum [313]), which is physiologically similar to M. thermoacetica [314].
A decade before it was shown to grow autotrophically, growth yields of M. thermoacetica on glucose and fructose were calculated to be higher than one would expect for ATP production by substrate level phosphorylation alone, and it was proposed that additional ATP is generated by an anaerobic respiratory process [315]. The high growth yields measured when A. woodii is cultured on fructose or caffeate also supported a chemiosmotic energy conservation mechanism for this acetogen [316]. Electron transport-linked ATP synthesis in M. thermoacetica received further support when b-type cytochromes and a menaquinone were identified in its membranes during growth on CO2 and glucose [317].
In A. woodii and M. thermoacetica, the F1F0 ATP synthases responsible for ATP synthesis have been isolated. The F1F0 ATPase from M. thermoacetica was shown to be similar in subunit composition, and pattern of inhibition and stimulation by metal cations, anions, and alcohols to ATPases from mitochondria, chloroplasts, and other bacteria that use them for energy generation [318]. Evidence for ATP formation dependent on a proton gradient in M. thermoacetica includes increased ATP formation following an extracellular drop in pH, which was inhibited by ATPase inhibitor dicyclohexylcarbodiimide (DCCD) [318].
The Wood-Ljungdahl pathway enzyme(s) that are linked to transmembrane proton gradient formation is not known. The enzymes needed for acetate synthesis have been found to be soluble after preparation of cell extracts with a French pressure cell, but after gentler osmotic lysis of M. thermoautotrophica cells, much of the CODH and methylenetetrahydrofolate reductase activities remained in the membrane fractions [319]. Incubation of CODH- and methylenetetrahydrofolate reductase-containing membranes with CO or with dithionite caused eduction of the b-type cytochromes present in the membranes (the redox potentials of these cytochromes are −200 mV and −48 mV). When CO-reduced membranes were treated with CO2, the lower potential cytochrome was partially re-oxidized [319]. Further studies with M. thermoautotrophica membranes showed that electrons from CO can reduce cytochrome b559, followed by menaquinone, followed by cytochrome b554. Based on these studies, and on the redox potential for the methylenetetrahydrofolate/methyltetrahydrofolate couple (−120 mV), Ljungdahl’s group proposed an electron transport chain (Figure 22) involving the following sequential steps: oxidation of CO (CO+H2O ⇌CO22e−+2H+, ΔEo’ = −540 mV) coupled to formation of H2 or NADH by a flavoprotein, reduction of cytochrome b559, which could then reduce methylenetetrahydrofolate or menaquinone and cytochrome b554, which would finally reduce rubredoxin (ΔEo’ ≈ 0 mV vs. SHE [320]) [321]. Driven by the oxidation of CO with ferricyanide, a proton motive force that could drive ATP synthesis and amino acid transport was measured in M. thermoacetica membrane vesicles [322]. However, which step would extrude protons in the proposed electron transport chain is still not known.
Figure 22.
Proposed electron transport chain from work done in Moorella species (modified from [98]). The specific proton translocating step and which specific electron carriers are coupled to the Wood-Ljungdahl pathway are not known. The cytochromes may be parts of larger protein complexes (i.e. cytochrome bd oxidase and formate dehydrogenase).
A cytochrome bd oxidase has recently been identified in M. thermoacetica [323]. Although M. thermoacetica is a strict anaerobe, it can tolerate traces of oxygen [324]. Furthermore, M. thermoacetica membranes catalyzed NADH-dependent O2 uptake that is apparently coupled to a membrane-associated electron transport chain. Duroquinol- and quinol-dependent O2 uptake activities increased in membrane preparations that were enriched for cytochrome bd oxidase, and it was proposed that the cytochrome bd oxidase is involved in protection from oxidative stress. Based on the genome sequence, the cytochrome b in this complex could be identified as cytb559, which had been identified by Ljungdahl’s group many years earlier [323]. Another cytochrome (b554) is part of a gene cluster annotated as formate dehydrogenase, although the substrate specificity of this protein is not clear from the sequence. Both of these annotations support the possible role of these cytochromes in electron transport coupled to energy conservation.
While some acetogens like M. thermoacetica seem to use a cytochrome-based proton-coupled electron transfer pathway, others, like A. woodii lack membrane-bound cytochromes, but, contain membrane-bound corrinoids that have been suggested, in analogy to the corrinoid-containing, Na+ -pumping methyltetrahydromethanopterin: coenzyme M methyltransferases of methanogenic archaea, to be involved in energy conservation ([325] and reviewed in [162]). A. woodii growth on fructose, methanol, or H2 + CO2, is dependent on the sodium concentration in the growth medium, and growth on fructose at low sodium levels decreased acetate production from 2.7 to 2.1 moles of acetate per mole of fructose, which is consistent with inhibition of synthesis of the third molecule of acetate from fructose by the Wood-Ljungdahl pathway (as described for acetogenesis from glucose in section II B1) [161]. Experiments with resting A. woodii cells showed that acetate production from H2 + CO2 is also dependent on sodium ions and that these cells can produce a strong Na+ gradient in which concentrations inside cells are as much as forty-fold lower than outside, corresponding to a transmembrane chemical potential of −90 mV [161]. Furthermore, Na+-dependent ATP formation by A. woodii cells is inhibited by the F1F0 ATPase inhibitor DCCD [326, 327]. Na+ is taken up into inverted membrane vesicles, coupled with acetogenesis [328]. The Na+ dependent ATPase was purified and shown to be an unusual F1F0 ATPase [329], with two types of rotor subunits, e.g., two bacterial F(0)-like c subunits and an 18 kDa eukaryal V(0)-like c subunit [330, 331]. The Na+ dependent step was shown to be in the methyl branch of the Wood-Ljungdahl pathway, and it has suggested that the responsible enzyme is either methylenetetrahydrofolate reductase or the methyltransferase/corrinoid protein involved in methyl group transfer from tetrahydrofolate to ACS.
A possible respiratory pathway for growth on caffeate as an electron acceptor has been recently identified in A. woodii [332] (reviewed in [333]). Membrane-bound ferredoxin:NAD+ oxidoreductase activity was found, and is postulated to be the activity of a protein complex homologous to the Rnf complex of Rhodobacter capsulatus. Genetic evidence for such a complex in A. woodii has been shown [332]. The possible Rnf complex has also been hypothesized as the Na+-translocating protein for growth by the Wood-Ljungdahl pathway. Reduced ferredoxin, from hydrogenase or PFOR would be used by Rnf to reduce NAD+, which could provide electrons to the methyl branch of the pathway.
In summary, both H+- and Na+-dependent acetogens produce electrochemical gradients that can be used for ATP synthesis by F1F0 ATPases. In A. woodii, ATP synthesis from a transmembrane Na+ gradient is linked to acetogenesis by some step in the methyl branch of the Wood-Ljungdahl pathway, possibly at methylenetetrahydrofolate reductase, or via an Rnf ferredoxin:NAD+ oxidoreductase. In M. thermoacetica and M. thermoautotrophica, methylenetetrahydrofolate reductase and CO dehydrogenase could be linked to energy conserving steps in the Wood-Ljungdahl pathway. The possible electron transport chain in Moorella is summarized in Figure 22, which was modified from [322].
The genome studies of M. thermoacetica give some insight into possible energy conserving pathways [98]. Homologs of NADH dehydrogenase I and archaeal heterodisulfide reductase are present in the genome. NADH dehydrogenase in E. coli, as in other bacteria, has 13 subunits. As discussed in the genome paper [98], the genes encoding NADH dehydrogenase are indicated in Figure 23 to be encoded by three gene clusters: Moth_0977-87, which encodes ten of the thirteen NADH dehydrogenase subunits; and Moth_1717-9 and Moth_1886-6, which appear to be redundant and encode the other three subunits (NuoE, F, and G in E. coli). Moth_0979, which is in the middle of the first gene cluster, has no homolog in E. coli. Other genes identified that may play important roles in electron transfer linked proton translocation are Moth_2184-90, with significant homology to the genes encoding the subunits of E. coli’s hydrogenase 4, which has been proposed to catalyze proton translocation [334]. These various sets of genes could encode partial or complete membrane-bound complexes that would function in anaerobic respiration by generating a transmembrane proton gradient for use by the F1F0ATP synthase. Localization of genes encoding one homolog of heterodisulfide reductase, a hydrogenase, and 5,10-methylene-H4folate reductase near the acs gene cluster are consistent with a suggestion that the reductase and a hydrogenase might link to CODH to generate a proton motive force, driving anaerobic respiration [160], as suggested in Figure 23. This figure is depicted as a speculative and highly incomplete scheme to underline our lack of knowledge in this area. Hopefully, the M. thermoacetica genome sequence [98], which contains a number of potential electron transfer components with as yet unknown functions, will aid in the identification of the various electron acceptors in the pathways shown in this scheme. Furthermore, there is evidence that in M. thermoautotrophica, CO dehydrogenase and methyl-H4folate reductase, which were isolated from the soluble fraction of the cell extract (above in the Wood-Ljungdahl pathway enzyme section), can associate with membranes [319]. The presence of multiple gene clusters encoding possible membrane-bound respiratory complexes could be explained by the ability of M. thermoacetica to use many electron acceptors as different respiratory proteins could be induced by different growth substrates.
Figure 23.
Protein complexes possibly involved in proton transfer in M. thermoacetica.
VIII. Prospective: Questions that remain unanswered
Many questions that remain about the biochemistry and bioenergetics of acetogenesis and of the Wood-Ljungdahl pathway have become more tractable with the sequencing of the first acetogenic genome. Some of these questions are reiterated here, with the details provided above.
A number of questions remain about the enzymology of the Wood-Ljungdahl pathway. Although it has been clearly shown that general acid catalysis is involved in the methyl transfer reactions of the MeTr involved in the Wood-Ljungdahl pathway and in methionine synthase, it needs to be firmly established whether there is a generally applicable mechanism of proton transfer (i.e., in the binary or ternary complex, or in the transition state for the methyl transfer reaction), or if perhaps these enzymes use different mechanisms. Biochemical, biophysical, and structural studies of the various proposed conformational states of the MeTr:CFeSP and CFeSP:CODH/ACS complexes. Although the structure of the CFeSP clearly defined the two TIM barrel domains including the corrinoid binding site, it is important to determine the protein structure around the [4Fe-4S] cluster, which was not resolved.
Although there is ample spectroscopic evidence for Ni-CO intermediates on CODH and ACS, neither of these has been observed by X-ray crystallography. Furthermore, no crystal structures have been reported for the methylated or the CoA-bound states of ACS. It is important to resolve the apparently conflicting results supporting the binding of the CODH competitive inhibitor CN− to Ni or Fe. Evidence for a spin-coupled state on ACS was recently provided by Mossbauer spectroscopy and there are proposals for a Ni(0) state. Although the Ni(0) seems rather unlikely, it is important to further characterize and to establish whether or not there is any physiological relevance for the spin-coupled state.
The role of one CODH, which is part of the acs gene cluster as part of the CODH/ACS machine is well understood; however, enzymatic and gene expression studies are required to elucidate the function of the other CODH (Moth_1972), which is unlinked to this gene cluster.
A number of questions remain about the coupling of acetyl-CoA formation by the Wood-Ljungdahl pathway to the reactions and pathways that are coupled to it. The phosphotransacetylase associated with the Wood-Ljungdahl pathway needs to be unambiguously identified by sequencing the active protein. It is not clear how binding of CoA accelerates electron transfer among the internal iron-sulfur clusters by ~100,000-fold. With respect to the pyruvate synthase reaction, it is not clear how acetogens accomplish the energetically unfavorable carboxylation of acetyl-CoA; for example, if reverse electron transfer is involved in driving this reaction and, if so, what is the mechanism by which this is accomplished. In addition, whether M. thermoacetica has a complete or incomplete reductive TCA cycle is unclear.
It is unknown what sequence of reactions M. thermoacetica uses for growth on the twocarbon substrates oxalate, glyoxylate, and glycolate, because only partial pathways known to be involved in glycolate metabolism in other organisms are present in M. thermoacetica; thus, biochemical and perhaps gene expression studies will be required to characterize these pathways.
Perhaps the most important unsolved question related to acetogenesis and the Wood-Ljungdahl pathway is how acetogens conserve energy for growth. A proposed mechanism of energy conservation and proton translocation involving the 5,10-methylene-H4folate reductase and the membrane-associated hydrogenase and/or heterodisulfide reductase. It is likely that the acetogens that contain membrane-bound cytochromes (like M. thermoacetica) will have some components of the energy generation system in common with those that lack cytochromes (like A. woodii) and some that are distinct. For the non-cytochrome containing acetogens, there are important questions about the role of the Rnf complex described above [332]. Is it an ion (Na+) pump? Is it a coupling site in acetogens, and, if so, how is it coupled to the Wood-Ljungdahl pathway? What is the role of the membrane bound corrinoids in the acetogens that lack cytochromes, and does the corrinoid-dependent methyltransferase catalyze Na+ translocation?
