Introduction
More than half a century ago, Linus Pauling wrote: “enzymes are molecules that are complementary in structure to the activated complexes of the reactions that they catalyze, ···, [rather than] entering into reactions.”1 This model has had a profound impact on drug design. Structure-based drug design usually involves the conceptualization and synthesis of molecules that have shapes and binding surfaces that are highly complementary to a protein receptor or enzyme binding site.2, 3 The goal is to achieve high binding affinity and selectivity. A drug must also have appropriate ADMET properties, but affinity is the first step.
Affinity and selectivity are generally improved by ensuring more perfect geometric and noncovalent interactions with a binding site. Crystal structures of a protein-ligand complex suggest structural modifications to better occupy a hydrophobic pocket. Such modifications can improve potency from the millimolar to the nanomolar range,4 and have helped lead to clinically approved compounds such as the HIV protease inhibitor nelfinavir.5, 6
Kuntz et al. have shown that small molecule affinity for protein binding sites resulting from noncovalent interactions generally peaks at 10 picomolar (10−11 M), corresponding to ΔGbinding of 15 kcal/mol.7 In contrast, binding constants for enzymes with transition states correspond to average Δ Gbinding of 22 kcal/mol, and up to 38 kcal/mol—many orders of magnitude stronger than can be attributed to noncovalent factors alone.8, 9 Earlier studies in our group have shown how these and other data point to the generality of covalent and partial covalent bonding in transition states of enzyme-catalyzed reactions.8, 9 This strength of binding may be achieved by fully covalent bonding such as Schiff base or acylenzyme intermediate formation, but partial covalent bonds that take place in general acid/base catalysis and interactions with metal cofactors can partially share electrons with the substrate or other reactants such as water molecules in the transition state.9
The harnessing of such strong covalent interactions could help provide the high potency that is needed at early stages of drug development. There has been a tendency to avoid covalent drugs, going back to studies in the early 1970s demonstrating hepatotoxicity as a result of covalent binding of compounds such as [14C]bromobenzene and acetaminophen.10, 11 At the same time, however, there are many examples of highly successful covalently acting drugs on the market, from proton pump inhibitors omeprazole and related compounds,12, 13 to the entire class of β-lactam antibiotics.14 The toxicity attributed to covalent binding is not an inherent feature of these interactions, per se, but rather a result of the specific spectrum of off-target modifications that may be made by the drug or even by metabolites of drugs whose primary mechanism of action is noncovalent.15, 16 Advances in chemical biology, along with bioinformatics data analysis methods, are increasingly able to unravel which covalent modifications are tolerable and which are toxic, suggesting a reevaluation of the role of covalent binding in drugs and drug leads.16–19 In this Perspective, the limits achievable by noncovalent binding in enzyme–inhibitor complexes, and the greater affinity achieved by covalent bonding of the drug to the receptor, are discussed. Selected examples of covalently acting drugs will be presented (for more comprehensive reviews see references 20, 21), as well as opportunities for their future structure-based design with advances in molecular modeling or screening with target-specific libraries.
Factors Involved in Ligand-Receptor Binding
Drug-receptor binding has been discussed in terms of an equation for ΔGbinding that is sometimes referred to as the “master equation”:22
ΔGsolvent encompasses the desolvation of both ligand and receptor upon complex formation; this often involves favorable removal of hydrophobic molecules from aqueous solution and displacement of water molecules that would otherwise be held in a pocket in the protein. ΔGint is the interaction free energy involving direct contact of ligand with protein, and includes noncovalent interactions such as van der Waals interactions, electrostatic interactions, and hydrogen bonds. These can further improve the binding, but generally only to 15 kcal/mol, according to the study by Kuntz et al.7 The few cases which exceed the maximal noncovalent binding of 15 kcal/mol include the biotin-avidin and biotin–streptavidin complexes.7, 23 In these cases, the high affinity is believed to be due to enhanced hydrogen-bonding cooperativity, among other things.24, 25 A small molecule host-guest complex with such a large affinity has also recently been reported.26
ΔGconf and ΔGmotion represent unfavorable terms for conformational strain and entropy loss upon complexation, respectively. These two factors are important to include but their calculation is difficult to quantify; nevertheless, research in these areas is active.27, 28 In this Perspective, relative magnitudes and limits of ΔGbinding are discussed, with a focus on how they might be enhanced in order to lead to more potent compounds for the inhibition of therapeutically relevant targets.
Distributions of Experimental Binding Constants: The 15 kcal/mol Limit of Noncovalent Binding Energy
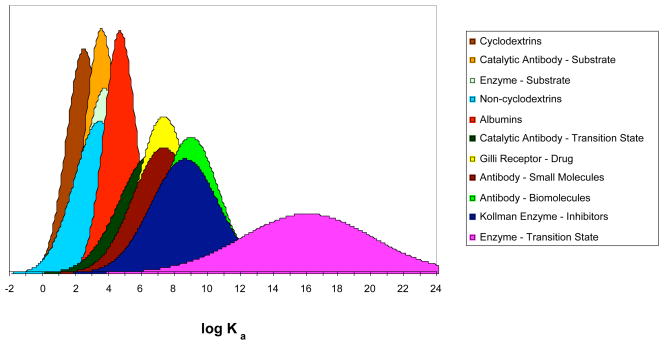
Previously, we surveyed the aqueous binding constants for all types of organic and protein hosts, including cyclodextrins, various synthetic organic hosts, albumins, antibodies, enzymes, and other nonenzymatic drug receptors.8 The binding constants fall into three regions (Figure 1): weak-binding, medium-binding, and strong-binding. The weak-binding region corresponds to binding constants in the decimolar to hundreds of micromolar range (log Ka of 1–4) and covers cyclodextrins and synthetic hosts binding organic guests, catalytic antibodies and enzymes binding their substrates, and albumins binding organic ligands. The medium-binding region corresponds to binding constants in the micromolar to nanomolar range (log Ka of 6–9) and contains antibody–antigen complexes (including antibody–small molecule and antibody–biomolecule complexes), catalytic antibody–TS (transition state) complexes, receptor–drug complexes, and enzyme–inhibitor complexes. The strong-binding region is for the enzyme–TS complexes, whose proficiencies suggest binding constants in the sub-picomolar range (log Ka values of 16±4).
Figure 1.
Summary of the typical binding constants for host–guest complexes. For each class, the binding distribution is represented by an idealized normal distribution. Each curve is normalized to have the same area, the maximum occurs at the average value of Ka, and the standard deviation is used to set the width of the curve. Reprinted with permission from reference 9. Copyright 2005 Americal Chemical Society.
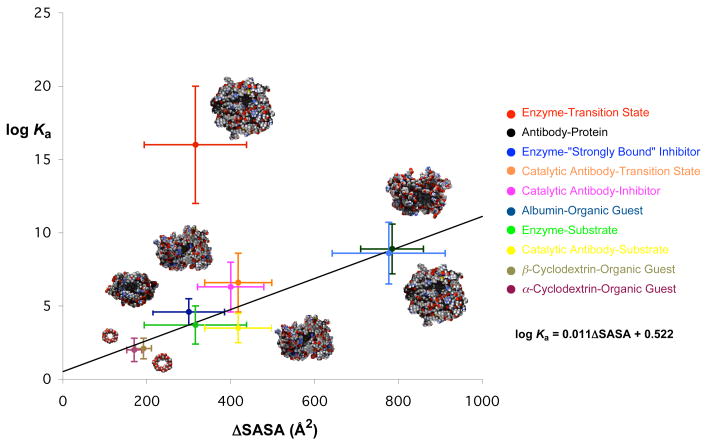
As shown in Figure 2, a rough correlation between log Ka and ΔSASA (Å2) is found for all the complexes in the weak- and medium-binding systems.8, 9 This suggests that weak to medium bindings are strongly influenced by desolvation of hydrophobic surfaces and bringing them together upon binding. A linear fit gives a slope of 0.011, corresponding to a γ value of 0.007 kcal/mol/Å2 in the equation ΔG = γΔSASA.8 Other noncovalent factors including van der Waals, hydrogen bonding, and electrostatic interactions, which are commonly involved in the binding process, contribute variations of as much as 3 orders of magnitude from this rough correlation.8 These noncovalent interactions appear to achieve an upper limit for association constants, Kas, of ~1011.7 With this limit in mind, it is sensible that most commercial drugs or prospective drug candidates which rely merely on noncovalent binding interactions have a binding strength in the sub-nanomolar to nanomolar range.
