Abstract
The human Polybromo-1 protein (Pb1) was recently identified as a unique subunit of the PBAF (Polybromo, Brg1-Associated Factors) chromatin remodeling complex required for kinetochore localization during mitosis and the transcription of estrogen-responsive genes. Pb1 coordinates key features common to all remodeling complexes, including chromatin localization, recruitment of protein subunits and alteration of chromatin architecture. A comprehensive analysis of individual domains composing Pb1 is used to propose new information regarding the function of Pb1 in the PBAF chromatin remodeling complex. The newly identified regulatory role of this important protein is also examined to explain both native function and the emerging role of Pb1 as a tumor suppressor found to be mutated in breast cancer.
Keywords: Polybromo, BAF180, PBAF, bromodomain
Introduction
The importance of the Polybromo-1 (Pb1) protein and PBAF (Polybromo, Brg1-Associated Factors) chromatin remodeling complex in regulating eukaryotic gene transcription is only beginning to be understood. It has been clear for many years that such remodeling complexes contribute to transcriptional regulation by altering the structure of chromatin and controlling the accessibility of DNA [1-8]. Recent studies have led to the striking observation that individual protein subunits act by localizing these complexes to specific chromatin sites in order to execute specific biological functions [9-17]. This review will focus on the native and mutant Pb1 protein to explain its role in the PBAF chromatin remodeling complex. We provide a description of the domain organization as it is currently understood and introduce new information regarding the function of Pb1 in the PBAF complex. We also review the data available with regard to variations in domain composition for the six isoforms of Pb1 to explain functional variations in PBAF. Finally, we examine the biological features and newly identified regulatory role of this important protein to explain current observations implicating Pb1 mutants in breast cancer.
Discovery of the PBAF chromatin remodeling complex
The human polybromo protein (originally termed BAF180) was originally discovered in screens identifying subunits homologous to the yeast SWI/SNF (SWItching/Sucrose Non-Fermentable) complex from several mammalian cell lines using antibodies to Brg1 [18, 19]. Consequently, the proteins identified in these screens were termed Brg1-Associated Factors. The PBAF chromatin-remodeling complex, originally termed SWI/SNF-B due to its similarity with SWI/SNF, contains nine common subunits and four interchangeable subunits depending on the subclass (described in detail below). The minimal catalytic core required for chromatin remodeling activity in vitro requires four subunits: Brg1, BAF155, BAF170, and BAF47, although Brg1 alone exhibits some remodeling activity [20]. In vivo, however, stable association of the BAF complex with chromatin requires the presence of actin and BAF53, two subunits that are strongly associated with Brg1 [21]. Because these BAF subunits are also constituents of PBAF and the ATPase activity of Brg1 is optimal when bound to actin and BAF53, it can be argued that these subunits are essential constituents of the active core complex [22]. BAF155 and BAF170 show high sequence homology and, depending on the tissue type, may form homo- or heterodimers in the complex [19, 23]. BAF57 interfaces with various nuclear receptors including, glucocorticoid, estrogen and androgen receptors, by direct association to recruit Brg1-based complexes to receptor-responsive promoters thereby altering transcriptional activity [15, 24-27]. Interestingly, protein-protein interactions between the leucine zipper motifs of BAF155 and BAF170 seem to regulate steady-state levels of the BAF57 subunit and, consequently, the overall stoichiometry of the complex. BAF47, a human homologue of yeast SNF5 protein, triggers the remodeling activity of Brg1 in vitro [20], interacts with transcription factors such as c-Myc [28] and induces cell cycle arrest in G0/G1 [29, 30]. The recently discovered BAF200 subunit serves to regulate certain interferon-responsive genes and acts to stabilize Pb1 in the higher-order PBAF complex [31]. Three highly homologous BAF60 genes (BAF60a, BAF60b, and BAF60c) have been identified in different tissue types, suggesting a tissue-specific role for this nuclear receptor binding protein [32, 33]. This is also the case for BAF45 subunits, which associates with BAF53 to control proliferation and differentiation of neuronal stem cells [34]. BAF53, the closest homolog of Arp3, is one of the actin-related proteins necessary for the ATPase activity of Brg1 in vivo [21]. Considering the observation that actin co-purifies with Pb1 [35] and the Pb1 subunit is required to localize PBAF at kinetochores of mitotic chromosomes [36], it is possible that Pb1 mediates both nucleosome targeting and actin-dependent migration of chromosomal regions within the nucleus.
Differences in the two highly conserved subclasses of human SWI/SNF are defined by the presence of either Polybromo-1 and BAF200 or BAF250 and BRM subunits in the complex [31, 36, 37]. When considering that the majority of subunits are shared between the complexes, chromatin localization is likely a function of these distinctive subunits. In fact, it is thought that the Pb1 and BAF200 subunits act to target PBAF to different gene regions via protein-protein interactions. Yeast proteins homologous to BAF250 (Swi1) and Polybromo-1 (Rsc1, Rsc2 and Rsc4) are known as the signature subunits of ySWI/SNF or yRSC (Remodels the Structure of Chromatin), respectively. No homolog to Swi1 exists in the yRSC complex [12, 13, 38] and similarly, the Rsc1, Rsc2 and Rsc4 proteins lack a counterpart in ySWI/SNF [36]. As a result of the observed compositional variations, the complexes were divided into two subfamilies; one comprising yeast SWI/SNF and the mammalian homolog BAF containing BAF250 and BRM, and a second subfamily including yeast RSC and mammalian PBAF complexes. A key relationship between the human PBAF and yeast RSC complex is based on the subunit composition of Pb1, which is homologous to the RSC subunits Rsc1, Rsc2, and Rsc4 [39]. RSC is more abundant in comparison to ySWI/SNF which may reflect the broader role of RSC in chromatin maintenance and structural regulation. Recently, RSC has been shown to play a role in sister chromatid cohesion and chromosome segregation [40-42]. Because PBAF was shown to localize to kinetochores during mitosis [36], it has been proposed that PBAF, like RSC, is required for cell cycle progression through mitosis.
Polybromo-1: a unique PBAF subunit
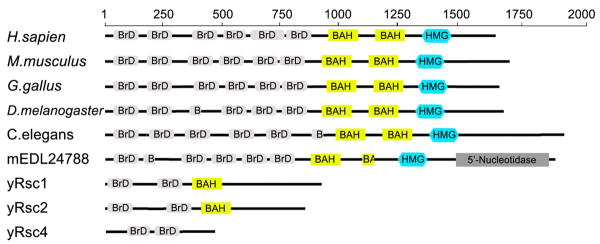
The full length 1689-amino acid Pb1 protein contains six tandem bromodomains (BrDs), two bromo-adjacent homology domains (BAH), and a high-mobility group (HMG) (Fig. 1). Taken together, the fact that BrDs bind acetylated histones [43-45], BAH domains are protein-interaction modules [46, 47], and HMGs have been shown to bind nucleosomal DNA [48-50], the Pb1 protein serves as an important PBAF subunit coordinating several roles central to the function of most known remodeling complexes: [1] targeting chromatin sites, [2] recruiting specific effector proteins, and [3] altering the histone-DNA interactions to control genetic functions. It should be noted that polybromo proteins from different organisms show high sequence homology for all known and computationally predicted polybromo proteins. When only considering mammalian polybromo proteins, sequence identities approach 90%. Sequence alignments between polybromo proteins from H.sapien, D.melanogaster and C.elegans, and Rsc1, Rsc2 and Rsc4 from S.cerevisiae show the three highly homologous Rsc proteins have similar domain organization and share multiple conserved structural motifs [51]. Because the Rsc proteins have a total of six BrDs and two BAH domains, it has been suggested that Pb1 may be a product of gene fusions [36], conferring features in human PBAF that the three Rsc proteins impart upon the yeast RSC complex.
