Abstract
Peroxiredoxin 6 (Prdx6) is a unique antioxidant enzyme that can reduce phospholipid and other hydroperoxides. A549 cells, a human lung-derived cell line, express both Prdx6 and Nrf2, a transcription factor that binds to antioxidant response elements (AREs) and promotes expression of antioxidant genes. Treatment of A549 cells with 500μM H2O2 increased Prdx6 mRNA levels 2.5 fold while treatment with 400μM H2O2 or 200μM tert-butylhydroquinone (tBHQ) triggered a corresponding 2.5 fold increase in reporter gene activity in A549 cells transfected with the pSEAP2-Basic vector (BD, Bioscience), containing 1524 nucleotides of the human Prdx6 promoter region. Deletion of a consensus ARE sequence present between positions 357 and 349 before the start of transcription led to a striking decrease in both basal and H2O2 or tBHQ-induced activation in A549 cells and H2O2-induced activation in primary rat alveolar type II cells. Co-transfection with Nrf2 stimulated the Prdx6 promoter in an ARE-dependent manner, while it was negatively regulated by Nrf3. siRNA targeting Nrf2 down-regulated reporter gene expression whereas siRNA targeting the Nrf2 repressor, Keap1, up-regulated it. Binding of Nrf2 to the ARE sequence in chromatin was confirmed by PCR following chromatin immunoprecipitation. These data demonstrate that the ARE within the Prdx6 promoter is a key regulator of basal transcription of the Prdx6 gene and of its inducibility under conditions of oxidative stress.
Keywords: Prdx6 promoter, Transcription factor, Nrf2, Keap1, lung cells, reporter gene assay
Introduction
Lung tissue is unique as it is directly exposed to high oxygen tension and oxidants. Reactive oxygen species (ROS) that are produced as byproducts of cellular metabolism are potentially toxic, although at low concentrations they are known to function as messengers in signal transduction processes [1]. Previous investigators have studied the relative importance of antioxidant enzymes in the ability of lung cells to tolerate oxidative stress, focusing mainly on classical antioxidant enzymes such as catalase, glutathione peroxidase (GPx) [2], and superoxide dismutases [3]. Recently, a new family of antioxidant genes, the peroxiredoxin (Prdx) family has been identified. Peroxiredoxins, a family of 20–30-kDa peroxidases, are represented in organisms from all kingdoms [4]. The mammalian representatives include six enzymes (Prdx 1–6) that degrade H2O2 and other peroxides using the thiol groups of their cysteines (Cys) as catalytical centers [5, 6]. Prdx 1–4 contain two conserved catalytic cysteines and utilize thioredoxin as the reductant [7]. Prdx5 also uses thioredoxin as reductant and is classified as an atypical 2-cys Prdx [7]. Prdx6 contains a single conserved cysteine [8] and utilizes GSH but not thioredoxin to catalyze the reduction of peroxides; it is the only Prdx reported so far to reduce phospholipid hydroperoxides (PLOOH) [9]. Interestingly, Prdx6 also exhibits phospholipase A2 activity that has a catalytic serine as the reactive center [10, 11]. The ability to reduce phospholipid hydroperoxides could allow Prdx6 to repair membrane damage caused by oxidative stress. Prdx6 is highly enriched in lung compared to other organs [6, 12]. Altered expression of Prdx6 has been shown in several human lung diseases including malignant mesothelioma [13] and high-grade squamous cell carcinoma [14]. Overexpression of Prdx6 in cells confers protection against membrane damage associated with phospholipid peroxidation [15], while lipid peroxidation and apoptosis occurred when Prdx6 expression was blocked by treatment with an antisense oligonucleotide [16]. Consistent with a key role for Prdx6 in antioxidant defense, mice in which the Prdx6 gene had been deleted exhibited increased sensitivity to oxidative stress [17–21], while mice overexpressing Prdx6 were more resistant than wild-type to oxygen exposure [21].