Figure 2.
Plot of log Ka versus buried solvent accessible surface area (ΔSASA). Reprinted with permission from reference 9. Copyright 2005 Americal Chemical Society.
The Kuntz et al. survey of experimental data on 160 of the strongest-binding drugs and inhibitors to receptors and enzymes was referred to earlier.7 It indicates that the binding energies of small molecules increase by about 1.5 kcal/mol with each non-hydrogen atom.7 There is a 15 kcal/mol maximum affinity of drugs and inhibitors to receptors and enzymes, and for ligands that contain more than 10 non-hydrogen atoms, the binding energy increases very little. While up to 21 kcal/mol binding has been observed for a few cases, 92% of cases from the Kuntz et al. studies have binding energies of 15 kcal/mol or less. Their analysis of the dominant interactions suggests that van der Waals interactions, solvation and desolvation contribute to the binding affinities across the entire set of ligands. It is noteworthy that the outliers that bound unusually strongly include metal ions, covalently attached ligands, and a few well-known complexes such as biotin–avidin. In addition, the 15 kcal/mol maximum affinity (Ka = 1011 M−1) of drugs with receptors has also been noted by Gilli et al., which they explained by entropy–enthalpy compensation.29, 30 Combining our survey8 with those of Kuntz et al.7 and Gilli et al.,29, 30 the strength of binding of drugs and inhibitors to receptors and enzymes rarely exceeds 15 kcal/mol by noncovalent binding interactions (log Ka <11).9
Covalent Interactions with Transition States as the Origins of Enzyme Proficiency
Enzymes are exceptional catalysts that have evolved naturally over millions of years. A quantitative measurement of enzyme catalysis is proficiency, defined by Wolfenden as Ktx−1, where Ktx−1 = (kcat/KM/)kuncat.31 Proficiency has been interpreted as the hypothetical equilibrium constant for conversion of the transition state (TS) of the uncatalyzed reaction plus the enzyme, both in water, into the enzyme–TS complex—the apparent binding constant of the TS to the enzyme. This interpretation of the proficiency is somewhat controversial but the conclusions about enzymes that show particularly high proficiencies and about how enzyme transition state binding compares to general non-covalent interactions drawn below do not depend on the exact numerical interpretation of these values. While abundant kcat and KM values have been measured and are available in the BRENDA enzyme database,32 few kuncat values have been documented due to the slow rates of the uncatalyzed reactions in water. Additionally, many enzyme-catalyzed reactions do not have an equivalent in pure aqueous solution; redox reactions are good examples of this. Wolfenden and coworkers have measured the kuncat of twenty-four hydrolases, isomerases, and lyases,31, 33, 34 which enabled plotting of the binding constant data for enzyme–TS complexes, shown in Figure 1.9 The 1016 average binding constant (Figure 1) is much greater than expected from noncovalent interactions. We have proposed that such exceptional binding is a result of covalent or partial covalent bond formation. This may involve the formation of an intermediate covalently bound to the enzyme or cofactor, proton transfer (general acid/base catalysis and possibly low-barrier hydrogen bonds) occurring in the transition state, or bonding to metal cations in the transition state.9
As discussed above and in previous publications,7–9, 29, 30 noncovalent interactions are likely to contribute no more than 15 kcal/mol for binding, corresponding to a binding constant of 1011 M−1, for typical small molecules binding to proteins. The twenty-four Wolfenden enzymes with known proficiencies31, 33, 34 were divided into two groups using 1011 M−1 as the dividing line.9 Twenty-one enzymes (88%) had proficiencies over 1011 M−1 and only three enzymes had proficiencies below 1011 M−1.9 A literature survey of the catalytic mechanisms of the twenty-four enzymes showed that catalysis of the twenty-one enzymes with proficiencies over 1011 M−1 all involve partial covalent bond-breaking or bond-forming processes, often general acid/base catalysis, as well as the participation of metals and organic cofactors.9 Our use of the terms covalent bonding or covalent catalysis does not require irreversible binding, but only the formation of partial bonds. Catalysis of two of the enzymes, chorismate mutase and cyclophilin, with proficiencies below 1011 M−1 involves hydrogen bonds and electrostatic interactions that are noncovalent in nature.9, 35, 36 The remaining enzyme, carbonic anhydrase, with a proficiency of 109 M−1, involves general acid/base catalysis and metal binding.9, 37 This demonstrates that covalent catalysis is not excluded from the low proficiency class, as there is no real limit to how weak covalent forces can be, but it is not necessary to achieve the modest acceleration therein. This survey emphasizes that enzymes with proficiencies over 1011 M−1 always involve some type of covalent catalysis.9
While the covalent hypothesis has been checked with twenty-four enzymes with known proficiencies, this is a tiny number compared to the tens of thousands of enzymes with unknown proficiencies. To establish the proficiencies of more enzymes, the twenty-four kuncat values measured by Wolfenden were used to estimate kuncat values for related reactions.9 For example, the rates of amide hydrolysis or glycolysis fluctuate across a small range. Therefore, the available kuncat values were employed to estimate the proficiencies of enzymes that catalyze related reactions. By this method, the proficiencies of 1017 enzymes were estimated.9 These results show that 97% of the enzymes have proficiencies over 1011 M−1, and therefore involve covalent catalysis.9
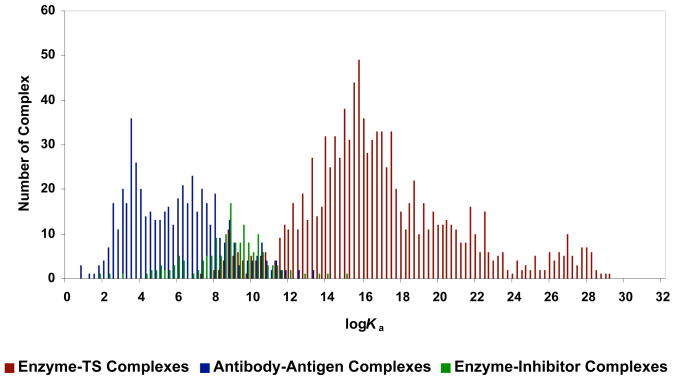
The binding constants of 507 antibody–antigen complexes (excluding catalytic antibodies) from our survey,8 160 enzyme–inhibitor complexes from Kuntz’s study,7 and 1017 enzyme–TS complexes from our proficiency estimation were compared.9 The results are shown in Figure 3.9 The naturally evolved antibodies show micromolar binding constant, Ka = 106±2 M−1, while enzyme–inhibitor complexes exhibit nanomolar binding constant, Ka = 109±2 M−1, both of which are well within the domain of noncovalent binding. Enzyme–TS complexes achieve femtomolar binding, Ka = 1016±4 M−1, which must originate from covalent interactions. The 15 kcal/mol dichotomy between noncovalent binding of antigens by antibodies and inhibitors by enzymes and covalent binding of transition states by enzymes is apparent.9
Figure 3.
Frequency plot of association constant for 507 antibody–antigen complexes, 160 enzyme–inhibitor complexes, and 1017 measurements of enzyme–transition state complex. Reprinted with permission from reference 9. Copyright 2005 Americal Chemical Society.
In summary, proficiency greater than 1011 M−1 signals covalent or partially covalent catalysis by enzymes. When we refer to “covalent catalysis” we include electrophilic catalysis such as the participation of metals, nucleophilic catalysis such as Schiff base formation, general acid/base catalysis, or the covalent participation of cofactors. Most enzymes perform covalent catalysis, employing different mechanisms to those available in aqueous solution. Proficiency or binding affinity of 1011 M−1 is found empirically to be the usual limit of noncovalent binding comprised of hydrogen bonds, electrostatics, van der Waals interactions, and hydrophobic effects.7–9, 29, 30
Potential Quantitive Advantages of Covalent Enzyme Inhibitors and Covalent Drugs
The development of a new drug can cost about a billion dollars and take almost 13 years from development to approval.38 The first step is often to find a “hit”, a drug-like molecule with detectable affinity for a clinically relevant target. Presently, there are various strategies for finding a hit, including the use of natural products,39, 40 high-throughput screening,41 virtual screening or rational design,42, 43 or a combination of techniques. The second step is the optimization of the hit to generate a lead compound with increased affinity and selectivity.
