Abstract
The Drosophila eag gene has been shown to regulate neuronal excitability (Wu et al., 1983), olfaction (Dubin et al., 1998), associative learning (Griffith et al., 1994) and larval locomotion (Wang et al., 2002a). Not all of the roles of this gene in these processes can be explained by its function as a voltage-gated potassium channel (e.g. Zhong and Wu, 1991). In this study, we show that the eag gene is spliced in a PKA- and PKC-regulated manner to produce a protein lacking channel domains. This protein, in the context of activated PKA, can engage cellular signaling pathways that alter cell structure. Nuclear localization is necessary for C-terminal-mediated effects, which also require MAPK. The requirement for PKA/PKC activation in the synthesis and function of this novel protein suggests that it may couple membrane events to nuclear signaling to regulate neuronal function on long time scales.
Introduction
The ether-à-go-go (eag) gene was first identified as a locus that caused leg-shaking in Drosophila in response to ether anesthesia (Kaplan and Trout, 1969). Subsequent cloning of the gene revealed that eag encodes a voltage-gated potassium channel (Robertson et al., 1996; Warmke et al., 1991) which defines a novel family of potassium channels that includes the human long Q-T syndrome gene HERG (Curran et al., 1995; Warmke and Ganetzky, 1994).
Recordings from animals that have mutations in eag demonstrate an increase in presynaptic release and spontaneous firing (Wu et al., 1983), consistent with loss of a potassium conductance. Closer examination of an array of eag alleles, however, paints a more complex picture of the role of eag in regulation of excitability. Voltage clamp recordings from larval muscle showed that, in eag mutants, several other identifiable potassium conductances were affected, and more interestingly, there were allele-specific interactions between eag and Shaker, a gene encoding a fast-inactivating potassium channel (Zhong and Wu, 1991; Zhong and Wu, 1993). These data indicate that eag can either directly or indirectly affect the activity of other channels.
Several mechanisms could be invoked to explain these effects of eag mutations. The most direct is that the Eag protein interacts or coassembles with other potassium channel subunits to form unique conductances. Testing this in heterologous expression systems has yielded conflicting results (Chen et al., 1996; Chen et al., 2000; Tang et al., 1998). A second (but not mutually exclusive) possibility is that the Eag channel could serve as a scaffold and regulator for signal transduction molecules at the cell membrane. Eag is known to bind a variety of signaling molecules including calmodulin (Sun et al., 2004), dCASK (Marble et al., 2005), and CaMKII (Wang et al., 2002b), which it directly activates (Sun et al., 2004). Eag also has a cyclic nucleotide binding motif, and can be regulated by these second messengers (Bruggemann et al., 1993), although it is not known if this is due to direct binding. Recently, Eag has been shown to have a role in activation of MAPK pathways in a voltage-dependent, but conductance-independent manner (Hegle et al., 2006).
In this study we document a third mechanism for the regulation of cellular signaling processes by Eag. We show that eag transcripts can be alternatively spliced to produce a message encoding an 80kDa protein containing both N- and C-terminal sequences but lacking all channel-forming transmembrane domains. Production of this alternative splice form can be stimulated by calcium influx or activation of either PKA or PKC. In transfected cells, C-terminal fragments of the Eag protein can enter the nucleus and activate a MAPK pathway that alters cell morphology. The intact Eag80 splice product can also alter cell morphology, but only in the presence of activated PKA or PKC. These data demonstrate a non-channel role for the eag gene that may be important in long-term regulation of cellular function.
Experimental Procedures
Plasmids and construction
For COS cell expression, full length Eag cDNAs were cloned into pCDNA3. The pYFP-C1 vector was generated by replacing CFP of pCFP-C1(Clontech) with YFP from pYFP-N1(Clontech). Eag was cloned into pYFP-C1 to get pYFP-Eag, which expresses an Eag fusion protein with YFP at its N-terminal. To obtain endogenous Eag splice form cDNAs, nested PCR products from RT-PCR were cloned into the TOPO cloning vector (Invitrogen), and spliced clones verified by sequencing, resulting in TOPO-Eag80. Two XmnI sites flanking the splicing sites were used to swap the sequence from TOPO-Eag80 into pYFP-Eag to produce pYFP-Eag80. pCDNA3-80 kDa were produced by swapping the Eag sequence using EcoRI and BstEII sites between pCDNA3-Eag and pYFP-Eag80. PCR products for Δ1-675, Δ1-662, Δ1-700 were cloned into pYFP-C1 using EcoRI and XmaI sites. PCR based site-directed mutagenesis was done using the QuickChange kit (Stratagene) to produce point mutations. All clones and mutations were verified by sequencing. Activated PKA and PKC in pCDNA3 were gifts from Yi Zhou (Florida State University, Tallahassee). Myc-Eag plasmids were gifts from Gisela Wilson.
Materials and chemicals
TPA was from Sigma. Ionomycin, PKC/PKA inhibitor (bisindolylmalemide I), MAPK inhibitor (PD98059), translational inhibitor (Anisomycin) and transcriptional inhibitor (Actinomycin D) were from Calbiochem (La Jolla, CA). The specificity of bisindolylmaleimide I is dependent on the amount used; at low concentrations it is relatively specific for PKC, but at higher concentrations also inhibits PKA and GSK-3 (http://www.emdbiosciences.com/product/203290). Anti-Eag rabbit polyclonal antibodies were raised against the last 143 amino acids of Eag (aa 1032-1174) fused to GST. To express the C-terminal Eag fragment for antibody production, PCR product containing the sequence between aa 1032-1174 was cloned between EcoRI and XbaI sites of pGEX-2T. GST-1032-1174, purified as described previously (Sun et al., 2004) was used to immunize rabbits to produce polyclonal antibody. Other antibodies include FITC- and HRP-conjugated secondary antibodies (anti rabbit and anti-mouse, Jackson ImmunoResearch Laboratories), anti-Myc (Boehringer Mannheim), anti-GFP (Roche Applied Biosciences) and rabbit anti-Eag made against aa 755-771 of Eag (a gift from Gisela Wilson, UW Madison).
