Abstract
The p53 tumor suppressor plays pivotal role in the organism by supervising strict compliance of individual cells to needs of the whole organisms. It has been widely accepted that p53 acts in response to stresses and abnormalities in cell physiology by mobilizing the repair processes or by removing the diseased cells through initiating the cell death programs. Recent studies, however, indicate that even under normal physiological conditions certain activities of p53 participate in homeostatic regulation of metabolic processes and that these activities are important for prevention of cancer. These novel functions of p53 help to align metabolic processes with the proliferation and energy status, to maintain optimal mode of glucose metabolism and to boost the energy efficient mitochondrial respiration in response to ATP deficiency. Additional activities of p53 in non-stressed cells tune up the antioxidant defense mechanisms reducing the probability of mutations caused by DNA oxidation under conditions of daily stresses. The deficiency in the p53-mediated regulation of glycolysis and mitochondrial respiration greatly accounts for the deficient respiration of the predominance of aerobic glycolysis in cancer cells (the Warburg effect), while the deficiency in the p53-modulated antioxidant defense mechanisms contributes to mutagenesis and additionally boosts the carcinogenesis process.
1. Introduction
The p53 tumor suppressor plays a key role in securing genetic stability. The importance of p53 is underscored by the fact that its activity is lost in the vast majority of human cancers [1]. While in nearly half of all human cancers the p53 gene itself is mutated leading to accumulation of dysfunctional protein, in the other half there can be found other abnormalities within the p53 pathway that compromise its tumor suppressor functions [2]. Germline mutations of the p53 gene in Li-Fraumeni syndrome are associated with tremendous susceptibility to cancer [3]. Similarly, p53 −/− mice demonstrate a cancer-prone phenotype and severe karyotype instability [4].
Decades of intense studies have established a role for p53 as a stress-induced protein that protects genetic stability by restricting proliferation, motility and viability of abnormal or stress-exposed cells [5]. As the p53 knockout mice look and develop normally, it was initially concluded that functions of the p53 gene are dispensable for normal physiology. Recent studies however, while not questioning the importance of stress-induced p53 in protection against cancer, suggest additional important roles for p53 in physiology of normal non-stressed cells. The p53 is tightly involved in the homeostatic regulation of energy-producing processes, coordination of overall rate of biosynthesis, and mobilization of defenses against reactive oxygen species (ROS). Together with the stress-induced functions of p53, which eliminate the effects of existing damage, the novel functions enforce preventive mechanisms that reduce probability of mutations. These functions address daily hazards to which a cell is exposed under charge of normal physiological processes. Indeed, excluding extreme conditions such as excessive radiation and treatment with genotoxic drugs, the damages generated by normal physiological processes constitute major hazards leading to cancer and driving the ageing process. In the present review we will describe recently identified mechanisms by which p53 affects the overall rate of biosynthesis, regulates energy metabolism and controls the intracellular redox status.
2. Modulation of p53 activity in response to stresses
In non-stressed cells the level of p53 is low, owing to both the ubiquitin-mediated degradation in 26S proteasomes, through processes controlled by Mdm2 and some other E3 ubiquitin ligases [6], and by the ubiquitin-independent degradation by default in 20S proteasomes [7]. p53 is induced in response to genotoxic influences [8], such as γ-radiation, UV, genotoxic drugs and oxidative stress. The p53 response can be also triggered by a variety conditions that challenge genome integrity indirectly, such as defective cytoskeleton, altered cell adhesion, depleted pools of intermediates, activated oncogenes, etc.[9, 10].
p53 acts mainly as a transcription factor that activates or represses several functional clusters of genes [11]. The p53 inhibitor Mdm2 is induced by p53, forming a negative autoregulatory loop, which forces degradation of p53, as its activity increases [12]. Some signals activate enzymes that modify p53 and prevent its interaction with Mdm2 protein [13], while other signals prevent degradation of p53 by inducing p14ARF, which blocks Mdm2 [14]. In addition, stabilization of p53 can be mediated through the induction of NADPH: quinone oxidoreductases NQO1 and NQO2, which prevent degradation of p53 in 20S proteasomes [15]. The transcriptional activity of p53 itself can be modulated by a wide array of modifying enzymes and interacting proteins that modify DNA-binding preferences and affect the consequences of p53 binding to DNA [16].
The p53 activation leads to certain functional outcomes providing optimal solution to the particular condition that initiated the p53 response [9]. The outcomes are mediated by products of the genes that p53 controls as a transcription factor. Several lines of studies identify more than 500 genes that are either stimulated, or repressed by p53 [11, 17–19], although the spectrum of the genes is strongly cell and tissue specific [20].
The p53 regulated genes can be divided into several functional groups. There are scores of p53 responsive genes that participate in intrinsic and extrinsic cell death pathways [21]. A large group of p53 regulated genes act to generate high levels of ROS, which potentiate the apoptotic response [22]. Certainly, upon grave insults and damages, the best solution would be safe elimination of irretrievable yet dangerous cells. However, the outcome of p53 response greatly depends on cell context: in certain cell types lethal stresses induce terminal senescence rather than apoptosis [23]. A large group of p53 regulated genes is capable of arresting the cells at specific points of the cell cycle, thus giving the stressed cell time to repair a damage. Another group of p53-regulated genes controls DNA repair factors. In combination with reversible cell cycle arrest the induced DNA repair genes help in recovering from damage before resuming normal cell divisions. The recovering process is facilitated by several p53-regulated genes (such as MDM2, cyclin G, Siah1 etc.) participating in negative autoregulatory loops [12]. Products of these genes simultaneously communicate with other pathways in the signaling network, and coordinate responses to stress conditions.
Communication of a stressed cell with its environment is achieved through a large set of p53-regulated genes encoding secreted proteins. The proteins can prevent spreading and dissemination of abnormal cells by inhibiting angiogenesis, modifying extracellular matrix, or affecting cell motility [20]. The p53-regulated activation of exosomes [24] contributes to communication between affected cells and the environment and assists in presenting antigens of abnormal cells to immune system, thus helping in immunizing the host against cancer antigens [20].
3. p53 is supervising pathways that control biosynthesis rate
Recent studies reveal that p53 has much broader capacities in controlling basic processes within the cell. Under conditions of mild physiological stresses or even without any stresses at all p53 acts to adjust overall rate of biosynthesis with energy status of the cell and the availability of nutrients, growth factors and hormones. Apparently, these activities relate to pro-survival functions of p53 that act to maintain healthy homeostasis and to delay the ageing process.
Cell growth, proliferation and specific cellular functions require sufficient supply of oxygen and nutrients. Both the external supply of nutrients and the functional demands of the organism vary in great extent. In times of food limitations or extensive energy spending the viability of an organism depends on homeostatic mechanisms that mobilize internal resources by shutting down dispensable processes and by switching certain metabolic reactions.
The mTOR pathway plays critical role in switching between catabolic and anabolic processes. Stimulation of the mTOR by availability of nutrients, growth factors and hormones leads to a more robust protein synthesis that boosts cell growth and proliferation. In contrast, ATP depletion and limited nutrients switches metabolism to a slower but more energy efficient aerobic respiration and mobilizes catabolic processes, such as macroautophagy, thus lowering cell’ dependence from external energy sources. The mTOR pathway is regulated by sensing and integrating signals that measure levels of nutrients, growth factors and hormones, and overall energy status of the cell. The signals are delivered through the activity of two opposing upstream kinases, the Akt (or protein kinase B), which depends on availability of nutrients and growth factors and the AMP-dependent kinase (AMPK), which is activated by the depleted energy status. The p53 tumor suppressor exerts additional level of regulation over the mTOR pathway, thereby integrating the metabolic signals to a broader context and resolving cell’ fate according to needs of the whole organism.
3.1. The AMP-dependent kinase (AMPK) mediates a p53-dependent metabolic checkpoint
Limited availability of glucose leads to exhaustion of intracellular energy resources, which is characterized by low ATP:AMP ratio. During intense consumption of ATP, adenylate cyclase compensates the deficiency by converting two molecules of ADP to AMP and ATP. Therefore, ratio of AMP to ATP is the true indicator of the energy charge.