To date, relatively little is known about the control of Prdx6 expression. Prdx6 mRNA and protein expression were induced by H2O2 and paraquat in a rat lung epithelial (L2) cell line as well as by hyperoxia in rat lungs [22], although, the mechanism underlying this response remains unclear. The goals of this study were to identify the human Prdx6 promoter and the mechanism for induction of Prdx6 by oxidant stress. We identified a functional anti-oxidant response element (ARE) [23], also called an electrophile-response element (EpRE) [24], in the Prdx6 promoter. This transcription factor binds to the ARE and appears to be involved in regulation of Prdx6 expression.
Materials and methods
Chemicals and reagents
MEM was purchased from Life Technologies (Grand Island, NY), OptiMEM was from Invitrogen, (Carlsbad, CA), H2O2 and tert-butylhydroquinone (t-BHQ) were from (Sigma). pSEAP2 alkaline phosphatase reporter vectors and an alkaline phosphatase activity assay kit were from BD Bioscience, competent cells, restriction enzymes and the Beta-glo assay system were from Promega (Madison, WI). All chemicals used were at least analytical grade.
Cell culture
A549 cells are a cell line derived from a malignancy originating from human broncho-alvelolar epithelial cells [25]. They have been shown to express all six peroxiredoxins [26]. A549 cells [American Type Culture Collection (ATCC), Manassas, VA] were cultured in MEM (Life Technologies, Grand Island, NY) supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin in a humidified incubator containing 5% CO2 at 37°C. Type II pneumocytes were isolated from the lungs of anesthetized male Sprague-Dawley rats according to our previously published methods [27]. Procedures involving animals were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Rats were maintained under filtered-air conditions. Briefly, lungs were perfused through the pulmonary artery until free of blood, excised, and lavaged through the trachea. The tissue was digested with elastase, and the lobes were separated and minced with a tissue chopper. The preparation was sequentially filtered through nylon screens (160-, 37-, and 10-μm mesh sizes) and enriched for type II cells by panning on rat immunoglobulin G (Sigma, St. Louis, MO)-coated Petri dishes for 1 h at 37°C to remove contaminating macrophages. The pneumocytes were plated on 12 well tissue-culture dishes at 1.5 × 106 cells/dish (2.2 cm in diameter). Cells were cultured overnight in 10% fetal calf serum (FCS) in Eagle’s minimal essential medium (MEM) at 37° C in a humidified incubator with 5% CO2 in air.
Western blot analysis
To determine the presence of Prdx6 in A549 cells, whole cell lysates were prepared and protein concentrations were determined using the Bio-Rad Bradford protein assay. Lysate (10μg protein) was subjected to 12% SDS-PAGE and transferred to nitrocellulose membranes (Amersham, Piscataway, NJ). Western blotting was carried out using a polyclonal anti-Prdx6 peptide antibody (1:3,000 dilution) and peroxidase-congugated secondary antibody (1:5,000 dilution) as described previously [28]. Recombinant rat Prdx6 was used as a standard [10].
RT-PCR
RT (reverse transcription)-PCR was performed with Prdx6 and Nrf2 primers in order to detect expression of Prdx6 and Nrf2 in A549 cells. Briefly, 1μg total RNA was reverse-transcribed into cDNA using a 1st Strand cDNA Synthesis kit for RT-PCR (AMV, Roche, Foster City, CA). PCR was performed on a GeneAmp 2400 (Perkin-Elmer, Waltham, MA) for 40 cycles for Prdx6 and Nrf2 and 27 cycles for GAPDH as a control. Each cycle included denaturation at 95°C for 30s, annealing at 58°C for 30 s, and elongation at 72°C for 30 S. The primers used for Prdx6 were: 5′-GGACGTGGCTCCCAACTTT-3′ and 5′-CGAGGGTGGGAGAAGAGAATG-3′. Those used for Nrf2 were: 5′-TACTCCCAGGTTGCCCACA-3′ and 5′-CATCTACAAACGGGAATGTCTGC-3′. The primers used for GAPDH were:5′-AGCCACATCGCTCAGACAC-3′ and 5′-TGGACTCCACGACGTACTC-3′. The size of the PCR products was determined by 2% (Prdx6 and Nrf2) or 1% (GAPDH) agarose gel electrophoresis with visualization by ethidium bromide staining.