Fig. 1.
Schematic representation of human Polybromo-1 domain organization. Known and predicted domains for Pb1 are compared to polybromo proteins from the indicated organisms and the homologous yeast Rsc proteins. The proteins shown here share similar domain organization of bromodomains (BrD), bromo-adjacent homology domains (BAH), and the high-mobility group (HMG). Partial domains are indicated.
The tandem bromodomains
The ‘histone code’ hypothesis suggests that posttranslational modification of histone proteins can alter local and global protein architecture and generate novel interaction interfaces capable of recruiting effector proteins [52, 53]. Consistent with this notion, site-specific acetylation patterning on conserved histone tail regions plays an important role for multiple signaling pathways which converge on histones to affect chromatin structure and regulate transcription [54-60]. The ability to selectively recognize and bind acetylated histones is a feature of a family of evolutionarily conserved protein modules roughly 100 amino acids in length called bromodomains [61-63]. Bromodomains are acetyllysine (AcK) binding domains found in subunits of chromatin remodeling complexes and in many histone acetyltransferases (HATs) [64-67]. New findings reveal that bromodomain-acetyllysine recognition serves as a pivotal mechanism for regulating protein–protein interactions in different cellular processes such as chromatin remodeling and transcriptional activation [68-70]. Essentially, the BrDs unique role as interpreters of the histone acetylation code centers on the ability to target specific histone acetylation sites [62, 71-73] making these domains key targeting elements of chromatin remodeling complexes such as RSC, SWI/SNF and PBAF [36, 64, 74-78].
Solved structures, including the bromodomain of human SWI/SNF related matrix associated actin-dependent regulator of chromatin subfamily A member 2 (PDB ID: 2DAT), hRING3 (PDB ID: 2G4A), hKAP1 (PDB ID: 2RO1) [79], hBrd2 (PDB ID: 1XOJ), hBrd3 (PDB ID: 2E7N), hBrd4 (PDB ID: 2I8N), hBrd7 (PDB ID: 2I7K) [80], Brg1 (PDB ID: 2GRC), hGcn5 (PDB ID: 1F68) [81], hBRDT (PDB ID: 2RFJ), yGcn5 (PDB ID: 1E6I) [67], peregrine (PDB ID: 2D9E), TAF(II)250 dibromodomain (PDB ID: 1EQF) [82], yRSC4 (PDB ID: 2R0S) [83], and fifth BrD from mouse polybromo-1 (PDB ID: 2YQD), reveal a four-helix bundle with a pronounced hydrophobic pocket formed by two loops. Comparison with structures of BrDs bound to acetylated peptides, including P/CAF (PDB ID: 1JM4) [84], CBP (PDB ID: 1JSP) [85, 86], a HAT bromodomain (PDB ID: 1N72) [66], hBrd2 (PDB ID: 2E3K), and hBrg1 (PDB ID: 2H60) [87] indicate that the acetyllysine peptide binds at one end of the helical bundle within the hydrophobic pocket formed by the two loop segments. Structural data further indicates the BrD-acetylhistone interface spans several side chains encompassing the acetyllysine and, in fact, the acetyllysine alone is insufficient to confer protein interaction. For example, Gcn5 shows negligible binding to the free amino acid form of lysine, acetyllysine or N-acetyl-histamine whereas, Gcn5, Bdf1 and TAF(II)250 show strong binding to acetylated histone tail peptides from H4 [67, 81, 82, 88, 89].
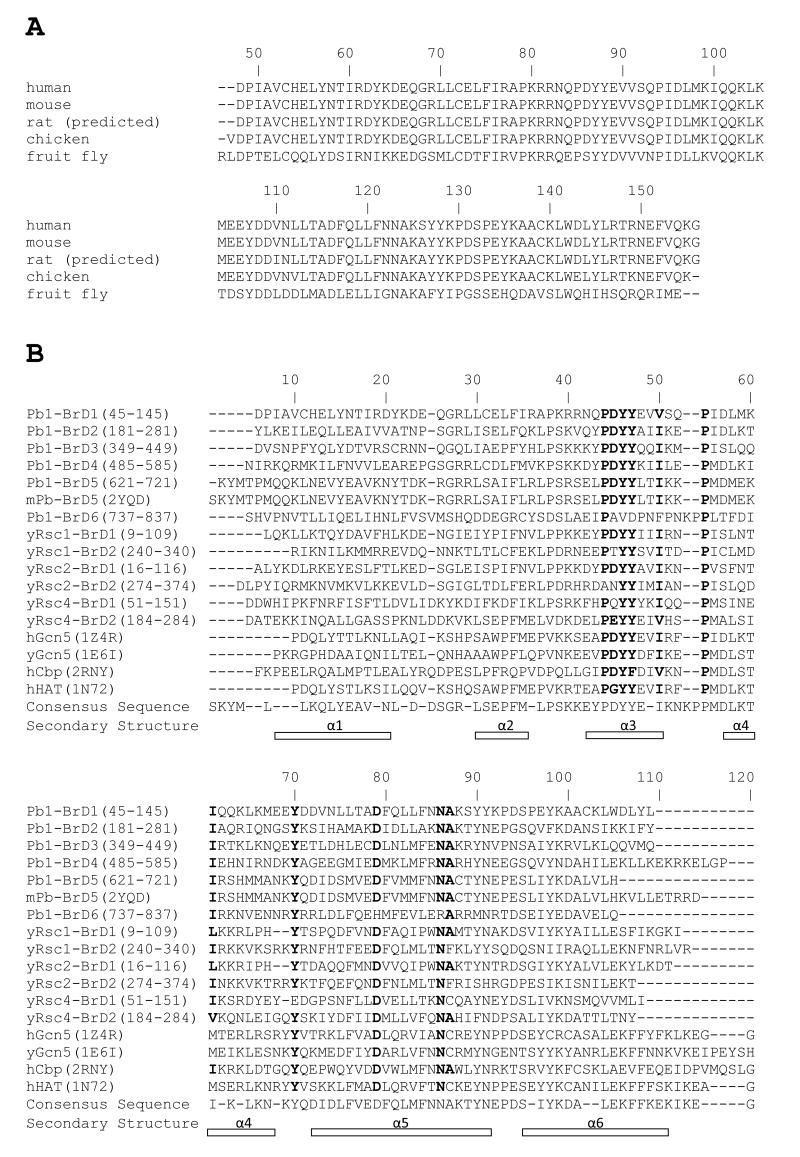
Computational analysis of bromodomains from human and mouse Pb1, yeast Rsc and multiple structures from the Protein Database (PDB) suggests the amino acid sequences of bromodomains are highly conserved in regions that directly contact the acetyllysine, yet less conserved in regions that contact histone side chains surrounding the modified lysine (Fig. 2) [51, 90, 91]. The tyrosine-rich binding pocket contains the highly conserved -YY- and -YN- (these amino acids occur at positions 46, 47 and 90, 91 in Fig. 2B) sequences that contribute to the selection for acetylated lysine side chains. The highly conserved tyrosine side-chains form a hydrophobic pocket that surrounds the acetyllysine. The lack of side-chains in the pocket capable of stabilizing a positive charge may discriminate the acetylation-state and act to select against unmodified lysine [39]. Interestingly, significant BrD sequence variations are found in regions that form the acetyllysine interface, suggesting a molecular level explanation for the observed site-specificity of many bromodomain proteins. For example, comparison of sequence alignments with secondary structures indicate that bromodomain side chains -EEV- shown to be at the binding interface in Gcn5 correspond to -KKY- in Pb1-BrD3 and -SEL- in Pb1-BrD5 (these amino acids occur at positions 41-43 in Fig. 2B). The loop regions of bromodomains from P/CAF and Gcn5 appear to undergo conformational changes to accommodate peptide binding [67, 84].