AMPK keeps energy balance within the cell, as well as the whole-body energy metabolism [25]. Through phosphorylation of multiple substrates, AMPK turns off ATP-consuming pathways and simultaneously activates energy production. Particularly, AMPK phosphorylates and inhibits HMG-Co reductase and acetyl CoA carboxylase (ACC), thereby suppressing cholesterol and fatty acid synthesis, inhibits protein synthesis through phosphorylation of TSC2, an upstream repressor of mTOR, inhibits glucose synthesis in the liver and attenuates insulin secretion. Simultaneously, AMPK stimulates glucose uptake, fatty acid oxidation and mitochondrial respiration [26].
The AMPK consists of a catalytic (α) and two regulatory (β and γ) subunits [27]. AMPK is slightly activated by binding of AMP to its γ subunit. However, a dramatic activation is achieved by phosphorylation of the α-subunit at Thr172 [28] by Ca2+/calmodulin sensitive kinase and by a constitutively active kinase LKB1. Loss of function of the LKB1 kinase is implicated in a variety of human cancers, placing it among major tumor suppressor genes [29]. Due to the stimulation by the upstream kinases the AMPK is active by default. The modulation of AMPK in response to AMP is mediated mainly through the activity of protein phosphatase 2C (PP2C), which inhibits AMPK by dephosphorylating Thr172. Binding of AMP to AMPK makes it a worse substrate for PP2C, thereby increasing its enzymatic activity [28, 30].
Functions of AMPK and p53 are tightly linked. In response to a DNA-damage p53 activates transcription of the genes for β subunits of AMPK [31]. The β-subunits function as a scaffold for binding of α and γ subunits, and modulate localization and activity of AMPK [32]. Furthermore, p53 itself is controlled by AMPK and by its upstream kinase LKB1. The LKB1 kinase forms complex with p53 and directly or indirectly phosphorylates its Ser15 and Ser392 [33]. In the complex with p53 LKB1 binds to control regions of the p21/CDKN1A and other p53-regulated genes, and participates in transcriptional activation by phosphorylating components of chromatin or the general transcriptional machinery [33]. Activated AMPK can modify p53, and in addition it somehow induces activation of the p53 gene promoter [34], which further enhanced the p53 response.
Normal fibroblasts display a reversible p53-dependent cell-cycle arrest at the G1/S boundary when placed into low-glucose medium. The cell-cycle arrest depends on the activity of AMPK, but it occurs even under conditions when the mTOR pathway is not inhibited. The delay in cell cycle progression represents a checkpoint that restricts further divisions when energy resources are exhausted. This metabolic checkpoint is absent in the p53−/− cells. Moreover, the p53 +/+ fibroblasts that grow in low glucose are relatively resistant to apoptosis upon complete removal of glucose, as opposed to p53−/− fibroblasts, which are not protected [35]. Therefore, p53 is required for the adaptive response to limited glucose availability that enhances the viability of cells under conditions of glucose deprivation.
The metabolic checkpoint involves direct phosphorylation of p53 by AMPK at Ser15, leading to stabilization and transcriptional activation of p53 [35]. Noteworthy, AMPK- and p53-dependent responses to low glucose differ substantially depending on the cell type. In normal thymocytes and in a human osteosarcoma cell line glucose deprivation is associated with phosphorylation of p53 at Ser46 leading to apoptosis [34]. Similarly, apoptotic cell death prevails in oncogene-transformed cells that have inactive pRB and defective checkpoints, pointing to a role of cell-cycle arrest in the pro-survival function of p53 [31]. Meanwhile, in cell types that sustain cell cycle arrest the viability of starved cells may be promoted by the stimulation of p53-dependent autophagy [36], which mobilizes nutrients by digesting part of their cytoplasm [37] (see below).
3.2. p53-mediated control of the IGF-1 - Akt pathway
Protein kinase B or Akt delivers survival signals in response to growth factors and hormones. The pathway is regulated by insulin-like growth factor (IGF-1) or by insulin, which signal the availability of nutrients in the organism. Binding of IGF-1 to its tyrosine kinase receptor IGF-1R induces recruitment of active phosphoinositide 3-kinase (PI3K), which increases local concentration of PIP3 at the plasma membrane. The PIP3 tethers to plasma membrane proteins containing the pleckstrin-homology (PH) domain, such as Akt and PDK1. As the result of co-localization the PDK1 phosphorylates Akt at Thr308, which stimulates its kinase activity. However, full activation of Akt is achieved when it is phosphorylated at Ser473 by the kinase complex mTORC2 [38].
The induction of Akt by PI3K is reversed by dephosphorylation of PIP3 by lipid phosphatase PTEN. Without the activity of PTEN the pathway remains permanently active, explaining why PTEN is a frequently mutated tumor suppressor gene in cancer [39].
The Akt modulates a variety of cellular functions through phosphorylation of different proteins [38, 40]. Particularly, Akt stimulates cell proliferation by blocking p27kip1 and by activating c-myc and cyclin D1. It counteracts-apoptosis through the inhibition of Bad, Mdm2 and FoxO proteins and through activation of NFκB. Akt can stimulate aerobic glycolysis, by increasing concentration of glucose transporters Glut1 and Glut4 at the plasma membrane [41, 42] and by stimulating the mitochondria-associated hexokinase [43], which initiates glycolysis and pentose-phosphate pathway (PPP). Akt activates ATP citrate lyase, thereby enhancing the de novo synthesis of fatty acids [44] necessary for building membranes in rapidly growing cells. The Akt kinase phosphorylates FoxO transcription factors, leading to their inhibition by nuclear exclusion and to downregulation of the enzymes involved in gluconeogenesis – PEPCK and glucose-6-phosphatase. Finally, Akt is a strong activator of the mTOR pathway, which is described below.
p53 can negatively regulate the IGF-1-Akt pathway. One of the p53-regulated genes is IGF-BP3 [45], which encodes a major IGF-1 carrying protein and a modulator of its bioactivity in the circulation. IGF-BP3 is a strong inhibitor of IGF-1 in extravascular tissue compartment, where it is locally secreted in a controlled manner [46]. By inducing the IGF-BP3 expression p53 attenuates the IGF-1 signaling, not only in the same cell, but also in the neighborhood, producing a bystander effect. The PTEN gene is a p53-activated gene [47]. Therefore, conditions leading to activation of p53 attenuate the activity of IGF-1/Akt pathway and induce substantial changes to overall metabolism of the cell. By stimulating PTEN p53 downregulates Akt, leading to reduced glycolysis and fatty acids synthesis and enhanced β-oxidation of lipids.
3.3. p53 and the mTOR pathway
Mammalian target of rapamycin (mTOR) serves as a major integrator of signals measuring intracellular energy level, availability of nutrients and growth factors. It stimulates cell growth promoting ribosome biogenesis, mRNA translation, and by inhibiting autophagy [38, 48, 49]. mTOR acts as a catalytic subunit of two distinct Ser/Thr kinase complexes– mTORC1 and mTORC2 [48]. Of these complexes only the mTORC1 is responsible for the control of protein synthesis and cell growth [38], while the mTORC2 regulates cytoskeleton through the stimulation of F-actin stress fibers, paxillin, RhoA, Rac1, Cdc42, and PKCα [50]. The mTORC2, along with the PDK1, also serves as an upstream kinase in the activation of Akt [38].
The mTORC1 is positively regulated by growth factors [48, 51] and inhibited by nutrient limitation and stresses, such as hypoxia, oxidative stress, DNA damage [51, 52]. In addition to mTOR, the mTORC1 complex includes PRAS40, mLST8 and Raptor. The most important effectors downstream of mTORC1 are the p70 ribosomal protein S6 kinase1 (S6K1) and the eIF4E binding protein 1 (4EBP1). Phosphorylation by the mTORC1 activates the S6K1, and inactivates the 4EBP1 inhibitor of cap-dependent translation. The phosphorylation of these two proteins stimulates ribosome assembly and protein translation and promotes cell growth [51].