Northern blot analysis
Total RNA was prepared from cells using the RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacture’s instructions. 12μg of total RNA was electrophoresed on a 1% agarose gel with 0.66 M formaldehyde. The size-fractionated RNAs were transferred to a positively-charged nylon filter (Schleicher and Schuell, Keene, NH) by capillary action. Prdx6 specific cDNA was labeled with 32P-dCTP using the Rad Prime DNA labeling system (GibcoBRL, Bethesda, MD) and separated from unincorporated nucleotides using a Microspin 6 column (BioRad, Hercules, CA). Hybridizations were performed with quick-Hyb solution (Stratagene, LaJolla, CA). Membranes were washed and then exposed to BioMax film (Kodak, Rochester, NY) at −80°C. The bands were scanned and relative densitometric units were measured with a densitometer (Fluor-S-MultiImager; Bio-Rad).
Plasmid constructs
BAC clones were obtained from the Children’s Hospital Oakland Research Institute (Oakland, CA). 1524 bp of human Prdx6 promoter region obtained by PCR from BAC clone 638L8 was cloned into the Mlu I and Xho I sites of the pSEAP2-Basic vector (BD, Bioscience) which contains alkaline phosphatase as a reporter gene, creating pSEAP2:Prdx6-wild type (wt). The BAC clone 638L8 contains a single A to T transversion at position −273 compared to the Genbank sequence. Truncations of the Prdx6 promoter were produced by digesting pSEAP2:Prdx6-wt with Mlu I that cuts 5′ to the translational start and a series of enzymes that cut within the Prdx6 coding region to create deletions. Spe I was used to generate a pSEAP2 clone with 1182 base pairs upstream of the transcriptional start, pSEAP2:Prdx6 −1182, BsiW I was used to generate pSEAP2:Prdx6 −410, Pml I was used to generate pSEAP2:Prdx6 −338 and Age I to generate pSEAP2:Prdx6 −44. All plasmid sequences were confirmed by sequencing analysis at the DNA sequencing facility of University of Pennsylvania. Full-length expression plasmids of human Nrf2 and Nrf3 in PCDNA3 were kindly provided by Dr Anil K. Jaiswal (University of Maryland School of Medicine).
Site-directed mutagenesis
Deletion of the ARE utilized the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Due to the high GC content of the Prdx6 promoter, we performed the mutagenesis on a limited region of the promoter. The promoter region between the BsiW I and the Pml I site was cloned by PCR amplification into plasmid pCR 2.1 using the topoTA cloning kit (Invitrogen). ARE deletion was performed using primers in which the ARE sequence was omitted. The forward primer for deleting ARE had the sequence: 5′TCCAGGGGGCAACGCCCGCATCACGTGTGC-3′ and the reverse primer had the sequence: 5′-GCACACGTGATGCGGGCGTTGCCCCCTGGA-3′ for PCR amplification of the Prdx6 promoter. The mutagenic oligonucleotide primer pair was added to 50 μl of reaction mixture containing 25 ng of DNA template, and the reaction was amplified in a thermal cycler for 18 cycles. Then 10 units of Dpn I restriction enzyme was added to the reaction, mixed and incubated at 37°C for 1 h to digest the parental dsDNA. One microliter of Dpn I-treated DNA was used to transform XL1-Blue competent cells. Clones were selected, and mutagenesis was confirmed by DNA sequencing. The promoter fragment with the deleted ARE was then excised by digestion with BsiW I and Pml I from the pCR 2.1 plasmid and inserted back into the pSEAP2-basic vector containing the wild type clone, replacing the wild-type DNA between these sites, which had been deleted previously by digestion with these same enzymes. The final product contained the sequence shown above in the forward primer, with the deletion occurring between the nucleotides 14 and 15.