Optimization of hits normally improves their potency by 100–1000 fold, which corresponds to 2.8–4.2 kcal/mol binding energy. After optimization, the lead compounds are typically expected to achieve nano- to picomolar binding affinity, which corresponds to 12.6–16.8 kcal/mol binding energy. Valid hits are below or around micromolar inhibitors and contribute at least two-thirds of the net binding energy of fully optimized lead compounds. Because of the ultimate importance of finding a highly potent hit, huge drug-like compound libraries have been built within companies or provided by chemical vendors to allow their identification.44 Subsequent optimization is against other, non-efficacy parameters such as physical and ADMET properties.
For drug lead compounds, a Ki of 1 nM—within the realm of the strongest noncovalent binding—is considered quite good by consensus. Is there an advantage to exploit covalent interactions in search of sub-picomolar affinities? The answer depends upon the importance of the drug’s residence time in their binding sites.45, 46 Generally, drugs exert their effects when they are bound to their receptors, and therefore anything that keeps them bound longer is seen as beneficial. This has both kinetic and thermodynamic components. Kinetics and thermodynamics of a simple one-step binding and one-step dissociation mechanism are related by the expression for the inhibition equilibrium constant, Ki = koff/kon. This is related to the occupancy time for such a drug complex by the half life of occupancy, t1/2 = 0.693/koff.46 The expressions for the equilibrium constants and half lives of complexes that involve an induced fit mechanism are slightly more complicated but similar.46 The rate constant for association of antibodies with their haptens has been determined to be in the region of 106 to 107 M−1 s−1.47 Using this range for kon, and 1 nM as a hypothetical Ki, the half life for first order decomposition of the enzyme/inhibitor complex would be between 1.2 and 12 minutes. Results at this level are very sensitive, however, and a modest change of Ki to 5 nM produces a new range of half-lives of 14 to 140 seconds. As Ki gets smaller, corresponding to stronger binding, the koff rates get slower, and the occupancy time is longer. The koff rate is also determined by the barrier to dissociation of the drug from the receptor, and this represents another way of optimizing residence times.
If inhibitors could capture enough TS binding affinity to reliably give Kis of 1 pM, this would change the half life range to 19 to 190 hours. For comparison, half lives of 12 drugs and drug candidates have been measured to be between 8 minutes and 40 hours.45 Covalent interactions can lead to long dissociation half lives, as in the case of finasteride, which forms a covalent bond with the NADPH cofactor in the active site of the steroid 5α-reductase enzyme as treatment for benign prostatic hyperplasia.48 The half life for dissociation of this complex is greater than 30 days.45 Noncovalent interactions can also lead to long dissociation half lives, as with natural protease inhibitors with half lives of up to 4 months.46 The increased half lives of drug binding represent a way to achieve what Schramm has termed the “ultimate physiological goal,” where efficacy is maintained until the target is physically replaced by the body.49 The benefits of long residency times would be seen in reduced dosages, resulting in fewer drug-drug interactions and less off-target toxicity due to the lower concentration of free drug in serum (assuming that the covalent interactions are not able to contribute to off-target effects).45, 46
Another way to address the place of covalent interactions in pharmaceutical agents is through the analyses of which small molecules make the best drugs. This question is of fundamental importance for drug discovery. In recent years, databases of successful compounds have been the subject of intense statistical scrutiny in attempts to discern any empirical metrics by which candidate molecules can be judged.50–52 A number of trends have emerged, including a premium on a high potency to molecular weight ratio.50 In order to design lead compounds that meet this and other criteria, our analysis and hypothesis suggest the use of covalent bonds. With the strength of covalent interactions ensuring high potency, more effort could be placed on optimizing moieties for ADMET and specificity properties.
Covalent approaches are also of utility for bringing challenging biological targets into the range of small molecule inhibitors. Many biologically important molecules are large and well structured and therefore may distribute interactions over a broad surface; each interaction need only contribute a small amount of binding to generate a large cumulative affinity. This is not easily matched by small molecules with acceptable ADMET properties. For such molecules, every interaction must count as there will only be a limited number available. A covalent interaction can provide the binding energy needed to compete. Such challenges are presented by protein-protein and protein-DNA interactions but also by enzymes that process proteins such as kinases and proteases. It is not coincidental that known kinase inhibitors bind at the ATP pocket rather at the binding site of the protein substrate despite the daunting challenges that competition with ATP presents in terms of selectivity.53
A recent example of inhibiting the interaction of the thyroid hormone receptor and coregulator proteins demonstrates that even this challenging area may be made more amenable by use of a covalent interaction.54 In this instance, it is suggested that the requisite α,β-unsaturated ketone is liberated at the protein by Mannich elimination of a β-amino ketone. The irreversible inhibitor then reacts with a specific surface cysteine residue making this compound act as a mechanism based inhibitor.
An area of particular current interest, particularly in the area of cancer therapy is that of HDAC inhibitors.55 These enzymes catclyze the deacetylation of histones which are involved in the control of access to nuclear DNA. The active site is an 11Å long tube with a zinc ion at the active site at the far end. The tube like nature of this active site limits the surface area available for non-covalent interactions without also capitalizing on interactions with the surface surrounding the entrance to the binding site which is used by the enzyme to selectively bind its protein substrate. Small molecules with the same shape, surface area and even increased lipophilicity as the recently approved vorinostat show dramatic drops in binding when the covalent metal binding hydroxamate group is removed.56
These latter two examples highlight a key challenge for covalent inhibitors – achieveing selectivity. Reaction with any free cysteine or any zinc containing enzyme would be catastrophic in vivo. While the covalent interaction can confer potency, the remainder of the molecule must be used to judiciously control selectivity through non-covalent interactions. Ideally, this involves them being involved in the initial molecular recognition which is followed by covalent bond formation. This may require careful selection of a suitable reactive group (a number of these so-called “warheads” and the type of interactions they make have been mentioned) followed by tuning of the reactivity of the moiety that will form the covalent bond. The kinetics of the binding event as well as the strength of the interaction finally achieved must both be examined carefully during the design and optimization of covalent inhibitors.
Selected Examples of Covalent Inhibitors and Drugs: Transition State Analogues, Irreversible and Reversible Binders, and Metal-Coordinators
The proposal of using covalent interactions is implicit in the rational design of inhibitors that are transition state analogues. Drugs that have been classified as TSAs include the anticancer agent pentostatin (deoxycoformycin),57 the HIV protease inhibitor saquinavir,58 and the influenza drugs oseltamivir, and zanamivir.59 As we have shown, the vast majority of enzymes involve covalent or partially covalent interactions to effect such strong binding affinities to transition states. By employing covalent interactions, the inhibition constants of TSAs could in principle become comparable to the TS binding constants, Ktx. Unfortunately, this has not proven to be the case. The field has a history of nearly 40 years,60 yet even the best TSAs have Kis in the picomolar range.61–63 A previous comparison of 8 enzymes’ Ktxs to the Kis of their TSAs showed that, while Ktxs are between 10−11 and 10−24, the Kis are only between 10−6 and 10−12.63 The question of why TSAs are not able to take full advantage of the binding affinity reflected in Ktx constants has been put down to two main problems: 1) imperfect TS analogy in terms of geometry and electrostatics, and 2) an inherent inability of a ground state to reproduce certain of these parameters such as partial bonds and hyperpolarized states.63 There have been a number of approaches to convert theoretical information about TSs into design criteria for TS analogues.64, 65 Quantum mechanically calculated electrostatic potentials have been used along with experimental kinetic isotope effects to help design TSAs for human nucleoside phosphorylase, among others,65 and combined quantum mechanical/molecular mechanical modeling of a hepatitis C virus protease reaction has been carried out to suggest how TSAs might be designed to help treat this disease.66
Suicide inhibitors are compounds that irreversibly acylate or alkylate the appropriate active site nucleophiles (such as Ser, His, Cys etc.) with their electrophilic functional groups (such as β-lactams, alkyl fluorophosphates, halomethyl ketones, and sulfonyl fluorides).20, 21, 67–69 Because of their inherent specificity, stoichiometry, and irreversible nature, suicide inhibitors have wide applications in biochemical and pharmalogical research.20 Moreover, suicide inhibitors have shown great potential as therapeutic agents, as reviewed comprehensively by Sjoerdsma in 1981 and more recently by Robertson in 2005.21, 69 Since then, however, the perception of promise has waned, to the extent that a more recent review of irreversible protease inhibitors notes with dismay that, “A strong bias against irreversible inhibitors exists in the pharmaceutical industry, and it is an uphill battle to get irreversible inhibitors considered as potential clinical candidates.”70 However, many frequently used and non-toxic drugs were found to be irreversible inhibitors long after their use, such as penicillin14 and aspirin.71 This indicates that drugs acting in this manner should not be dismissed out of hand. As discussed in the Introduction to this Perspective, the outlook for covalent drugs may be more favorable in the near future.