The mTORC1 activity is modulated through the complex of TSC1 (hemartin) and TSC2 (tuberin) proteins, which represent products of tumor suppressor genes commonly mutated in tuberous sclerosis, a rare genetic disease characterized by multiple benign tumors, hemartomas, in brain and other vital organs [53]. Acting as a rheostat, the TSC1/2 complex regulates the mTOR, restraining cell growth under stress conditions, and releasing the inhibition when the conditions are favorable for growth. Mechanistically, the TSC2 subunit serves as a GTPase activating protein (GAP) for the small GTPase protein Rheb, which activates the mTOR when it is GTP-bound [48, 52].
The Akt kinase serves as a positive regulator of cell growth through activation of the mTORC1. Phosphorylation of TSC2 by Akt leads to the disruption of its complex with TSC1 and binding to 14-3-3 proteins [54]. As result of the inhibited GAP activity of TSC2 the GTP-bound Rheb stimulates the mTOR kinase. In addition, Akt stimulates the mTOR pathway through the phosphorylation of the mTORC1 inhibitor PRAS40 [55].
An opposite effect is induced by the AMPK, which is an upstream inhibitor of the mTORC1. The AMPK phosphorylates the TSC2 at the amino acid residue that is different from the one phosphorylated by the Akt, which serves as prerequisite for additional activating phosphorylation of the TSC2 at multiple sites by the GSK-3 [56]. This leads to stimulation of the Rheb-GAP activity and inhibition of mTOR [57]. Noteworthy, the TSC2-inhibiting effect of AMPK is dominant over the stimulating effects of Akt and Erk [26], which means that in the absence of energy resources the mechanism prevents stimulation of protein synthesis by positive signals. In addition to TSC2 the AMPK can also target the mTORC1 by phosphorylating Raptor [58]. Therefore, like Akt, the AMPK regulates mTOR by targeting both the TSC1/2 complex and the mTORC1, although the outcomes are opposing.
The mTOR pathway is negatively regulated by the p53 tumor suppressor. Low energy status induces AMPK, which activates p53. In response, p53 arrests the cell cycle, and shuts down the unnecessary protein synthesis through the inhibition of mTOR.
There are several components of the mTOR pathway that are affected by p53. It has been mentioned that p53 upregulates the β subunits of AMPK, which translates into the AMPK-induced activation of TSC2, inhibition of Raptor, and downregulation of the mTOR kinase. Another mechanism involves activation of AMPK by p53-modulated Sesn1 and Sesn2 [59]. In addition to their role in the p53-mediated antioxidant activity [60](see below) sestrins interact with the catalytic α-subunit of AMPK and promote its AMP and LKB1 independent phosphorylation. Apparently, binding of sestrins to AMPK inhibits the dephosphorylation of Thr172 by PP2C, and maintains high activity of the AMPK. Upregulation of the AMPK increases phosphorylation of p53, thereby amplifying the effect. Moreover, sestrins bind to TSC2 and promote its interaction with AMPK, leading to selective inactivation of the mTOR pathway. This mechanism suggests that during metabolic stress the AMPK-induced activation of p53 not only amplifies the effect, but also assists in more focused targeting of the mTOR pathway.
Finally, the TSC2 itself is a p53-regulated gene, being induced by the conditions that increase activity of p53 [31, 61]. It should however be kept in mind that although p53 can inhibit the mTOR pathway at many levels, the effect has strong tissue specificity, suggesting that final effects of the p53 induction on activity of the mTOR pathway will depend both on the cell type and the degree of p53 induction [31].
3.4. Role of p53 in autophagy
Prolonged nutrient deprivation initiates a self-eating process, macroautophagy (autophagy), which provides cells with nutrients needed for their survival, through sacrificing some of the existing cell mass. Autophagy is a membrane-trafficking process that includes formation of autophagosomes, double-membrane vesicles that engulf certain volumes of cytoplasm - portions of endoplasmic reticulum, endosomes and mitochondria. The autophagosomes fuse with lysosomes to form autolysosomes in which the engulfed structures are digested to provide nutrients from the carbohydrate, lipid and protein turnover [37]. In addition, the autophagy helps to remove certain waste material, such as damaged organelles and oxidized proteins. Defective mitochondria elicit certain diffusible molecules and ROS that serve as signals to initiate the autophagy [62], leading to the removal of unwanted sources of mutations that fuel the ageing process [63, 64]. Although in many instances autophagy helps to maintain viability, in certain cell contexts its components are required for apoptosis [65].
Autophagy is a tightly regulated evolutionary conserved process [66]. The mTOR pathway is implicated in the negative regulation of autophagy, although the exact mechanisms are not completely understood [67]. The p53 has a rather complex relation to autophagy. First, p53 can activate autophagy through inhibition of the mTOR pathway [61]. Second, p53 can induce autophagy through a p53-activated gene DRAM. Forced expression of DRAM in p53-negative cells can stimulate autophagy, while the effect is inhibited by RNAi [68]. Remarkably, autophagy may have a pro-survival role [69], as ectopic expression of DRAM results in substantial increase in clonogenicity [70]. However, DRAM was also shown to be required for the p53 dependent apoptosis [68] suggesting the relation of autophagy to cell death pathways.
Recent studies suggest that other components of the p53 pathway may also contribute to the autophagy process. Transcriptionally active p53 family member TA-p73 can upregulate autophagy in a DRAM-independent manner [71]. Autophagy can be also triggered by ARF, which is a strong inducer of p53 in response to oncogene activation [72]. However, the process is only partially p53 dependent, as ARF can induce autophagy even in the absence of p53 [72]. The induction of autophagy is even greater in response to a shorter isoform of ARF (the smARF), which unlike the full-length nucleolar ARF resides in mitochondria [73].
Remarkably, the autophagy can be also triggered by the inhibition of p53 [74]. It was found that while the transcriptionally active nuclear p53 promotes autophagy, the cytoplasmic form of p53 is a strong inhibitor of autophagy [75]. Balanced action of the two forms of p53 makes the cells sensitive to autophagy induction in response to starvation or fasting, while in p53-knockout cells the level of autophagy is permanently high and cannot be modulated [74].
The mechanism for constitutively hyperactive autophagy in p53-deficient cells is not clear. Perhaps, the deficiency in p53 may compromise certain functions and create signals for autophagy. Particularly, p53 is required for normal mitochondrial biogenesis [76], base excision repair of mitochondrial DNA [77] and for maintaining transcription of AIF [78] and SCO2 [79] genes. The AIF gene encodes the “apoptosis inducing factor”, a NADH oxidase in the inner mitochondrial membrane [80], which participates in oxidative phosphorylation by assisting proper assembly of the mitochondrial respiratory complex I [81]. In response to strong insults there is a PARP-1 dependent translocated of AIF from mitochondria to nuclei, which has a pro-apoptotic effect [82]. Although, AIF may play a cytoprotective function under conditions of physiological stresses [78]. Similarly, the product of the SCO2 gene participates in oxidative phosphorylation [83] by assisting the assembly of mitochondrial respiratory complex IV [79]. Certainly, the deficiency in these p53 regulated proteins can compromise mitochondrial functions, leading to the induction of autophagy.
4. Functions of the p53 tumor suppressor control aerobic respiration and glycolysis
Energy demands of cells vary substantially depending on their tissue origin, current physiological condition, proliferation status etc. In normal cells glucose is the major external source of energy. Energy of glucose is converted into energy of ATP. Glycolysis, an ancient anaerobic process in the cytoplasm, produces two molecules of pyruvate and only two molecules of ATP. Aerobic mitochondrial respiration finalizes glucose oxidation by yielding nearly 30 molecules of ATP. Despite its high energy efficiency the aerobic respiration is a slow process, compared to glycolysis, which may be a robust source of energy. Also, in addition to providing energy the mitochondrial TCA cycle serves as major source of intermediates used for anabolic purposes. Continuous withdrawal of components from the process would stop its recycling, unless it is replenished with suitable metabolites [84]. Therefore, despite the apparently higher efficiency of aerobic respiration, glycolytic production of ATP may become preferable under certain conditions that require either rapid release of energy, such as intense contractions of skeletal muscular fibers [85], or massive biosynthesis of cellular structures (membranes, organelles) in rapidly proliferating cells [86]. Certainly, optimal balance between glycolytic and mitochondrial branches of energy metabolism is a subject to tight regulation [87].