Transfection procedure
A549 cells (70–80% confluence) were transfected with plasmids using Lipofectamine 2000 transfection reagent (Invitrogen). The pSEAP2:Control plasmid that expresses SEAP from an SV40 promoter was used as a positive control while pSEAP2:Basic, with no insert, served as a negative control. A plasmid expressing β-galactosidase from the SV40 promoter was used as a transfection control. For transfection, DNA, consisting of 1.4 μg of SEAP plasmid and 0.2 μg of β-galactosidase plasmid, was mixed with 200 μl of OptiMEM (Invitrogen) and 4 μl of Lipofectamine 2000, according to the supplier’s protocol, and incubated for 20 min, then added to each well of a 12 well dish (3.8 cm in diameter). In some experiments cells were co-transfected with the full length promoter construct and the Nrf2 or Nrf3 expression plasmid. For siRNA transfection, cells in 6 well dishes (9.6 cm in diameter) were transfected with 40 pmoles of siRNA duplex using Lipofectamine 2000 and OPTI-MEM medium, containing reduced serum, according to the manufacture’s protocol. 48h after siRNA transfection with Lipofectamine 2000, 2.8μg of SEAP plasmid and 0.4μg of β-galactosidase plasmid was added to each well for another 24h. For type II cells, we have optimized a high-efficiency transfection technique using the Nucleofector kit for normal human bronchial epithelial cells (Amaxa Biosystems, Cologne, Germany), in which plasmid DNA is transfected directly into the cell nucleus. Transfection protocols were performed using program W-001 following the manufacturer’s instructions. For all studies with A549 and type II cells, the medium was replaced 6 hours after transfection and cells were studied 18–24h later.
Exposure of cells to oxidant stress and assay of alkaline phosphatase and β-galactosidase activities
Cells (A549 or type II) were exposed to H2O2 or tert-butyl hydroquinone (t-BHQ) while attached to plastic dishes. Cells were rinsed with cold PBS before harvest. In some experiments, cell viability was determined by flow cytometry analysis following incubation for 15 min with propidium iodide. To determine optimal conditions, A549 cells were exposed to varying concentrations of H2O2 for 12 h or for varying times to 400μM H2O2. Based on these results, for subsequent studies, A549 cells were treated with 400 μM H2O2 for 8 hours. Based on a published protocol [29], A549 cells were exposed to 200μM t-BHQ for 6 h. Type II cells, because they began to detach from the culture dish during prolonged incubation, were treated with 400μM H2O2 for 4 hours. Following exposure to oxidants, samples of medium were taken in order to assay for SEAP activity using a chemiluminescence SEAP assay kit (BD, Bioscience). The cell pellet was lysed with assay buffer II (Promega, Madison, WI) and centrifuged at 12,500g for 5 min. β-galactosidase was measured in the supernatant using the Beta-glo assay kit (Promega, Madison, WI). Readings were performed on a luminometer and the specific SEAP activity of each promoter construct was normalized to the β-galactosidase activity.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed by using a commercially available kit (ChIP-IT™, Active Motif, Carlsbad, CA). Briefly, cells attached to the culture dish after incubation with H2O2 (400μM, 8h) were fixed for 10 min with formaldehyde that was directly added to the medium at room temperature and then isolated by scraping and sonicated. Immunoprecipitation of DNA was carried out using control rabbit IgG or anti-Nrf2 antibody (Santa Cruz Biotechnology). The immunoprecipitated DNA fragments were used as templates for PCR with primers designed to amplify 120 nucleotides of the Prdx6 promoter region surrounding the ARE. The forward primer sequence was: 5′-GCATCCTCAAGCTTCCAGGGGGC-3′; the reverse primer sequence was: 5′-GGACTACCGCGGTCGCTGTTGG-3′.
Statistical analysis
Results are expressed as mean ± SE. Statistical significance was determined by one-way ANOVA or t-test as appropriate using SigmaStat (Jandell Scientific, San Rafael, CA). P <0.05 was considered statistically significant.