Great effort has also been devoted to the development of reversible covalent inhibitors, especially for proteases, which by their nature have a nucleophilic residue already primed for addition.67, 70, 72, 73 Inhibitors of this kind usually have an electrophilic functional group such as an aldehyde which can reversibly form a hemiacetal with, for example, serine hydroxyl or cysteine thiol groups. The reversible nature of boronic ester formation has also been exploited to replace aldehydes in such an inhibitor, and this has led to the clinically approved compound bortezomib, which is being used as a treatment for multiple myeloma.74, 75 Formation of reversible covalent bonds increases the binding affinity to proteases by 10–1000 times.73 Although much work has been devoted to the design of reversible covalent inhibitors of serine proteases, translation of this information into marketable drugs has proven difficult.76 Metabolic instability, low selectivity, and slow kinetics of binding have been invoked as explanations.76 However, Andrews et al. have shown that reactive 5,5-trans-lactams can have their pharmacokinetic profile tuned to make acceptable drug candidates.77 Removing the entire electrophilic isostere drops the affinity from the picomolar to nanomolar range, indicating the importance of the reactive species for binding.73
The use of metals in pharmaceutical pursuits has abundant precedent, with compounds known to interact with DNA and enzymes.78–81 The potential of metal ions to form covalent and ionic bonds makes them appealing, and already several prominent compounds that either contain a metal themselves—such as the antineoplastic agent cisplatin82—or interact with a metal complexed by their target—such as the antihypertensive angiotensin converting enzyme inhibitor captopril83—have proven to be successful drugs. As with TSAs, however, these inhibitors do not appear to have access to the full strength of covalent interactions that produces sub-picomolar Kis. The reasons for this are likely to be the same as those for the inadequacies of TSAs, especially an inability to reproduce a hyperpolarized transition state with a stable ground state. Metals are still an exciting prospect for therapeutic compounds, since both thermodynamic and kinetic benefits have been observed. In a class of zinc-chelating inhibitors which complex with the active site Ser and His of serine proteases, inclusion of the zinc was found to give an added 4–7 kcal/mol of binding affinity, raising the binding constant by 3 to 5 orders of magnitude.78 This led to Kis in the tens of picomolar to nanomolar range, but these compounds also exhibited relatively long half lives for the EI complex, from 11 to 32 hours. These compounds can also be highly specific, with the best ones having Kis four orders of magnitude smaller for tryptase compared to two other serine proteases trypsin and thrombin.78
Prospects for the Design of Covalent Drugs
The strength of binding afforded by covalent drugs may not be advantageous in all cases and for all targets. Specifically, it has been suggested that infectious disease targets may replicate too quickly for the increased residence time of covalent drugs to be a factor.45 However, others see bacterial, viral, and parasitic diseases as a golden opportunity for irreversible inhibitor design, in part because the drug is only administered for a short period of time, thereby reducing any off-target toxicity.70 Viral proteases have been recognized as excellent drug targets because they are essential for the virus to replicate, provide an opportunity for specificity by their exogenous origins, and their mechanism-based inhibitors are believed to be less susceptible to resistance since they target the catalytically essential residues.84, 85 Transition state analogues such as saquinavir are anti-HIV protease drugs,58 and a covalently acting SARS coronavirus 3CL protease inhibitor has also shown promising efficacy.86
While covalently acting drugs do represent a path towards “ultimate physiological” inhibition,49 the benefits of this will have to be evaluated on a case-by-case basis. They appear to be excellent candidates for blocking protein-protein interactions,87 but may be less ideal for other targets. For example, while G-protein coupled receptors represent a significant proportion of drug targets,88 they are subject to a trafficking process by which they are internalized from their place in the cell membrane and either recycled back or degraded.89 This turnover may make some GPCRs unattractive candidates for covalent drugs, yet the anticoagulant clopidogrel (Plavix®) acts by irreversible inhibition of the P2Y12 GPCR on platelets.90 As noted earlier, some enzyme-TS binding that uses covalent interactions does not achieve binding constants that exceed those that are readily attained by non-covalent interactions. In such cases (which may be numerous) it is unreasonable to expect that even inhibitors that also use covalent bonds might achieve substantially tighter binding.
If it is decided that a particular target is suited to covalent inhibition, there are a variety of developing tools, both experimental and computational, for arriving at such an inhibitor. Cravatt and colleagues have reviewed the use of active site-directed covalent probes, or activity based probes, for activity based protein profiling.91, 92 In ABPP, a covalently interacting core group based on the mechanism of a given enzyme may be used, for example, to probe the proteomic phenotype of that enzyme subtype in disease states or to assign a function to an unclassified protein.93, 94 In the field of enzyme inhibitors, ABPP has been used to investigate the selectivity profiles of covalently acting inhibitors such as bortezomib.95 The ABPs would form an ideal nucleus for the creation of a structure-based, target-specific compound library for screening,96 as has been done for a family of quinic acid derivatives that coordinate with a calcium ion among other groups in the binding site of E-selectin.97 Similar libraries could be created with metal-chelating groups such as sulfonamides to access the strong binding potential of metal-enzyme interactions. The ABP or metal would help ensure potency by virtue of covalent or partially covalent interactions, allowing for a concerted focus on specificity through optimization of the rest of the molecule.
Computational chemistry tools such as SkelGen98 and BOMB99 can be used to grow functional groups off of a covalently interacting group in silico and evaluate their binding energies to provide another route to rational design. Covalent docking programs are also being developed,100 and it may be that the binding orientations of covalently interacting drugs are easier to predict given the constraint of a known interaction at the binding site. Combined quantum mechanical/molecular mechanical calculations have been used to calculate the preferred binding orientation of a covalent inhibitor of fatty acid amide hydrolase, URB524.101 Pure quantum mechanics methods have been used to calculate the reactivities of substituted nitriles with thiols, which was shown to correlate with the formation of covalent adducts with an active site cysteine residue of cathepsin K.102 All of these tools would be equally applicable to fully covalent interactions and partial covalent interactions such as metal chelation. Although the prospect of a rationally designed, covalently interacting core with subsequent optimization of specificity has been mentioned, these two steps need not occur in that order. The reverse has already been done for both antibodies and small molecules.103, 104 In these cases, rational design of covalent bond formation came after specificity was ensured by working with a pre-optimized antibody, in one case, and a known drug scaffold in the other.
Conclusions
Pauling’s finding that enzymes provide binding by complementing the shapes and characteristics of transition states provides the principles for the design of reversible noncovalent inhibitors in the pharmaceutical industry. The aim of this perspective article has been to present and discuss the limit of noncovalent binding and our recent discovery about the origins of the enormous catalytic acceleration that is usually manifested in enzyme catalysis.8, 9 While complementarity proposed by Pauling can account for acceleration up to 11 orders of magnitude, most enzymes exceed that proficiency. Enzymes with proficiency > 1011 M−1 achieve > 15 kcal/mol of “transition state binding” not merely by a concatenation of noncovalent effects but by partial covalent bond formation between enzyme or cofactor and transition state, involving a change in mechanism from that in aqueous solution. The involvement of partially covalent bonds does not require that a proficient enzyme form an actual covalent intermediate with a substrate. The bonding can be partially covalent or ionic, such as in cases of a metal ion associated with the enzyme coordinating with the substrate. Or it can be partly covalent, such as in cases of proton transfer between enzyme and substrate (general acid/base catalysis).