Fig. 2.
Sequence alignments of Pb1 bromodomains. (A) Bromodomain 1 (the amino-terminal bromodomain) for the indicated species from amino acids 45-155 are compared. (B) Bromodomains from Pb1 and Rsc are compared to those with known structures. Highly homologous amino acids based on identity or chemical features (i.e. hydrophobicity or charge) are shown in bold. Annotation of secondary structural elements is based on the structure of the fifth bromodomain (BrD5) of mouse polybromo protein (PDB ID: 2YQD). ClustalW was used to perform the alignment.
The emerging picture of BrD-acetylhistone interactions support the concept that functional diversity of a conserved protein modular structure is achieved by evolutionary changes of amino acid sequences in regions composing the binding interface. Considering that histone tail sequences are nearly invariant from yeast to human, it is possible that bromodomains have evolved to discriminate the subtle molecular differences in the tail sequences. Bromodomain proteins are highly specific for a particular acetyllysine, as indicated by in vitro binding studies [62, 71, 73]. Recently, it was shown that individually cloned and expressed bromodomains from Pb1 were able to discriminate the lysine acetylation state on histone tail peptides [39]. Studies employing a peptide library composed of short acetylhistone tail segments were the first proof that individual bromodomains were targeting specific acetyllysines [73]. The data indicate bromodomain-acetylhistone interactions require the lysine to be acetylated and exhibit variable affinities and specificities as observed for DNA-binding proteins. Kinetic and thermodynamic analyses have shown that acetyllysines located at different positions bind with a range of affinities due to differential effects of ligand-induced folding – consistent with observations from NMR structures [71, 72, 81]. In vivo studies reveal strong connections between biological function and the requirement by bromodomain proteins for acetylation at certain histone side-chains. The yeast RSC targeting protein Rsc4, for example, requires acetylation at Lys14 of histone H3 [75]. Mutant derivatives of Rsc4 were unable to bind H3-AcK14 and were unable to activate certain genes. It remains to be determined whether the variations in site-specificity observed in vitro translate into biologically relevant acetylation patterns.
The overall role of the polybromo region, which contains six tandem bromodomains, may be to selectively target Pb1, and consequently the PBAF complex, to certain chromatin sites defined by discreet nucleosome acetylation patterns. Results from our lab indicate that several Pb1-BrDs target specific acetyllysines within the histone tail regions. Quantitative screening of individual bromodomains from Pb1 against an acetylhistone peptide library has revealed a hypothetical histone acetylation pattern that may act as the nucleosome binding site for the Pb1 protein. If we assume each bromodomain targets an acetyllysine in vivo, then there would be more than a million potential arrangements of the acetylation sites that could act as the pattern targeted by the native Pb1 protein. Our library screens identified several potential high-affinity combinations based on the lowest KD values for the six Pb1 bromodomains. The amino-terminal or first bromodomain (BrD1) preferentially targets H3-K4 and H2A-K5, which share an -RXKQX- motif. BrD3 exhibits high affinity and high-specificity for H3-K9, having an equilibrium dissociation constant of approximately 1 μM. The second BrD also interacts with histone H3-K9, raising speculation that Pb1 is able to contact two H3-K9 sites within the same or adjacent nucleosomes. BrD4 preferentially targets the -XTKAX- motifs of H3-K23 and H2B-K20. The two remaining bromodomains (BrD5 and BrD6) target several sites and we speculate that these domains bind in a non-specific manner to enhance the thermodynamic stabilization of the polybromo-nucleosome complex (unpublished results). These observations support our hypothesis that Pb1 targets a discreet histone acetylation pattern (or subset of patterns) defined by the sum of the individual bromodomain binding events. Examining the relevance of a combinatorial histone acetylation code by Pb1 will require testing our hypothetical binding site in the nucleosomal context to understand the cooperativity of multiple bromodomains and the mechanisms used to discriminate among different acetylation patterns.
The bromo-adjacent homology domains
The bromo-adjacent homology (BAH) domain is a 130 amino acid region first identified in the polybromo protein of vertebrates and later found to be present in a variety of proteins involved in transcriptional regulation [47]. Although relatively little is known about the BAH domains, the function seems to center around its ability to act as a protein-protein interaction module [46, 47, 92]. For example, proteins containing BAH domains have been found to interact with the silent information regulator Sir1p within yeast Orc1p [93]. The interaction interface formed by the BAH domain may coordinate the spatial organization of proteins and, in the case of Pb1, the tandem BAH domains may serve to anchor the subunit within the PBAF complex.
Solved structures for BAH domains include yeast Orc1 (PDB ID: 1M4Z) [93], ySir3 (PDB ID: 2FL7) [94], and the N-terminal BAH domain from chicken polybromo (PDB ID: 1W4S) [92]. Structural elements of the globular BAH domain are composed primarily of β-strands creating a rather distorted β-barrel [92]. The ‘open-barrel’ shape of the BAH domain is completely filled by hydrophobic side chains from short β-strands and a 310 helix that creates an extended loop fold. A co-crystal structure of yeast Sir1 and Orc1 (PDB IDs: 1ZBX, 1ZHI) gives details regarding the interface formed between the Orc1 BAH domain and Sir1 [95, 96].