Results
Expression of Prdx6 and Nrf2 in lung epithelial A549 cells
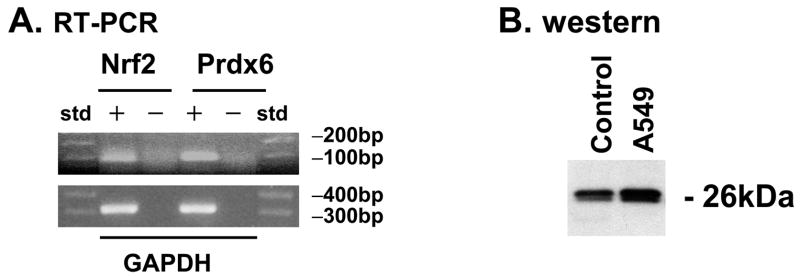
A549 cells were examined to determine whether Prdx6 and the transcription factor, Nrf2, are expressed in this cell line. RNA was detected using RT PCR and clearly shows expression of both Prdx6 and Nrf2 (Fig. 1A). Prdx6 protein expression was demonstrated by western blot, using recombinant Prdx6 as a control (Fig. 1B).
Fig. 1.
(A) RT-PCR for expression of mRNA for Nrf2 and Prx6 (upper panel) in human lung epithelial A549 cells. GAPDH (lower panel) was used as a control. PCR products were separated on a 2% agarose gel for Nrf2 and Prdx6 and 1% agarose for GAPDH. +, enzyme (reverse transcriptase) added; −, negative control (no enzyme), std, standards for molecular size. (B) Western blot using polyclonal anti-Prdx6 peptide antibody; Control: recombinant rat Prdx6 protein. A549:10 μg cell protein.
Prdx6 mRNA levels and promoter activity of Prdx6 are increased by H2O2
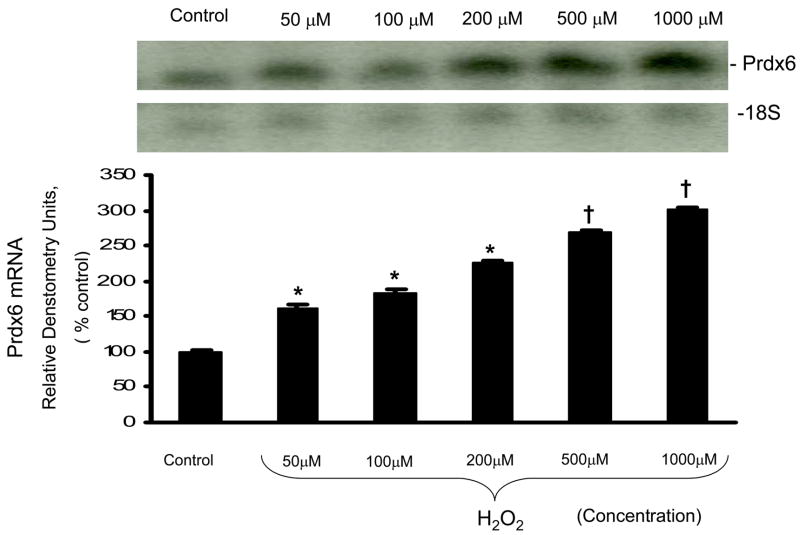
A549 cells were exposed to H2O2 at concentrations ranging from 50–1000μM for 12 h. There was a concentration-dependent increase in Prdx6 mRNA expression with H2O2 treatment as measured by northern blot (Fig. 2). Analysis by propidium iodide staining for cell membrane permeability indicated that dye exclusion was >90% for 400 μM H2O2 but decreased to <80% with 1000 μM H2O2 (not shown). Therefore, 400 μM H2O2 was used for most subsequent studies. A549 cells that had been transfected with the full length Prdx6 promoter: SEAP construct and incubated with 400μM H2O2 for varying times (not shown) showed a time-dependent increase of expression but no change between 8 and 12 h of incubtion; therefore, subsequent experiments used an 8 h incubation period for A549 cells.