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barker HA, Kamen MD. Carbon dioxide utilization in the synthesis of acetic acid by Clostridium thermoaceticum. Proc. Natl. Acad. Sci. USA. 1945;31:219–225. doi: 10.1073/pnas.31.8.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drake HL, Daniel SL, Matthies C, Küsel K. Acetogenesis, acetogenic bacteria, and the acetyl-CoA pathway: Past and current perspectives. In: Drake HL, editor. Acetogenesis. New York: Chapman and Hall; 1994. pp. 3–60. [Google Scholar]
- 3.Müller V, Imkamp F, Rauwolf Andreas, Küsel K, Drake HL. Molecular and Cellular Biology of Acetogenic Bacteria. In: Nakano MM, Zuber P, editors. Strict and Facultative Anaerobes: Medical and Environmental Aspects. Wymondham, UK: Horizon Bioscience; 2004. pp. 251–281. [Google Scholar]
- 4.Fischer F, Lieske R, Winzer K. Biologische gasreaktionen. II. Gber die bildung von essigs ure bei der biologischen umsetzung von kohlenoxyd und kohlens ure mit wasserstoff zu methan. Biochem. Z. 1932;245:2–12. [Google Scholar]
- 5.Wieringa KT. Over het verdwinjhnen van waterstof en koolzuur onder anaerobe voorwaarden. Antonie van Leeuwenhoek. 1936;3:263–273. [Google Scholar]
- 6.Wieringa KT. The formation of acetic acid from carbon dioxide and hydrogen by anaerobic spore-forming bacteria. Antonie van Leeuwenhoek. 1940;6:251–262. [Google Scholar]
- 7.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 8.Fontaine FE, Peterson WH, McCoy E, Johnson MJ, Ritter GJ. A new type of glucose fermentation by Clostridium thermoaceticum. J. Bacteriol. 1942;43:701–715. doi: 10.1128/jb.43.6.701-715.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breznak JA. Acetogenesis from carbon dioxide in termite guts. In: Drake HL, editor. Acetogenesis. New York: Chapman and Hall; 1994. pp. 303–330. [Google Scholar]
- 10.Mackie RI, Bryant MP. Acetogenesis and the rumen: syntrophic relationships. In: Drake HL, editor. Acetogenesis. New York: Chapman and Hall; 1994. pp. 331–364. [Google Scholar]
- 11.Rosencrantz D, Rainey FA, Janssen PH. Culturable Populations of Sporomusa spp. and Desulfovibrio spp. in the Anoxic Bulk Soil of Flooded Rice Microcosms. Appl. Environ. Microbiol. 1999;65:3526–3533. doi: 10.1128/aem.65.8.3526-3533.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ollivier B, Caumette P, Garcia J-L, Mah RA. Anaerobic Bacteria from Hypersaline Environments. Microbiol. Rev. 1994;58:27–38. doi: 10.1128/mr.58.1.27-38.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters V, Conrad R. Methanogenic and Other Strictly Anaerobic Bacteria in Desert Soil and Other Oxic Soils. Appl. Environ. Microbiol. 1995;61:1673–1676. doi: 10.1128/aem.61.4.1673-1676.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kusel K, Wagner C, Drake HL. Enumeration and metabolic product profiles of the anaerobic microflora in the mineral soil and litter of a beech forest. FEMS Microbiology Ecology. 1999;29:91–103. [Google Scholar]
- 15.Liu S, Suflita JM. H2-CO2-Dependent Anaerobic O-Demethylation Activity in Subsurface Sediments and by an Isolated Bacterium. Appl. Environ. Microbiol. 1993;59:1325–1331. doi: 10.1128/aem.59.5.1325-1331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adrian NR, Arnett CM. Anaerobic Biodegradation of Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by Acetobacterium malicum Strain HAAP-1 Isolated from a Methanogenic Mixed Culture. Current Microbiology. 2004;48:332–340. doi: 10.1007/s00284-003-4156-8. [DOI] [PubMed] [Google Scholar]
- 17.Macbeth TW, Cummings DE, Spring S, Petzke LM, Sorenson KS., Jr Molecular Characterization of a Dechlorinating Community Resulting from In Situ Biostimulation in a Trichloroethene-Contaminated Deep, Fractured Basalt Aquifer and Comparison to a Derivative Laboratory Culture. Appl. Environ. Microbiol. 2004;70:7329–7341. doi: 10.1128/AEM.70.12.7329-7341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greening RC, Leedle JAZ. Enrichment and isolation of Acetitomaculum ruminis, gen. nov., sp. nov.: acetogenic bacteria from the bovine rumen. Arch. Microbiol. 1989;151:399–406. doi: 10.1007/BF00416597. [DOI] [PubMed] [Google Scholar]
- 19.Tholen A, Brune A. Localization and In Situ Activities of Homoacetogenic Bacteria in the Highly Compartmentalized Hindgut of Soil-Feeding Higher Termites (Cubitermes spp.) Appl. Environ. Microbiol. 1999;65:4497–4505. doi: 10.1128/aem.65.10.4497-4505.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chassard C, Bernalier-Donadille A. H2 and acetate transfers during xylan fermentation between a butyrate-producing xylanolytic species and hydrogenotrophic microorganisms from the human gut. FEMS Microbiol. Lett. 2006;254:116–122. doi: 10.1111/j.1574-6968.2005.00016.x. [DOI] [PubMed] [Google Scholar]
- 21.Pester M, Brune A. Hydrogen is the central free intermediate during lignocellulose degradation by termite gut symbionts. ISME J. 2007;1:551–565. doi: 10.1038/ismej.2007.62. [DOI] [PubMed] [Google Scholar]
- 22.Odelson DA, Breznak JA. Volatile fatty acid production by the hindgut microbiota of xylophagous termites. Appl. Environ. Microbiol. 1983;45:1602–1613. doi: 10.1128/aem.45.5.1602-1613.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Van TD, Robinson JA, Ralph J, Greening RC, Smolenski WJ, Leedle JAZ, Schaefer DM. Assessment of Reductive Acetogenesis with Indigenous Ruminal Bacterium Populations and Acetitomaculum ruminis. Appl. Environ. Microbiol. 1998;64:3429–3436. doi: 10.1128/aem.64.9.3429-3436.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 1997;61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ragsdale SW. The Eastern and Western branches of the Wood/Ljungdahl pathway: how the East and West were won. BioFactors. 1997;9:1–9. doi: 10.1002/biof.5520060102. [DOI] [PubMed] [Google Scholar]
- 27.Ljungdahl LG. The acetyl-CoA pathway and the chemiosmotic generation of ATP during acetogenesis. In: Drake HL, editor. Acetogenesis. New York: Chapman and Hall; 1994. pp. 63–87. [Google Scholar]
- 28.Drake HL, Kusel K, Matthies C. Ecological consequences of the phylogenetic and physiological diversities of acetogens. Antonie Van Leeuwenhoek. 2002;81:203–213. doi: 10.1023/a:1020514617738. [DOI] [PubMed] [Google Scholar]
- 29.Stupperich E, Hammel KE, Fuchs G, Thauer RK. Carbon monoxide fixation into the carboxyl group of acetyl coenzyme A during autotrophic growth of Methanobacterium. FEBS Lett. 1983;152:21–23. doi: 10.1016/0014-5793(83)80473-6. [DOI] [PubMed] [Google Scholar]
- 30.Lapado J, Whitman WB. Method for isolation of auxotrophs in the methanogenic archaebacteria: role of the acetyl-CoA pathway of autotrophic CO2 fixation in Methanococcus maripauludis. Proc. Natl. Acad. Sci. USA. 1990;87:5598–5602. doi: 10.1073/pnas.87.15.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferry JG. Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol. Ecol. 1999;23:13–38. doi: 10.1111/j.1574-6976.1999.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 32.Ingram-Smith C, Gorrell A, Lawrence SH, Iyer P, Smith K, Ferry JG. Characterization of the acetate binding pocket in the Methanosarcina thermophila acetate kinase. J. Bacteriol. 2005;187:2386–2394. doi: 10.1128/JB.187.7.2386-2394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorrell A, Lawrence SH, Ferry JG. Structural and kinetic analyses of arginine residues in the active site of the acetate kinase from Methanosarcina thermophila. J. Biol. Chem. 2005;280:10731–10742. doi: 10.1074/jbc.M412118200. [DOI] [PubMed] [Google Scholar]
- 34.Iyer PP, Lawrence SH, Luther KB, Rajashankar KR, Yennawar HP, Ferry JG, Schindelin H. Crystal structure of phosphotransacetylase from the methanogenic archaeon Methanosarcina thermophila. Structure. 2004;12:559–567. doi: 10.1016/j.str.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Spormann AM, Thauer RK. Anaerobic acetate oxidation to CO2 by Desulfotomaculum acetoxidans - Demonstration of enzymes required for the operation of an oxidative acetyl-CoA/carbon monoxide dehydrogenase pathway. Arch. Microbiol. 1988;150:374–380. [Google Scholar]
- 36.Schauder R, Preuβ A, Jetten MS, Fuchs G. Oxidative and reductive acetyl CoA/carbon monoxide dehydrogenase pathway in Desulfobacterium autotrophicum. Arch. Microbiol. 1988;151:84–89. [Google Scholar]
- 37.Ferry JG. Biochemistry of methanogenesis. Crit. Rev. Biochem. Mol. Biol. 1992;27:473–502. doi: 10.3109/10409239209082570. [DOI] [PubMed] [Google Scholar]
- 38.Fontaine FE, Peterson WH, McCoy E, Johnson MJ, Ritter GJ. A new type of glucose fermentation by Clostridium thermoaceticum n. sp. Journal of Bacteriology. 1942;43:701–715. doi: 10.1128/jb.43.6.701-715.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braun M, Mayer F, Gottschalk G. Clostridium aceticum (Wieringa), a microorganism producing acetic acid from molecular hydrogen and carbon dioxide. Arch. Microbiol. 1981;128:288–293. doi: 10.1007/BF00422532. [DOI] [PubMed] [Google Scholar]
- 40.Adamse AD. New isolation of Clostridium aceticum (Wieringa) Antonie van Leeuwenhoek. 1980;46:523–531. doi: 10.1007/BF00394009. [DOI] [PubMed] [Google Scholar]
- 41.Kerby R, Zeikus JG. Growth of Clostridium thermoaceticum on H2/CO2 or CO as energy source. Current Microbiology. 1983;8:27–30. [Google Scholar]
- 42.Bassham JA, Benson AA, Calvin M. The path of carbon in photosynthesis. J. Biol. Chem. 1950;185:781–787. [PubMed] [Google Scholar]
- 43.Walker DA. From Chlorella to chloroplasts: a personal note. Photosynth Res. 2007;92:181–185. doi: 10.1007/s11120-007-9139-3. [DOI] [PubMed] [Google Scholar]
- 44.Wood HG. A study of carbon dioxide fixation by mass determination on the types of C13-acetate. J. Biol. Chem. 1952;194:905–931. [PubMed] [Google Scholar]
- 45.Wood HG. Fermentation of 3,4-C14- and 1-C14-labeled glucose by Clostridium thermoaceticum. J. Biol. Chem. 1952;199:579–583. [PubMed] [Google Scholar]
- 46.Utter MF, Wood HG. Mechanisms of fixation of carbon dioxide by heterotrophs and autotrophs. Adv. Enzymol. Relat. Areas Mol. Biol. 1951;12:41–151. doi: 10.1002/9780470122570.ch2. [DOI] [PubMed] [Google Scholar]
- 47.Balch WE, Schoberth S, Tanner RS, Wolfe RS. Acetobacterium, a new genus of hydrogen-oxidizing, carbon-dioxide-reducing, anaerobic bacteria. International Journal of Systematic Bacteriology. 1977;27:355–361. [Google Scholar]
- 48.Balch WE, Wolfe RS. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere. Appl. Environ. Microbiol. 1976;32:781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drake HL, Gossner AS, Daniel SL. Old Acetogens, New Light. Ann. N. Y. Acad. Sci. 2008;1125:100–128. doi: 10.1196/annals.1419.016. [DOI] [PubMed] [Google Scholar]
- 50.Ljungdahl L, Wood HG. Incorporation of C14 from carbon dioxide into sugar phosphates, carboxylic acids, and amino acids by Clostridium thermoaceticum. J. Bacteriol. 1965;89:1055–1064. doi: 10.1128/jb.89.4.1055-1064.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whipple GH, Robscheit-Robbins FS. Blood regeneration in severe anemia. II. Favorable influence of liver, heart and skeletal muscle in diet. Am. J. Physiol. 1925;72:408–418. [Google Scholar]
- 52.Minot GR, Murphy WP. Treatment of pernicious anaemia by a special diet. J. Am Med. Assn. 1926;87:470–476. [PMC free article] [PubMed] [Google Scholar]
- 53.Smith EL. Purification of Anti-pernicious Anæmia Factors from Liver. Nature. 1948;161:638–639. doi: 10.1038/161638a0. [DOI] [PubMed] [Google Scholar]
- 54.Rickes EL, Brink NG, Koniuszy FR, Wood TR, Folkers K. Crystalline Vitamin B12. Science. 1948;107:396–397. doi: 10.1126/science.107.2781.396. [DOI] [PubMed] [Google Scholar]