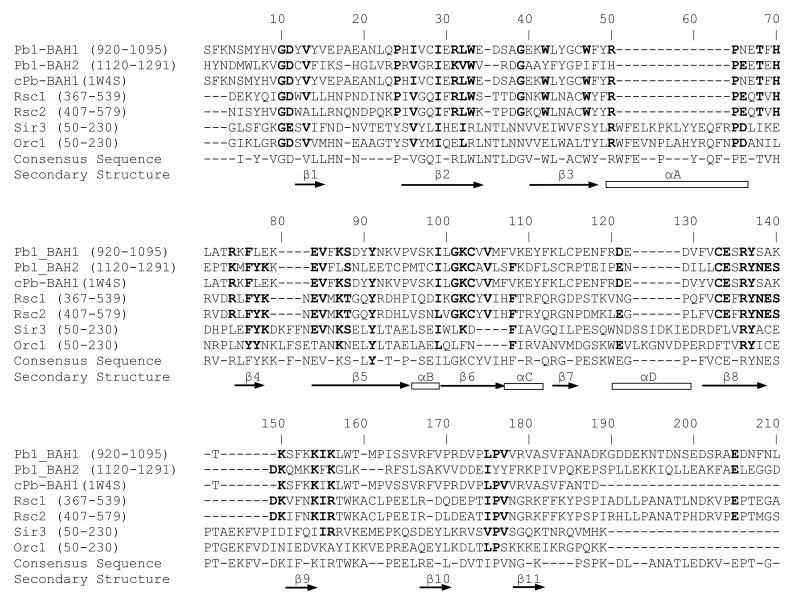
Sequence alignments comparing BAH domains from different organisms indicate a high degree of homology throughout the region and illustrate several interesting points (Fig. 3). First, the Pb1-BAH domains are highly homologous to each other and to yeast Rsc1 and Rsc2 BAH domains (approx. 30% identical). Second, the yeast Sir1 and Orc1 proteins share greater than 50% identity with each other, yet significantly less homology with Pb1 and yRsc BAH domains. In fact, the Rsc1 and Rsc2 BAH domains are more similar in sequence to Pb1 BAH domains than to the two other yeast proteins. Three amino acids, Pro65, Phe76 and Glu84 (numbering based on Fig. 3) are absolutely conserved in all polybromo and Rsc BAH domains. Previous examinations of BAH domains indicated that these three amino acids are clustered at a single region forming a small pocket or depression in the surface of the BAH domain [92]. Speculation that these three amino acids might permit binding of a partner protein was further supported by the observation that mutation of the glutamic acid to a lysine in yRsc2 disrupted the proper segregation of daughter cells [97]. The primary sequence and secondary structure annotation derived from structural data from the nearly identical BAH1 domain from chicken polybromo suggest that these proteins have comparable function. Compare this observation with proteins such as the D.melanogaster Ash1 and A.thaliana Orc1, which have unique regions within the canonical BAH region that may confer a regulatory role unique to these proteins [46]. Computational analyses suggests, but does not prove, that seemingly diverse roles are based on the highly variable carboxy-terminal 50 or so amino acids. In fact, the connection between these strategically located sequence variations was offered as an explanation into the diverse roles of BAH domains in transcriptional activation, replication and silencing [98]
Fig. 3.
Sequence alignment comparing BAH domains from human and yeast proteins. Highly homologous amino acids based on identity or chemical properties are shown in bold. Annotation of secondary structural elements is based on the structure of the first BAH domain from the chicken polybromo protein (PDB ID: 1W4S). ClustalW was used to perform the alignment.
Although the genes for several BAH-containing proteins have shown severe phenotypic defects, no data existed to prove the BAH domain itself is directly involved. This was until Cairns and coworkers showed that the BAH domains for Rsc1 and 2 were required for the assembly of the RSC complex but not the bromodomains [78]. This observation led to speculation that the BAH domain had functions that center around protein-protein interactions required for the multi-subunit assembly of RSC and potentially the interaction with DNA-binding transcription factors. Conversely, the BAH domain of yOrc1 protein has been linked to silencing of HML and HMR mating-type loci via the interaction of its N-terminal BAH domain [99]. Subsequent studies showing that Sir1 interacts with the N-terminal region of Orc1 in replication-dependent repression further support the requirement of the BAH domain in forming critical protein-protein interactions [98, 100, 101].
The role of the BAH domains in Pb1 is expected to be comparable to that of the homologous BAH domains in the two Rsc proteins found in the RSC complex. The two BAH domains of Pb1 may selectively bind to one or more binding partners in the service of forming the multi-subunit PBAF. It is important to identify the binding partner(s) to understand the assembly mechanism of the complex. For example, the observation that BAF200 is required to anchor Pb1 within the PBAF complex [31] makes a strong argument for a BAH-dependent interaction between the two proteins. This raises the further question about the loss of BAH regions observed in different Pb1 isoforms (discussed below) that might deactivate the domain and adversely impact the ability to form protein-protein interactions or assemble into PBAF. This positions Pb1 as a central protein connecting the acetylation pattern present on nucleosomes that act as the binding target of the polybromo region with the proteins recruited by the BAH domains.
High-mobility group
The high mobility group domain, or HMG-box (HMGB), is defined by a region roughly 80 amino acids in length and found in a variety of eukaryotic chromosomal proteins and nucleosome binding proteins [48, 50, 102]. HMG-boxes bind in the minor groove of DNA and are grouped into three major classes based on their mode of binding. Class I consist of proteins containing a single HMG box that interact with specific DNA sequences and structures, including four-way DNA junctions and duplex DNA targets. For example, Sox proteins are transcriptional regulators associated with developmental processes that act by altering DNA conformation and nucleosomal architecture [103-105]. Class II and III are proteins bind DNA in a non-specific manner and oftentimes contain two or more tandem HMG-boxes. Examples of Class II proteins include upstream binding factors (UBF) and non-histone chromosomal proteins that bind to bent and distorted DNA. Because HMGB1 and 2 proteins are involved in the assembly of nucleoprotein complexes in various biological processes, including, V(D)J recombination, transcriptional initiation, and DNA repair, they are both thought to play architectural roles [49]. Class III proteins also bind four-way DNA junctions and include nucleolar and mitochondrial transcription factors.
Solved structures of HMG-boxes are quite abundant and include the yeast non-histone protein Nhp6A (PDB ID: 1J5N) [106], rat HMG1 (PDB IDs: 1HME) [107], human HMG-box transcription factor 1 (PDB ID: 2E6O), mouse HMGX2 (PDB ID: 2CRJ), the second HMG-box from human B3 (PDB ID: 2YQI), and the HMG-box domain of thymus high mobility group box protein TOX from mouse (PDB ID: 2CO9). HMG-boxes exhibit a variety of functional roles and the exact role of the domain in Pb1 is currently unknown. Because several types of HMGB proteins interact with nucleosomes in the absence of histone H1, it is possible that the HMG-box from Pb1 displaces H1 to loosen chromatin structure. To clarify some of the speculation, we performed BLAST searches and secondary structure analysis with known domains and structures to propose possible functions for the HMG-box in Pb1.
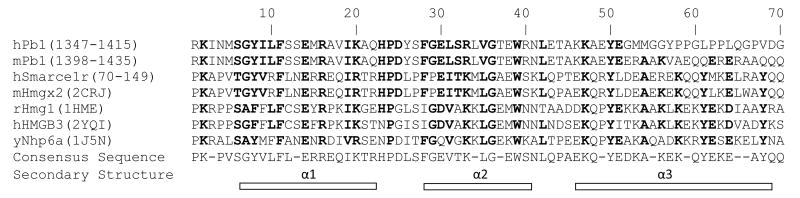
The HMG-box found in Pb1 exhibits some striking similarities to the mammalian HMG box-containing protein 20B, which is a SMARCE1-related protein. This protein is SWI/SNF-related matrix-associated actin-dependent regulator of chromatin which has a Sox-like transcriptional factor [108]. This is a nonspecific DNA-binding HMG-box. This protein, also termed BRAF35, was highly expressed in proliferating tissues and nuclear staining revealed a close association with chromatin containing histone H3 phosphorylation. Similar to Pb1, the BRAF35 protein may play a role in modulating progression of the cell cycle. It is interesting to note that the highly homologous mouse and human polybromo proteins, which show >90% identity, have identical HMG-box sequences until amino acid 52 (corresponds to amino acid 1399 in human and 1444 in mouse). Sequences beyond this point show negligible homology raising the questions about the evolution of these regions and the potential for alternative function. The regions of HMG expected to be required for DNA binding based on structural modeling are intact (Fig. 4) and no adverse effect on binding would be expected. Further comparison indicates Pb1 is highly homologous to yeast NHP6A, a non-sequence-specific DNA-binding protein which belongs to the HMGB protein family. Nhp6a was shown to increase V(D)J cleavage efficiency by binding distorted DNA structures and promoting RAG 1/2-mediated chemical reactions on the DNA [109].