Tissue culture
COS cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% bovine serum at 37°C in an atmosphere containing 5% CO2. Plasmids were transfected into 50-80% confluent COS cells using FuGENE6 Transfection Regent (Roche Molecular Biochemicals) according to the manufacturer’s instructions.
Immunoblotting
After 24-29 hours transfection, cells incubated with or without inhibitor and/or TPA as indicated in figure legends were washed with PBS buffer and then directly lysed with 1X SDS sample buffer (2% SDS, 10% glycerol, 0.1 M dithiothreitol, 120 mM Tris-HCl, pH6.8, 0.02% bromphenol blue) and boiled for 15 min for immunoblot analysis. Cell lysates were resolved by 7.5 or 8.0% SDS-PAGE. The gel was electrophoretically transferred to nitrocellulose at 230 mA for 60 min and probed with anti-Eag, and anti-Myc in the case of detecting Myc-Eag. Bands were visualized with HRP-conjugated secondary antibodies and enhanced chemiluminescence reagents (ECL, Amersham-Pharmacia Biotech).
mRNA purification and reverse transcription
RT-PCR was carried out on cDNA made from both transfected COS cells and from Canton S wild type fly heads. COS cells were transfected using pCDNA3-Eag and treated with TPA for 5 hours to induce the production of Eag80. Total RNA from both COS cells and fly heads was extracted and purified using a commercial kit according to manufacturer’s instructions (TRIzol, Invitrogen). First-strand synthesis was conducted using the SuperScript kit (Invitrogen) by using an Eag specific primer (the last 18 bp of Eag coding sequence: 5’CAGGAAGTCCAGCAGCTTGCC3’). To amplify the Eag80 cDNA, the first-strand cDNA from reverse transcription was used as template with nested primers from regions we knew were present in Eag80 based on its antigenicity (outer primer pair: 5’CACATTCCTCGAGAACATCATCC3’ and 5’TTCTCGACATCACTTTGGCCGGAA3’ß; inner primer pair: 5’TCGATTTCCCGATCGTCTAC3’ and 5’GAGTGCGACGAAATTTGGAGA3’). Cycling parameters were 98°C for 2 min, followed by 35 cycles of 98°C for 12 s, 58°C for 30 s and 72°C for 10 s. PCR products were separated on agarose gels and extracted for cloning and sequencing. An alternative message formed by a splice donor in the coding sequence from genomic exon 2 attaching to a splice acceptor that is in coding sequence from genomic exon 14 excises a cryptic intron that contains exons 3-13. Splicing guidance is provided by information (branchpoint and pyrimidine-rich tract) that is present in coding sequences from genomic exon 14.
Co-immunoprecipitation from fly head extracts
Flies were grown on standard medium at 25°C. Canton-S was used as the wild type strain. Fly head extraction and co-immunoprecipitation were basically performed as described (Sun et al., 2004), with the exception that there was no addition of calcium or EDTA/EGTA in the buffer. Proteins bound to beads were resolved by 7.5% SDS-PAGE. The gel was electrophoretically transferred to nitrocellulose at 230 mA for 60 min and probed with anti-Eag. Bands were visualized with HRP-conjugated secondary antibodies and enhanced chemiluminescence reagents (ECL, Amersham-Pharmacia Biotech).
Immunostaining and immunohistochemistry
After transfection for 24-29 hours, COS cells plated and grown on 12 mm poly-D-Lysine-coated glass slides (Lab-Tek, Flaskette), were fixed in 4% paraformaldehyde in PBS at RT for 20 min. After washing 3 X with PBS, cells were permeabilized with 0.1% Triton in PBS for 15 min at room temperature and blocked for 20 min in PBS containing 10% normal goat serum. Cells were then incubated with a 1:200 dilution of anti-Eag in PBS with 5% normal goat serum overnight at 4°C, washed with PBS, and incubated with FITC-conjugated secondary antibody for 1 h at room temperature. After washing with PBS, coverslips were mounted in Vectashield-diamidino-phenylindole (DAPI) solution.
Results
Immunoblot analysis of Drosophila head extracts immunoprecipitated with an antibody generated against the distal C-terminal of the protein (aa 1032-1175) consistently identifies both the 130 kDa full length channel protein and an 80 kDa band (Figure 1, left panel). Both of these bands can also be detected using an antibody generated against a peptide (aa 755-771) that lies between the CaM- and CaMKII-binding domains of the channel (data not shown), suggesting that the 80 kDa protein is a genuine Eag product. The ability of this “Eag80” fragment to be recognized by these antibodies also suggests that it contains most of the C-terminus of the full length Eag protein, a region rich in binding sites for signal transduction molecules (Marble et al., 2005; Sun et al., 2004). A similar 80 kDa band is found in extracts of COS cells which have been transfected with a full length Eag cDNA (Figure 1, right panel). The regulated production of soluble fragments from transmembrane proteins has been shown to be an important form of signal transduction in many contexts (Ehrmann and Clausen, 2004). Since the transfected tissue culture system recapitulated the in vivo production of Eag80, we used it to study the generation and function of this channel fragment.
Figure 1. An 80 kDa Eag protein is present in both fly head extracts and COS cells transfected with a full length eag cDNA.