- 55.Hodgkin DC, Kamper J, Mackay M, Pickworth J, Trueblood KN, White JG. Structure of vitamin B12. Nature. 1956;178:64–66. doi: 10.1038/178064a0. [DOI] [PubMed] [Google Scholar]
- 56.Poston JM, Kuratomi K, Stadtman ER. Methyl-vitamin B12 as a source of methyl groups for the synthesis of acetate by cell-free extracts of Clostridium thermoaceticum. Ann. N.Y. Acad. Sci. 1964;112:804–806. doi: 10.1111/j.1749-6632.1964.tb45057.x. [DOI] [PubMed] [Google Scholar]
- 57.Ljungdahl LG, Irion E, Wood HG. Total synthesis of acetate from CO2. I. Co-methylcobyric acid and Co-(methyl)-5-methoxybenzimidazolylcobamide as intermediates with Clostridium thermoaceticum. Biochem. 1965;4:2771–2780. doi: 10.1021/bi00888a030. [DOI] [PubMed] [Google Scholar]
- 58.Ljungdahl L, Irion E, Wood HG. Role of corrinoids in the total synthesis of acetate from CO2 by Clostridium thermoaceticum. Fed. Proc. 1966;25:1642–1648. [PubMed] [Google Scholar]
- 59.Larrabee AR, Rosenthal S, Cathou RE, Buchanan JM. Enzymatic synthesis of the methyl group of methionine. Isolation, characterization, and role of 5-methyl tetrahydrofolate. J. Biol. Chem. 1963;238:1025. [Google Scholar]
- 60.Gibson F, Woods DD. The synthesis of methionine by suspensions of Escherichia coli. Biochem. J. 1960;74:160. doi: 10.1042/bj0740160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeyama S, Hatch FT, Buchanan JM. Enzmatic synthesis of the methyl group of methionine. Involvement of vitamin B12. J. Biol. Chem. 1961;236:1102. [PubMed] [Google Scholar]
- 62.Ljungdahl L, Wood HG. Total synthesis of acetate from CO2 by heterotrophic bacteria. Ann. Rev. Microbiol. 1969;23:515. doi: 10.1146/annurev.mi.23.100169.002503. [DOI] [PubMed] [Google Scholar]
- 63.Rabinowitz JC. Folic acid. In: Boyer, Lardy, Myrback, editors. The Enzymes. 2nd Ed. Vol. 2. 1960. p. 185. [Google Scholar]
- 64.Parker DJ, Wu T-F, Wood HG. Total synthesis of acetate from CO2: methyltetrahydrofolate, an intermediate, and a procedure for separation of the folates. J. Bacteriol. 1971;108:770–776. doi: 10.1128/jb.108.2.770-776.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghambeer RK, Wood HG, Schulman M, Ljungdahl LG. Total synthesis of acetate from CO2. III. Inhibition by alkylhalides of the synthesis from CO2, methyltetrahydrofolate, and methyl-B12 by Clostridium thermoaceticum. Arch. Biochem. Biophys. 1971;143:471. doi: 10.1016/0003-9861(71)90232-3. [DOI] [PubMed] [Google Scholar]
- 66.Ljungdahl LG, Wood HG. Acetate biosynthesis. In: Dolphin D, editor. Vitamin B12, vol. Vol. 2. New York: Wiley; 1982. pp. 166–202. [Google Scholar]
- 67.Hu S-I, Pezacka E, Wood HG. Acetate synthesis from carbon monoxide by Clostridium thermoaceticum. Purification of the corrinoid protein. J. Biol. Chem. 1984;259:8892–8897. [PubMed] [Google Scholar]
- 68.Ragsdale SW, Lindahl PA, Münck E, Mössbauer EPR, and optical studies of the corrinoid/Fe-S protein involved in the synthesis of acetyl-CoA by Clostridium thermoaceticum. J. Biol. Chem. 1987;262:14289–14297. [PubMed] [Google Scholar]
- 69.Drake HL, Hu S-I, Wood HG. Purification of five components from Clostridium thermoacticum which catalyze synthesis of acetate from pyruvate and methyltetrahydrofolate. Properties of phosphotransacetylase. J. Biol. Chem. 1981;256:11137–11144. [PubMed] [Google Scholar]
- 70.Diekert GB, Thauer RK. Carbon monoxide oxidation by Clostridium thermoaceticum and Clostridium formicoaceticum. J. Bacteriol. 1978;136:597–606. doi: 10.1128/jb.136.2.597-606.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Diekert GB, Graf EG, Thauer RK. Nickel requirement for carbon monoxide dehydrogenase formation in Clostridium thermoaceticum. Arch. Microbiol. 1979;122:117–120. [Google Scholar]
- 72.Drake HL, Hu S-I, Wood HG. Purification of carbon monoxide dehydrogenase, a nickel enzyme from Clostridium thermoaceticum. J. Biol. Chem. 1980;255:7174–7180. [PubMed] [Google Scholar]
- 73.Ragsdale SW, Clark JE, Ljungdahl LG, Lundie LL, Drake HL. Properties of purified carbon monoxide dehydrogenase from Clostridium thermoaceticum a nickel, iron-sulfur protein. J. Biol. Chem. 1983;258:2364–2369. [PubMed] [Google Scholar]
- 74.Ragsdale SW. Life with carbon monoxide. CRC Crit Rev Biochem and Mol Biol. 2004;39:165–195. doi: 10.1080/10409230490496577. [DOI] [PubMed] [Google Scholar]
- 75.Hu S-I, Drake HL, Wood HG. Synthesis of acetyl coenzyme A from carbon monoxide, methyltetrahydrofolate, and coenzyme A by enzymes from Clostridium thermoaceticum. J. Bacteriol. 1982;149:440–448. doi: 10.1128/jb.149.2.440-448.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clark JE, Ragsdale SW, Ljungdahl LG, Wiegel J. Levels of enzymes involved in the synthesis of acetate from CO2 in Clostridium thermoaceticum. J. Bacteriol. 1982;151:507–509. doi: 10.1128/jb.151.1.507-509.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diekert G, Ritter M. Purification of the nickel protein carbon monoxide dehydrogenase of Clostridium thermoaceticum. FEBS Lett. 1983;151:41–44. doi: 10.1016/0014-5793(83)80338-x. [DOI] [PubMed] [Google Scholar]
- 78.Ragsdale SW, Ljungdahl LG, DerVartanian DV. Isolation of the carbon monoxide dehydrogenase from Acetobacterium woodii and comparison of its properties with those of the Clostridium thermoaceticum enzyme. J. Bacteriol. 1983;155:1224–1237. doi: 10.1128/jb.155.3.1224-1237.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ingraham LL. B12 coenzymes: biological Grignard reagents. Ann. N.Y. Acad. Sci. 1964;112:713. doi: 10.1111/j.1749-6632.1964.tb45049.x. [DOI] [PubMed] [Google Scholar]
- 80.Banerjee R, Ragsdale SW. The Many Faces of Vitamin B12: Catalysis by Cobalamin-dependent Enzymes. Ann Rev. Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 81.Parker DJ, Wood HG, Ghambeer RK, Ljungdahl LG. Total synthesis of acetate from carbon dioxide. Retention of deuterium during carboxylation of trideuteriomethyltetrahydrofolate or trideuteriomethylcobalamin. Biochem. 1972;11:3074. doi: 10.1021/bi00766a021. [DOI] [PubMed] [Google Scholar]
- 82.Kräutler B. Acetyl-cobalamin from photoinduced carbonylation of methyl-cobalamin. Helv. Chim. Acta. 1984;67:1053. [Google Scholar]
- 83.Ragsdale SW, Wood HG. Acetate biosynthesis by acetogenic bacteria: evidence that carbon monoxide dehydrogenase is the condensing enzyme that catalyzes the final steps of the synthesis. J. Biol. Chem. 1985;260:3970–3977. [PubMed] [Google Scholar]
- 84.Kumar M, Lu W-P, Liu L, Ragsdale SW. Kinetic evidence that CO dehydrogenase catalyzes the oxidation of CO and the synthesis of acetyl-CoA at separate metal centers. J. Am. Chem. Soc. 1993;115:11646–11647. [Google Scholar]
- 85.Ragsdale SW, Kumar M. Ni containing carbon monoxide dehydrogenase/acetyl-CoA synthase. Chem. Rev. 1996;96:2515–2539. doi: 10.1021/cr950058+. [DOI] [PubMed] [Google Scholar]
- 86.Menon S, Ragsdale SW. Evidence that carbon monoxide is an obligatory intermediate in anaerobic acetyl-CoA synthesis. Biochem. 1996;35:12119–12125. doi: 10.1021/bi961014d. [DOI] [PubMed] [Google Scholar]
- 87.Drennan CL, Doukov TI, Ragsdale SW. The Metalloclusters of Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase: A Story in Pictures. J. Biol. Inorg. Chem. 2004;9:511–515. doi: 10.1007/s00775-004-0563-y. [DOI] [PubMed] [Google Scholar]
- 88.Lindahl PA. Acetyl-Coenzyme A Synthase: The Case for a Nip0-Based Mechanism of Catalysis. J. Biol. Inorg. Chem. 2004;9:516–524. doi: 10.1007/s00775-004-0564-x. [DOI] [PubMed] [Google Scholar]
- 89.Brunold TC. Spectroscopic and Computational Insights into the Geometric and Electronic Properties of the A Cluster of Acetyl-Coenzyme A Synthase. J. Biol. Inorg. Chem. 2004;9:533–541. doi: 10.1007/s00775-004-0566-8. [DOI] [PubMed] [Google Scholar]
- 90.Charles G, Riordan Acetyl coenzyme A synthase: new insights into one of Natures bioorganometallic catalysts. Journal of Biological Inorganic Chemistry. 2004;9:509–510. [Google Scholar]
- 91.Ragsdale SW, Ljungdahl LG, DerVartanian DV. EPR evidence for nickel substrate interaction in carbon monoxide dehydrogenase from Clostridium thermoaceticum. Biochem. Biophys. Res. Commun. 1982;108:658–663. doi: 10.1016/0006-291x(82)90880-4. [DOI] [PubMed] [Google Scholar]
- 92.Ragsdale SW, Ljungdahl LG, DerVartanian DV. 13C and 61Ni isotope substitution confirm the presence of a nickel(III)-carbon species in acetogenic CO dehydrogenases. Biochem. Biophys. Res. Commun. 1983;115:658–665. doi: 10.1016/s0006-291x(83)80195-8. [DOI] [PubMed] [Google Scholar]
- 93.Ragsdale SW, Wood HG, Antholine WE. Evidence that an iron-nickel-carbon complex is formed by reaction of CO with the CO dehydrogenase from Clostridium thermoaceticum. Proc. Natl. Acad. Sci. USA. 1985;82:6811–6814. doi: 10.1073/pnas.82.20.6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Riordan C. Synthetic Chemistry and Chemical Precedents for Understanding the Structure and Function of Acetyl Coenzyme A Synthase. J. Biol. Inorg. Chem. 2004;9:542–549. doi: 10.1007/s00775-004-0567-7. [DOI] [PubMed] [Google Scholar]
- 95.Schidlowski M, Hayes JM, Kaplan IR. Earth's Earliest Biosphere: Its Origin and Evolution. In: Schopf JW, editor. Princeton, NJ: Princeton Univ. Press; 1983. pp. 149–186. [Google Scholar]
- 96.Brock TD. Evolutionary relationships of the autotrophic bacteria. In: Schlegel HG, Bowien B, editors. Autotrophic Bacteria. Madison, WI: Science Tech Publishers; 1989. pp. 499–512. [Google Scholar]
- 97.Martin W, Russell MJ. On the origin of biochemistry at an alkaline hydrothermal vent. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007;362:1887–1925. doi: 10.1098/rstb.2006.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pierce E, Xie G, Barabote RD, Saunders E, Han CS, Detter JC, Richardson P, Brettin TS, Das A, Ljungdahl LG, Ragsdale SW. The Complete Genome Sequence of Moorella thermoacetica. Environ. Microbiol. 2008 doi: 10.1111/j.1462-2920.2008.01679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bao Q, Tian Y, Li W, Xu Z, Xuan Z, Hu S, Dong W, Yang J, Chen Y, Xue Y, Xu Y, Lai X, Huang L, Dong X, Ma Y, Ling L, Tan H, Chen R, Wang J, Yu J, Yang H. A Complete Sequence of the Thermoanaerobacter tengcongensis Genome. Genome Res. 2002;12:689–700. doi: 10.1101/gr.219302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wiegel J, Ljungdahl LG. Thermoanaerobacter ethanolicus gen. nov., spec. nov., a new, extreme thermophilic, anaerobic bacterium. Archives of Microbiology. 1981;128:343–348. [Google Scholar]
- 101.Lundie LL, Jr, Drake HL. Development of a minimally defined medium for the acetogen Clostridium thermoaceticum. Journal of Bacteriology. 1984;159:700–703. doi: 10.1128/jb.159.2.700-703.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ljungdahl LG, Irion E, Wood HG. Role of corrinoids in the total synthesis of acetate from CO2 by Clostridium thermoaceticum. Federal Proceedings. 1966;25:1642. [PubMed] [Google Scholar]
- 103.Stupperich E, Eisinger H-J, Schurr S. Corrinoids in anaerobic bacteria. FEMS Microbiology Letters. 1990;87:355–360. [Google Scholar]
- 104.Ragsdale SW. The Eastern and Western branches of the Wood/Ljungdahl pathway: how the East and West were won. BioFactors. 1997;6:3–11. doi: 10.1002/biof.5520060102. [DOI] [PubMed] [Google Scholar]
- 105.Ljungdahl LG. The autotrophic pathway of acetate synthesis in acetogenic bacteria. Ann. Rev. Microbiol. 1986;40:415–450. doi: 10.1146/annurev.mi.40.100186.002215. [DOI] [PubMed] [Google Scholar]