Fig. 4.
Sequence alignment comparing the Pb1 HMG-box with proteins from other organisms. Native sequence or PDB ID is shown in parenthesis. Highly homologous amino acids based on identity or chemical features (i.e. hydrophobicity or charge) are shown in bold. Annotation of secondary structural elements is based on the structure of the HMG domain from mouse HMGX2 (PDB ID: 2CRJ). ClustalW was used to perform the alignment.
The HMG-box domains are often found in chromatin-associated proteins and induce DNA structural changes upon binding to regulate chromatin function and gene expression. The sequence analysis suggests that the HMG-box in Pb1 is homologous to human SMARCE1-related protein. The SMARCE1r high mobility group protein was found to be associated with the nuclear matrix and to have DNA binding activity [110]. Because the HMG-box family is quite diverse it is unclear if the domain in Pb1 acts to loosen the chromatin by histone H1 displacement or put topological constraints on the DNA, both of which might facilitate transcriptional activation. Although highly speculative, its similarity to the closely related BAF57 (Smarce1), also a subunit of PBAF, raises the question of whether the HMG-box in Pb1 provides complementary DNA binding activity or nucleosome association to enhance the activity of the complex.
Isoforms of Polybromo-1
The human polybromo-1 protein has six isoforms generated as a result of alternative splicing products, five of which alter the number and type of domains present in the Pb1 protein [111]. Considering the role of these domains, Pb1 variants may target different histone acetylation patterns and alternately, recruit distinct binding partners to different chromatin sites based on the composition of BrD and BAH domains present. These statements are highly speculative, but the following analysis permits us to propose alternate roles for the different isoforms and presumably the function of PBAF.
The full length human Polybromo-1 protein has 1689 amino acids composing all the functional domains described above. The six tandem bromodomains are found in the amino-terminal 900 amino acids. The number of amino acids between adjacent bromodomains is 60-70 amino acids for all but BrD2 and BrD3, which are separated by 130 amino acids (Fig. 5). The two protein-interaction modules, BAH1 (956 – 1074) and BAH2 (1156 – 1272), both of which are roughly 120 amino acids in length occur in tandem to the carboxy-side of the polybromo region. A putative HMG domain (1379 – 1447), roughly 70 amino acids in length, is found at the carboxy-region of Pb1.
Fig. 5.
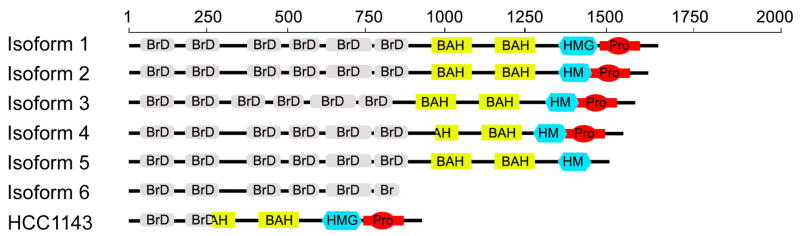
Schematic representation of variations in domain organization for Pb1 isoforms. The HCC1143 mutation of the PB1 gene, as observed in breast tumors, is shown for comparison.
Isoform 2 is a 1634 amino acid protein that differs from the full protein (canonical sequence) in that amino acids 1430-1484 in the native sequence are missing. In fact, this region is only present in isoform 1 and absent in all other isoforms. All the six BrDs and both BAH domains remain conserved as in the full length polybromo protein. Fifty-five amino acids are deleted which includes the carboxy-18 amino acids of the HMG box (1430-1447). This deletion brings a proline rich region, which was approximately 30 amino acids separated from the HMG domain, into contact with the HMG-box. Isoform 3 is a 1602 amino acid protein with two major sections deleted for an overall deletion of 87 amino acids. In addition to the deletion of amino acids 1430-1484, 33 residues are deleted from the region between Brd2 and Brd3 (300-332 from the full spacer region 271-399) and one Serine residue is added at 300th position bringing BrD2 and Brd3 closer by a net of 32 residues. As stated above, the shorter separation distance between adjacent BrDs may alter the cooperativity of Pb1 binding to acetylated nucleosomes. Isoform 4 generates a 1582 amino acid protein with three major sections deleted for a total deletion of 107 amino acids (989-1013, 1336-1362, 1430-1484). All six bromodomains are conserved but 25 amino acids (989-1013) are deleted from BAH1 shortening it from 119 amino acids to 94 amino acids. Considering sequence alignments results, we speculate that a substantial portion of the secondary structure and consequently, the binding interface will be disrupted. Furthermore, twenty seven amino acids are deleted (1336-1362) in the region between BAH2 and HMG box which spans 1273-1378 in the canonical isoform. Isoform 5 is a 1582 amino acid proteins with native polybromo and BAH regions. Amino acids 1430-1536 are deleted, disrupting the HMG domain and removing the proline-rich region. Isoform 6 contains only the first 856 amino acids. Bromodomains 1-5 are unaffected and BrD6 is shortened by seven amino acids. It is unclear what affect this will have on BrD6 binding, as prior examination of binding data for BrD6, which was taken from amino acids 740-864 in the native sequence, did include the amino acids lost in this isoform [73]. Examination of sequence alignments suggests that all secondary structural elements required to form the hydrophobic core are present (Fig. 2).
Several key questions regarding the impact of the presence of different Pb1 isoforms on the function of PBAF are raised. If the BAH and HMG domains in various isoforms are inactive, it is likely that the PBAF complex will not properly form. Alternatively, if there is a change in domain composition, there could be different PBAF sub-classes that have differential regulatory effects. To some degree this has been observed in neural stem cells, where different developmental stages of neurogenesis exhibit a switch in the expression of BAF45 and BAF53 family proteins [34]. The presence of only one BAH domain may select or limit the type of partner proteins or even prevent proper assembly of Pb1 in the PBAF complex. In the case of the HMG-box partial deletion, a loss or decrease in activity may result in a lower affinity of the complex to nucleosomal locations or limit architectural alterations in chromatin that might facilitate transcription. Rapid dissociation may be an advantage in regions of high transcriptional activity, but may adversely impact PBAF anchoring at kinetochores; a trait required in segregation of sister chromatids. An interesting observation from our bromodomain-acetylhistone studies was that BrD2 and BrD3 have a strong preference for binding histone H3 acetylated at lysine 9. This led to speculation that it is possible for each domain to target a copy of H3-AcK9 on the same nucleosome or adjacent nucleosomes and consequently, the spatial arrangement of the two BrDs may require greater separation in Pb1 [73]. The shorter separation distance between these two bromodomains in isoform 3 raises the question of whether alternate patterns can act as the binding target or if Pb1 itself has primary and secondary chromatin binding targets. Finally, the most extreme isoform loses all but the polybromo region, suggesting it is able to target the same acetyl-nucleosome sites, but imparts no nucleation of PBAF via the BAH domains. In effect, this form of the protein would block PBAF assembly at chromatin.