Fig. 2.
Up-regulation of Prdx6 mRNA by oxidative stress. A549 cells were treated with 50μM-1 mM H2O2 for 12 hours. Total RNA was isolated and subjected to northern blot hybridization analysis. Individual data were quantified as densitometry units and normalized to 18S RNA as a loading control. The values shown in the bar graph are the means ±SE (n=3).*P<0.05, †P<0.01, compared with the control (no H2O2).
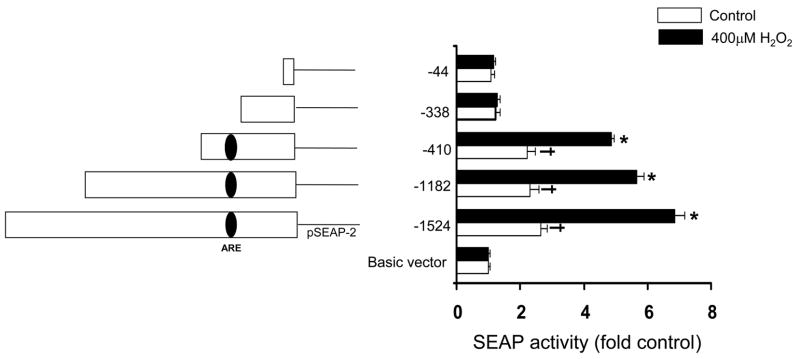
Promoter activity of Prdx6 was measured using the Secreted Alkaline Phosphatase (SEAP) reporter system. A 1.5-kb region of the human Prdx6 promoter was obtained from a BAC clone, amplified by PCR, and cloned into the promoterless pSEAP2 vector. Initial testing indicated that this construct contained the ability to direct transcription as well as the ability to be stimulated by H2O2. The full-length promoter construct pSEAP2:wt and a series of Prdx6 promoter deletions were generated in the pSEAP2 reporter plasmid by digestion with restriction enzymes. A549 cells were transiently transfected with these constructs and SEAP activity was determined in the presence and absence of 400 μM H2O2. The full length construct (−1524 bp) induced a 2.5 fold increase in the basal SEAP activity, which was further increased by almost 3-fold in response to H2O2 treatment (Fig. 3). The −1182 bp and −410 bp constructs exhibited the same ability to direct transcription as the full-length construct in the presence and absence of H2O2 (Fig. 3). However, SEAP activity of cells transfected with the Prdx6 promoter construct containing only 338 bpbefore the start of transcription (−338) was the same as that detected in cells transfected with pSEAP2-basic vector and this construct did not respond to H2O2 (Fig. 3). Since the −338 construct showed no significant increase in basal promoter activity when compared with the empty vector control and did not show any induction with H2O2, the elements involved in basal expression and in H2O2 induction must lie upstream of this region. Reporter gene expression and induction with H2O2 from the −410 bp construct was similar to that obtained with the largest construct. Thus, these data indicate that the DNA fragment from −410 to −338 contains regulatory elements controlling basal and H2O2-induced expression of Prdx6 in A549 cells.
Fig. 3.
Basal and H2O2-inducible activity of the Prdx6 promoter. A series of constructs was generated by restriction enzyme digestion of the 1.5 kb DNA fragment derived from the 5′-flanking region of Prdx6 into the promoterless pSEAP-2 basic vector. The approximate location of the putative ARE is shown within the promoter (left panel). Right panel, A549 cells were transiently transfected with reporter vectors and then incubated for 24 h, followed by 400 μM H2O2 treatment (or no treatment in control samples). SEAP activity was determined 8 h later. Values were normalized to the activity of cotransfected β-galactosidase. Means ±SE (n=3) are shown. *P<0.05 compared with vehicle control, †P<0.05 compared with Basic vector (no insert).