- 106.Drake HL, Daniel SL. Acetogenic bacteria: what are the in situ consequences of their diverse metabolic versitilities? BioFactors. 1997;6:13–24. doi: 10.1002/biof.5520060103. [DOI] [PubMed] [Google Scholar]
- 107.Li L-F, Ljungdahl L, Wood HG. Properties of nicotinamide adenine dinucleotide phosphate-dependent formate dehydrogenase from Clostridium thermoaceticum. Journal of Bacteriology. 1966;92:405–412. doi: 10.1128/jb.92.2.405-412.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Andreesen JR, Ljungdahl LG. Formate dehydrogenase of Clostridium thermoaceticum: incorporation of selenium-75, and the effects of selenite, molybdate, and tungstate on the enzyme. J. Bacteriol. 1973;116:867–873. doi: 10.1128/jb.116.2.867-873.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yamamoto I, Saiki T, Liu S-M, Ljungdahl LG. Purification and Properties of NADP-dependent formate dehydrogenase from Clostridium thermoaceticum, a tungsten-selenium-iron protein. J. Biol. Chem. 1983;258:1826–1832. [PubMed] [Google Scholar]
- 110.Ruschig U, Muller U, Willnow P, Hopner T. CO2 reduction to formate by NADH catalysed by formate dehydrogenase from Pseudomonas oxalaticus. Eur. J. Biochem. 1976;70:325–330. doi: 10.1111/j.1432-1033.1976.tb11021.x. [DOI] [PubMed] [Google Scholar]
- 111.Thauer RK. CO2 reduction to formate in Clostridium acidi-urici. J. Bacteriol. 1973;114:443. doi: 10.1128/jb.114.1.443-444.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thauer RK, Kaufer B, Fuchs G. The active species of 'CO2' utilized by reduced ferredoxin CO2 oxidoreductase from Clostridium pasteurianum. Eur. J. Biochem. 1975;55:111–117. doi: 10.1111/j.1432-1033.1975.tb02143.x. [DOI] [PubMed] [Google Scholar]
- 113.Ljungdahl LG, Andreesen JR. Tungsten, a component of active formate dehydrogenase from Clostridium thermoacetium. FEBS Lett. 1975;54:279–282. doi: 10.1016/0014-5793(75)80092-5. [DOI] [PubMed] [Google Scholar]
- 114.Latimer WM. The Oxidation States of the Elements and Their Potentials in Aqueous Solution. 2 ed. New York: 1961. [Google Scholar]
- 115.Fuchs G. CO2 fixation in acetogenic bacteria: variations on a theme. FEMS Microbiol. Rev. 1986;39:181–213. [Google Scholar]
- 116.Kisker C, Schindelin H, Rees D. Molybdenum-cofactor-containing enzymes: structure and mechanism. Annu. Rev. Biochem. 1997;66:233–267. doi: 10.1146/annurev.biochem.66.1.233. [DOI] [PubMed] [Google Scholar]
- 117.Schwarz G, Mendel RR. Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu. Rev. Plant Biol. 2006;57:623–647. doi: 10.1146/annurev.arplant.57.032905.105437. [DOI] [PubMed] [Google Scholar]
- 118.Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC, Sun PD. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science. 1997;275:1305–1308. doi: 10.1126/science.275.5304.1305. [DOI] [PubMed] [Google Scholar]
- 119.Raaijmakers HC, Romao MJ. Formate-reduced E. coli formate dehydrogenase H: The reinterpretation of the crystal structure suggests a new reaction mechanism. J Biol Inorg Chem. 2006;11:849–854. doi: 10.1007/s00775-006-0129-2. [DOI] [PubMed] [Google Scholar]
- 120.MacKenzie RE. Biogenesis and interconversion of substituted tetrahydrofolates. In: Blakley RL, Benkovic SJ, editors. Folates and pterins. New York, N.Y: John Wiley & Sons, Inc.; 1984. pp. 256–306. [Google Scholar]
- 121.Blakeley RL, Benkovic SJ. Folates and Pterins. New York: John Wiley and Sons, Inc.; 1984. [Google Scholar]
- 122.Himes RH, Harmony JA. Formyltetrahydrofolate synthetase. CRC Crit Rev Biochem. 1973;1:501–535. doi: 10.3109/10409237309105441. [DOI] [PubMed] [Google Scholar]
- 123.Lovell CR, Przybyla A, Ljungdahl LG. Cloning and expression in Escherichia coli of the Clostridium thermoaceticum gene encoding thermostable formyltetrahydrofolate synthetase. Arch. Microbiol. 1988;149:280–285. doi: 10.1007/BF00411642. [DOI] [PubMed] [Google Scholar]
- 124.Lovell CR, Przybyla A, Ljungdahl LG. Primary structure of the thermostable formyltetrahydrofolate synthetase from Clostridium thermoaceticum. Biochem. 1990;29:5687–5694. doi: 10.1021/bi00476a007. [DOI] [PubMed] [Google Scholar]
- 125.McGuire JJ, Rabinowitz JC. Studies on the mechanism of formyltetrahydrofolate synthetase. The Peptococcus aerogenes enzyme. J. Biol. Chem. 1978;253:1079. [PubMed] [Google Scholar]
- 126.O'Brien WE, Brewer JM, Ljungdahl LG. Chemical, physical and enzymatic comparisons of formyltetrahydrofolate synthetases from thermo- and mesophilic clostridia. In: Zuber H, editor. Proceedings of the International Symposium on Enzymes and Proteins from Thermophilic Microorganisms, Structure and Functions. Basel, Switzerland: Birkhauser-Verlag; 1975. p. 249. [Google Scholar]
- 127.Himes RH, Wilder T. Formyltetrahydrofolate synthetase. Effect of pH and temperature on the reaction. Arch. Biochem. Biophys. 1968;124:230. doi: 10.1016/0003-9861(68)90323-8. [DOI] [PubMed] [Google Scholar]
- 128.Radfar R, Shin R, Sheldrick GM, Minor W, Lovell CR, Odom JD, Dunlap RB, Lebioda L. The crystal structure of N(10)-formyltetrahydrofolate synthetase from Moorella thermoacetica. Biochemistry. 2000;39:3920–3926. doi: 10.1021/bi992790z. [DOI] [PubMed] [Google Scholar]
- 129.Shannon KW, Rabinowitz JC. Isolation and characterization of the Saccharomyces cerevisiae MIS1 gene encoding mitochondrial C1-tetrahydrofolate synthase. J. Biol. Chem. 1988;263:7717. [PubMed] [Google Scholar]
- 130.Staben C, Rabinowitz JC. Nucleotide sequence of the Saccharomyces cerevisiae ADE3 gene encoding C1-tetrahydrofolate synthase. J. Biol. Chem. 1986;261:2986. [PubMed] [Google Scholar]
- 131.Lovell CR, Przybyla A, Ljungdahl LG. Primary structure of the thermostable formyltetrahydrofolate synthetase from Clostridium thermoaceticum. Biochemistry. 1990;29:5687–5694. doi: 10.1021/bi00476a007. [DOI] [PubMed] [Google Scholar]
- 132.Majillano MR, Jahansouz H, Matsunaga TO, Kenyon GL, Himes RH. Formation and utilization of formyl phosphate by N10-formyltetrahydrofolate synthetase: Evidence for formyl phosphate as an intermediate in the reaction. Biochem. 1989;28:5136–5145. doi: 10.1021/bi00438a034. [DOI] [PubMed] [Google Scholar]
- 133.Leaphart AB, Trent Spencer H, Lovell CR. Site-directed mutagenesis of a potential catalytic and formyl phosphate binding site and substrate inhibition of N10-formyltetrahydrofolate synthetase. Arch Biochem Biophys. 2002;408:137–143. doi: 10.1016/s0003-9861(02)00552-0. [DOI] [PubMed] [Google Scholar]
- 134.Moore MR, O'Brien WE, Ljungdahl LG. Purification and characterization of nicotinamide adenine dinucleotide- dependent methylenetetrahydrofolate dehydrogenase from Clostridium formicoaceticum. J. Biol. Chem. 1974;249:5250–5253. [PubMed] [Google Scholar]
- 135.Ragsdale SW, Ljungdahl LG. Purification and properties of NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Acetobacterium woodii. J. Biol. Chem. 1984;259:3499–3503. [PubMed] [Google Scholar]
- 136.Clark JE, Ljungdahl LG. Purification and properties of 5,10-methenyltetrahydrofolate cyclohydrolase. J. Biol. Chem. 1982;257:3833–3836. [PubMed] [Google Scholar]
- 137.Poe M, Benkovic SJ. 5-formyl- and 10-formyl-5,6,7,8-tetrahydrofolate. Conformation of the tetrahydropyrazine ring and formyl group in solution. Biochem. 1980;19:4576–4582. doi: 10.1021/bi00561a006. [DOI] [PubMed] [Google Scholar]
- 138.Moncrief MBC, Hausinger RP. Purification and activation properties of UreD-UreF-urease apoprotein. J. Bacteriol. 1996;178:5417–5421. doi: 10.1128/jb.178.18.5417-5421.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pawelek PD, Allaire M, Cygler M, MacKenzie RE. Channeling efficiency in the bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase domain: the effects of site-directed mutagenesis of NADP binding residues. Biochim Biophys Acta. 2000;1479:59–68. doi: 10.1016/s0167-4838(00)00058-3. [DOI] [PubMed] [Google Scholar]
- 140.Pelletier JN, MacKenzie RE. Binding and interconversion of tetrahydrofolates at a single site in the bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase. Biochem. 1995;34:12673–12680. doi: 10.1021/bi00039a025. [DOI] [PubMed] [Google Scholar]
- 141.Rios-Orlandi EM, MacKenzie RE. The activities of the NAD-dependent methylenetetrahydrofolate dehydrogenase-methylenetetrahydrofolate cyclohydrolase from ascites tumor cells are kinetically independent. J. Biol. Chem. 1988;263:4662. [PubMed] [Google Scholar]
- 142.Monzingo AF, Breksa A, Ernst S, Appling DR, Robertus JD. The X-ray structure of the NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Saccharomyces cerevisiae. Protein Sci. 2000;9:1374–1381. doi: 10.1110/ps.9.7.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shen BW, Dyer DH, Huang JY, D'Ari L, Rabinowitz J, Stoddard BL. The crystal structure of a bacterial, bifunctional 5,10 methylene-tetrahydrofolate dehydrogenase/cyclohydrolase. Protein Sci. 1999;8:1342–1349. doi: 10.1110/ps.8.6.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schmidt A, Wu H, MacKenzie RE, Chen VJ, Bewly JR, Ray JE, Toth JE, Cygler M. Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase. Biochemistry. 2000;39:6325–6335. doi: 10.1021/bi992734y. [DOI] [PubMed] [Google Scholar]
- 145.O'Brien WE, Brewer JM, Ljungdahl LG. Purification and characterization of thermostable 5,10- methylenetetrahydrofolate dehydrogenase from Clostridium thermoaceticum. J. Biol. Chem. 1973;248:403–408. [PubMed] [Google Scholar]
- 146.Clark JE, Ljungdahl LG. Purification and properties of 5,10-methylenetetrahydrofolate reductase, an iron-sulfur flavoprotein from Clostridium formicoaceticum. J. Biol. Chem. 1984;259:10845–10889. [PubMed] [Google Scholar]
- 147.Park EY, Clark JE, DerVartanian DV, Ljungdahl LG. 5,10-methylenetetrahydrofolate reductases: iron-sulfur-zinc flavoproteins of two acetogenic clostridia. In: Miller F, editor. Chemistry and Biochemistry of Flavoenzymes. vol. 1. Boca Raton, Fl: CRC Press; 1991. pp. 389–400. [Google Scholar]
- 148.Wohlfarth G, Geerligs G, Diekert G. Purification and properties of a NADH-dependent 5,10- methylenetetrahydrofolate reductase from Peptostreptococcus productus. Eur. J. Biochem. 1990;192:411–417. doi: 10.1111/j.1432-1033.1990.tb19242.x. [DOI] [PubMed] [Google Scholar]
- 149.Sheppard CA, Trimmer EE, Matthews RG. Purification and properties of NADH-dependent 5, 10- methylenetetrahydrofolate reductase (MetF) from escherichia coli [In Process Citation] J. Bacteriol. 1999;181:718–725. doi: 10.1128/jb.181.3.718-725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Matthews RG, Sheppard C, Goulding C. Methylenetetrahydrofolate reductase and methionine synthase: biochemistry and molecular biology. Eur J Pediatr. 1998;157 Suppl 2:S54–S59. doi: 10.1007/pl00014305. [DOI] [PubMed] [Google Scholar]
- 151.Yamada K, Chen Z, Rozen R, Matthews RG. Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. Proc. Natl. Acad. Sci. U S A. 2001;98:14853–14858. doi: 10.1073/pnas.261469998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Katzen HM, Buchanan JM. Enzymatic synthesis of the methyl group of methionine. VIII. Repression-derepression, purification, and properties of 5,10-methylenetetrahydrofolate reductase from Escherichia coli. J. Biol. Chem. 1965;240:825. [PubMed] [Google Scholar]
- 153.Vanoni MA, Lee S, Floss HG, Matthews RG. Stereochemistry of reduction of methylenetetrahydrofolate to methyltetrahydrofolate catalyzed by pig liver methylenetetrahydrofolate reductase. J. Am. Chem. Soc. 1990;112:3987. [Google Scholar]
- 154.Guenther BD, Sheppard CA, Tran P, Rozen R, Matthews RG, Ludwig ML. The structure and properties of methylenetetrahydrofolate reductase from Escherichia coli: a model for the role of folate in ameliorating hyperhomocysteinemia in humans. Nat. Struct. Biol. 1999;6:359–365. doi: 10.1038/7594. [DOI] [PubMed] [Google Scholar]