Regulatory roles of Polybromo-1
The details about which genes are under direct regulation by Pb1 and the PBAF complex are only beginning to emerge and already roles are being discovered in cell division, transcription and, under aberrant conditions, cancer. Initial examination of mammalian systems indicate that Pb1 is required for cardiac and placental development via Pb1 gene targets, including the retinoic acid-induced genes RARβ2 and CRABPII (cellular retinoic acid binding protein II, found in mouse) [112]. Retinoic acid receptors (RARs) activate transcription by recruiting coactivator complexes such as histone acetyltransferases (HAT) and the mediator complex, to increase chromatin accessibility by general transcription factors and to promote transcription initiation. Ablation of the PB1 gene led to a partial decrease of the expression of RARβ2 gene, whereas CRABPII expression was totally abolished [112]. It was determined that transcriptional regulation by RARs involved distinct coactivator complexes, including the hSWI/SNF and ISWI complexes, the mediator complex, HATs, such as CBP/p300, pCAF and p160 proteins. The role of Pb1 in retinoid-mediated transcription suggested that PBAF is an important coactivator for RARs. Recently, it was shown that Pb1 binds to the p21 promoter to regulate transcription [113].
Pb1 mutations occur in approximately 4% of sporadic breast cancers. The observation that different breast tumor samples contained mutations in the PB1 gene, resulting in a truncated form of polybromo-1 has led to speculation regarding the tumor suppressor properties of Pb1 [113]. The mechanism remains unclear, but the mutant protein has been shown to redirect the estrogen-stimulated estrogen receptor pathway thereby preventing transcriptional activity [114]. Because wild-type Pb1 plays a central role in cell cycle arrest in response to growth inhibitory signals, it has been proposed that cell cycle exit is adversely impacted by the mutation [113]. The observation that the mutant PB1 gene has a deletion resulting in Pb1 proteins missing four bromodomains and a portion of the first BAH domain (Fig. 5) might cause speculation that if the tandem bromodomains do in fact target acetyl-nucleosome sites for proper localization, their deletion should be detrimental to Pb1 function. Recently, these mutants were shown to be incorrectly localized in the cell [114] further supporting the notion that Pb1 and the PBAF complex require the targeting of specific chromatin sites to execute proper function
Acknowledgments
We thank the Thompson lab for helpful discussions and contributions. This research was supported in part by a Faculty Scholarship Grant from Michigan Technological University and National Institutes of Health Grant GM076055.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kornberg RD. Structure of chromatin. Ann Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg R, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg RD, Lorch Y. Chromatin structure and transcription. Ann Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- 4.Widom J. Structure, dynamics, and function of chromatin in vitro. Ann Rev Biophys Biomol Str. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 5.Becker P, Hörz W. Atp-dependent nucleosome remodeling. Ann Rev Biochem. 2002;71:243–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 6.Workman J, Kingston R. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Ann Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 7.McGhee JD, Felsenfeld G. Nucleosome structure. Ann Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- 8.Ramakrishnan V. Histone structure and the organization of the nucleosome. Ann Rev Biophys Biomol Str. 1997;26:83–112. doi: 10.1146/annurev.biophys.26.1.83. [DOI] [PubMed] [Google Scholar]
- 9.Yoon HG, Choi Y, Cole PA, Wong JM. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol Cell Biol. 2005;25:324–335. doi: 10.1128/MCB.25.1.324-335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng Z, Chen CJ, Chamberlin M, Lu F, Blobel GA, Speicher D, Cirillo LA, Zaret KS, Lieberman PM. The cbp bromodomain and nucleosome targeting are required for zta-directed nucleosome acetylation and transcription activation. Mol Cell Biol. 2003;23:2633–2644. doi: 10.1128/MCB.23.8.2633-2644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winston F, Allis C. The bromodomain: A chromatin-targeting module? Nat Struct Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 12.Nie Z, Xue Y, Yang D, Zhou S, Deroo B, Archer T, Wang W. A specificity and targeting subunit of a human swi/snf family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins R, Furukawa T, Tanese N, Treisman J. Osa associates with the brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 1999;18:7029–7040. doi: 10.1093/emboj/18.24.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson C, Workman J. Promoter targeting and chromatin remodeling by the swi/snf complex. Curr Opin Genet Dev. 2000;10:187–92. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 15.Belandia B, Orford R, Hurst H, Parker M. Targeting of swi/snf chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 2002;21:4094–4103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montecino M, Stein J, Stein G, Lian J, van Wijnen A, Cruzat F, Gutierrez S, Olate J, Marcellini S, Gutierrez J. Nucleosome organization and targeting of swi/snf chromatin-remodeling complexes: Contributions of the DNA sequence. Biochem Cell Biol. 2007;85:419–425. doi: 10.1139/O07-070. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Cote J, Xue Y, Zhou S, Khavari P, Biggar S, Muchardt C, Kalpana G, Goff S, Yaniv M, Workman J, Crabtree G. Purification and biochemical heterogeneity of the mammalian swi-snf complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Xue Y, Zhou S, Kuo A, Cairns B, Crabtree G. Diversity and specialization of mammalian swi/snf complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 20.Phelan M, Sif S, Narlikar G, Kingston R. Reconstitution of a core chromatin remodeling complex from swi/snf subunits. Mol Cell. 1999;3:247–53. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 21.Zhao K, Wang W, Rando O, Xue Y, Swiderek K, Kuo A, Crabtree G. Rapid and phosphoinositol-dependent binding of the swi/snf-like BAF complex to chromatin after t lymphocyte receptor signaling. Cell. 1998;95:625–36. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 22.Rando OJ, Zhao KJ, Janmey P, Crabtree GR. Phosphatidylinositol-dependent actin filament binding by the swi/snf-like baf chromatin remodeling complex. Proc Natl Acad Sci USA. 2002;99:2824–2829. doi: 10.1073/pnas.032662899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Archer T. Regulating swi/snf subunit levels via protein-protein interactions and proteasomal degradation. Mol Cell Biol. 2005;25:9016–9027. doi: 10.1128/MCB.25.20.9016-9027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall T, Link K, Petre-Draviam C, Knudsen K. Differential requirement of swi/snf for androgen receptor activity. J Biol Chem. 2003;278:30605–13. doi: 10.1074/jbc.M304582200. [DOI] [PubMed] [Google Scholar]
- 25.García-Pedrero J, Kiskinis E, Parker M, Belandia B. The swi/snf chromatin remodeling subunit baf57 is a critical regulator of estrogen receptor function in breast cancer cells. J Biol Chem. 2006;281:22656–64. doi: 10.1074/jbc.M602561200. [DOI] [PubMed] [Google Scholar]
- 26.Hsiao P, Fryer C, Trotter K, Wang W, Archer T. Baf60a mediates critical interactions between nuclear receptors and the brg1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23:6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Link K, Burd C, Williams E, Marshall T, Rosson G, Henry E, Weissman B, Knudsen K. Baf57 governs androgen receptor action and androgen-dependent proliferation through swi/snf. Mol Cell Biol. 2005;25:2200–2215. doi: 10.1128/MCB.25.6.2200-2215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Davies K, Allen J, Zhu L, Pestell R. Cell cycle arrest and repression of cyclin d1 transcription by ini1/hsnf5. Mol Cell Biol. 2002;22:5975–5988. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oruetxebarria I, Venturini F, Kekarainen T, Houweling A, Zuijderduijn L. P16ink4a is required for hsnf5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cell. J Biol Chem. 2004;279:3807–3816. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- 30.Versteege I, Medjkane S, Rouillard D, Delattre O. A key role of the hsnf5/ini1 tumour suppressor in the control of the g1-s transition of the cell cycle. Oncogene. 2002;21:6403–6412. doi: 10.1038/sj.onc.1205841. [DOI] [PubMed] [Google Scholar]