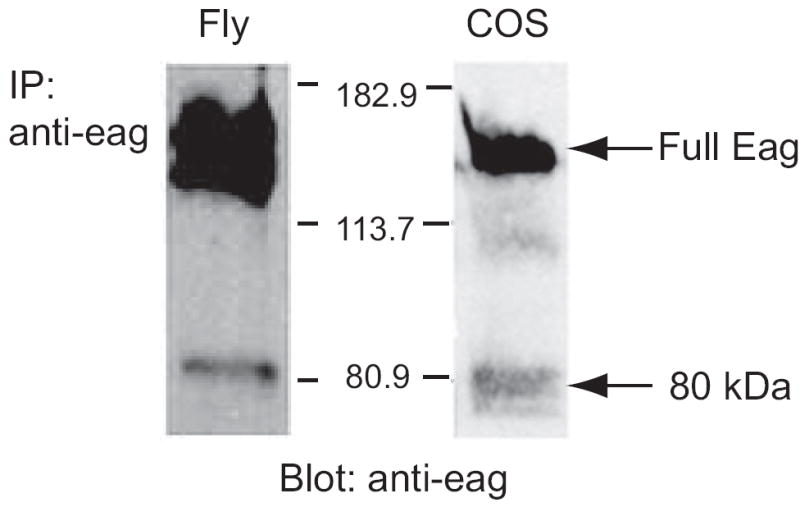
Left lane, immunoprecipitation of Eag from fly head extracts. Fly head extracts from adult wild type Canton-S flies were immunoprecipitated with anti-Eag antiserum generated against the distal C-terminal of the protein (aa 1032-1175) and protein A/G sepharose beads. Right lane, Eag-transfected COS cell lysates. Proteins of both lanes were resolved by 7.5% SDS-PAGE and transferred to nitrocellulose. Eag was detected by anti-Eag (1:2000) immunoblot. Full length and 80 kDa proteins are indicated by arrows.
Production of Eag80 in COS cells was induced by activation of multiple signal transduction cascades. Treatment of transfected cells with 162 nM TPA (Figure 2A), a phorbol activator of PKC, co-transfection with the catalytic subunit of PKA or activated PKC (Figure 2B) or treatment with 10 μM ionomycin (Figure 2C), a calcium ionophore, all significantly increase the amount of Eag80. There is also an increase in variety of anti-Eag-reactive bands of both higher and lower molecular weight (discussed below). With TPA treatment, accumulation of Eag80 begins within an hour and peaks at about 5 hours. Maximal induction with PKA or PKC occludes further TPA-induced increases, suggesting that the induction pathway is the same for all these agents. Inhibition of PKC and PKA with bisindolylmalemide or inhibition of MAP kinase by PD98059 can block the ability of TPA to induce Eag80 production in cells transfected with Eag alone, and significantly suppresses basal expression of the 80 kDa protein (Figure 2D and E).
Figure 2. The production of Eag80 is activity-dependent.
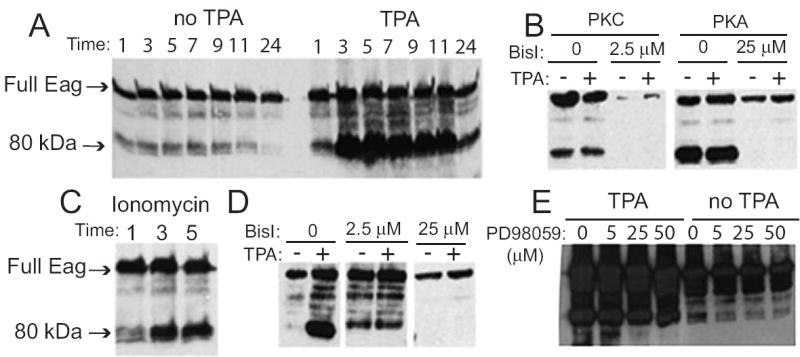
COS cells were transfected with Eag alone (A, C-E) or co-transfected with PKA or PKC (B). Cell lysates were separated by SDS-PAGE and Eag detected by immunoblot. TPA (162 nM, A) and ionophore (10 μM, C) can induce the production of Eag80 protein. (B) Cotransfection with PKA or PKC induces Eag80 and occludes TPA induction. Both effects are blocked by bisindolylmalemide I (BisI), an inhibitor of PKC (at low concentration) and PKA (at high concentration). (D) BisI blocks TPA-induced production of Eag80 in cells transfected with Eag alone. (E) MAPK inhibitor inhibits the production of Eag80 in cells transfected with Eag alone. Kinase inhibitors, at indicated concentrations, were added ca. 23 h after transfection, TPA was added 1 h later and cells harvested after an additional 5 hours.
Soluble signaling fragments of transmembrane proteins have been shown in a number of cases to be generated by proteolysis, for example Notch (Ye and Fortini, 2000) and L-type calcium channels (Gomez-Ospina et al., 2006). We investigated this possibility by testing the ability of various classes of protease inhibitors to block TPA-induced Eag80 production. In no case did we find a consistent reduction in TPA-induced Eag80, although higher molecular weight bands were attenuated (data not shown). In contrast, Anisomycin, an inhibitor of protein synthesis and Actinomycin D, an transcriptional inhibitor, were both potent blockers of Eag80 generation, but had little effect on the level of the full length Eag protein and bands with molecular weights greater than 80kDa (Figure 3A,B). These data suggest that Eag80 is produced from a novel, TPA-induced, transcript.
Figure 3. Eag80 is produced by alternative splicing of eag.