- 155.Vanoni MA, Daubner SC, Ballou DP, Matthews RG. Methylenetetrahydrofolate reductase. Steady state and rapid reaction studies on the NADPH-methylenetetrahydrofolate, NADPH-menadione, and methytethydrofolate-menadione oxidoreductase activities of the enzyme. J. Biol. Chem. 1983;258:11510. [PubMed] [Google Scholar]
- 156.Matthews RG. Studies on the methylene/methyl interconversion catalyzed by methylenetetrahydrofolate reductase from pig liver. Biochem. 1982;21:4165. doi: 10.1021/bi00260a038. [DOI] [PubMed] [Google Scholar]
- 157.Matthews RG, Drummond JT. Providing one-carbon units for biological methylations: mechanistic studies on serine hydroxymethyltransferase, methylenetetrahydrofolate reductase, and methyltetrahydrofolate-homocysteine methyltransferase. Chem. Rev. 1990;90:1275. [Google Scholar]
- 158.Kallen RG, Jencks WP. The mechanism of the condensation of formaldehyde with tetrahydrofolic acid. J. Biol. Chem. 1966;241:5851–5863. [PubMed] [Google Scholar]
- 159.Matthews RG, Haywood BJ. Inhibition of pig liver methylenetetrahydrofolate reductase by dihydrofolate: some mechanistic and regulatory implications. Biochem. 1979;18:4845. doi: 10.1021/bi00589a012. [DOI] [PubMed] [Google Scholar]
- 160.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriological Reviews. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Heise R, Muller V, Gottschalk G. Sodium dependence of acetate formation by the acetogenic bacterium Acetobacterium woodii. J Bacteriol. 1989;171:5473–5478. doi: 10.1128/jb.171.10.5473-5478.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Muller V. Energy conservation in acetogenic bacteria. Appl. Environ. Microbiol. 2003;69:6345–6353. doi: 10.1128/AEM.69.11.6345-6353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Roberts DL, James-Hagstrom JE, Garvin DK, Gorst CM, Runquist JA, Baur JR, Haase FC, Ragsdale SW. Cloning and expression of the gene cluster encoding key proteins involved in acetyl-CoA synthesis in Clostridium thermoaceticum: CO dehydrogenase, the corrinoid/FeS protein, and methyltransferase. Proceedings of the National Academy of Sciences. 1989;86:32–36. doi: 10.1073/pnas.86.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jeon WB, Cheng J, Ludden PW. Purification and characterization of membrane-associated CooC protein and its functional role in the insertion of nickel into carbon monoxide dehydrogenase from Rhodospirillum rubrum. Journal of Biological Chemistry. 2001;42:38602–38609. doi: 10.1074/jbc.M104945200. [DOI] [PubMed] [Google Scholar]
- 165.Wu M, Ren Q, Durkin AS, Daugherty SC, Brinkac LM, Dodson RJ, Madupu R, Sullivan SA, Kolonay JF, Haft DH, Nelson WC, Tallon LJ, Jones KM, Ulrich LE, Gonzalez JM, Zhulin IB, Robb FT, Eisen JA. Life in Hot Carbon Monoxide: The Complete Genome Sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genetics. 2005;1:e65. doi: 10.1371/journal.pgen.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lu W-P, Schiau I, Cunningham JR, Ragsdale SW. Sequence and expression of the gene encoding the corrinoid/iron-sulfur protein from Clostridium thermoaceticum and reconstitution of the recombinant protein to full activity. J. Biol. Chem. 1993;268:5605–5614. [PubMed] [Google Scholar]
- 167.Matthews RG. Cobalamin-dependent methyltransferases. Acc. Chem. Res. 2001;34:681–689. doi: 10.1021/ar0000051. [DOI] [PubMed] [Google Scholar]
- 168.Ragsdale SW. Catalysis of Methyl Group Transfers Involving Tetrahydrofolate and B12. In: Litwack G, editor. Vitamins and Hormones. vol. 79. Amsterdam, The Netherlands: Folic Acid and Folates, Elsevier, Inc.; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hagemeier CH, Krer M, Thauer RK, Warkentin E, Ermler U. Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex. Proc. Natl. Acad. Sci. U S A. 2006;103:18917–18922. doi: 10.1073/pnas.0603650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Evans JC, Huddler DP, Hilgers MT, Romanchuk G, Matthews RG, Ludwig ML. Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase. Proc. Natl. Acad. Sci. U S A. 2004;101:3729–3736. doi: 10.1073/pnas.0308082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 172.Doukov T, Seravalli J, Stezowski J, Ragsdale SW. Crystal structure of a methyltetrahydrofolate and corrinoid dependent methyltransferase. Structure. 2000;8:817–830. doi: 10.1016/s0969-2126(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 173.Doukov TI, Hemmi H, Drennan CL, Ragsdale SW. Structural And Kinetic Evidence For An Extended Hydrogen Bonding Network In Catalysis Of Methyl Group Transfer: Role Of An Active Site Asparagine Residue In Activation Of Methyl Transfer By Methyltransferases. J. Biol. Chem. 2007;282:6609–6618. doi: 10.1074/jbc.M609828200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hilhorst E, Iskander AS, Chen TBRA, Pandit U. Alkyl transfer from quaternary ammonium salts to cobalt(I): model for the cobalamin-dependent methionine synthase reaction. Tetrahedron. 1994;50:8863–8870. [Google Scholar]
- 175.Pratt JM, Norris PR, Hamza SA, Bolton R. Methyl Transfer from Nitrogen to Cobalt: Model for the B12-Dependent Methyl Transfer Enzymes. J. Chem. Soc., Chem. Commun. 1994:1333–1334. [Google Scholar]
- 176.Zheng D, Darbre T, Keese R. Methanol and dimethylaniline as methylating agents of Co(I) corrinoids under acidic conditions. J. Inorg. Biochem. 1999;73:273–275. [Google Scholar]
- 177.Wedemeyer-Exl C, T D, R K. A model for the cobalamin-dependent methionine synthase. Helv. Chim. Acta. 1999;82:1173–1184. [Google Scholar]
- 178.Seravalli J, Shoemaker RK, Sudbeck MJ, Ragsdale SW. Binding of (6R,S)-methyltetrahydrofolate to methyltransferase from Clostridium thermoaceticum: role of protonation of methyltetrahydrofolate in the mechanism of methyl transfer. Biochem. 1999;38:5736–5745. doi: 10.1021/bi9824745. [DOI] [PubMed] [Google Scholar]
- 179.Smith AE, Matthews RG. Protonation state of methyltetrahydrofolate in a binary complex with cobalamin-dependent methionine synthase. Biochem. 2000;39:13880–13890. doi: 10.1021/bi001431x. [DOI] [PubMed] [Google Scholar]
- 180.Zhao S, Roberts DL, Ragsdale SW. Mechanistic studies of the methyltransferase from Clostridium thermoaceticum: origin of the pH dependence of the methyl group transfer from methyltetrahydrofolate to the corrinoid/iron-sulfur protein. Biochem. 1995;34:15075–15083. doi: 10.1021/bi00046a013. [DOI] [PubMed] [Google Scholar]
- 181.Rod TH, Brooks CL., 3rd How dihydrofolate reductase facilitates protonation of dihydrofolate. J. Am. Chem. Soc. 2003;125:8718–8719. doi: 10.1021/ja035272r. [DOI] [PubMed] [Google Scholar]
- 182.Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Laresec JZ, Schramm VL, Almo SC. Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis. Biochem. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]
- 183.Kicska GA, Tyler PC, Evans GB, Furneaux RH, Shi W, Fedorov A, Lewandowicz A, Cahill SM, Almo SC, Schramm VL. Atomic dissection of the hydrogen bond network for transition-state analogue binding to purine nucleoside phosphorylase. Biochem. 2002;41:14489–14498. doi: 10.1021/bi026636f. [DOI] [PubMed] [Google Scholar]
- 184.Jablonski PE, Lu WP, Ragsdale SW, Ferry JG. Characterization of the metal centers of the corrinoid/iron-sulfur component of the CO dehydrogenase enzyme complex from Methanosarcina thermophila by EPR spectroscopy and spectroelectrochemistry. J. Biol. Chem. 1993;268:325–329. [PubMed] [Google Scholar]
- 185.Holliger C, Regeard C, Diekert G. Dehalogenation by anaerobic bacteria. In: Häggblom MM, Bossert ID, editors. Dehalogenation - Microbial Processes and Environmental Applications. Boston, MA: Kluwer Academic Publishers; 2003. pp. 115–157. [Google Scholar]
- 186.Krasotkina J, Walters T, Maruya KA, Ragsdale SW. Characterization of the B12- and Iron-Sulfur Containing Reductive Dehalogenase from Desulfitobacterium chlororespirans. J. Biol. Chem. 2001;276:40991–40997. doi: 10.1074/jbc.M106217200. [DOI] [PubMed] [Google Scholar]
- 187.Svetlitchnaia T, Svetlitchnyi V, Meyer O, Dobbek H. Structural insights into methyltransfer reactions of a corrinoid iron-sulfur protein involved in acetyl-CoA synthesis. Proc. Natl. Acad. Sci. U S A. 2006;103:14331–14336. doi: 10.1073/pnas.0601420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Banerjee R, Frasca V, Ballou DP, Matthews RG. Participation of Cob(I)alamin in the Reaction Catalyzed by Methionine Synthase from Escherichia coli: A Steady-State and Rapid Reaction Kinetic Analysis. Biochem. 1990;29:11101–11109. doi: 10.1021/bi00502a013. [DOI] [PubMed] [Google Scholar]
- 189.Schrauzer GN, Deutsch EJ. Reactions of cobalt(I) supernucleophiles. The alkylation of vitamin B12s, cobaloximes (I), and related compounds. J. Am. Chem. Soc. 1969;91:3341–3350. doi: 10.1021/ja01040a041. [DOI] [PubMed] [Google Scholar]
- 190.Schrauzer GN, Deutsch E, Windgassen RJ. The nucleophilicity of vitamin B12s. J. Am. Chem. Soc. 1968;90:2441–2442. doi: 10.1021/ja01011a054. [DOI] [PubMed] [Google Scholar]
- 191.Tackett SL, Collat JW, Abbott JC. The formation of hydridocobalamin and its stability in aqueous solution. Biochem. 1963;2:919–923. doi: 10.1021/bi00905a004. [DOI] [PubMed] [Google Scholar]
- 192.Banerjee RV, Harder SR, Ragsdale SW, Matthews RG. Mechanism of reductive activation of cobalamin-dependent methionine synthase: an electron paramagnetic resonance spectroelectrochemical study. Biochem. 1990;29:1129–1135. doi: 10.1021/bi00457a005. [DOI] [PubMed] [Google Scholar]
- 193.Harder SA, Lu W-P, Feinberg BF, Ragsdale SW. Spectroelectrochemical studies of the corrinoid/iron-sulfur protein from Clostridium thermoaceticum. Biochem. 1989;28:9080–9087. doi: 10.1021/bi00449a019. [DOI] [PubMed] [Google Scholar]
- 194.Harder SA, Feinberg BF, Ragsdale SW. A spectroelectrochemical cell designed for low temperature electron paramagnetic resonance titration of oxygen-sensitive proteins. Anal. Biochem. 1989;181:283–287. doi: 10.1016/0003-2697(89)90244-3. [DOI] [PubMed] [Google Scholar]
- 195.Fujii K, Galivan JH, Huennekens FM. Activation of methionine synthase: further characterization of flavoprotein system. Arch. Biochem. Biophys. 1977;178:662–670. doi: 10.1016/0003-9861(77)90238-7. [DOI] [PubMed] [Google Scholar]
- 196.Drummond JT, Matthews RG. Cobalamin-dependent and cobalamin-independent methionine synthases in Escherichia coli: two solutions to the same chemical problem. Adv Exp Med Biol. 1993;338:687–692. doi: 10.1007/978-1-4615-2960-6_142. [DOI] [PubMed] [Google Scholar]
- 197.Menon S, Ragsdale SW. The role of an iron-sulfur cluster in an enzymatic methylation reaction: methylation of CO dehydrogenase/acetyl-CoA synthase by the methylated corrinoid iron-sulfur protein. J. Biol. Chem. 1999;274:11513–11518. doi: 10.1074/jbc.274.17.11513. [DOI] [PubMed] [Google Scholar]
- 198.Bouchev VF, Furdui CM, Menon S, Muthukumaran RB, Ragsdale SW, McCracken J. ENDOR studies of pyruvate: ferredoxin oxidoreductase reaction intermediates. J. Am. Chem. Soc. 1999;121:3724–3729. [Google Scholar]
- 199.Menon S, Ragsdale SW. Role of the [4Fe-4S] cluster in reductive activation of the cobalt center of the corrinoid iron-sulfur protein from Clostridium thermoaceticum during acetyl-CoA synthesis. Biochem. 1998;37:5689–5698. doi: 10.1021/bi9727996. [DOI] [PubMed] [Google Scholar]
- 200.Olteanu H, Banerjee R. Human methionine synthase reductase, a soluble P-450 reductase-like dual flavoprotein, is sufficient for NADPH-dependent methionine synthase activation. J. Biol. Chem. 2001;276:35558–35563. doi: 10.1074/jbc.M103707200. [DOI] [PubMed] [Google Scholar]