- 31.Yan ZJ, Cui KR, Murray DM, Ling C, Xue YT, Gerstein A, Parsons R, Zhao KJ, Wang WD. Pbaf chromatin-remodeling complex requires a novel specificity subunit, baf200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debril M, Gelman L, Fayard E, Annicotte J, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the swi/snf complex through the baf60c subunit. J Biol Chem. 2004;279:16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- 33.Lickert H, Takeuchi J, Both I, Walls J, McAuliffe F, Adamson S, Henkelman R, Wrana J, Rossant J, Bruneau B. Baf60c is essential for function of baf chromatin remodelling complexes in heart development. Nature. 2004;432:107–12. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 34.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bettinger B, Gilbert D, Amberg D. Actin up in the nucleus. Nat Rev Molec Cell Biol. 2004;5:410–415. doi: 10.1038/nrm1370. [DOI] [PubMed] [Google Scholar]
- 36.Xue Y, Canman J, Lee C, Nie Z, Yang D, Moreno G, Young M, Salmon E, Wang W. The human swi/snf-b chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci USA. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nature Rev Mol Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- 38.Dallas P, Pacchione S, Wilsker D, Bowrin V, Kobayashi R, Moran E. The human swi-snf complex protein p270 is an arid family member with non-sequence-specific DNA binding activity. Mol Cell Biol. 2000;20:3137–3146. doi: 10.1128/mcb.20.9.3137-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandrasekaran R, Thompson M. Expression, purification and characterization of individual bromodomains from human polybromo-1. Prot Expr Purif. 2006;50:111–117. doi: 10.1016/j.pep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Baetz K, Krogan N, Emili A, Greenblatt J, Hieter P. The ctf13-30/ctf13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex rsc, which is required for the establishment of sister chromatid cohesion. Mol Cell Biol. 2004;24:1232–1244. doi: 10.1128/MCB.24.3.1232-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu J, Huang J, Meluh P, Laurent B. The yeast rsc chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol Cell Biol. 2003;23:3202–3215. doi: 10.1128/MCB.23.9.3202-3215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, Hsu J, Laurent B. The rsc nucleosome-remodeling complex is required for cohesin's association with chromosome arms. Mol Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 43.Zeng L, Zhou M. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 44.Jeanmougin F, Wurtz J, Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 45.Filetici P, Ornaghi P, Ballario P. The bromodomain: A chromatin browser. Front Biosci. 2001;6:866–876. doi: 10.2741/filetici. [DOI] [PubMed] [Google Scholar]
- 46.Goodwin G, Nicolas R. The bah domain, polybromo and the rsc chromatin remodelling complex. Gene. 2001;268:1–7. doi: 10.1016/s0378-1119(01)00428-0. [DOI] [PubMed] [Google Scholar]
- 47.Callebaut I, Courvalin JC, Mornon JP. The bah (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446:189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 48.Thomas J, Travers A. Hmg1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 49.Thomas J. Hmg1 and 2: Architectural DNA-binding proteins. Biochem Soc Trans. 2001;29:395–401. doi: 10.1042/bst0290395. [DOI] [PubMed] [Google Scholar]
- 50.Štros M, Launholt D, Grasser K. The hmg-box: A versatile protein domain occurring in a wide variety of DNA-binding proteins. Cellular and Molecular Life Sciences (CMLS) 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul S, Madden T, Schäffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strahl BD, Allis CD. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 53.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Development. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 55.Cheung P, Allis CD, Sassone-Corsi P. Signalling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 56.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 57.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 58.Loyola A, LeRoy G, Wang YH, Reinberg D. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev. 2001;15:2837–2851. doi: 10.1101/gad.937401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 60.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 61.Staal A, Enserink JM, Stein JL, Stein GS, Van Wijnen AJ. Molecular characterization of celtix-1, a bromodomain protein interacting with the transcription factor interferon regulatory factor 2. J Cellular Physiology. 2000;185:269–279. doi: 10.1002/1097-4652(200011)185:2<269::AID-JCP12>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 62.Garzón JFG, Kupitz CJ, Bailey JD, Thompson M. Acetylation-dependent binding analysis of the yeast gcn5 bromodomain protein. Am J Undergrad Res. 2008;7:19–25. [Google Scholar]
- 63.Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Molecular Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 64.Cairns B, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R. Rsc, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 65.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Ann Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 66.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 67.Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone h4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE. Identification of transcription complexes that contain the double bromodomain protein brd2 and chromatin remodeling machines. J Proteome Res. 2006;5:502–511. doi: 10.1021/pr050430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horn P, Peterson C. The bromodomain: A regulator of atp-dependent chromatin remodeling? Front Biosci. 2001;6:1019–1023. doi: 10.2741/horn. [DOI] [PubMed] [Google Scholar]
- 70.Schweiger MR, You JX, Howley PM. Bromodomain protein 4 mediates the papillomavirus e2 transcriptional activation function. J Virol. 2006;80:4276–4285. doi: 10.1128/JVI.80.9.4276-4285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kupitz C, Chandrasekaran R, Thompson M. Kinetic analysis of acetylation-dependent Pb1 bromodomain-histone interactions. Biophys Chem. 2008;136:7–12. doi: 10.1016/j.bpc.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chandrasekaran R, Thompson M. Thermodynamic analysis of the acetylation dependence of bromodomain-histone interactions. Anal Biochem. 2008;374:304–312. doi: 10.1016/j.ab.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chandrasekaran R, Thompson M. Polybromo-1 bromodomains bind histone h3 at specific acetyl-lysine positions. Biochem Biophys Res Comm. 2007;355:661–666. doi: 10.1016/j.bbrc.2007.01.193. [DOI] [PubMed] [Google Scholar]
- 74.Mohrmann L, Verrijzer CP. Composition and functional specificity of swi2/snf2 class chromatin remodeling complexes. Biochimica Et Biophysica Acta-Gene Structure and Expression. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 75.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns B. Tandem bromodomains in the chromatin remodeler rsc recognize acetylated histone h3 lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hassan AH, Awad S, Prochasson P. The swi2/snf2 bromodomain is required for the displacement of saga and the octamer transfer of saga-acetylated nucleosomes. J Biol Chem. 2006;281:18126–18134. doi: 10.1074/jbc.M602851200. [DOI] [PubMed] [Google Scholar]