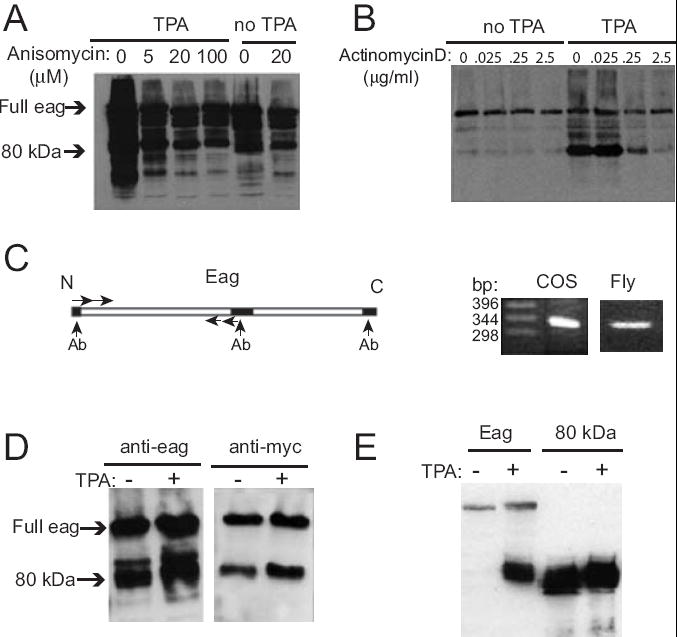
(A) Anisomycin, a translational inhibitor, inhibits the production of Eag80. 23 h after transfection, cells were incubated with anisomycin at indicated concentrations for 1 h, then for a further 5 h with or without addition of 162 nM TPA. (B) Actinomycin D, a translational inhibitor, significantly inhibits the production of Eag80. 23 h after transfection, cells were incubated with actinomycin D at indicated concentrations for 1 h, then for a further 5 h with or without addition of 162 nM TPA. (C) Reverse transcription identifies an Eag80 splice variant. mRNA was extracted from eag-transfected COS cells treated with TPA and wild type Canton-S heads. (D) Eag80 contains Eag N-terminal sequences. A full length eag cDNA with an N-terminal Myc tag was transfected into COS cells, which were then treated with or without 162 nM TPA for 5 h. Immunoblot of cell lysates were done using either anti-Myc or an anti-Eag antibody that recognizes the distal C-terminal. (E) A synthetic Δ67-698 eag cDNA produced a family of protein bands identical in size to Eag80 in the absence of TPA. Treatment of cells with TPA for 5 h did not alter the production of this protein, although it did induce production of identical bands from a full length cDNA.
There were two possible types of novel transcripts that could account for Eag80 production from a transfected full length Eag cDNA. The first would be the result of a cryptic transcriptional start that produced a truncated message in which the first methionine available for translation was distal to the Eag transmembrane domains. We tested this possibility by mutating methionine codons (M527, M536, M553 and M585) that could theoretically serve as a start to produce an 80 kDa protein. There was no effect on basal or TPA-induced Eag80 levels (data not shown).
The second type of novel transcript that could produce an 80 kDa protein from a full length cDNA would involve TPA induction of a cryptic in-frame splice event. Translation would start at the same methionine codon as for the full length protein, but removal of internal sequence from the mRNA would result in a smaller protein product that contained both N- and C-terminal Eag sequences. To test this, we transfected cells with an Eag cDNA that contained an N-terminal Myc tag (Wang et al., 2002b) and immunoblotted extracts with either anti-Myc or anti-Eag Ab that recognizes the distal C-terminal (Figure 3D). This experiment shows that Eag80 contains both the N- and C-terminal of full length Eag, consistent with the splicing model.
To determine where TPA-induced splicing was occurring, we carried out RT-PCR on mRNA extracted from transfected, TPA-treated, COS cells using nested primers from regions we knew were present in Eag80 based on its antigenicity (see above). Sequencing revealed a novel transcript that was generated by splicing between coding sequences of exons 2 and 14 (Figure 3C). Examination of the sequences of the eag cDNA and the eag genomic regions indicated that the sequences at the putative splice site, while contained in coding exons, conformed to canonical splice donor and acceptor sequences. Splice guidance is likely provided by putative branchpoint and pyrimidine-rich sequences found in exon 14. The fact that all of this splicing information is present in coding exons has allowed us to capture what is probably normally an event occurring in the pre-mRNA by transfection of a cDNA into heterologous cells.
RT-PCR on mRNA from fly heads using the same primers that were used on COS cell extracts produced an identical band, indicating that this splice also occurs in vivo from the genomic eag locus. The predicted protein product of this message would lack amino acids 66-699, which includes all the transmembrane domains of the channel. Consistent with this being the basis of Eag80 generation, a synthetic cDNA encoding the Δ66-699 mRNA constitutively produced a family of protein bands identical in size to Eag80 and was not TPA-inducible (Figure 3E). The multiple bands produced by the full length and Δ66-699 cDNAs are likely to be due to proteolysis of the N-terminal of Eag80, since anti-Myc recognizes only a single Eag80 band when the Myc-Eag cDNA is transfected (Figure 3D). To verify that splicing was responsible for generation of Eag80 in COS cells, the putative splice donor in exon 2 was mutated from AGGT to GCAA. Transfection of this cDNA produced only full length protein, even when TPA was added (Figure 4A).
Figure 4. Splicing of eag is required for production of Eag80 and alterations in COS cell morphology.
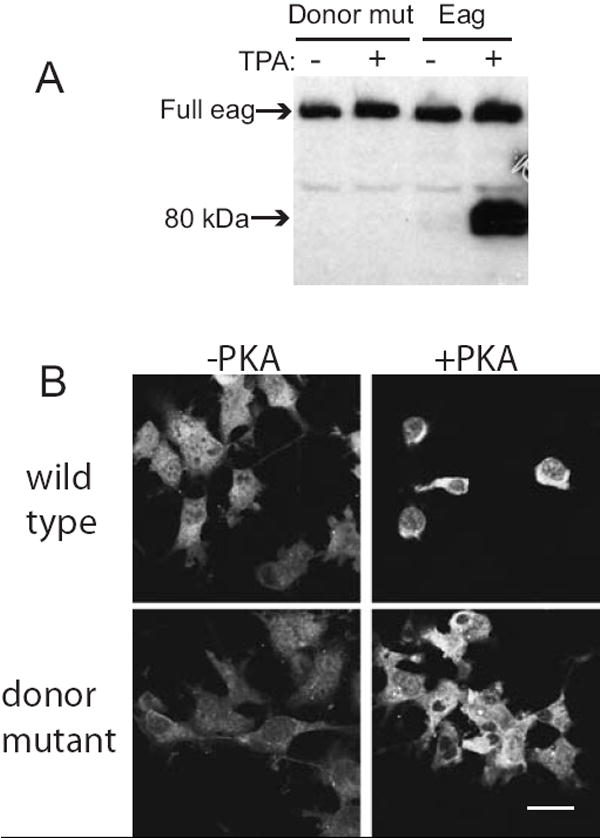
Full length eag cDNAs, either the wild type (panel A, right and panel B, top) or a non-splicing donor site mutant (panels A, left and panel B, bottom), were transfected into COS cells. (A) Cells were treated ± 16 μM TPA for 5 h and extracts immoblotted with anti-Eag. (B) Cells were cotransfected ± activated PKA and cell morphology assessed by staining with anti-Eag. Scale bar = 30 μm.