- 201.Lexa D, Savéant J-M. Electrochemistry of vitamin B12. I. Role of the base-on/base-off reaction in the oxidoreduction mechanism of the B12r-B12s system. J. Am. Chem. Soc. 1976;98:2652–2658. doi: 10.1021/ja00425a039. [DOI] [PubMed] [Google Scholar]
- 202.Stich TA, Seravalli J, Venkateshrao S, Spiro TG, Ragsdale SW, Brunold TC. Spectroscopic Studies of the Corrinoid/Iron-Sulfur Protein from Moorella thermoacetica. J. Am. Chem. Soc. 2006;128:5010–5020. doi: 10.1021/ja054690o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jarrett JT, Choi CY, Matthews RG. Changes in protonation associated with substrate binding and Cob(I)alamin formation in cobalamin-dependent methionine synthase. Biochem. 1997;36:15739–15748. doi: 10.1021/bi971987t. [DOI] [PubMed] [Google Scholar]
- 204.Ragsdale SW. Nickel and the Carbon Cycle. J Inorg Biochem. 2007;101:1657–1666. doi: 10.1016/j.jinorgbio.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ragsdale SW. Metals and their scaffolds to promote difficult enzymatic reactions. Chem Rev. 2006;106:3317–3337. doi: 10.1021/cr0503153. [DOI] [PubMed] [Google Scholar]
- 206.Ragsdale SW. Enzymology of the Wood-Ljungdahl Pathway of Acetogenesis. Ann. N. Y. Acad. Sci. 2008;1125:129–136. doi: 10.1196/annals.1419.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Svetlitchnyi V, Peschel C, Acker G, Meyer O. Two membrane-associated NiFeS-carbon monoxide dehydrogenases from the anaerobic carbon-monoxide-utilizing eubacterium Carboxydothermus hydrogenoformans. J. Bacteriol. 2001;183:5134–5144. doi: 10.1128/JB.183.17.5134-5144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lindahl PA, Münck E, Ragsdale SW. CO dehydrogenase from Clostridium thermoaceticum: EPR and electrochemical studies in CO2 and argon atmospheres. J. Biol. Chem. 1990;265:3873–3879. [PubMed] [Google Scholar]
- 209.Lindahl PA, Ragsdale SW, Münck E. Mössbauer studies of CO dehydrogenase from Clostridium thermoaceticum. J. Biol. Chem. 1990;265:3880–3888. [PubMed] [Google Scholar]
- 210.Bastian NR, Diekert G, Niederhoffer EG, Teo B-K, Walsh CP, Orme-Johnson WH. Nickel and iron EXAFS of carbon monoxide dehydrogenase from Clostridium thermoaceticum. J. Am. Chem. Soc. 1988;110:5581–5582. [Google Scholar]
- 211.Drennan CL, Heo J, Sintchak MD, Schreiter E, Ludden PW. Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase. Proc. Natl. Acad. Sci. U S A. 2001;98:11973–11978. doi: 10.1073/pnas.211429998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Dobbek H, Svetlitchnyi V, Gremer L, Huber R, Meyer O. Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster. Science. 2001;293:1281–1285. doi: 10.1126/science.1061500. [DOI] [PubMed] [Google Scholar]
- 213.Doukov TI, Iverson T, Seravalli J, Ragsdale SW, Drennan CL. A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Science. 2002;298:567–572. doi: 10.1126/science.1075843. [DOI] [PubMed] [Google Scholar]
- 214.Darnault C, Volbeda A, Kim EJ, Legrand P, Vernede X, Lindahl PA, Fontecilla-Camps JC. Ni-Zn-[Fe(4)-S(4)] and Ni-Ni-[Fe(4)-S(4)] clusters in closed and open alpha subunits of acetyl-CoA synthase/carbon monoxide dehydrogenase. Nat Struct Biol. 2003;10:271–279. doi: 10.1038/nsb912. [DOI] [PubMed] [Google Scholar]
- 215.Dobbek H, Svetlitchnyi V, Liss J, Meyer O. Carbon monoxide induced decomposition of the active site [Ni-4Fe-5S] cluster of CO dehydrogenase. J. Am. Chem. Soc. 2004;126:5382–5387. doi: 10.1021/ja037776v. [DOI] [PubMed] [Google Scholar]
- 216.Sun J, Tessier C, Holm RH. Sulfur ligand substitution at the nickel(II) sites of cubane-type and cubanoid NiFe3S4 clusters relevant to the C-clusters of carbon monoxide dehydrogenase. Inorg. Chem. 2007;46:2691–2699. doi: 10.1021/ic062362z. [DOI] [PubMed] [Google Scholar]
- 217.Feng J, Lindahl PA. Carbon monoxide dehydrogenase from Rhodospirillum rubrum: effect of redox potential on catalysis. Biochem. 2004;43:1552–1559. doi: 10.1021/bi0357199. [DOI] [PubMed] [Google Scholar]
- 218.Jeoung JH, Dobbek H. Carbon dioxide activation at the Ni,Fe-cluster of anaerobic carbon monoxide dehydrogenase. Science. 2007;318:1461–1464. doi: 10.1126/science.1148481. [DOI] [PubMed] [Google Scholar]
- 219.Kim EJ, Feng J, Bramlett MR, Lindahl PA. Evidence for a proton transfer network and a required persulfide-bond-forming cysteine residue in ni-containing carbon monoxide dehydrogenases. Biochem. 2004;43:5728–5734. doi: 10.1021/bi036062u. [DOI] [PubMed] [Google Scholar]
- 220.Heo J, Halbleib CM, Ludden PW. Redox-dependent activation of CO dehydrogenase from Rhodospirillum rubrum. Proc. Natl. Acad. Sci. U S A. 2001;98:7690–7693. doi: 10.1073/pnas.141230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Parkin A, Seravalli J, Vincent KA, Ragsdale SW, Armstrong FA. Rapid electrocatalytic CO2/CO interconversions by Carboxydothermus hydrogenoformans CO dehydrogenase I on an electrode. J. Am. Chem. Soc. 2007;129:10328–10329. doi: 10.1021/ja073643o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Seravalli J, Ragsdale SW. 13C NMR Characterization of an Exchange Reaction between CO and CO2 Catalyzed by Carbon Monoxide Dehydrogenase. Biochem. 2008;47:6770–6781. doi: 10.1021/bi8004522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Menon S, Ragsdale SW. Unleashing hydrogenase activity in pyruvate:ferredoxin oxidoreductase and acetyl-CoA synthase/CO dehydrogenase. Biochem. 1996;35:15814–15821. doi: 10.1021/bi9615598. [DOI] [PubMed] [Google Scholar]
- 224.Bhatnagar L, Krzycki JA, Zeikus JG. Analysis of hydrogen metabolism in Methanosarcina barkeri: regulation of hydrogenase and role of CO-dehydrogenase in H2 production. FEMS Microbiol. Lett. 1987;41:337–343. [Google Scholar]
- 225.Santiago B, Meyer O. Characterization of hydrogenase activities associated with the molybdenum CO dehydrogenase from Oligotropha carboxidovorans. FEMS Microbiol. Lett. 1996;136:157–162. [Google Scholar]
- 226.Chen J, Huang S, Seravalli J, G H, Jr, Swartz DJ, Ragsdale SW, Bagley KA. Infrared Studies of Carbon Monoxide Binding to Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase from Moorella thermoacetica. Biochem. 2003;42:14822–14830. doi: 10.1021/bi0349470. [DOI] [PubMed] [Google Scholar]
- 227.Ha SW, Korbas M, Klepsch M, Meyer-Klaucke W, Meyer O, Svetlitchnyi V. Interaction of Potassium Cyanide with the [Ni-4Fe-5S] Active Site Cluster of CO Dehydrogenase from Carboxydothermus hydrogenoformans. J. Biol. Chem. 2007;282:10639–10646. doi: 10.1074/jbc.M610641200. [DOI] [PubMed] [Google Scholar]
- 228.Hu ZG, Spangler NJ, Anderson ME, Xia JQ, Ludden PW, Lindahl PA, Münck E. Nature of the C-cluster in Ni-containing carbon monoxide dehydrogenases. J. Am. Chem. Soc. 1996;118:830–845. [Google Scholar]
- 229.DeRose VJ, Telser J, Anderson ME, Lindahl PA, Hoffman BM. A multinuclear ENDOR study of the C-cluster in CO dehydrogenase from Clostridium thermoaceticum: Evidence for HxO and histidine coordination to the [Fe4S4] center. J. Am. Chem. Soc. 1998;120:8767–8776. [Google Scholar]
- 230.Bertini I, Luchinat C. The reaction pathways of zinc enzymes and related biological catalysts. In: Bertini I, Gray HB, Lippard SJ, Valentine JS, editors. Bioinorganic Chemistry. Mill Valley, CA: University Science Books; 1994. pp. 37–106. [Google Scholar]
- 231.Svetlitchnyi V, Dobbek H, Meyer-Klaucke W, Meins T, Thiele B, Romer P, Huber R, Meyer O. A functional Ni-Ni-[4Fe-4S] cluster in the monomeric acetyl-CoA synthase from Carboxydothermus hydrogenoformans. Proc. Natl. Acad. Sci. U S A. 2004;101:446–451. doi: 10.1073/pnas.0304262101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bramlett MR, Stubna A, Tan X, Surovtsev IV, Munck E, Lindahl PA. Mossbauer and EPR study of recombinant acetyl-CoA synthase from Moorella thermoacetica. Biochem. 2006;45:8674–8685. doi: 10.1021/bi060003+. [DOI] [PubMed] [Google Scholar]
- 233.Seravalli J, Ragsdale SW. Channeling of Carbon Monoxide during Anaerobic Carbon Dioxide Fixation. Biochem. 2000;39:1274–1277. doi: 10.1021/bi991812e. [DOI] [PubMed] [Google Scholar]
- 234.Maynard EL, Lindah PA. Evidence of a molecular tunnel connecting the active sites for CO2 reduction and acetyl-CoA synthesis in acetyl-CoA synthase from Clostridium thermoaceticum. J. Am. Chem. Soc. 1999;121:9221–9222. [Google Scholar]
- 235.Doukov TI, Blasiak LC, Seravalli J, Ragsdale SW, Drennan CL. Xenon in and at the end of the tunnel of bifunctional Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase. Biochem. 2008;47:3474–3483. doi: 10.1021/bi702386t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Johnson BJ, Cohen J, Welford RW, Pearson AR, Schulten K, Klinman JP, Wilmot CM. Exploring molecular oxygen pathways in Hansenula polymorpha copper-containing amine oxidase. J. Biol. Chem. 2007;282:17767–17776. doi: 10.1074/jbc.M701308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wilce MC, Dooley DM, Freeman HC, Guss JM, Matsunami H, McIntire WS, Ruggiero CE, Tanizawa K, Yamaguchi H. Crystal structures of the copper-containing amine oxidase from Arthrobacter globiformis in the holo and apo forms: implications for the biogenesis of topaquinone. Biochem. 1997;36:16116–16133. doi: 10.1021/bi971797i. [DOI] [PubMed] [Google Scholar]
- 238.Seravalli J, Ragsdale SW. Pulse-chase studies of the synthesis of acetyl-CoA by carbon monoxide dehydrogenase/acetyl-CoA synthase: evidence for a random mechanism of methyl and carbonyl addition. J Biol Chem. 2008;283:8384–8394. doi: 10.1074/jbc.M709470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Gencic S, Grahame DA. Two Separate One-Electron Steps in the Reductive Activation of the A Cluster in Subunit beta of the ACDS Complex in Methanosarcina thermophila. Biochemistry. 2008;47:5544–5555. doi: 10.1021/bi7024035. [DOI] [PubMed] [Google Scholar]
- 240.Tan X, Martinho M, Stubna A, Lindahl PA, Munck E. Mossbauer Evidence for an Exchange-Coupled {[Fe4S4](1+) Nip(1+)} A-Cluster in Isolated alpha Subunits of Acetyl-Coenzyme A Synthase/Carbon Monoxide Dehydrogenase. J Am Chem Soc. 2008 doi: 10.1021/ja801981h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Seravalli J, Kumar M, Ragsdale SW. Rapid Kinetic Studies of Acetyl-CoA Synthesis: Evidence Supporting the Catalytic Intermediacy of a Paramagnetic NiFeC Species in the Autotrophic Wood-Ljungdahl Pathway. Biochem. 2002;41:1807–1819. doi: 10.1021/bi011687i. [DOI] [PubMed] [Google Scholar]
- 242.Shin W, Anderson ME, Lindahl PA. Heterogeneous nickel environments in carbon monoxide dehydrogenase from Clostridium thermoaceticum. J. Am. Chem. Soc. 1993;115:5522–5526. [Google Scholar]
- 243.Barondeau DP, Lindahl PA. Methylation of carbon monoxide dehydrogenase from Clostridium thermoaceticum and mechanism of acetyl coenzyme A synthesis. J. Am. Chem. Soc. 1997;119:3959–3970. [Google Scholar]
- 244.Seravalli J, Xiao Y, Gu W, Cramer SP, Antholine WE, Krymov V, Gerfen GJ, Ragsdale SW. Evidence That Ni-Ni Acetyl-CoA Synthase Is Active And That The Cu-Ni Enzyme Is Not. Biochem. 2004;43:3944–3955. doi: 10.1021/bi036194n. [DOI] [PubMed] [Google Scholar]
- 245.Ramer SE, Raybuck SA, Orme-Johnson WH, Walsh CT. Kinetic characterization of the [3'-32P]coenzyme A/acetyl coenzyme A exchange catalyzed by a three-subunit form of the carbon monoxide dehydrogenase/acetyl-CoA synthase from Clostridium thermoaceticum. Biochem. 1989;28:4675–4680. doi: 10.1021/bi00437a025. [DOI] [PubMed] [Google Scholar]