- 77.Carey M, Li B, Workman JL. Rsc exploits histone acetylation to abrogate the nucleosomal block to rna polymerase II elongation. Molecular Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cairns B, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg R, Winston F. Two functionally distinct forms of the rsc nucleosome-remodeling complex, containing essential at hook, bah and bromodomains. Molecular Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 79.Zeng L, Yap KL, Ivanov AV, Wang X, Mujtaba S, Plotnikova O, Rauscher FJ, Zhou MM. Structural insights into human kap1 phd finger-bromodomain and its role in gene silencing. Nat Struct Mol Biol. 2008;15:626–633. doi: 10.1038/nsmb.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun H, Liu J, Zhang J, Shen W, Huang H, Xu C, Dai H, Wu J, Shi Y. Solution structure of brd7 bromodomain and its interaction with acetylated peptides from histone h3 and h4. Biochem Biophys Res Commun. 2007;358:435–441. doi: 10.1016/j.bbrc.2007.04.139. [DOI] [PubMed] [Google Scholar]
- 81.Hudson B, Martinez-Yamout M, Dyson H, Wright P. Solution structure and acetyl-lysine binding activity of the gcn5 bromodomain. J Mol Biol. 2000;304:355–370. doi: 10.1006/jmbi.2000.4207. [DOI] [PubMed] [Google Scholar]
- 82.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human taf(ii)250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 83.VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Molecular Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mujtaba S, He Y, Zeng L, Farooq A, Carlson J, Ott M, Verdin E, Zhou M. Structural basis of lysine-acetylated hiv-1 tat recognition by pcaf bromodomain. Mol Cell. 2002;9:575–586. doi: 10.1016/s1097-2765(02)00483-5. [DOI] [PubMed] [Google Scholar]
- 85.Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le N, Ronai Z, Zhou M. Structural mechanism of the bromodomain of the coactivator cbp in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 86.Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structural basis of site-specific histone recognition by the bromodomains of human coactivators pcaf and cbp/p300. Structure. 2008;16:643–652. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shen WQ, Xu C, Huang W, Zhang JH, Carlson JE, Tu XM, Wu JH, Shi YY. Solution structure of human brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46:2100–2110. doi: 10.1021/bi0611208. [DOI] [PubMed] [Google Scholar]
- 88.Pamblanco M, Poveda A, Sendra R, Rodriguez-Navarro S, Perez-Ortin JE, Tordera V. Bromodomain factor 1 (bdf1) protein interacts with histones. Febs Letters. 2001;496:31–35. doi: 10.1016/s0014-5793(01)02397-3. [DOI] [PubMed] [Google Scholar]
- 89.Pizzitutti F, Giansanti A, Ballario P, Ornaghi P, Torreri P, Ciccotti G, Filetici P. The role of loop za and pro371 in the function of yeast gcn5p bromodomain revealed through molecular dynamics and experiment. J Mol Recognit. 2006;19:1–9. doi: 10.1002/jmr.748. [DOI] [PubMed] [Google Scholar]
- 90.Pearson W. Searching protein sequence libraries: Comparison of the sensitivity and selectivity of the smith-waterman and fasta algorithms. Proc Nat Acad Sci USA. 1988;85:2444–2448. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- 91.Thompson JD, Higgins DG, Gibson TJ. Clustal w: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oliver A, Jones S, Roe S, Matthews S, Goodwin G, Pearl L. Crystal structure of the proximal bah domain of the polybromo protein. Biochem J. 2005;389:657–64. doi: 10.1042/BJ20050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Z, Hayashi M, Merkel O, Stillman B, Xu R. Structure and function of the bah-containing domain of orc1p in epigenetic silencing. EMBO J. 2002;21:4600–4611. doi: 10.1093/emboj/cdf468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hou Z, Danzer JR, Fox CA, Keck JL. Structure of the sir3 protein bromo adjacent homology (bah) domain from s. Cerevisiae at 1.95 a resolution. Protein Sci. 2006;15:1182–1186. doi: 10.1110/ps.052061006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hsu HC, Stillman B, Xu RM. Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing. Proc Natl Acad Sci USA. 2005;102:8519–8524. doi: 10.1073/pnas.0502946102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hou Z, Bernstein DA, Fox CA, Keck JL. Structural basis of the sir1-origin recognition complex interaction in transcriptional silencing. Proc Natl Acad Sci USA. 2005;102:8489–8494. doi: 10.1073/pnas.0503525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wong M, Scott-Drew S, Hayes M, Howard P, Murray J. Rsc2 encoding a component of the rsc nucleosome remodeling complex is essential for 2 micron plasmid maintenance in saccharomyces cerevisiae. Mol Cell Biol. 2002;22:4218–4229. doi: 10.1128/MCB.22.12.4218-4229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Noguchi K, Vassilev A, Ghosh S, Yates J, DePamphilis M. The bah domain facilitates the ability of human orc1 protein to activate replication origins in vivo. EMBO J. 2006;25:5372–5382. doi: 10.1038/sj.emboj.7601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bell S, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of orc1p reveals similarity to regulators of DNA-replication and transcriptional silencing. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 100.Gardner K, Rine J, Fox C. A region of the sir1 protein dedicated to recognition of a silencer and required for interaction with the orc1 protein in saccharomyces cerevisiae. Genetics. 1999;151:31–44. doi: 10.1093/genetics/151.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and sir1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 102.Zhang Q, Wang Y. High mobility group proteins and their post-translational modifications. Biochim Biophys Acta. 2008;1784:1159–66. doi: 10.1016/j.bbapap.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bowles J, Schepers G, Koopman P. Phylogeny of the sox family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 104.Wegner M. From head to toes: The multiple facets of sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wiβmüller S, Kosian T, Wolf M, Finzsch M, Wegner* M. The high-mobility-group domain of sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res. 2006;34:1735–1744. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Masse JE, Wong B, Yen YM, Allain FHT, Johnson RC, Feigon J. The s. Cerevisiae architectural HMGB protein nhp6a complexed with DNA: DNA and protein conformational changes upon binding. J Mol Biol. 2002;323:263–284. doi: 10.1016/s0022-2836(02)00938-5. [DOI] [PubMed] [Google Scholar]
- 107.Weir HM, Kraulis PJ, Hill CS, Raine AR, Laue ED, Thomas JO. Structure of the hmg box motif in the b-domain of hmg1. EMBO J. 1993;12:1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marmorstein L, Kinev A, Chan G, Bochar D, Beniya H, Epstein J, Yen T, Shiekhattar R. A human brca2 complex containing a structural DNA binding component influences cell cycle progression. Cell. 2001;104:247–257. doi: 10.1016/s0092-8674(01)00209-4. [DOI] [PubMed] [Google Scholar]
- 109.Dai Y, Wong B, Yen Y, Oettinger M, Kwon J, Johnson R. Determinants of hmgb proteins required to promote rag1/2-recombination signal sequence complex assembly and catalysis during v(d)j recombination. Mol Cell Biol. 2005;25:4413–4425. doi: 10.1128/MCB.25.11.4413-4425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Leea Y, Shinb H, Choib W, Ahna S, Kim W. Characterization of human smarce1r high-mobility-group protein. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 2002;1574:269–276. doi: 10.1016/s0167-4781(01)00373-6. [DOI] [PubMed] [Google Scholar]
- 111.Horikawa I, Barrett J. Cdna cloning of the human polybromo-1 gene on chromosome 3p21. DNA Sequence. 2002;13:211–215. doi: 10.1080/1042517021000021590. [DOI] [PubMed] [Google Scholar]
- 112.Wang Z, Zhai WG, Richardson JA, Olson EN, Meneses JJ, Firpo MT, Kang CH, Skarnes WC, Tjian R. Polybromo protein baf180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18:3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xia W, Nagase S, Montia A, Kalachikov S, Keniry M, Su T, Memeo L, Hibshoosh H, Parsons R. Baf180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008;68:1667–74. doi: 10.1158/0008-5472.CAN-07-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.A. Montia (2007), Vol. PhD, pp. 104, Columbia University, New York.