In the course of examining the induction of Eag80, we noted that conditions that stimulated production of this protein caused COS cells to round up and eventually float off the substrate. To determine if this morphological change was due to Eag80, we compared the effects of the non-splicing cDNA to wild type full length Eag. Figure 4B shows COS cells transfected with wild type or non-splicing Eag ± activated PKA. Co-transfection of a non-splicing Eag with PKA did not lead to changes in cell morphology, suggesting that eag splicing was required to initiate changes in cell morphology. It is also notable that the effects on cell morphology are not an artifact of dosage. PKA is a potent stimulator of protein production from cDNAs cloned into the vector we used (this is most easily seen when cell morphology is not altered: compare fluorescence intensity ± PKA for Figure 4B, bottom panels), but even when the non-splicing Eag is present at very high levels, it does not affect cell structure.
The effect of Eag80 on COS cells did not appear to be due to non-specific toxicity, since even when rounded up, cells were alive and able to exclude trypan blue (data not shown). We suspected that Eag might be activating an endogenous COS cell signal transduction pathway that controls cell architecture and that understanding the ability of Eag80 to hijack the COS pathway might give us insight into the function of this Eag splice form. The Eag C-terminal contains a number of domains (shown in Figure 6A) that could be involved in its actions in COS cells, including two putative nuclear localization (NLS) signals, one putative nuclear export signal (NES), binding domains for calmodulin and CaMKII (Sun et al., 2004) and a binding domain for dCASK that overlaps with the second NLS (Marble et al., 2005).
Figure 6. NLS1 of the Eag C-terminal is required for morphological effects in COS cells.
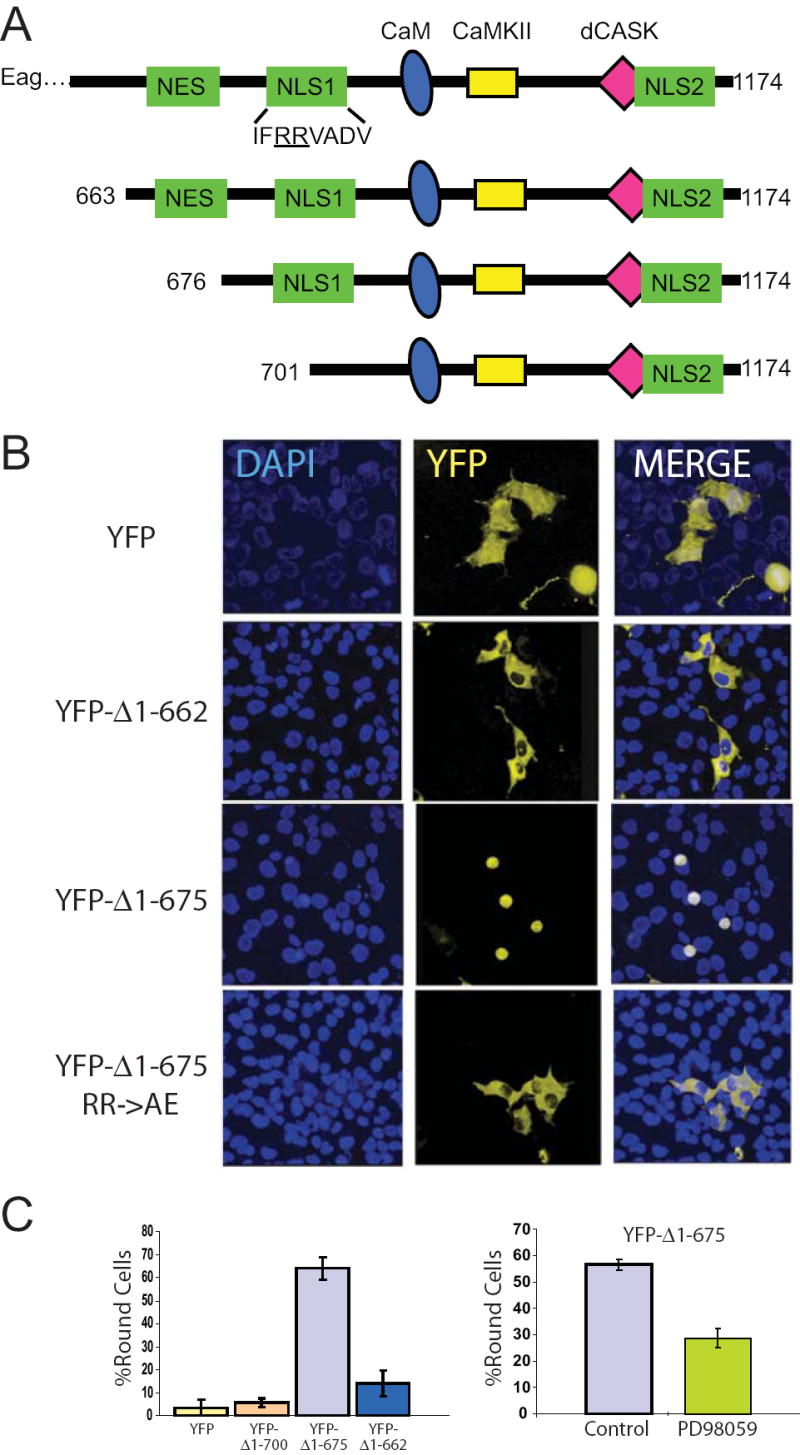
(A) Cartoon of Eag C-terminal domains and constructs. The Eag protein has multiple signaling modules including putative nuclear export (NES) and nuclear localization (NLS) sequences. It also contains binding sites for calmodulin (CaM), calcium/CaM-dependent protein kinase II (CaMKII) and dCASK. (B) Three YFP-tagged Eag C-terminal fragments (YFP-Δ1-662, which contains the putative NES, YFP-Δ1-675, which lacks the NES but contains both NLSs, and YFP-Δ1-662 with a point mutation in NLS1) were expressed in COS cell for 29 hours. Two examples of YFP-Δ1-675 are shown at different stages of the rounding process. YFP alone was expressed as control. DAPI staining shows nuclei. Scale bar = 30 μm. (C) Left panel, percentage round cells is shown for YFP, YFP-Δ1-700, which lacks both the NES and NLS1, YFP-Δ1-675 and YFP-Δ1-662. Right panel, MAPK inhibitor PD98059 (50 μM), added 6 h after transfection, inhibits YFP-Δ1-675-dependent cell rounding.