Recent studies suggest that the p53 tumor suppressor has multiple roles in aligning energy metabolism with current physiological condition and proliferation status of cells [83, 88], and that the effect of p53 is observed under apparently normal non-stressful conditions. Inactivation of p53 leads to greater dependence of cells from the glycolytic energy production and to substantial impairment of the aerobic mitochondrial functions [76, 89, 90]. As was described above, under the conditions of low energy or deficient growth stimulation the activity of p53 shuts down anabolic processes to complement the inhibitory effect of p53 on cell cycle progression. The effect of p53 on energy metabolism is seen even without any stresses. However, it does not exclude a possibility of certain p53-dependent modulation of energy metabolism by changing physiological conditions.
4.1. p53 affects mitochondrial energy production
Inhibition of p53 results in substantial deficiency in mitochondrial biogenesis [76], decrease in oxygen consumption [79], and stimulation of glycolysis, which is manifested by the increased lactate production. Although the overall level of ATP in the p53−/− cells does not change, it was found that in the p53+/+ cells the proportion of ATP produced from glycolysis vs. mitochondrial respiration is 1:3, while in the p53−/− cells it is increased to 3:1 [79], suggesting that the mitochondrial ATP synthesis depends on p53 functions.
Search for the candidate genes that could mediate these effects of p53 has identified SCO2 gene. The gene is apparently upregulated by basal levels of p53 [79]. It encodes a copper chaperon protein required for the assembly of mitochondrial cytochrome c oxidase complex IV [91]. The decreased aerobic respiration in the p53-deficient cells can be rescued by the reexpression of SCO2 at physiological levels. Similarly, targeted disruption of one allele of the SCO2 gene reduces the mitochondrial ATP production and increases the glycolytic activity.
The results confirm the involvement of SCO2 in the p53 mediated control of aerobic ATP production, although do not exclude contribution of additional p53 regulated genes in the mitochondrial energy production. One of the candidate genes is the p53-regulated AIF, encoding a protein required for the assembly of mitochondrial respiratory complex I [81], although its involvement in energy production is still elusive.
As deficient mitochondrial functions in the p53−/− cells are easily compensated by the increased glycolysis, it suggests the coordinate regulation of the glycolytic and aerobic pathways. In addition to the known mechanisms of feed-back regulation, an array of the p53 regulated gene products affecting glucose metabolism contribute to this balance. It should however be kept in mind that the p53 dependent effects may be highly cell type specific explaining why some of the mechanisms cannot be reproduced in a particular cell system.
4.2. p53 modifies glycolytic pathways
The p53 functions affect glucose catabolism at multiple levels. p53 can in some systems retard glucose uptake by repressing transcription of GLUT1 and GLUT4 genes encoding glucose transporters [92]. In other instances, p53 can stimulate glucose uptake by increasing the transcription of Hexokinase II gene [93], which converts glucose to glucose-6 phosphate and serves as a key enzyme in the glycolytic pathway. At the first glance, this activity of p53 contradicts its function as tumor suppressor, as upregulation of Hexokinase II is widely observed in cancer cells [94]. However, mild upregulation of Hexokinase II may be part of the pro-survival function of p53 in normal cells under conditions of mild stress, when p53 helps to recover from metabolic checkpoint by refueling energy resources [95]. Phosphoglycerate mutase (PGM), an enzyme acting at the later stages of the glycolytic pathway by performing reversible rearrangement of phosphoglycerates, is also regulated by p53. Upregulation of PGM increases substantially the glycolytic flux [96]. The PGM gene contains a p53-responsive element that mediates transcriptional activation of PGM, at least in cardiomyocytes [97]. However, p53 is also able to repress the expression of PGM through a posttranscriptional mechanism [96], adding additional level of complexity to the regulation of glycolysis.
Recently yet another enzyme indirectly connected with glycolysis was identified as a p53-upregulated product. TIGAR gene encodes a protein that is homologous to bisphosphatase domain of a bifunctional enzyme 6-phosphofructo-2-kinase, an enzyme converting glucose-6-phosphate to fructose-2,6-bisphosphate [98]. The reaction is reversible, as the bisphosphatase domain of the same enzyme converts the fructose-2,6-bisphosphate to fructose-6-phosphate. The fructose-2,6-bisphosphate is a strong inhibitor of fructose-1,6-bisphosphatase, a regulatory enzyme in biosynthesis of glucose [99], suggesting that the p53-regulated TIGAR can potentially stimulate gluconeogenesis. However, the fructose-2,6-bisphosphate is also a strong allosteric activator of 6-phosphofructo-1-kinase, a key enzyme of glycolysis. By this mechanism the product of the p53-induced TIGAR can block glycolysis at the fructose-6-phosphate stage and divert the glucose catabolism toward the oxidative branch of the PPP. Interestingly, glucose-6-phosphate dehydrogenase (G6PD), which catalyzes a rate-limiting step in the PPP [100], is also activated by p53 [19]. Stimulation of PPP leads to increased synthesis of the nucleotides needed for the DNA repair, and to the enhanced accumulation of NADPH, which boosts the antioxidant defense. Therefore, inhibition of glycolysis and stimulation of PPP by p53 serves as important pro-survival mechanism that helps the cell to recover from minor injuries [88].
Among the first identified p53 targets was muscular creatine kinase gene (MCK) [101]. Functional p53 responsive element is also contained in the gene encoding the brain isoform of creatine kinase [102]. Creatine kinase restores ATP level by phosphorylating ADP and by consuming phosphocreatine, which serves as energy reservoir in tissues that are actively spending ATP, such as skeletal muscle, brain and smooth muscles. Therefore, restoration of ATP levels can also be the function of p53 that acts to maintain intracellular homeostasis and to promote cell survival.
Regulation of homeostatic functions of p53 would require milder changes in its activity compared to the more robust and deadly activation of p53 observed in response to severe stresses and insults. Results of in vitro studies suggest that energy status of the cell and major metabolic units may affect transcriptional activities of p53 in a feed-back manner, similar to the regulation of glycolytic enzymes by their metabolites. Specifically, it was found that ADP can bind to p53 tetramer and promote its DNA binding [103], while ATP (or GTP) and NAD+ can act in opposite manner [103, 104]. However, regulation of p53-dependent homeostatic functions requires additional studies.
4.3. p53 and the Warburg Effect
One of the prominent hallmarks of cancer cells is the constitutive switch in their energy mechanism from oxidative phosphorylation to the predominance of aerobic glycolysis. This phenomenon was first described by Otto Warburg [105, 106] and is known as the Warburg effect. Warburg proposed that carcinogenesis process starts when cells acquire irreversible injury to respiration, leading to selection of less differentiated and primitive cells that “grow wildly” and “succeed in replacing the irretrievably lost respiration by fermentation energy” [105].