The discussion has illustrated that noncovalent interactions can be approximated by estimating surface area of the small ligands. In order to design drug-like inhibitors with very high receptor affinities, covalent interactions have to be considered. While these interactions alone do not automatically lead to sub-picomolar binding affinities, Kis of this magnitude cannot be achieved without them. The goal of this perspective has been to emphasize the potential role of covalent bonding in rational drug design.
Acknowledgments
We thank the National Institute of General Medical Sciences, National Institutes of Health, and the National Science Foundation for financial support of this research.
Biographies
Adam J. T. Smith is a graduate student in the lab of Prof. K. N. Houk at the University of California, Los Angeles, where he is a member of the Medical Scientist Training Program (MSTP), a dual-degree MD/Ph.D. program. He is a member of the Chemistry-Biology Interface Training Program. He received his Bachelor’s degree from Emory University in Atlanta, Georgia.
Xiyun Zhang is from Chongqing, China. She studied chemistry at Beijing Normal University (China) and remained at Beijing Normal University to pursue a M.S. in organic synthesis with Prof. Q. Chen. She received her Ph.D. from University of California, Los Angeles, with Prof. K. N. Houk in the area of computational chemistry and biochemistry. She was the recipient of the Thomas L. and Ruth Jacobs award for graduate research at UCLA. She has carried out postdoctoral studies with Professor Houk, Professor T. M. Handel at University of California, San Diego, and Professor M. Pellecchia at the Burnham Institute for Medical Research in La Jolla, California. Currently she is a Bioinformatics Scientist at Codexis in San Francisco, California.
Andrew G. Leach is a Computational Chemist with AstraZeneca at their Alderley Park UK site. He originates from Chester in the UK and obtained his PhD with Prof. S. V. Ley at the University of Cambridge. From 2000 until 2002, he was a Fulbright PostDoctoral Scholar at the University of California, Los Angeles in the lab of Prof. K. N. Houk.
K. N. Houk has been on the chemistry faculties at Louisiana State University, the University of Pittsburgh, and UCLA. He was the 2003 winner of the American Chemical Society Award for Computers in Chemical and Pharmaceutical Research.
References and Notes
- 1.Pauling L. The Nature of Forces Between Large Molecules of Biological Interest. Nature. 1948;161:707–709. doi: 10.1038/161707a0. [DOI] [PubMed] [Google Scholar]
- 2.Congreve M, Murray CW, Blundell TL. Structural Biology and Drug Discovery. Drug Discov Today. 2005;10(13):895–907. doi: 10.1016/S1359-6446(05)03484-7. [DOI] [PubMed] [Google Scholar]
- 3.Williams SP, Kuyper LF, Pearce KH. Recent Applications of Protein Crystallography and Structure-Guided Drug Design. Curr Opin Chem Biol. 2005;9(4):371–380. doi: 10.1016/j.cbpa.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Gill AL, Frederickson M, Cleasby A, Woodhead SJ, Carr MG, Woodhead AJ, Walker MT, Congreve MS, Devine LA, Tisi D, O’Reilly M, Seavers LC, Davis DJ, Curry J, Anthony R, Padova A, Murray CW, Carr RA, Jhoti H. Identification of Novel p38alpha MAP Kinase Inhibitors Using Fragment-Based Lead Generation. J Med Chem. 2005;48(2):414–426. doi: 10.1021/jm049575n. [DOI] [PubMed] [Google Scholar]
- 5.Kaldor SW, Kalish VJ, Davies JF, 2nd, Shetty BV, Fritz JE, Appelt K, Burgess JA, Campanale KM, Chirgadze NY, Clawson DK, Dressman BA, Hatch SD, Khalil DA, Kosa MB, Lubbehusen PP, Muesing MA, Patick AK, Reich SH, Su KS, Tatlock JH. Viracept (Nelfinavir Mesylate, AG1343): a Potent, Orally Bioavailable Inhibitor of HIV-1 Protease. J Med Chem. 1997;40(24):3979–3985. doi: 10.1021/jm9704098. [DOI] [PubMed] [Google Scholar]
- 6.Reich SH, Melnick M, Davies JF, 2nd, Appelt K, Lewis KK, Fuhry MA, Pino M, Trippe AJ, Nguyen D, Dawson H, et al. Protein Structure-Based Design of Potent Orally Bioavailable, Nonpeptide Inhibitors of Human Immunodeficiency Virus Protease. Proc Natl Acad Sci U S A. 1995;92(8):3298–3302. doi: 10.1073/pnas.92.8.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuntz ID, Chen K, Sharp KA, Kollman PA. The Maximal Affinity of Ligands. Proc Natl Acad Sci U S A. 1999;96(18):9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houk KN, Leach AG, Kim SP, Zhang X. Binding Affinities of Host-Guest, Protein-Ligand, and Protein-Transition-State Complexes. Angew Chem Int Ed Engl. 2003;42(40):4872–4897. doi: 10.1002/anie.200200565. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Houk KN. Why Enzymes Are Proficient Catalysts: Beyond the Pauling Paradigm. Acc Chem Res. 2005;38(5):379–85. doi: 10.1021/ar040257s. [DOI] [PubMed] [Google Scholar]
- 10.Brodie BB, Reid WD, Cho AK, Sipes G, Krishna G, Gillette JR. Possible Mechanism of Liver Necrosis Caused by Aromatic Organic Compounds. Proc Natl Acad Sci U S A. 1971;68(1):160–164. doi: 10.1073/pnas.68.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jollow DJ, Mitchell JR, Potter WZ, Davis DC, Gillette JR, Brodie BB. Acetaminophen-Induced Hepatic Necrosis. II. Role of Covalent Binding In Vivo. J Pharmacol Exp Ther. 1973;187(1):195–202. [PubMed] [Google Scholar]
- 12.Lindberg P, Nordberg P, Alminger T, Brandstrom A, Wallmark B. The Mechanism of Action of the Gastric Acid Secretion Inhibitor Omeprazole. J Med Chem. 1986;29(8):1327–1329. doi: 10.1021/jm00158a001. [DOI] [PubMed] [Google Scholar]
- 13.Lorentzon P, Bayati A, Lee H, Andersson K. Selective Inhibition of the Gastric H+,K(+)-ATPase by Omeprazole and Related Compounds. Ann N Y Acad Sci. 1997;834:592–599. doi: 10.1111/j.1749-6632.1997.tb52328.x. [DOI] [PubMed] [Google Scholar]
- 14.Waxman DJ, Strominger JL. Penicillin-Binding Proteins and the Mechanism of Action of Beta-Lactam Antibiotics. Annu Rev Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 15.Baillie TA. Future of Toxicology-Metabolic Activation and Drug Design: Challenges and Opportunities in Chemical Toxicology. Chem Res Toxicol. 2006;19(7):889–893. doi: 10.1021/tx060062o. [DOI] [PubMed] [Google Scholar]
- 16.Liebler DC. Protein Damage by Reactive Electrophiles: Targets and Consequences. Chem Res Toxicol. 2008;21(1):117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennehy MK, Richards KA, Wernke GR, Shyr Y, Liebler DC. Cytosolic and Nuclear Protein Targets of Thiol-Reactive Electrophiles. Chem Res Toxicol. 2006;19(1):20–29. doi: 10.1021/tx050312l. [DOI] [PubMed] [Google Scholar]
- 18.Said MR, Begley TJ, Oppenheim AV, Lauffenburger DA, Samson LD. Global Network Analysis of Phenotypic Effects: Protein Networks and Toxicity Modulation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101(52):18006–18011. doi: 10.1073/pnas.0405996101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huth JR, Song D, Mendoza RR, Black-Schaefer CL, Mack JC, Dorwin SA, Ladror US, Severin JM, Walter KA, Bartley DM, Hajduk PJ. Toxicological Evaluation of Thiol-Reactive Compounds Identified Using a LA Assay to Detect Reactive Molecules by Nuclear Magnetic Resonance. Chem Res Toxicol. 2007;20(12):1752–1759. doi: 10.1021/tx700319t. [DOI] [PubMed] [Google Scholar]