- 246.Lu WP, Ragsdale SW. Reductive activation of the coenzyme A/acetyl-CoA isotopic exchange reaction catalyzed by carbon monoxide dehydrogenase from Clostridium thermoaceticum and its inhibition by nitrous oxide and carbon monoxide. J. Biol. Chem. 1991;266:3554–3564. [PubMed] [Google Scholar]
- 247.Bhaskar B, DeMoll E, Grahame DA. Redox-dependent acetyl transfer partial reaction of the acetyl-CoA Decarbonylase/Synthase complex: kinetics and mechanism. Biochem. 1998;37:14491–14499. doi: 10.1021/bi9812423. [DOI] [PubMed] [Google Scholar]
- 248.Shanmugasundaram T, Kumar GK, Wood HG. Involvement of tryptophan residues at the coenzyme A binding site of carbon monoxide dehydrogenase from Clostridium thermoaceticum. Biochem. 1988;27:6499–6503. doi: 10.1021/bi00417a045. [DOI] [PubMed] [Google Scholar]
- 249.Drake HL, Hu SI, Wood HG. Purification of five components from Clostridium thermoaceticum which catalyze synthesis of acetate from pyruvate and methyltetrahydrofolate. Properties of phosphotransacetylase. J. Biol. Chem. 1981;256:11137–11144. [PubMed] [Google Scholar]
- 250.Liu Y, Leal NA, Sampson EM, Johnson CLV, Havemann GD, Bobik TA. PduL Is an Evolutionarily Distinct Phosphotransacylase Involved in B12-Dependent 1,2-Propanediol Degradation by Salmonella enterica Serovar Typhimurium LT2. J. Bacteriol. 2007;189:1589–1596. doi: 10.1128/JB.01151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hughes NJ, Chalk PA, Clayton CL, Kelly DJ. Identification of carboxylation enzymes and characterization of a novel four-subunit pyruvate:flavodoxin oxidoreductase from Helicobacter pylori. J. Bacteriol. 1995;177:3953–3959. doi: 10.1128/jb.177.14.3953-3959.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Cammack R, Kerscher I, Oesterhelt D. A stable free radical intermediate in the reaction of 2-oxoacid:ferredoxin oxidoreductases of Halobacterium halobium. FEBS Lett. 1980;118:271–273. [Google Scholar]
- 253.Brostedt E, Nordlund S. Purification and partial characterization of a pyruvate oxidoreductase from the photosynthetic bacterium Rhodospirillum rubrum grown under nitrogen-fixing conditions. Biochem. J. 1991;279(Pt 1):155–158. doi: 10.1042/bj2790155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kletzin A, Adams MWW. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J. Bacteriol. 1996;178:248–257. doi: 10.1128/jb.178.1.248-257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Kerscher L, Oesterhelt D. Pyruvate:ferredoxin oxidoreductase - new findings on an ancient enzyme. Trends in Biochemical Sciences. 1982;7:371–374. [Google Scholar]
- 256.Wahl RC, Orme-Johnson WH. Clostridial pyruvate oxidoreductase and the pyruvate oxidizing enzyme specific to nitrogen fixation in Klebsiella pneumoniae are similar enzymes. J. Biol. Chem. 1987;262:10489–10496. [PubMed] [Google Scholar]
- 257.Ragsdale SW. Pyruvate:ferredoxin oxidoreductase and its radical intermediate. Chem. Rev. 2003;103:2333–2346. doi: 10.1021/cr020423e. [DOI] [PubMed] [Google Scholar]
- 258.Cavazza C, Contreras-Martel C, Pieulle L, Chabriere E, Hatchikian EC, Fontecilla-Camps JC. Flexibility of thiamine diphosphate revealed by kinetic crystallographic studies of the reaction of pyruvate-ferredoxin oxidoreductase with pyruvate. Structure. 2006;14:217–224. doi: 10.1016/j.str.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 259.Pieulle L, Guigliarelli B, Asso M, Dole F, Bernadac A, Hatchikian EC. Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. Biochim. Biophys. Acta-Protein Struct Mol Enzym. 1995;1250:49–59. doi: 10.1016/0167-4838(95)00029-t. [DOI] [PubMed] [Google Scholar]
- 260.Chabriere E, Charon M-H, Volbeda A, Pieulle L, Hatchikian EC, Fontecilla-Camps J-C. Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat. Struct. Biol. 1999;6:182–190. doi: 10.1038/5870. [DOI] [PubMed] [Google Scholar]
- 261.Blamey JM, Adams MWW. Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim. Biophys. Acta. 1993;1161:19–27. doi: 10.1016/0167-4838(93)90190-3. [DOI] [PubMed] [Google Scholar]
- 262.Blamey JM, Adams MWW. Characterization of an ancestral type of pyruvate ferredoxin oxidoreductase from the hyperthermophilic bacterium, Thermotoga maritima. Biochem. 1994;33 doi: 10.1021/bi00170a019. [DOI] [PubMed] [Google Scholar]
- 263.Adams MWW, Kletzin A. Oxidoreductase-type enzymes and redox proteins involved in fermentative metabolisms of hyperthermophilic archaea. Adv. Prot. Chem. 1996;48:101–180. doi: 10.1016/s0065-3233(08)60362-9. [DOI] [PubMed] [Google Scholar]
- 264.Bock A-K, Prieger-Kraft A, Schönheit P. Pyruvate - a novel substrate for growth and methane mormation in Methanosarcina barkeri. Arch. Microbiol. 1994;161:33–46. [Google Scholar]
- 265.Tersteegen A, Linder D, Thauer RK, Hedderich R. Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur. J. Biochem. 1997;244:862–868. doi: 10.1111/j.1432-1033.1997.00862.x. [DOI] [PubMed] [Google Scholar]
- 266.Simpson PG, Whitman WB. Anabolic pathways in methanogens. In: Ferry JG, editor. Methanogenesis: ecology physiology, biochemistry & genetics. London: Chapman & Hall; 1993. pp. 445–472. [Google Scholar]
- 267.Horner DS, Hirt RP, Embley TM. A single eubacterial origin of eukaryotic pyruvate : ferredoxin oxidoreductase genes: Implications for the evolution of anaerobic eukaryotes. Mol Biol Evol. 1999;16:1280–1291. doi: 10.1093/oxfordjournals.molbev.a026218. [DOI] [PubMed] [Google Scholar]
- 268.Zhang Q, Iwasaki T, Wakagi T, Oshima T. 2-oxoacid:ferredoxin oxidoreductase from the thermoacidophilic archaeon, Sulfolobus sp strain 7. J. Biochem. Tokyo. 1996;120:587–599. doi: 10.1093/oxfordjournals.jbchem.a021454. [DOI] [PubMed] [Google Scholar]
- 269.Furdui C, Ragsdale SW. The Roles of Coenzyme A in the Pyruvate:Ferredoxin Oxidoreductase Reaction Mechanism: Rate Enhancement of Electron Transfer from a Radical Intermediate to an Iron-Sulfur Cluster. Biochemistry. 2002;41:9921–9937. doi: 10.1021/bi0257641. [DOI] [PubMed] [Google Scholar]
- 270.Breslow R. Rapid Deuterium Exchange in Thiazolium Salts. J. Am. Chem. Soc. 1957;79:1762–1763. [Google Scholar]
- 271.Kerscher L, Oesterhelt D. The catalytic mechanism of 2-oxoacid:ferredoxin oxidoreductases from Halobium halobium. One-electron transfer at two distinct steps of the catalytic cycle. Eur. J. Biochem. 1981;116:587–594. doi: 10.1111/j.1432-1033.1981.tb05377.x. [DOI] [PubMed] [Google Scholar]
- 272.Chabriere E, Vernede X, Guigliarelli B, Charon MH, Hatchikian EC, Fontecilla-Camps JC. Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase. Science. 2001;294:2559–2563. doi: 10.1126/science.1066198. [DOI] [PubMed] [Google Scholar]
- 273.Mansoorabadi SO, Seravalli J, Furdui C, Krymov V, Gerfen GJ, Begley TP, Melnick J, Ragsdale SW, Reed GH. EPR Spectroscopic and Computational Characterization of the Hydroxyethylidene-Thiamine Pyrophosphate Radical Intermediate of Pyruvate:Ferredoxin Oxidoreductase. Biochem. 2006;45:7122–7131. doi: 10.1021/bi0602516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Astashkin AV, Seravalli J, Mansoorabadi SO, Reed GH, Ragsdale SW. Pulsed Electron Paramagnetic Resonance Experiments Identify the Paramagnetic Intermediates in the Pyruvate Ferredoxin Oxidoreductase Catalytic Cycle. J. Am. Chem. Soc. 2006;128:3888–3889. doi: 10.1021/ja0585275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Menon S, Ragsdale SW. Mechanism of the Clostridium thermoaceticum pyruvate:ferredoxin oxidoreductase: Evidence for the common catalytic intermediacy of the hydroxyethylthiamine pyropyrosphate radical. Biochem. 1997;36:8484–8494. doi: 10.1021/bi970403k. [DOI] [PubMed] [Google Scholar]
- 276.Furdui C, Ragsdale SW. The Roles of Coenzyme A in the Pyruvate:Ferredoxin Oxidoreductase Reaction Mechanism: Rate Enhancement of Electron Transfer from a Radical Intermediate to an Iron-Sulfur Cluster. Biochem. 2002;41:9921–9937. doi: 10.1021/bi0257641. [DOI] [PubMed] [Google Scholar]
- 277.Meuer J, Kuettner HC, Zhang JK, Hedderich R, Metcalf WW. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5632–5637. doi: 10.1073/pnas.072615499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Bock AK, Kunow J, Glasemacher J, Schonheit P. Catalytic properties, molecular composition and sequence alignments of pyruvate: ferredoxin oxidoreductase from the methanogenic archaeon Methanosarcina barkeri (strain Fusaro) Eur. J. Biochem. 1996;237:35–44. doi: 10.1111/j.1432-1033.1996.0035n.x. [DOI] [PubMed] [Google Scholar]
- 279.Yoon KS, Hille R, Hemann C, Tabita FR. Rubredoxin from the green sulfur bacterium Chlorobium tepidum functions as an electron acceptor for pyruvate ferredoxin oxidoreductase. J. Biol. Chem. 1999;274:29772–29778. doi: 10.1074/jbc.274.42.29772. [DOI] [PubMed] [Google Scholar]
- 280.Evans MCW, Buchanan BB, Arnon DI. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc. Natl. Acad. Sci. USA. 1966;55:928–934. doi: 10.1073/pnas.55.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hugler M, Wirsen CO, Fuchs G, Taylor CD, Sievert SM. Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the epsilon subdivision of proteobacteria. J. Bacteriol. 2005;187:3020–3027. doi: 10.1128/JB.187.9.3020-3027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Hügler M, Huber H, Molyneaux SJ, Vetriani C, Sievert SM. Autotrophic CO2 fixation via the reductive tricarboxylic acid cycle in different lineages within the phylum Aquificae: evidence for two ways of citrate cleavage. Environ. Microbiol. 2007;9:81–92. doi: 10.1111/j.1462-2920.2006.01118.x. [DOI] [PubMed] [Google Scholar]
- 283.Wood AP, Aurikko JP, Kelly DP. A challenge for 21st century molecular biology and biochemistry: what are the causes of obligate autotrophy and methanotrophy? FEMS Microbiol. Ecol. 2004;28:335–352. doi: 10.1016/j.femsre.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 284.Fuchs G, Stupperich E. Evidence for an incomplete reductive carboxylic acid cycle in Methanobacterium thermoautotrophicum. Arch. Microbiol. 1978;118:121–125. doi: 10.1007/BF00406084. [DOI] [PubMed] [Google Scholar]
- 285.Stols L, Donnelly MI. Production of succinic acid through overexpression of NAD(+)-dependent malic enzyme in an Escherichia coli mutant. Appl. Environ. Microbiol. 1997;63:2695–2701. doi: 10.1128/aem.63.7.2695-2701.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Dorn M, Andreesen JR, Gottschalk G. Fermentation of fumarate and L-malate by Clostridium formicoaceticum. J. Bacteriol. 1978;133:26–32. doi: 10.1128/jb.133.1.26-32.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Bache R, Pfennig N. Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch. Microbiol. 1981;130:255–261. [Google Scholar]
- 288.Daniel SL, Keith ES, Yang H, Lin YS, Drake HL. Utilization of methoxylated aromatic compounds by the acetogen Clostridium thermoaceticum: expression and specificity of the co- dependent O-demethylating activity. Biochem. Biophys. Res. Commun. 1991;180:416–422. doi: 10.1016/s0006-291x(05)81309-9. [DOI] [PubMed] [Google Scholar]
- 289.Naidu D, Ragsdale SW. Characterization of a Three-Component Vanillate O-Demethylase from Moorella thermoacetica. J. Bacteriol. 2001;183:3276–3281. doi: 10.1128/JB.183.11.3276-3281.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Kaufmann F, Wohlfarth G, Diekert G. O-demethylase from Acetobacterium dehalogenans--substrate specificity and function of the participating proteins. Eur. J. Biochem. 1998;253:706–711. doi: 10.1046/j.1432-1327.1998.2530706.x. [DOI] [PubMed] [Google Scholar]
- 291.Kaufmann F, Wohlfarth G, Diekert G. Isolation of O-demethylase, an ether-cleaving enzyme system of the homoacetogenic strain MC. Arch. Microbiol. 1997;168:136–142. doi: 10.1007/s002030050479. [DOI] [PubMed] [Google Scholar]