After more than six decades we can argue that many different factors contribute to the Warburg effect in cancer. First, the switch from the apparently more energy saving oxidative phosphorylation to the more wasteful glycolytic fermentation is characteristic to many normal rapidly dividing cells, such as lymphocytes, hematopoietic, embryonic and other cell types [84, 107]. Apparently, the high glycolytic rate provides certain advantages for proliferating cells. Although the yield of ATP per glucose consumed is low, the rate of ATP production during glycolysis is much higher compared to oxidative phosphorylation [108]. Second, cell divisions require intermediates for biosynthetic pathways, including NADPH, citrate and glycerol for lipids, ribose sugars for nucleotides etc. While some of the intermediates originate from glycolysis and PPP, others are taken from the TCA cycle. Therefore, in proliferating cells much of the carbon that enters the TCA cycle is used for the biosynthetic processes, which consume rather than produce ATP. The shortage in ATP is covered by the excessive glycolysis. As the rate of glycolysis outpaces the maximal velocity of pyruvate oxidation, its excess is converted to lactate, which is secreted from the cell and can be used by other cells as an energy source. Recent findings suggest that p53 might play a role in adjusting metabolic processes in normal proliferating cells by diverting some of the glucose catabolites to PPP, which yields additional intermediates for nucleic acid biosynthesis, and NADPH for conversion of excessive pyruvate to lactate, synthesis of glutathione etc. By sensing decreased level of ATP p53 could stimulate oxidative phosphorylation and the production of ATP through the upregulation of the SCO2 gene. Similarly, the accumulation of NAD+ could allosterically activate p53 and stimulate the production of NADPH through the induction of TIGAR.
Although, the Warburg effect in cancer is not entirely explained by the normal mechanisms that regulate metabolism. Mutations leading to the irreversible breakage of different signaling pathways in the cell create distortions in the whole regulatory network. The abnormalities induce p53 response that restrict proliferation or induce apoptosis of affected cells. The eventual disruption of the p53 pathway occurring upon cancer progression lifts the ban for uncontrollable proliferation and permits the competition among the abnormal cells for the supply of nutrients and oxygen. The increased production and secretion of lactate from rapidly growing cells relying mostly on glycolysis confers additional advantage for cancer cells, as the acidification by lactic acid is toxic for normal cells but nontoxic for cancer cells [109]. Hence, there is further selection of the cells that maintain high level of aerobic glycolysis. Simultaneously, elimination of the p53 gene activity leads to the deficiency in oxidative phosphorylation due to the decreased levels of SCO2 and, possibly, of some other p53-regulated components of the respiratory chain [78, 90, 102, 110]. Therefore, the former suggestion made by Otto Warburg regarding the apparent functional deficiency of mitochondria in cancer cells appears to receive its final confirmation.
The deficiency in p53 in cancer cells abrogates the metabolic checkpoint that shuts down the mTOR pathway in response to depleted energy resources. The p53-dependent repression of the genes encoding glucose transporters, PGM and hexokinase is also relieved, while the TIGAR gene is downregulated, which collectively results in stimulation of the glycolytic flux. Hypoxia, which is one of the p53-inducing factors, can no longer inhibit proliferation of cancer cells, all the more so because the p53 dependent inhibition of angiogenesis is abrogated [111]. However, hypoxia induces HIF-1α transcription factor, which further stimulates glycolysis by boosting expression of many glycolytic enzymes and by promoting angiogenesis. Clearly, while many different factors contribute to the phenomenon of aerobic glycolysis predominance in cancer, the disruption of the p53 tumor suppressor function plays one of the central roles in the Warburg effect.
5. The antioxidant function of the p53 tumor suppressor
Organisms living in aerobic conditions utilize oxygen not only for the energy production through carbon oxidation. Oxygen can be metabolized into reactive oxygen species (ROS) that are highly reactive intermediates capable of modifying numerous biological substrates. Oxidation of lipids, proteins and nucleic acids by ROS damages cellular structures and represents a major hazard that fuels the aging process and leads to numerous pathologies. Despite the formerly widespread beliefs regarding ROS as mostly byproducts of mitochondrial respiration, detoxification by cytochrome P450, oxidation of fatty acids and other processes that involve redox reactions, recent studies indicate that substantial amounts of ROS are generated endogenously in response to various signals [112]. Hydrogen peroxide is an important signaling molecule that is produced by combined action of membrane bound NADPH oxidases and superoxide dismutase (SOD) following interaction of different membrane bound receptors with their extracellular ligands. The produced H2O2 performs its signaling role by oxidizing certain redox sensitive components of signaling pathways, such as protein phosphatases, proteinases, some transcription factors etc.
Several antioxidant systems participate in metabolism and homeostasis of ROS. Catalase and glutathione peroxidase are essential for rapid elimination of large quantities of ROS, which usually have exogenous sources. However, genetic studies with knockout models suggest that these highly efficient antioxidant enzymes are not critical for the protection against endogenously produced ROS, which represent major source for mutations and an important contributor to the aging process [112–114]. The latter function is performed by peroxiredoxins, a family of antioxidant thiol enzymes relying on thioredoxin system for their regeneration [115].
Changes in intracellular ROS mediate various responses, which depend greatly on cell type context and the level of ROS [116]. Exposure of cells to high levels of ROS leads to oxidative stresses, which among many other influences induce the p53 response. The induced p53 response leads to inhibition of cell proliferation, premature senescence or apoptotic cell death [88, 116]. Therefore, this function of p53 restricts further proliferation or viability of cells exposed to potentially mutagenic environment.
It is remarkable, that approximately one third of the genes that are highly responsive to H2O2 treatment represent transcriptional targets of p53 [117]. Also, among the genes induces by the p53 activation there is a number of genes, such as quinine oxidoreductases homologue PIG3 [22], proline oxidase PIG6 [118], ferredoxin reductase FDXR [119] whose products increase intracellular ROS and sensitize the cells to oxidative stress. These observations suggest that the pro-oxidant activity of p53 may facilitate apoptotic cell death through the oxidative degradation of mitochondrial components [22]. As the induction of apoptosis itself is accompanied by massive release of ROS, the other p53 regulated genes that are related to apoptosis induction can be also regarded as potentially pro-oxidant genes.
Unlike the condition of oxidative stress, mild increases in ROS may have stimulatory effect for certain functions, such as cell proliferation, motility etc. Abrogation of p53 functions in cancer may further contribute to the ROS-induced acceleration of proliferation and to stimulation of angiogenesis [120]. Following oncogenic transformation the permanent activation of growth promoting pathways can additionally boost the intracellular ROS level. In the absence of functional p53 the elevated ROS leads to increased oxidation of DNA, rapid accumulation of mutations and substantial genetic instability that further accelerates the oncogenic progression [121]. These results suggest that elimination of functional p53 may lead to progressive increase in intracellular ROS.
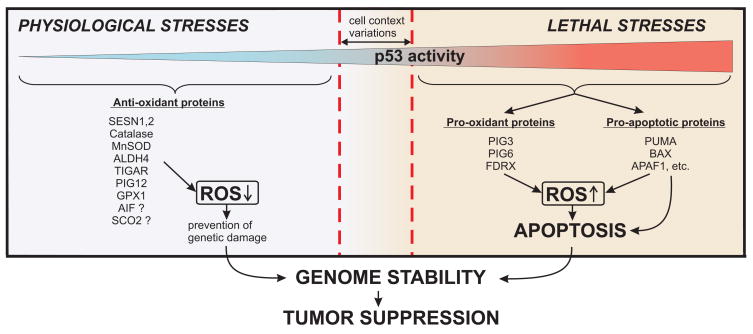
Recently a new model for the p53-dependent regulation of intracellular ROS levels has started to emerge that acknowledges a qualitatively different mode of p53 function in unstressed cells (Figure 2). Among the identified targets of p53 there are several genes with apparent anti-oxidant function. These are microsomal glutathione transferase homologue PIG12 [22], aldehyde dehydrogenase ALDH4A1 [122], glutathione peroxidase GPX1, Mn-superoxide dismutase SOD2 [123] and catalase [124]. In addition, two members of the sestrin family SESN1 (PA26) and SESN2 (Hi95) were also found to be regulated by p53 [125, 126]. Sestrins act as components of the peroxiredoxin regeneration system [60]. In tight cooperation with sulfiredoxin (Srx) the sestrins act as subunits of cysteinic sulfinyl reductase by regenerating inactive peroxiredoxins that overoxidize in response to massive bursts of H2O2 occurring during signal transduction.
Figure 2.
Levels of Reactive Oxygen Species (ROS) depend on p53 activity. Broken red line depicts critical level of p53 activity, which induces cell death pathways (varies in different tissues).