To investigate the function of Eag80, we transfected both N-terminal YFP-tagged Eag80 and untagged Eag80 into COS cells. In the absence of signal transduction activators, neither of these constructs altered cell morphology (Figure 5A, left). When cells were cotransfected with activated PKA (Figure 5A, right) or treated with TPA (data not shown), however, we saw cell rounding equivalent to that seen with full length Eag in the case of untagged Eag80, and a somewhat weaker effect with YFP-Eag80. Concurrent with the rounding of the cells, Eag80 moved to the plasma membrane in the presence of PKA. YFP-Eag80 showed some enrichment at the membrane, but not nearly to the same degree seen with untagged Eag80. Immunoblotting of extracts made from these COS cells showed that addition of PKA stimulates proteolysis of YFP-Eag80 which could lead to exposure of N-terminal Eag domains (Figure 5B) and release of soluble cytosolic YFP. Arrows indicate large fragments containing the intact C-terminal (recognized by anti-Eag, Figure 5B, left) and smaller fragments containing the N-terminal YFP tag (recognized by anti-GFP, Figure 5B, right). It is interesting to note that N-terminal proteolysis of the native Eag80 also occurs (see Figure 3D). In aggregate, these data suggest that activation of kinase is required for both the production and the function of Eag80. Residues in the N-terminal of Eag80 may be important for its ability to localize to the membrane and affect cell structure since blocking the N-terminal with YFP appears to decrease the ability of PKA to “activate” Eag80.
Figure 5. Kinase activation is required for Eag80 actions in COS cells.
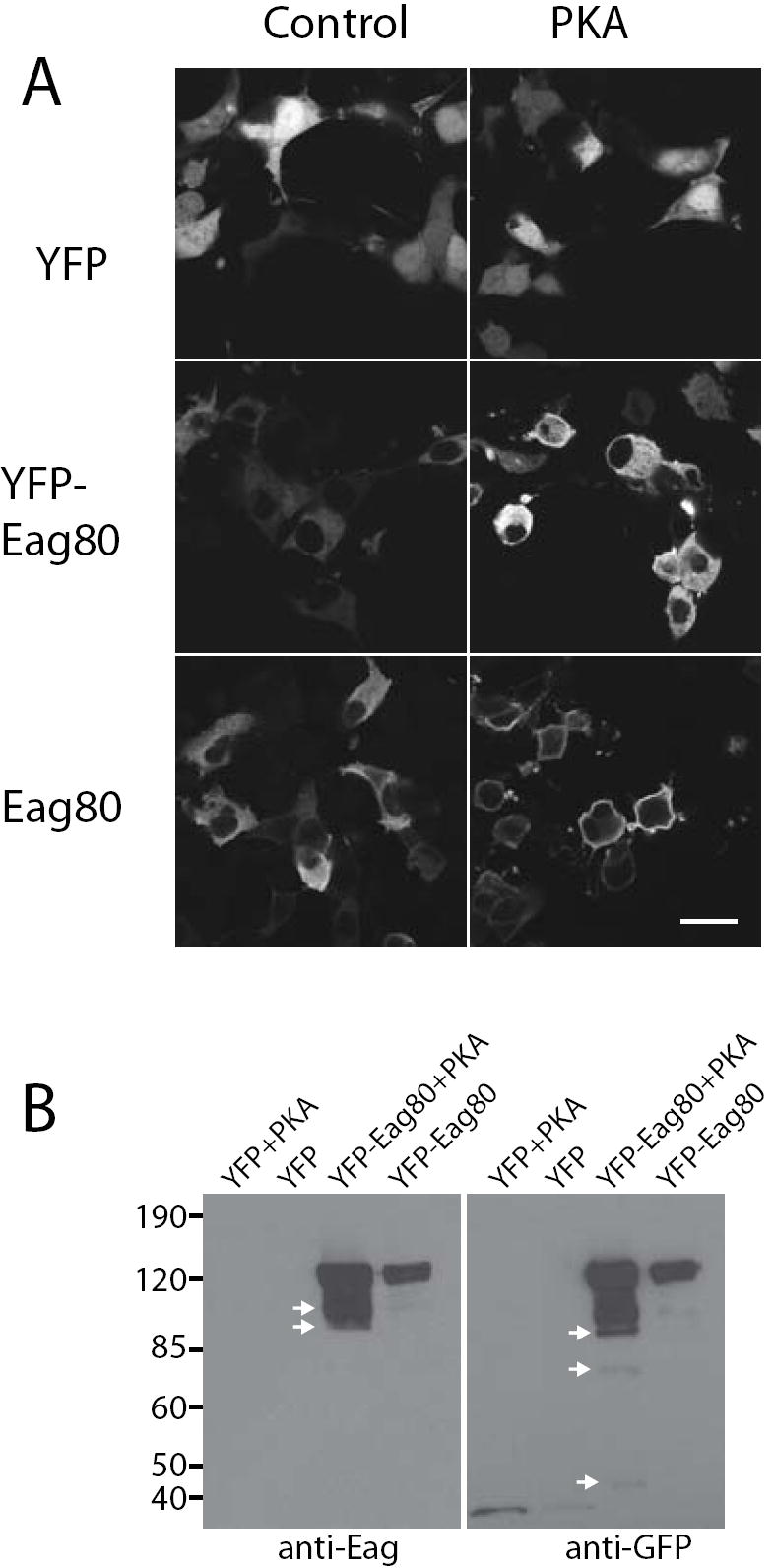
COS cells were transfected with a plasmid encoding YFP, YFP-Eag80 or untagged Eag-80, ± a plasmid containing cDNA for activated PKA. (A) YFP and YFP-Eag80 were imaged directly and Eag-80 was visualized by staining with anti-Eag. Scale bar = 30 μm. (B) Extracts of transfected cells were immunoblotted with anti-Eag (left) or anti-GFP (right).