- 292.Das A, Fu Z-Q, Tempel W, Liu Z-J, Chang J, Chen L, Lee D, Zhou W, Xu H, Shaw N, Rose JP, Ljungdahl LG, Wang B-C. Characterization of a corrinoid protein involved in the C1 metabolism of strict anaerobic bacterium Moorella thermoacetica. Proteins: Structure, Function, and Bioinformatics. 2007;67:167–176. doi: 10.1002/prot.21094. [DOI] [PubMed] [Google Scholar]
- 293.Barker HA, Kamen MD. Carbon dioxide utilization in the synthesis of acetic acid by Clostridium thermoaceticum. Proceedings of the National Academy of Sciences. 1945;31:219–225. doi: 10.1073/pnas.31.8.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Beaty SP, Ljungdahl LG. Growth of Clostridium thermoaceticum on methanol, ethanol, propanol, and butanol in medium containing either thiosulfate or dimethylsulfoxide. Abstracts of the Annual Meeting of the American Society for Microbiology. 1991:236. [Google Scholar]
- 295.Daniel SL, Drake HL. Oxalate- and glyoxylate-dependent growth and acetogenesis by Clostridium thermoaceticum. Applied and Environmental Microbiology. 1993;59:3062–3069. doi: 10.1128/aem.59.9.3062-3069.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Seifritz C, Fröstl JM, Drake HL, Daniel SL. Glycolate as a metabolic substrate for the acetogen Moorella thermoacetica. FEMS Microbiology Letters. 1999;170:399–405. [Google Scholar]
- 297.Göβner A, Devereux R, Ohnemüller N, Acker G, Stackebrandt E, Drake HL. Thermicanus aegyptius gen. nov., sp. nov., isolated from oxic soil, a fermentative microaerophile that grows commensally with the thermophilic acetogen Moorella thermoacetica. Applied and Environmental Microbiology. 1999;65:5124–5133. doi: 10.1128/aem.65.11.5124-5133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Andreesen JR, Schaupp A, Neurauter C, Brown A, Ljungdahl LG. Fermentation of glucose, fructose, and xylose by Clostridium thermoaceticum: effect of metals on growth yield, enzymes, and the synthesis of acetate from CO2. Journal of Bacteriology. 1973;114:743–751. doi: 10.1128/jb.114.2.743-751.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Seifritz C, Daniel SL, Gossner A, Drake HL. Nitrate as a preferred electron sink for the acetogen Clostridium thermoaceticum. J. Bacteriol. 1993;175:8008–8013. doi: 10.1128/jb.175.24.8008-8013.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Fröstl JM, Seifritz C, Drake HL. Effect of nitrate on the autotrophic metabolism of the acetogens Clostridium thermoautotrophicum and Clostridium thermoaceticum. J. Bacteriol. 1996;178:4597–4603. doi: 10.1128/jb.178.15.4597-4603.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Arendsen AF, Soliman MQ, Ragsdale SW. Nitrate-dependent regulation of acetate biosynthesis and nitrate respiration by Clostridium thermoaceticum. J. Bacteriol. 1999;181:1489–1495. doi: 10.1128/jb.181.5.1489-1495.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Seifritz C, Drake HL, Daniel SL. Nitrite as an Energy-Conserving Electron Sink for the Acetogenic Bacterium Moorella thermoacetica. Curr. Microbiol. 2003;46:329–333. doi: 10.1007/s00284-002-3830-6. [DOI] [PubMed] [Google Scholar]
- 303.Seifritz C, Fröstl JM, Drake HL, Daniel SL. Influence of nitrate on oxalate- and glyoxylate-dependent growth and acetogenesis by Moorella thermoacetica. Arch. Microbiol. 2002;178:457–464. doi: 10.1007/s00203-002-0475-6. [DOI] [PubMed] [Google Scholar]
- 304.Dilling S, Imkamp F, Schmidt S, Muller V. Regulation of caffeate respiration in the acetogenic bacterium Acetobacterium woodii. Appl. Environ. Microbiol. 2007;73:3630–3636. doi: 10.1128/AEM.02060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Misoph M, Drake HL. Effect of CO2 on the fermentation capacities of the acetogen Peptostreptococcus productus U-1. J. Bacteriol. 1996;178:3140–3145. doi: 10.1128/jb.178.11.3140-3145.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Matthies C, Freiberger A, Drake HL. Fumarate dissimilation and differential reductant flow by Clostridium formicoaceticum and Clostridium aceticum. Arch. Microbiol. 1993;160:273–278. [Google Scholar]
- 307.Andreesen JR, Gottschalk G, Schlegel HG. Clostridium formicoaceticum nov. spec. isolation, description and distinction from C. aceticum and C. thermoaceticum. Arch. Mikrobiol. 1970;72:154–174. doi: 10.1007/BF00409521. [DOI] [PubMed] [Google Scholar]
- 308.Winter JU, Wolfe RS. Methane formation from fructose by syntrophic associations of Acetobacterium woodii and different strains of methanogens. Arch. Microbiol. 1980;124:73–79. doi: 10.1007/BF00407031. [DOI] [PubMed] [Google Scholar]
- 309.Winter J, Wolfe RS. Complete degradation of carbohydrate to carbon dioxide and methane by syntrophic cultures of Acetobacterium woodii and Methanosarcina barkeri. Arch. Microbiol. 1979;121:97–102. doi: 10.1007/BF00409211. [DOI] [PubMed] [Google Scholar]
- 310.Lee MJ, Zinder SH. Isolation and Characterization of a Thermophilic Bacterium Which Oxidizes Acetate in Syntrophic Association with a Methanogen and Which Grows Acetogenically on H(2)-CO(2) Appl. Environ. Microbiol. 1988;54:124–129. doi: 10.1128/aem.54.1.124-129.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Zinder SH, Koch M. Non-aceticlastic methanogenesis from acetate: acetate oxidation by a thermophilic syntrophic coculture. Arch. Microbiol. 1984;138:263–272. [Google Scholar]
- 312.McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJ, Schink B, Rohlin L, Gunsalus RP. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. 2008;1125:58–72. doi: 10.1196/annals.1419.005. [DOI] [PubMed] [Google Scholar]
- 313.Collins M, Lawson P, Willems A, Cordoba J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 314.Savage MD, Drake HL. Adaptation of the acetogen Clostridium thermoautotrophicum to minimal medium. J. Bacteriol. 1986;165:315–318. doi: 10.1128/jb.165.1.315-318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Andreesen JR, Schaupp A, Neurater C, Brown A, Ljungdahl LG. Fermentation of glucose, fructose, and xylose by Clostridium thermoaceticum: effect of metals on growth yield, enzymes, and the synthesis of acetate from CO2. J. Bacteriol. 1973;114:743–751. doi: 10.1128/jb.114.2.743-751.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Tschech A, Pfennig N. Growth yield increase linked to caffeate reduction in Acetobacterium woodii. Arch. Microbiol. 1984;137:163–167. [Google Scholar]
- 317.Gottwald M, Andreesen JR, LeGall J, Ljungdahl LG. Presence of cytochrome and menaquinone in Clostridium formicoaceticum and Clostridium thermoaceticum. J. Bacteriol. 1975;122:325–328. doi: 10.1128/jb.122.1.325-328.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Ivey DM, Ljungdahl LG. Purification and characterization of the F1-ATPase from Clostridium thermoaceticum. J. Bacteriol. 1986;165:252–257. doi: 10.1128/jb.165.1.252-257.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Hugenholtz J, Ivey DM, Ljungdahl LG. Carbon monoxide-driven electron transport in Clostridium thermoautotrophicum membranes. J. Bacteriol. 1987;169:5845–5847. doi: 10.1128/jb.169.12.5845-5847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Yang SS, Ljungdahl LG, Dervartanian DV, Watt GD. Isolation and characterization of two rubredoxins from Clostridium thermoaceticum. Biochim. Biophys. Acta. 1980;590:24–33. doi: 10.1016/0005-2728(80)90143-7. [DOI] [PubMed] [Google Scholar]
- 321.Das A, Hugenholtz J, Van Halbeek H, Ljungdahl LG. Structure and function of a menaquinone involved in electron transport in membranes of Clostridium thermoautotrophicum and Clostridium thermoaceticum. J. Bacteriol. 1989;171:5823–5829. doi: 10.1128/jb.171.11.5823-5829.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Hugenholtz J, Ljungdahl LG. Electron transport and electrochemical proton gradient in membrane vesicles of Clostridium thermoautotrophicum. J. Bacteriol. 1989;171:2873–2875. doi: 10.1128/jb.171.5.2873-2875.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Das A, Silaghi-Dumitrescu R, Ljungdahl LG, Kurtz DM., Jr Cytochrome bd oxidase, oxidative stress, and dioxygen tolerance of the strictly anaerobic bacterium Moorella thermoacetica. J. Bacteriol. 2005;187:2020–2029. doi: 10.1128/JB.187.6.2020-2029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Karnholz A, Kusel K, Gossner A, Schramm A, Drake HL. Tolerance and metabolic response of acetogenic bacteria toward oxygen. Appl. Environ. Microbiol. 2002;68:1005–1009. doi: 10.1128/AEM.68.2.1005-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Dangel W, Schulz H, Diekert G, König H, Fuchs G. Occurrence of corrinoid-containing membrane proteins in anaerobic bacteria. Arch. Microbiol. 1987;148:52–56. [Google Scholar]
- 326.Heise R, Reidlinger J, Muller V, Gottschalk G. A sodium-stimulated ATP synthase in the acetogenic bacterium Acetobacterium woodii. FEBS Lett. 1991;295:119–122. doi: 10.1016/0014-5793(91)81400-3. [DOI] [PubMed] [Google Scholar]
- 327.Heise R, Muller V, Gottschalk G. Presence of a sodium-translocating ATPase in membrane vesicles of the homoacetogenic bacterium Acetobacterium woodii. Eur. J. Biochem. 1992;206:553–557. doi: 10.1111/j.1432-1033.1992.tb16959.x. [DOI] [PubMed] [Google Scholar]
- 328.Heise R, Müller V, Gottschalk G. Acetogenesis and ATP synthesis in Acetobacterium woodii are coupled via a transmembrane primary sodium ion gradient. FEMS Microbiol. Lett. 1993;112:261–268. [Google Scholar]
- 329.Reidlinger J, Müller V. Purification of ATP synthase from Acetobacterium woodii and identification as a Na+-translocating F1F0-type enzyme. Eur. J. Biochem. 1994;223:275–283. doi: 10.1111/j.1432-1033.1994.tb18992.x. [DOI] [PubMed] [Google Scholar]
- 330.Fritz M, Klyszejko AL, Morgner N, Vonck J, Brutschy B, Muller DJ, Meier T, Muller V. An intermediate step in the evolution of ATPases: a hybrid F(0)-V(0) rotor in a bacterial Na(+) F(1)F(0) ATP synthase. FEBS J. 2008;275:1999–2007. doi: 10.1111/j.1742-4658.2008.06354.x. [DOI] [PubMed] [Google Scholar]
- 331.Fritz M, Muller V. An intermediate step in the evolution of ATPases--the F1F0-ATPase from Acetobacterium woodii contains F-type and V-type rotor subunits and is capable of ATP synthesis. FEBS J. 2007;274:3421–3428. doi: 10.1111/j.1742-4658.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 332.Imkamp F, Biegel E, Jayamani E, Buckel W, Müller V. Dissection of the Caffeate Respiratory Chain in the Acetogen Acetobacterium woodii: Identification of an Rnf-Type NADH Dehydrogenase as a Potential Coupling Site. J. Bacteriol. 2007;189:8145–8153. doi: 10.1128/JB.01017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Müller V, Imkamp F, Biegel E, Schmidt S, Dilling S. Discovery of a Ferredoxin:NAD+-Oxidoreductase (Rnf) in Acetobacterium woodii: A Novel Potential Coupling Site in Acetogens. Ann. N. Y. Acad. Sci. 2008;1125:137–146. doi: 10.1196/annals.1419.011. [DOI] [PubMed] [Google Scholar]
- 334.Andrews SC, Berks BC, McClay J, Ambler A, Quail MA, Golby P, Guest JR. A 12-cistron Escherichia coli operon (hyf) encoding a putative proton- translocating formate hydrogenlyase system. Microbiology. 1997;143:3633–3647. doi: 10.1099/00221287-143-11-3633. [DOI] [PubMed] [Google Scholar]
- 335.Lindahl PA. Implications of a Carboxylate-Bound C-Cluster Structure of Carbon Monoxide Dehydrogenase. Angew. Chem. Int. Ed. Engl. 2008;47:4054–4056. doi: 10.1002/anie.200800223. [DOI] [PubMed] [Google Scholar]
- 336.Drake HL, Göβner AS, Daniel SL. Old Acetogens, New Light. Annals of the New York Academy of Sciences. 2008;1125:100–128. doi: 10.1196/annals.1419.016. [DOI] [PubMed] [Google Scholar]