The contribution of these antioxidant products to p53 functions was elusive until it was found that in unstressed cells a p53 function is required for reducing the intracellular ROS levels [127]. Abrogation of p53 functions by means of RNAi, or by overexpression of dominant negative p53, Mdm2 or papilloma virus E6 gene product resulted in a substantial increase in intracellular ROS. Similar increases in ROS were observed in tissues of the p53 −/− mice. The increases in ROS in the p53-deficient cells correlated with substantial downregulation of the p53 regulated genes GPX1, SESN1 and SESN2, suggesting that basal physiological levels of p53 are sufficient for maintaining functional state of the genes [127, 128]. Basal levels of p53 were also found sufficient for maintaining the expression of catalase [124] and TIGAR [98]. As described above, TIGAR encodes a homologue of bisphosphatase domain of the 6-phosphofructo-2-kinase which shifts glucose catabolism from glycolysis to an alternative PPP. Stimulation of PPP boosts the production of NADPH, leading to accumulation of reduced glutathione, which additionally contributes to the antioxidant activity of p53 [16].
The antioxidant activity plays an important role in overall tumor suppressor function of p53. It was found that ROS elevation connected with the deficiency in p53 increases dramatically oxidation of nuclear DNA, and the rate of mutagenesis. These effects could be substantially reversed by the overexpression of sestrins or by the application of antioxidant N-acetylcysteine [127]. Moreover, dietary supplementation with N-acetylcysteine substantially improves karyotype stability and prevents malignant lymphomas in p53 −/− mice, which suggests that the deficiency in the antioxidant function represents the major cause of frequent tumors connected with the p53 deficiency, at least in the mouse system [127].
Although the antioxidant function of p53 is observed under apparently physiological conditions, it does not exclude certain level of regulation in response to changing intracellular redox status. In fact, p53 contains several redox-active cysteines in the DNA binding domain that contribute to the regulation [129]. A p53 interacting protein ARE/Ref-1 mediates a redox-dependent modification of the domain that affects DNA-binding specificity and transcriptional activity of p53 [130]. In addition, the activity of p53 can be regulated by reversible redox-dependent S-glutathionylation of the oxidation-sensitive cysteines [131]. However, more studies are needed to reveal details of the redox-dependent regulation of the p53 activity, which would help to understand the mechanisms controlling homeostatic functions of p53 in non-stressed cells.
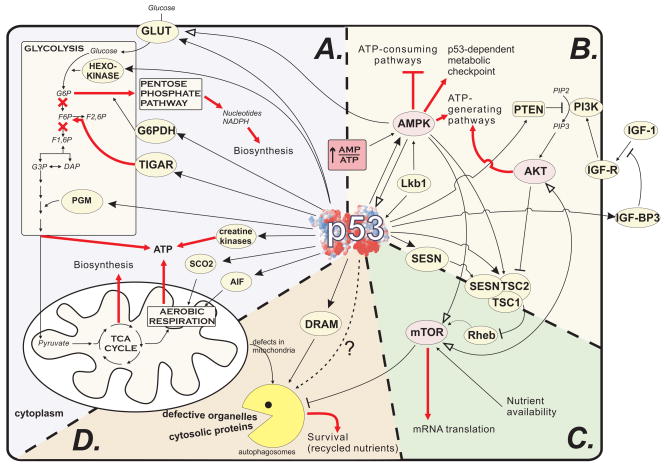
Figure 1.
P53 involvement in metabolism. A. Direct regulation of genes, which participate in major energetic pathways. Anabolic enzymes, regulated by p53. are not shown. GLUT – glucose transporters; G6PDH – glucose-6-phospate dehydrogenase; TIGAR – TP53-induced glycolysis and apoptosis regulator; PGM – phosphoglycerate mutase; SCO2 – synthesis of cytochrome c oxidase 2; AIF – apoptosis inducing factor; F2,6P – fructose-2,6-bisphosphate; B. Indirect modulation of cell metabolic activity through AMP-activated kinase (AMPK) and AKT kinase. IGF-1 – insulin-like growth factor 1; IGF-BP – IGF-binding protein; IGF-R – IGF-1 receptor; PI3K –phosphoinositol-3-kinase; PIP2/3 – phosphoinositol di/triphosphate; PTEN – phosphatase and tensin homolog, a PIP3 phosphatase; SESN –Sestrin 1 or 2; TSC1/2 – tuberous sclerosis protein 1/2, major regulators of mTOR activity; C. Indirect regulation of mTOR (mammalian target o rapamycin), a central cell growth, survival and motility regulator, by p53. RHEB – Ras homolog enriched in brain, a cell membrane anchored protein, the main regulator of mTOR activity. D. Control of autophagy by p53. DRAM – damage-regulated autophagy modulator. Bold red arrows indicate main outcomes of p53 metabolic activity. Arrows with large filled arrowheads depict direct transcriptional activation by p53. Arrows with large empty arrowheads show phosphorylation.
Acknowledgments
This work was supported by grants from the US National Institutes of Health R01CA10490 and R01AG025278 to P.M.C., from the Russian Basic Research Fund to P.M.C and J.E.K., from the Program on Molecular and Cellular Biology by the Russian Academy of Sciences to P.M.C. and J.E.K., and from the Howard Hughes Medical Institute to P.M.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Royds JA, Iacopetta B. p53 and disease: when the guardian angel fails. Cell Death Differ. 2006;13:1017–1026. doi: 10.1038/sj.cdd.4401913. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Lozano G, Zambetti GP. What have animal models taught us about the p53 pathway? J Pathol. 2005;205:206–220. doi: 10.1002/path.1704. [DOI] [PubMed] [Google Scholar]
- 4.Donehower LA. Genetic instability in animal tumorigenesis models. Cancer Surv. 1997;29:329–352. [PubMed] [Google Scholar]
- 5.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 6.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asher G, Reuven N, Shaul Y. 20S proteasomes and protein degradation “by default”. Bioessays. 2006;28:844–849. doi: 10.1002/bies.20447. [DOI] [PubMed] [Google Scholar]
- 8.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 9.Chumakov PM. Versatile functions of p53 protein in multicellular organisms. Biochemistry (Mosc) 2007;72:1399–1421. doi: 10.1134/s0006297907130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 11.Zhao R, Gish K, Murphy M, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes & development. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 12.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 13.Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 14.Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 15.Gong X, Kole L, Iskander K, Jaiswal AK. NRH:quinone oxidoreductase 2 and NAD(P)H:quinone oxidoreductase 1 protect tumor suppressor p53 against 20s proteasomal degradation leading to stabilization and activation of p53. Cancer Res. 2007;67:5380–5388. doi: 10.1158/0008-5472.CAN-07-0323. [DOI] [PubMed] [Google Scholar]
- 16.Vousden KH. Outcomes of p53 activation--spoilt for choice. Journal of cell science. 2006;119:5015–5020. doi: 10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- 17.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 18.Wei CL, Wu Q, Vega VB, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 19.Lyakhov IG, Krishnamachari A, Schneider TD. Discovery of novel tumor suppressor p53 response elements using information theory. Nucleic Acids Res. 2008;36:3828–3833. doi: 10.1093/nar/gkn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 21.Vousden KH. p53: death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 22.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 23.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 24.Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 25.Carling D. The AMP-activated protein kinase cascade--a unifying system for energy control. Trends Biochem Sci. 2004;29:18–24. doi: 10.1016/j.tibs.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Shaw RJ. Glucose metabolism and cancer. Curr Opin Cell Biol. 2006;18:598–608. doi: 10.1016/j.ceb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Carling D, Sanders MJ, Woods A. The regulation of AMP-activated protein kinase by upstream kinases. Int J Obes (Lond) 2008;32(Suppl 4):S55–59. doi: 10.1038/ijo.2008.124. [DOI] [PubMed] [Google Scholar]
- 28.Suter M, Riek U, Tuerk R, Schlattner U, Wallimann T, Neumann D. Dissecting the role of 5′-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. J Biol Chem. 2006;281:32207–32216. doi: 10.1074/jbc.M606357200. [DOI] [PubMed] [Google Scholar]