One model for Eag80 regulation suggested by this result is that the N-terminal might be an inhibitory domain that gates the ability of functional domains in the C-terminal to interact with cellular pathways. This model would predict that truncation of the N-terminal of Eag80 would eliminate the need for concurrent kinase activation. To determine if Eag C-terminal signal transduction domains were responsible for these cellular events, we transfected constructs containing YFP-Eag C-terminal fusions into COS cells in the absence of N-terminal sequences and signal transduction activators. YFP-Δ1-663, which contains all of the C-terminal domains, failed to cause cells to round up (Figure 6B and C). A truncation that removed the NES (YFP-Δ1-675) but left NLS1 intact phenocopied the effects of TPA treatment on cells transfected with full length Eag. Removal (YFP-Δ1-701) or mutation (YFP-Δ1-675 RR->AE) of NLS1 blocked the effects on cell morphology, as did treatment of cells with a MAPK inhibitor. Both mutation of NLS1 (Figure 6C, bottom panels) and treatment with MAPK inhibitor (data not shown) blocked nuclear localization. These data suggest that nuclear localization is required for the Eag C-terminal to activate a MAPK signal transduction pathway that regulates cell morphology, but that other domains within the C-terminal (e.g. aa 663-674) may be inhibitory.
Discussion
In this study we demonstrate that the Drosophila eag potassium channel has a non-channel splice form that can function as an activator of signal transduction pathways that alter cell structure. This splice form is generated by a regulated splicing event that can be stimulated by calcium and both PKC and PKA (Figure 2). The protein product, Eag80, contains both N- and C-terminal domains of the canonical Eag channel protein, but does not have any of the channel forming domains and thus cannot conduct ions or act as a membrane voltage sensor.
While it is clear that the major role of voltage-gated channels in the brain is to conduct ions and regulate the excitability and firing of neurons, in the past several years many channels have been shown to have functions that are independent of their ion conducting ability (Kaczmarek, 2006). Most of these non-conducting functions involve domains of the ion channel protein that have enzymatic activity or which provide a scaffolding function for other proteins. The full length Drosophila eag potassium channel has been shown to have several non-conducting functions. The intracellular C-terminal contains a number of binding sites for signal transduction molecules, some of which (CASK, CaM and CaMKII) regulate Eag channel activity (Marble et al., 2005; Wang et al., 2002b). The binding site for CaMKII is unique in that it can serve both as a scaffold to bring the kinase to the channel and as an activator of the kinase (Sun et al., 2004).
The roles of the C-terminal scaffolding modules in Eag are likely to be affected by the localization of that domain, i.e. when attached to the full length channel, they are in close proximity to the membrane, but when they are present in Eag80, they are no longer obligatorily membrane bound. The localization of soluble Eag80 appears to be both dynamic and complex. The complete Eag80 protein localizes to the membrane in a PKA-dependent manner (Figure 5), but the free C-terminal (Δ1-675, Figure 6) localizes to the nucleus. The nuclear localization of this fragment is required for MAPK-dependent morphological effects, although it does not appear to be a direct transcriptional activator (fusion of this fragment to GAL4 does not activate a UAS reporter, data not shown). How Eag80, which does not appear to transit to the nucleus at high levels, functions to affect cell structure remains to be determined, but its dynamic localization pattern suggests that it may have interactions in multiple cellular compartments.
Eag has also been shown to activate p38 MAPK and stimulate proliferation of several cell lines in a voltage-dependent manner that does not require ion flux (Hegle et al., 2006). Activation is regulated by channel conformation; constitutively “open” channels are defective for the proliferative response. This non-conducting function of the Eag channel does not appear to be related to the Eag80 splice form since the “open” mutants that block proliferation do not appreciably alter the levels of Eag80 (data not shown). It will be interesting to determine if these two forms of Eag use a common molecular interaction with MAPK or its regulators.
Once synthesized, the ability of Eag80 to effect changes in cell morphology in transfected cells is dependent on activation of PKA (Figure 5). The dual requirement for kinase activation in synthesis and function of this protein suggests that it could function as a biochemical coincidence detector in neurons. Synthesis of Eag80 could be stimulated by activation of PKC and not be able to carry out its signaling function unless PKA was concurrently activated. Another scenario for Eag80 function would involve it serving as a marker of previous kinase activation that can respond to a subsequently elevation in PKA activity more quickly.
Activity-dependent splicing of ion channels gene is a common finding (McKee et al., 2007), and can serve to fine tune the kinetic properties of channels to meet changes in activity (Chaudhuri et al., 2004; Xie and Black, 2001). To our knowledge this is the first example of an activity-dependent splicing event in a voltage-gated ion channel gene that generates a non-conducting signaling protein. These data suggest that the eag gene may have roles in cell regulation that are unrelated to membrane ion flux and could serve as part of nuclear loop for regulation of neuronal properties.