- 20.Metcalf BW. Recent Progress in the Design of Suicide Enzyme Inhibitors. Ann Rep Med Chem. 1981:289–297. [Google Scholar]
- 21.Robertson JG. Mechanistic Basis of Enzyme-Targeted Drugs. Biochemistry. 2005;44(15):5561–5571. doi: 10.1021/bi050247e. [DOI] [PubMed] [Google Scholar]
- 22.Ajay; Murcko MA. Computational Methods to Predict Binding Free Energy in Ligand-Receptor Complexes. J Med Chem. 1995;38(26):4953–4967. doi: 10.1021/jm00026a001. [DOI] [PubMed] [Google Scholar]
- 23.Weber PC, Wendoloski JJ, Pantoliano MW, Salemme FR. Crystallographic and Thermodynamic Comparison of Natural and Synthetic Ligands Bound to Streptavidin. J Am Chem Soc. 1992;114:3197–3200. [Google Scholar]
- 24.DeChancie J, Houk KN. The Origins of Femtomolar Protein-Ligand Binding: Hydrogen-Bond Cooperativity and Desolvation Energetics in the Biotin-(Strept)Avidin Binding Site. J Am Chem Soc. 2007;129(17):5419–5429. doi: 10.1021/ja066950n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei Y, Li H, Zhang R, Han S. Theoretical Study of Cooperativity in Biotin. J Phys Chem B. 2007;111(51):14370–14377. doi: 10.1021/jp076914q. [DOI] [PubMed] [Google Scholar]
- 26.Rekharsky MV, Mori T, Yang C, Ko YH, Selvapalam N, Kim H, Sobransingh D, Kaifer AE, Liu S, Isaacs L, Chen W, Moghaddam S, Gilson MK, Kim K, Inoue Y. A Synthetic Host-Guest System Achieves Avidin-Biotin Affinity by Overcoming Enthalpy-Entropy Compensation. Proc Natl Acad Sci U S A. 2007;104(52):20737–20742. doi: 10.1073/pnas.0706407105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruvinsky AM, Kozintsev AV. New and Fast Statistical-Thermodynamic Method for Computation of Protein-Ligand Binding Entropy Substantially Improves Docking Accuracy. J Comput Chem. 2005;26(11):1089–1095. doi: 10.1002/jcc.20246. [DOI] [PubMed] [Google Scholar]
- 28.Tirado-Rives J, Jorgensen WL. Contribution of Conformer Focusing to the Uncertainty in Predicting Free Energies for Protein-Ligand Binding. J Med Chem. 2006;49(20):5880–5884. doi: 10.1021/jm060763i. [DOI] [PubMed] [Google Scholar]
- 29.Gilli P, Ferretti V, Gilli G, Borea PA. Enthalpy-Entropy Compensation in Drug-Receptor Binding. J Phys Chem. 1994;98:1515–1518. [Google Scholar]
- 30.Borea PA, Dalpiaz A, Varani K, Gilli P, Gilli G. Can Thermodynamic Measurements of Receptor Binding Yield Information on Drug Affinity and Efficacy? Biochem Pharmacol. 2000;60(11):1549–1556. doi: 10.1016/s0006-2952(00)00368-3. [DOI] [PubMed] [Google Scholar]
- 31.Miller BG, Wolfenden R. Catalytic Proficiency: The Unusual Case of OMP Decarboxylase. Annu Rev Biochem. 2002;71:847–885. doi: 10.1146/annurev.biochem.71.110601.135446. [DOI] [PubMed] [Google Scholar]
- 32.Barthelmes J, Ebeling C, Chang A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA: The Enzyme Information System in 2007. Nucleic Acids Res. 2007;35(Database issue):D511–514. doi: 10.1093/nar/gkl972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lad C, Williams NH, Wolfenden R. The Rate of Hydrolysis of Phosphomonoester Dianions and the Exceptional Catalytic Proficiencies of Protein and Inositol Phosphatases. Proc Natl Acad Sci U S A. 2003;100(10):5607–5610. doi: 10.1073/pnas.0631607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snider MJ, Wolfenden R. The Rate of Spontaneous Decarboxylation of Amino Acids. J Am Chem Soc. 2000;122(46):11507–11508. [Google Scholar]
- 35.Fanghänel J. Enzymatic Catalysis of the Peptidyl-Prolyl Bond Rotation: Are Transition State Formation and Enzyme Dynamics Directly Linked? Angew Chem Int Ed Engl. 2003;42(5):490–492. doi: 10.1002/anie.200390149. [DOI] [PubMed] [Google Scholar]
- 36.Lee YS, Worthington SE, Krauss M, Brooks BR. Reaction Mechanism of Chorismate Mutase Studied by the Combined Potentials of Quantum Mechanics and Molecular Mechanics. J Phys Chem B. 2002;106:12059–12065. [Google Scholar]
- 37.Mauksch M, Brauer M, Weston J, Anders E. New Insights into the Mechanistic Details of the Carbonic Anhydrase Cycle as Derived From the Model System [(NH3)3Zn(OH)+/CO2: How Does the H2O/HCO3− Replacement Step Occur? ChemBioChem. 2001;2:190–198. doi: 10.1002/1439-7633(20010302)2:3<190::AID-CBIC190>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 38.Kola I, Landis J. Can the Pharmaceutical Industry Reduce Attrition Rates? Nat Rev Drug Discov. 2004;3(8):711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 39.Clardy J, Walsh C. Lessons from Natural Molecules. Nature. 2004;432(7019):829–837. doi: 10.1038/nature03194. [DOI] [PubMed] [Google Scholar]
- 40.Lesney MS. Nature’s Pharmaceuticals: Natural Products From Plants Remain at the Core of Modern Medicinal Chemistry. Today’s Chemist at Work. 2004 July;:26–32. [Google Scholar]
- 41.Gribbon P, Sewing A. High-Throughput Drug Discovery: What Can We Expect From HTS? Drug Discov Today. 2005;10(1):17–22. doi: 10.1016/S1359-6446(04)03275-1. [DOI] [PubMed] [Google Scholar]
- 42.Guido RV, Oliva G, Andricopulo AD. Virtual Screening and its Integration With Modern Drug Design Technologies. Curr Med Chem. 2008;15(1):37–46. doi: 10.2174/092986708783330683. [DOI] [PubMed] [Google Scholar]
- 43.Shoichet BK, McGovern SL, Wei B, Irwin JJ. Lead Discovery Using Molecular Docking. Curr Opin Chem Biol. 2002;6(4):439–446. doi: 10.1016/s1367-5931(02)00339-3. [DOI] [PubMed] [Google Scholar]
- 44.Erlanson DA, McDowell RS, O’Brien T. Fragment-Based Drug Discovery. J Med Chem. 2004;47(14):3463–3482. doi: 10.1021/jm040031v. [DOI] [PubMed] [Google Scholar]
- 45.Copeland RA, Pompliano DL, Meek TD. Drug-Target Residence Time and its Implications for Lead Optimization. Nat Rev Drug Discov. 2006;5(9):730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 46.Tummino PJ, Copeland RA. Residence Time of Receptor-Ligand Complexes and Its Effect on Biological Function. Biochemistry. 2008;47:5481–5492. doi: 10.1021/bi8002023. [DOI] [PubMed] [Google Scholar]
- 47.Foote J, Milstein C. Kinetic Maturation of an Immune Response. Nature. 1991;352(6335):530–532. doi: 10.1038/352530a0. [DOI] [PubMed] [Google Scholar]
- 48.Bull HG, Garcia-Calvo M, Andersson S, Baginsky WF, Chan HK, Ellsworth DE, Miller RR, Stearns RA, Bakshi RK, Rasmusson GH, Tolman RL, Myers RW, Kozarich JW, Harris GS. Mechanism-Based Inhibition of Human Steroid 5 -Reductase by Finasteride: Enzyme-Catalyzed Formation of NADP-Dihydrofinasteride, a Potent Bisubstrate Analog Inhibitor. J Am Chem Soc. 1996;118:2359–2365. [Google Scholar]
- 49.Lewandowicz A, Tyler PC, Evans GB, Furneaux RH, Schramm VL. Achieving the Ultimate Physiological Goal in Transition State Analogue Inhibitors for Purine Nucleoside Phosphorylase. J Biol Chem. 2003;278(34):31465–31468. doi: 10.1074/jbc.C300259200. [DOI] [PubMed] [Google Scholar]