- 29.Yoo LI, Chung DC, Yuan J. LKB1--a master tumour suppressor of the small intestine and beyond. Nature reviews. 2002;2:529–535. doi: 10.1038/nrc843. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen SB, Rose AJ. How is AMPK activity regulated in skeletal muscles during exercise? Front Biosci. 2008;13:5589–5604. doi: 10.2741/3102. [DOI] [PubMed] [Google Scholar]
- 31.Feng Z, Hu W, de Stanchina E, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 32.Warden SM, Richardson C, O’Donnell J, Jr, Stapleton D, Kemp BE, Witters LA. Post-translational modifications of the beta-1 subunit of AMP-activated protein kinase affect enzyme activity and cellular localization. Biochem J. 2001;354:275–283. doi: 10.1042/0264-6021:3540275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng PY, Berger SL. LKB1 is recruited to the p21/WAF1 promoter by p53 to mediate transcriptional activation. Cancer Res. 2006;66:10701–10708. doi: 10.1158/0008-5472.CAN-06-0999. [DOI] [PubMed] [Google Scholar]
- 34.Okoshi R, Ozaki T, Yamamoto H, et al. Activation of AMP-activated protein kinase induces p53-dependent apoptotic cell death in response to energetic stress. J Biol Chem. 2008;283:3979–3987. doi: 10.1074/jbc.M705232200. [DOI] [PubMed] [Google Scholar]
- 35.Jones RG, Plas DR, Kubek S, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 36.Buzzai M, Jones RG, Amaravadi RK, et al. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67:6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- 37.Lum JJ, Bauer DE, Kong M, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 38.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 40.Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27:5527–5541. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- 41.Young CD, Anderson SM. Sugar and fat - that’s where it’s at: metabolic changes in tumors. Breast Cancer Res. 2008;10:202. doi: 10.1186/bcr1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill MM, Clark SF, Tucker DF, Birnbaum MJ, James DE, Macaulay SL. A role for protein kinase Bbeta/Akt2 in insulin-stimulated GLUT4 translocation in adipocytes. Mol Cell Biol. 1999;19:7771–7781. doi: 10.1128/mcb.19.11.7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robey RB, Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–4696. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- 44.Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem. 2002;277:33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- 45.Buckbinder L, Talbott R, Velasco-Miguel S, et al. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 46.Ferry RJ, Jr, Cerri RW, Cohen P. Insulin-like growth factor binding proteins: new proteins, new functions. Horm Res. 1999;51:53–67. doi: 10.1159/000023315. [DOI] [PubMed] [Google Scholar]
- 47.Stambolic V, MacPherson D, Sas D, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 48.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 49.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 50.Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 51.Yang Q, Guan KL. Expanding mTOR signaling. Cell Res. 2007;17:666–681. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]
- 52.Corradetti MN, Guan KL. Upstream of the mammalian target of rapamycin: do all roads pass through mTOR? Oncogene. 2006;25:6347–6360. doi: 10.1038/sj.onc.1209885. [DOI] [PubMed] [Google Scholar]
- 53.Kwiatkowski DJ. Tuberous sclerosis: from tubers to mTOR. Ann Hum Genet. 2003;67:87–96. doi: 10.1046/j.1469-1809.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 54.Cai SL, Tee AR, Short JD, et al. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sancak Y, Thoreen CC, Peterson TR, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Inoki K, Ouyang H, Zhu T, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 57.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 58.Gwinn DM, Shackelford DB, Egan DF, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 61.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 63.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Droge W, Kinscherf R. Aberrant insulin receptor signaling and amino acid homeostasis as a major cause of oxidative stress in aging. Antioxid Redox Signal. 2008;10:661–678. doi: 10.1089/ars.2007.1953. [DOI] [PubMed] [Google Scholar]
- 65.Gozuacik D, Kimchi A. Autophagy and cell death. Curr Top Dev Biol. 2007;78:217–245. doi: 10.1016/S0070-2153(06)78006-1. [DOI] [PubMed] [Google Scholar]
- 66.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 68.Crighton D, Wilkinson S, Ryan KM. DRAM links autophagy to p53 and programmed cell death. Autophagy. 2007;3:72–74. doi: 10.4161/auto.3438. [DOI] [PubMed] [Google Scholar]
- 69.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nature reviews. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 70.Kerley-Hamilton JS, Pike AM, Hutchinson JA, Freemantle SJ, Spinella MJ. The direct p53 target gene, FLJ11259/DRAM, is a member of a novel family of transmembrane proteins. Biochim Biophys Acta. 2007;1769:209–219. doi: 10.1016/j.bbaexp.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Crighton D, O’Prey J, Bell HS, Ryan KM. p73 regulates DRAM-independent autophagy that does not contribute to programmed cell death. Cell Death Differ. 2007;14:1071–1079. doi: 10.1038/sj.cdd.4402108. [DOI] [PubMed] [Google Scholar]
- 72.Abida WM, Gu W. p53-Dependent and p53-independent activation of autophagy by ARF. Cancer Res. 2008;68:352–357. doi: 10.1158/0008-5472.CAN-07-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reef S, Zalckvar E, Shifman O, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–475. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 74.Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tasdemir E, Chiara Maiuri M, Morselli E, et al. A dual role of p53 in the control of autophagy. Autophagy. 2008;4:810–814. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- 76.Ibrahim MM, Razmara M, Nguyen D, Donahue RJ, Wubah JA, Knudsen TB. Altered expression of mitochondrial 16S ribosomal RNA in p53-deficient mouse embryos revealed by differential display. Biochim Biophys Acta. 1998;1403:254–264. doi: 10.1016/s0167-4889(98)00066-4. [DOI] [PubMed] [Google Scholar]
- 77.Achanta G, Sasaki R, Feng L, et al. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. The EMBO journal. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stambolsky P, Weisz L, Shats I, et al. Regulation of AIF expression by p53. Cell Death Differ. 2006;13:2140–2149. doi: 10.1038/sj.cdd.4401965. [DOI] [PubMed] [Google Scholar]
- 79.Matoba S, Kang JG, Patino WD, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 80.Miramar MD, Costantini P, Ravagnan L, et al. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J Biol Chem. 2001;276:16391–16398. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- 81.Vahsen N, Cande C, Briere JJ, et al. AIF deficiency compromises oxidative phosphorylation. The EMBO journal. 2004;23:4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heeres JT, Hergenrother PJ. Poly(ADP-ribose) makes a date with death. Curr Opin Chem Biol. 2007;11:644–653. doi: 10.1016/j.cbpa.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 83.Ma W, Sung HJ, Park JY, Matoba S, Hwang PM. A pivotal role for p53: balancing aerobic respiration and glycolysis. J Bioenerg Biomembr. 2007;39:243–246. doi: 10.1007/s10863-007-9083-0. [DOI] [PubMed] [Google Scholar]
- 84.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 85.Narkar VA, Downes M, Yu RT, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bauer DE, Harris MH, Plas DR, et al. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. FASEB J. 2004;18:1303–1305. doi: 10.1096/fj.03-1001fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yeung SJ, Pan J, Lee MH. Roles of p53, Myc and HIF-1 in Regulating Glycolysis - the Seventh Hallmark of Cancer. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 89.Ramanathan A, Wang C, Schreiber SL. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci U S A. 2005;102:5992–5997. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou S, Kachhap S, Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003;18:287–292. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]