Acknowledgments
This work was supported by NIH grant R01 GM54408 to L.C.G. We would like to thank Gisela Wilson for helpful discussion and for providing anti-Eag antiserum. We thank Ed Dougherty for imaging assistance. The Brandeis Biology confocal facility was supported by NIH grants P30 NS045713 and S10 RR16780.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bruggemann A, Pardo L, Stuhmer W, Pongs O. Ether-à-go-go encodes a voltage-gated channel permeable to K+ and Ca2+ and modulated by cAMP. Nature. 1993;365:445–448. doi: 10.1038/365445a0. [DOI] [PubMed] [Google Scholar]
- Chaudhuri D, Chang SY, DeMaria CD, Alvania RS, Soong TW, Yue DT. Alternative splicing as a molecular switch for Ca2+/calmodulin-dependent facilitation of P/Q-type Ca2+ channels. J Neurosci. 2004;24:6334–6342. doi: 10.1523/JNEUROSCI.1712-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M-L, Hoshi T, Wu C-F. Heteromeric interactions among K+ channel subunits from Shaker and eag families in Xenopus oocytes. Neuron. 1996;17:535–542. doi: 10.1016/s0896-6273(00)80185-3. [DOI] [PubMed] [Google Scholar]
- Chen ML, Hoshi T, Wu CF. Sh and eag K(+) channel subunit interaction in frog oocytes depends on level and time of expression. Biophys J. 2000;79:1358–1368. doi: 10.1016/S0006-3495(00)76388-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- Dubin AE, Liles MM, Harris GL. The K+ channel gene ether a go-go is required for the transduction of a subset of odorants in adult Drosophila melanogaster. J Neurosci. 1998;18:5603–5613. doi: 10.1523/JNEUROSCI.18-15-05603.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrmann M, Clausen T. Proteolysis as a regulatory mechanism. Annu Rev Genet. 2004;38:709–724. doi: 10.1146/annurev.genet.38.072902.093416. [DOI] [PubMed] [Google Scholar]
- Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LC, Wang J, Zhong Y, Wu CF, Greenspan RJ. Calcium/calmodulin-dependent protein kinase II and potassium channel subunit Eag similarly affect plasticity in Drosophila. Proc Natl Acad Sci USA. 1994;91:10044–10048. doi: 10.1073/pnas.91.21.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegle AP, Marble DD, Wilson GF. A voltage-driven switch for ion-independent signaling by ether-a-go-go K+ channels. Proc Natl Acad Sci U S A. 2006;103:2886–2891. doi: 10.1073/pnas.0505909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek LK. Non-conducting functions of voltage-gated ion channels. Nat Rev Neurosci. 2006;7:761–771. doi: 10.1038/nrn1988. [DOI] [PubMed] [Google Scholar]
- Kaplan WD, Trout WE. The behavior of four neurological mutants of Drosophila. Genetics. 1969;61:399–409. doi: 10.1093/genetics/61.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marble DD, Hegle AP, Snyder ED, 2nd, Dimitratos S, Bryant PJ, Wilson GF. Camguk/CASK enhances Ether-a-go-go potassium current by a phosphorylation-dependent mechanism. J Neurosci. 2005;25:4898–4907. doi: 10.1523/JNEUROSCI.4566-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AE, Neretti N, Carvalho LE, Meyer CA, Fox EA, Brodsky AS, Silver PA. Exon expression profiling reveals stimulus-mediated exon use in neural cells. Genome biology. 2007;8:R159. doi: 10.1186/gb-2007-8-8-r159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Coleman MJ, Hodge JJL, Budnik V, Griffith LC. Regulation of neuronal excitability in Drosophila by constitutively active CaMKII. J Neurobiol. 2002;52:24–42. doi: 10.1002/neu.10066. [DOI] [PubMed] [Google Scholar]
- Robertson GA, Warmke JM, Ganetzky B. Potassium currents expressed from Drosophila and mouse eag cDNAs in Xenopus oocytes. Neuropharmacology. 1996;35:841–850. doi: 10.1016/0028-3908(96)00113-x. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillips RW, Johnson-Schlitz D, Benz WK, Engels WR. A stable genomic source of P-element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Hodge JJ, Zhou Y, Nguyen M, Griffith LC. The eag potassium channel binds and locally activates calcium/calmodulin-dependent protein kinase II. J Biol Chem. 2004;279:10206–10214. doi: 10.1074/jbc.M310728200. [DOI] [PubMed] [Google Scholar]
- Tang CY, Schulteis CT, Jimenez RM, Papazian DM. Shaker and ether-a-go-go K+ channel subunits fail to coassemble in Xenopus oocytes. Biophys J. 1998;75:1263–1270. doi: 10.1016/S0006-3495(98)74046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Soll DR, Wu CF. Morphometric description of the wandering behavior in Drosophila larvae: a phenotypic analysis of K+ channel mutants. J Neurogenet. 2002a;16:45–63. doi: 10.1080/01677060213106. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wilson GF, Griffith LC. Calcium/Calmodulin-dependent Protein Kinase II Phosphorylates and Regulates the Drosophila Eag Potassium Channel. J Biol Chem. 2002b;277:24022–24029. doi: 10.1074/jbc.M201949200. [DOI] [PubMed] [Google Scholar]
- Warmke J, Drysdale R, Ganetzky B. A distinct potassium channel polypeptide encoded by the Drosophila eag locus. Science. 1991;252:1560–1562. doi: 10.1126/science.1840699. [DOI] [PubMed] [Google Scholar]
- Warmke JW, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci U S A. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CF, Ganetzky B, Haugland FN, Liu AX. Potassium currents in Drosophila: different components affected by mutations of two genes. Science. 1983;220:1076–1078. doi: 10.1126/science.6302847. [DOI] [PubMed] [Google Scholar]
- Xie J, Black DL. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature. 2001;410:936–939. doi: 10.1038/35073593. [DOI] [PubMed] [Google Scholar]
- Ye Y, Fortini ME. Proteolysis and developmental signal transduction. Semin Cell Dev Biol. 2000;11:211–221. doi: 10.1006/scdb.2000.0167. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Wu C-F. Alteration of four identified K+ currents in Drosophila muscles by mutations in eag. Science. 1991;252:1562–1564. doi: 10.1126/science.2047864. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Wu CF. Modulation of different K+ currents in Drosophila: A hypothetical role for the Eag subunit in multimeric K+ channels. J Neurosci. 1993;13:4669–4679. doi: 10.1523/JNEUROSCI.13-11-04669.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]