- 50.Abad-Zapatero C, Metz JT. Ligand Efficiency Indices as Guideposts for Drug Discovery. Drug Discov Today. 2005;10(7):464–469. doi: 10.1016/S1359-6446(05)03386-6. [DOI] [PubMed] [Google Scholar]
- 51.Ajay A, Walters WP, Murcko MA. Can We Learn to Distinguish Between “Drug-Like” and “Nondrug–Like” Molecules? J Med Chem. 1998;41(18):3314–3324. doi: 10.1021/jm970666c. [DOI] [PubMed] [Google Scholar]
- 52.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Developmental Settings. Adv Drug Delivery Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 53.Noble MEM, Endicott JA, Johnson LN. Protein Kinase Inhibitors: Insights into Drug Design from Structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 54.Arnold LA, Estébanez-Perpiñá E, Togashi M, Jouravel N, Shelat A, McReynolds AC, Mar E, Nguyen P, Baxter JD, Fletterick RJ, Webb P, Guy RK. Discovery of Small Molecule Inhibitors of the Interaction of the Thyroid Hormone Receptor with Transcriptional Regulators. J Biol Chem. 2005;280:43048–43055. doi: 10.1074/jbc.M506693200. [DOI] [PubMed] [Google Scholar]
- 55.Körner M, Tibes U. In: Progress in Medicinal Chemistry. Lawton G, Witty DR, editors. Vol. 40. Elsevier; 2008. pp. 205–280. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki T, Nagano Y, Kouketsu A, Matsuura A, Maruyama S, Kurotaki M, Nakagawa H, Miyata N. Novel Inhibitors of Human Histone Deacetylases: Design, Synthesis, Enzyme Inhibition, and Cancer Cell Growth Inhibition of SAHA-based Non-hydroxyamates. J Med Chem. 2005;48:1019–1032. doi: 10.1021/jm049207j. [DOI] [PubMed] [Google Scholar]
- 57.Agarwal RP, Spector T, Parks RE., Jr Tight-Binding Inhibitors--IV. Inhibition of Adenosine Deaminases by Various Inhibitors. Biochem Pharmacol. 1977;26(5):359–367. doi: 10.1016/0006-2952(77)90192-7. [DOI] [PubMed] [Google Scholar]
- 58.Roberts NA, Martin JA, Kinchington D, Broadhurst AV, Craig JC, Duncan IB, Galpin SA, Handa BK, Kay J, Krohn A, et al. Rational Design of Peptide-Based HIV Proteinase Inhibitors. Science. 1990;248(4953):358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 59.von Itzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B, Van Phan T, Smythe ML, White HF, Oliver SW, et al. Rational Design of Potent Sialidase-Based Inhibitors of Influenza Virus Replication. Nature. 1993;363(6428):418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 60.Wolfenden R. Transition State Analogues for Enzyme Catalysis. Nature. 1969;223(5207):704–705. doi: 10.1038/223704a0. [DOI] [PubMed] [Google Scholar]
- 61.Guo H, Rao N, Xu Q, Guo H. Origin of Tight Binding of a Near-Perfect Transition-State Analogue by Cytidine Deaminase: Implications for Enzyme Catalysis. J Am Chem Soc. 2005;127(9):3191–3197. doi: 10.1021/ja0439625. [DOI] [PubMed] [Google Scholar]
- 62.Wolfenden R. Conformational Aspects of Inhibitor Design: Enzyme-Substrate Interactions in the Transition State. Bioorg Med Chem. 1999;7(5):647–652. doi: 10.1016/s0968-0896(98)00247-8. [DOI] [PubMed] [Google Scholar]
- 63.Mader MM, Bartlett PA. Binding Energy and Catalysis: The Implications for Transition-State Analogs and Catalytic Antibodies. Chem Rev. 1997;97(5):1281–1302. doi: 10.1021/cr960435y. [DOI] [PubMed] [Google Scholar]
- 64.Richards WG. Quantum Chemistry in Drug Design. Pure & Appl Chem. 1988;60(2):227–279. [Google Scholar]
- 65.Schramm VL. Enzymatic Transition State Theory and Transition State Analogue Design. J Biol Chem. 2007;282(39):28297–28300. doi: 10.1074/jbc.R700018200. [DOI] [PubMed] [Google Scholar]
- 66.Oliva C, Rodriguez A, Gonzalez M, Yang W. A Quantum Mechanics/Molecular Mechanics Study of the Reaction Mechanism of the Hepatitis C Virus NS3 Protease with the NS5A/5B Substrate. Proteins. 2007;66(2):444–455. doi: 10.1002/prot.21190. [DOI] [PubMed] [Google Scholar]
- 67.Edwards PD, Bernstein PR. Synthetic Inhibitors of Elastase. Med Res Rev. 1994;14(2):127–194. doi: 10.1002/med.2610140202. [DOI] [PubMed] [Google Scholar]
- 68.Palfreyman MG, McDonald IA, Bey P, Danzin C, Zreika M, Lyles GA, Fozard JR. The Rational Design of Suicide Substrates of Amine Oxidases. Biochem Soc Trans. 1986;14(2):410–413. doi: 10.1042/bst0140410. [DOI] [PubMed] [Google Scholar]
- 69.Sjoerdsma A. Suicide Enzyme Inhibitors as Potential Drugs. Clin Pharmacol Ther. 1981;30(1):3–22. doi: 10.1038/clpt.1981.121. [DOI] [PubMed] [Google Scholar]
- 70.Powers JC, Asgian JL, Ekici OD, James KE. Irreversible Inhibitors of Serine, Cysteine, and Threonine Proteases. Chem Rev. 2002;102(12):4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 71.Van Der Ouderaa FJ, Buytenhek M, Nugteren DH, Van Dorp DA. Acetylation of Prostaglandin Endoperoxide Synthetase with Acetylsalicylic Acid. Eur J Biochem. 1980;109:1–8. doi: 10.1111/j.1432-1033.1980.tb04760.x. [DOI] [PubMed] [Google Scholar]
- 72.Bajusz S, Barabas E, Tolnay P, Szell E, Bagdy D. Inhibition of Thrombin and Trypsin by Tripeptide Aldehydes. Int J Pept Protein Res. 1978;12(4):217–221. doi: 10.1111/j.1399-3011.1978.tb02889.x. [DOI] [PubMed] [Google Scholar]
- 73.Leung D, Abbenante G, Fairlie DP. Protease Inhibitors: Current Status and Future Prospects. J Med Chem. 2000;43(3):305–341. doi: 10.1021/jm990412m. [DOI] [PubMed] [Google Scholar]
- 74.Kane RC, Farrell AT, Sridhara R, Pazdur R. United States Food and Drug Administration Approval Summary: Bortezomib for the Treatment of Progressive Multiple Myeloma After One Prior Therapy. Clin Cancer Res. 2006;12(10):2955–2960. doi: 10.1158/1078-0432.CCR-06-0170. [DOI] [PubMed] [Google Scholar]
- 75.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Bortezomib: Proteasome Inhibition as an Effective Anticancer Therapy. Annu Rev Med. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 76.Sanderson PE. Small, Noncovalent Serine Protease Inhibitors. Med Res Rev. 1999;19(2):179–197. doi: 10.1002/(sici)1098-1128(199903)19:2<179::aid-med4>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 77.Andrews DM, Chaignot HM, Coomber BA, Dowle MD, Hind SL, Johnson MR, Jones PS, Mills G, Patikis A, Pateman TJ, Robinson JE, Slater MJ, Trivedi N. The Design of Potent, Non-Peptidic Inhibitors of Hepatitis C Protease. Eur J Med Chem. 2003;38(4):339–343. doi: 10.1016/s0223-5234(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 78.Janc JW, Clark JM, Warne RL, Elrod KC, Katz BA, Moore WR. A Novel Approach to Serine Protease Inhibition: Kinetic Characterization of Inhibitors Whose Potencies and Selectivities are Dramatically Enhanced by Zinc(II) Biochemistry. 2000;39(16):4792–4800. doi: 10.1021/bi992182j. [DOI] [PubMed] [Google Scholar]
- 79.Louie AY, Meade TJ. Metal Complexes as Enzyme Inhibitors. Chem Rev. 1999;99(9):2711–2734. doi: 10.1021/cr9804285. [DOI] [PubMed] [Google Scholar]
- 80.So rich is the subject of medicinal inorganic chemistry that the entire issue of Chem. Rev. containing reference 79 (Vol. 99, Iss. 9) is dedicated to just this subject.