- 91.Jaksch M, Paret C, Stucka R, et al. Cytochrome c oxidase deficiency due to mutations in SCO2, encoding a mitochondrial copper-binding protein, is rescued by copper in human myoblasts. Hum Mol Genet. 2001;10:3025–3035. doi: 10.1093/hmg/10.26.3025. [DOI] [PubMed] [Google Scholar]
- 92.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 93.Mathupala SP, Heese C, Pedersen PL. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem. 1997;272:22776–22780. doi: 10.1074/jbc.272.36.22776. [DOI] [PubMed] [Google Scholar]
- 94.Smith TA. Mammalian hexokinases and their abnormal expression in cancer. Br J Biomed Sci. 2000;57:170–178. [PubMed] [Google Scholar]
- 95.Corcoran CA, Huang Y, Sheikh MS. The regulation of energy generating metabolic pathways by p53. Cancer Biol Ther. 2006;5:1610–1613. doi: 10.4161/cbt.5.12.3617. [DOI] [PubMed] [Google Scholar]
- 96.Kondoh H, Lleonart ME, Gil J, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 97.Ruiz-Lozano P, Hixon ML, Wagner MW, et al. p53 is a transcriptional activator of the muscle-specific phosphoglycerate mutase gene and contributes in vivo to the control of its cardiac expression. Cell Growth Differ. 1999;10:295–306. [PubMed] [Google Scholar]
- 98.Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 99.Okar DA, Manzano A, Navarro-Sabate A, Riera L, Bartrons R, Lange AJ. PFK-2/FBPase-2: maker and breaker of the essential biofactor fructose-2,6-bisphosphate. Trends Biochem Sci. 2001;26:30–35. doi: 10.1016/s0968-0004(00)01699-6. [DOI] [PubMed] [Google Scholar]
- 100.Tian WN, Braunstein LD, Apse K, et al. Importance of glucose-6-phosphate dehydrogenase activity in cell death. Am J Physiol. 1999;276:C1121–1131. doi: 10.1152/ajpcell.1999.276.5.C1121. [DOI] [PubMed] [Google Scholar]
- 101.Weintraub H, Hauschka S, Tapscott SJ. The MCK enhancer contains a p53 responsive element. Proc Natl Acad Sci U S A. 1991;88:4570–4571. doi: 10.1073/pnas.88.11.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Araki N, Morimasa T, Sakai T, et al. Comparative analysis of brain proteins from p53-deficient mice by two-dimensional electrophoresis. Electrophoresis. 2000;21:1880–1889. doi: 10.1002/(SICI)1522-2683(20000501)21:9<1880::AID-ELPS1880>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 103.Okorokov AL, Milner J. An ATP/ADP-dependent molecular switch regulates the stability of p53-DNA complexes. Mol Cell Biol. 1999;19:7501–7510. doi: 10.1128/mcb.19.11.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McLure KG, Takagi M, Kastan MB. NAD+ modulates p53 DNA binding specificity and function. Mol Cell Biol. 2004;24:9958–9967. doi: 10.1128/MCB.24.22.9958-9967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 106.Warburg O, Posener K, Negelein E. Ueber den Stoffwechsel der Tumoren. Biochemische Zeitschrift. 1924;152:319–344. [Google Scholar]
- 107.Wang T, Marquardt C, Foker J. Aerobic glycolysis during lymphocyte proliferation. Nature. 1976;261:702–705. doi: 10.1038/261702a0. [DOI] [PubMed] [Google Scholar]
- 108.Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- 109.Gillies RJ, Gatenby RA. Adaptive landscapes and emergent phenotypes: why do cancers have high glycolysis? J Bioenerg Biomembr. 2007;39:251–257. doi: 10.1007/s10863-007-9085-y. [DOI] [PubMed] [Google Scholar]
- 110.Okamura S, Ng CC, Koyama K, et al. Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system. Oncol Res. 1999;11:281–285. [PubMed] [Google Scholar]
- 111.Ren B, Yee KO, Lawler J, Khosravi-Far R. Regulation of tumor angiogenesis by thrombospondin-1. Biochim Biophys Acta. 2006;1765:178–188. doi: 10.1016/j.bbcan.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 112.Miwa S, Muller FL, Beckman KB. The basics of oxidative biochemistry. In: Miwa SKBB, Muller FL, editors. Aging Medicine: Oxidative stress in aging: From model systems to human diseases. Totowa, N.J: Humana Press; 2008. pp. 11–35. [Google Scholar]
- 113.Ho YS, Xiong Y, Ma W, Spector A, Ho DS. Mice lacking catalase develop normally but show differential sensitivity to oxidant tissue injury. J Biol Chem. 2004;279:32804–32812. doi: 10.1074/jbc.M404800200. [DOI] [PubMed] [Google Scholar]
- 114.Andziak B, O’Connor TP, Buffenstein R. Antioxidants do not explain the disparate longevity between mice and the longest-living rodent, the naked mole-rat. Mech Ageing Dev. 2005;126:1206–1212. doi: 10.1016/j.mad.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 115.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 116.Martindale JL, Holbrook NJ. Cellular response to oxidative stress: signaling for suicide and survival. J Cell Physiol. 2002;192:1–15. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- 117.Desaint S, Luriau S, Aude JC, Rousselet G, Toledano MB. Mammalian antioxidant defenses are not inducible by H2O2. J Biol Chem. 2004;279:31157–31163. doi: 10.1074/jbc.M401888200. [DOI] [PubMed] [Google Scholar]
- 118.Rivera A, Maxwell SA. The p53-induced gene-6 (proline oxidase) mediates apoptosis through a calcineurin-dependent pathway. J Biol Chem. 2005;280:29346–29354. doi: 10.1074/jbc.M504852200. [DOI] [PubMed] [Google Scholar]
- 119.Liu G, Chen X. The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene. 2002;21:7195–7204. doi: 10.1038/sj.onc.1205862. [DOI] [PubMed] [Google Scholar]
- 120.Halliwell B. Oxidative stress and cancer: have we moved forward? Biochem J. 2007;401:1–11. doi: 10.1042/BJ20061131. [DOI] [PubMed] [Google Scholar]
- 121.Kopnin PB, Agapova LS, Kopnin BP, Chumakov PM. Repression of sestrin family genes contributes to oncogenic Ras-induced reactive oxygen species up-regulation and genetic instability. Cancer Res. 2007;67:4671–4678. doi: 10.1158/0008-5472.CAN-06-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yoon KA, Nakamura Y, Arakawa H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. Journal of human genetics. 2004;49:134–140. doi: 10.1007/s10038-003-0122-3. [DOI] [PubMed] [Google Scholar]
- 123.Hussain SP, Amstad P, He P, et al. p53-induced up-regulation of MnSOD and GPx but not catalase increases oxidative stress and apoptosis. Cancer Res. 2004;64:2350–2356. doi: 10.1158/0008-5472.can-2287-2. [DOI] [PubMed] [Google Scholar]
- 124.O’Connor JC, Wallace DM, O’Brien C, Cotter TG. A novel antioxidant function for the tumour suppressor gene p53 in the retinal ganglion cell. Investigative ophthalmology & visual science. 2008 doi: 10.1167/iovs.08-1963. [DOI] [PubMed] [Google Scholar]
- 125.Budanov AV, Shoshani T, Faerman A, et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene. 2002;21:6017–6031. doi: 10.1038/sj.onc.1205877. [DOI] [PubMed] [Google Scholar]
- 126.Velasco-Miguel S, Buckbinder L, Jean P, et al. PA26, a novel target of the p53 tumor suppressor and member of the GADD family of DNA damage and growth arrest inducible genes. Oncogene. 1999;18:127–137. doi: 10.1038/sj.onc.1202274. [DOI] [PubMed] [Google Scholar]
- 127.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ding B, Chi SG, Kim SH, et al. Role of p53 in antioxidant defense of HPV-positive cervical carcinoma cells following H2O2 exposure. Journal of cell science. 2007;120:2284–2294. doi: 10.1242/jcs.002345. [DOI] [PubMed] [Google Scholar]
- 129.Liu B, Chen Y, St Clair DK. ROS and p53: a versatile partnership. Free Radic Biol Med. 2008;44:1529–1535. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fischer JL, Lancia JK, Mathur A, Smith ML. Selenium protection from DNA damage involves a Ref1/p53/Brca1 protein complex. Anticancer research. 2006;26:899–904. [PubMed] [Google Scholar]
- 131.Velu CS, Niture SK, Doneanu CE, Pattabiraman N, Srivenugopal KS. Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry. 2007;46:7765–7780. doi: 10.1021/bi700425y. [DOI] [PMC free article] [PubMed] [Google Scholar]