- 81.Hall MD, Mellor HR, Callaghan R, Hambley TW. Basis for Design and Development of Platinum(IV) Anticancer Complexes. J Med Chem. 2007;50(15):3403–3411. doi: 10.1021/jm070280u. [DOI] [PubMed] [Google Scholar]
- 82.Higby DJ, Wallace HJ, Jr, Albert D, Holland JF. Diamminodichloroplatinum in the Chemotherapy of Testicular Tumors. J Urol. 1974;112(1):100–104. doi: 10.1016/s0022-5347(17)59652-4. [DOI] [PubMed] [Google Scholar]
- 83.Cushman DW, Ondetti MA, Gordon EM, Natarajan S, Karanewsky DS, Krapcho J, Petrillo EW., Jr Rational Design and Biochemical Utility of Specific Inhibitors of Angiotensin-Converting Enzyme. J Cardiovasc Pharmacol. 1987;10(Suppl 7):S17–30. doi: 10.1097/00005344-198706107-00004. [DOI] [PubMed] [Google Scholar]
- 84.Byrd CM, Hruby DE. Viral Proteinases: Targets of Opportunity. Drug Dev Res. 2006;67:501–510. [Google Scholar]
- 85.Hsu JT, Wang HC, Chen GW, Shih SR. Antiviral Drug Discovery Targeting to Viral Proteases. Curr Pharm Des. 2006;12(11):1301–1314. doi: 10.2174/138161206776361110. [DOI] [PubMed] [Google Scholar]
- 86.Yang S, Chen SJ, Hsu MF, Wu JD, Tseng CT, Liu YF, Chen HC, Kuo CW, Wu CS, Chang LW, Chen WC, Liao SY, Chang TY, Hung HH, Shr HL, Liu CY, Huang YA, Chang LY, Hsu JC, Peters CJ, Wang AH, Hsu MC. Synthesis, Crystal Structure, Structure-Activity Relationships, and Antiviral Activity of a Potent SARS Coronavirus 3CL Protease Inhibitor. J Med Chem. 2006;49(16):4971–4980. doi: 10.1021/jm0603926. [DOI] [PubMed] [Google Scholar]
- 87.Way JC. Covalent Modification as a Strategy to Block Protein-Protein Interactions With Small-Molecule Drugs. Curr Opin Chem Biol. 2000;4(1):40–46. doi: 10.1016/s1367-5931(99)00049-6. [DOI] [PubMed] [Google Scholar]
- 88.Overington JP, Al-Lazikani B, Hopkins AL. How Many Drug Targets Are There? Nat Rev Drug Discov. 2006;5(12):993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 89.Marchese A, Chen C, Kim YM, Benovic JL. The Ins and Outs of G Protein-Coupled Receptor Trafficking. Trends Biochem Sci. 2003;28(7):369–376. doi: 10.1016/S0968-0004(03)00134-8. [DOI] [PubMed] [Google Scholar]
- 90.Storey RF. Biology and Pharmacology of the Platelet P2Y12 Receptor. Curr Pharm Des. 2006;12(10):1255–1259. doi: 10.2174/138161206776361318. [DOI] [PubMed] [Google Scholar]
- 91.Drahl C, Cravatt BF, Sorensen EJ. Protein-Reactive Natural Products. Angew Chem Int Ed Engl. 2005;44(36):5788–5809. doi: 10.1002/anie.200500900. [DOI] [PubMed] [Google Scholar]
- 92.Evans MJ, Cravatt BF. Mechanism-Based Profiling of Enzyme Families. Chem Rev. 2006;106(8):3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 93.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme Activity Profiles of the Secreted and Membrane Proteome that Depict Cancer Cell Invasiveness. Proc Natl Acad Sci U S A. 2002;99(16):10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jessani N, Young JA, Diaz SL, Patricelli MP, Varki A, Cravatt BF. Class Assignment of Sequence-Unrelated Members of Enzyme Superfamilies by Activity-Based Protein Profiling. Angew Chem Int Ed Engl. 2005;44(16):2400–2403. doi: 10.1002/anie.200463098. [DOI] [PubMed] [Google Scholar]
- 95.Berkers CR, Verdoes M, Lichtman E, Fiebiger E, Kessler BM, Anderson KC, Ploegh HL, Ovaa H, Galardy PJ. Activity Probe for In Vivo Profiling of the Specificity of Proteasome Inhibitor Bortezomib. Nat Methods. 2005;2(5):357–362. doi: 10.1038/nmeth759. [DOI] [PubMed] [Google Scholar]
- 96.Orry AJ, Abagyan RA, Cavasotto CN. Structure-Based Development of Target-Specific Compound Libraries. Drug Discov Today. 2006;11(5–6):261–266. doi: 10.1016/S1359-6446(05)03717-7. [DOI] [PubMed] [Google Scholar]
- 97.Kaila N, Somers WS, Thomas BE, Thakker P, Janz K, DeBernardo S, Tam S, Moore WJ, Yang R, Wrona W, Bedard PW, Crommie D, Keith JC, Jr, Tsao DH, Alvarez JC, Ni H, Marchese E, Patton JT, Magnani JL, Camphausen RT. Quinic Acid Derivatives as Sialyl Lewis(x)-Mimicking Selectin Inhibitors: Design, Synthesis, and Crystal Structure in Complex With E-Selectin. J Med Chem. 2005;48(13):4346–4357. doi: 10.1021/jm050049l. [DOI] [PubMed] [Google Scholar]
- 98.Firth-Clark S, Willems HM, Williams A, Harris W. Generation and Selection of Novel Estrogen Receptor Ligands Using the De Novo Structure-Based Design Tool, SkelGen. J Chem Inf Model. 2006;46(2):642–647. doi: 10.1021/ci0502956. [DOI] [PubMed] [Google Scholar]
- 99.Barreiro G, Kim JT, Guimaraes CR, Bailey CM, Domaoal RA, Wang L, Anderson KS, Jorgensen WL. From Docking False-Positive to Active Anti-HIV Agent. J Med Chem. 2007;50(22):5324–5329. doi: 10.1021/jm070683u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Katritch V, Byrd CM, Tseitin V, Dai D, Raush E, Totrov M, Abagyan R, Jordan R, Hruby DE. Discovery of Small Molecule Inhibitors of Ubiquitin-Like Poxvirus Proteinase I7L Using Homology Modeling and Covalent Docking Approaches. J Comput Aided Mol Des. 2007;21(10–11):549–558. doi: 10.1007/s10822-007-9138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lodola A, Mor M, Rivara S, Christov C, Tarzia G, Piomelli D, Mulholland AJ. Identification of Productive Inhibitor Binding Orientation in Fatty Acid Amide Hydrolase (FAAH) by QM/MM Mechanistic Modelling. Chem Commun (Camb) 2008;2:214–216. doi: 10.1039/b714136j. [DOI] [PubMed] [Google Scholar]
- 102.Oballa RM, Truchon JF, Bayly CI, Chauret N, Day S, Crane S, Berthelette C. A Generally Applicable Method for Assessing the Electrophilicity and Reactivity of Diverse Nitrile-Containing Compounds. Bioorg Med Chem Lett. 2007;17(4):998–1002. doi: 10.1016/j.bmcl.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 103.Chmura AJ, Orton MS, Meares CF. Antibodies with Infinite Affinity. Proc Natl Acad Sci U S A. 2001;98(15):8480–8484. doi: 10.1073/pnas.151260298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural Bioinformatics-Based Design of Selective, Irreversible Kinase Inhibitors. Science. 2005;308(5726):1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]