Role of the ARE
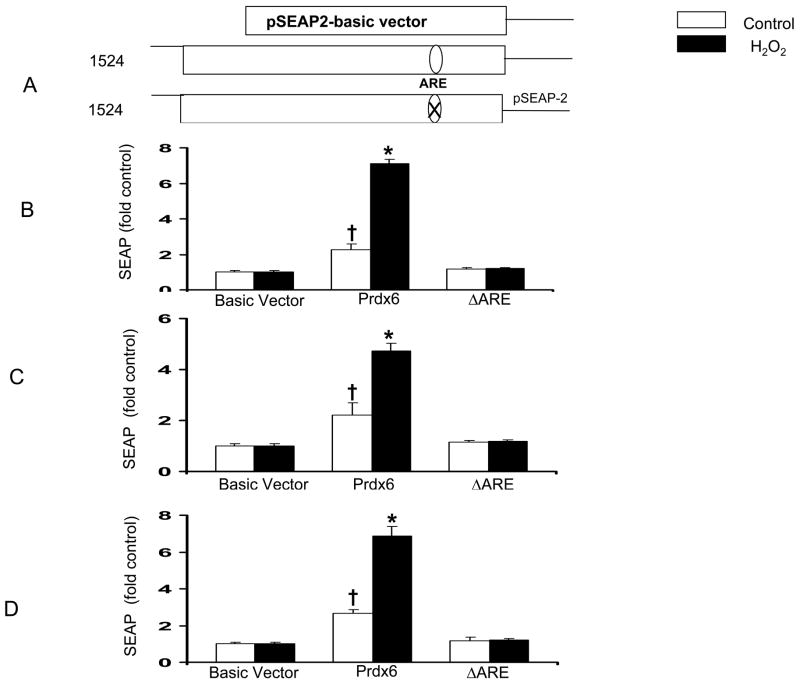
In analyzing nucleotides between −410 and −338 bp, we found an ARE consensus sequence between positions −357 and −349 in the Prdx6 gene (Fig 3). To examine whether this ARE isnecessary for H2O2-mediated activation, reporter gene constructs in which the ARE was deleted(ΔARE) were generated (Fig. 4A) and transfected into A549 cells. The ΔARE transfected cells showed markedly diminished basal and H2O2 mediated SEAP activity, suggesting that the ARE is indispensable for Prdx6 promoter activity and activation by H2O2 (Fig. 4B). To evaluate whether the ARE within the Prdx6 promoter would respond to a traditional ARE inducer, we treated A549 cells with t-BHQ, a phenolic oxidant used as a model inducing agent for ARE-driven transcription [29]. A significant, three fold increase in SEAP activity was noted with the full-length promoter, whereas transcription driven by the deleted ARE promoter did not change following tBHQ treatment (Fig. 4C).
Fig. 4.
Role of the ARE in Prdx6 gene expression. Deletion of the ARE was generated by site-directed mutagenesis of the full-length promoter in the pSEAP-2 basic vector. A549 or rat alveolar type II lung epithelial cells were transiently transfected with reporter vectors plus the β-galactosidase plasmid; after 24 h, cells were incubated with H2O2 or tBHQ. SEAP activity was determined at 4–8 h after treatment with oxidants. Cells were harvested and reporter gene assays performed; results for SEAP were normalized to cell β-galactosidase activity. A, Diagram showing vectors used in these experiments. Approximate location of the ARE within the promoter is indicated. X indicates ARE deletion. B, Transfection and treatment of A549 cells for 8 h with 400 μM H2O2. C, Transfection and treatment of A549 cells for 6 h with 200 μM tBHQ. D, Transfection and treatment of rat lung type II cells for 4 h with 400 μM H2O2. Values shown are means ± SE (n=3). *P<0.05 compared with untreated control, †P<0.05compared with Basic vector.
We also tested these constructs in rat alveolar primary type II cells and we obtained similar results to those found in A549 cells; namely, the ARE is essential for Prdx6 promoter activity (Fig. 4D). This indicates that A549 cells are a reasonable model for type II cells in these Prdx6 promoter studies.
Co-transfection of Nrf2 and Nrf3 exert opposite effects on Prdx6 promoter activity
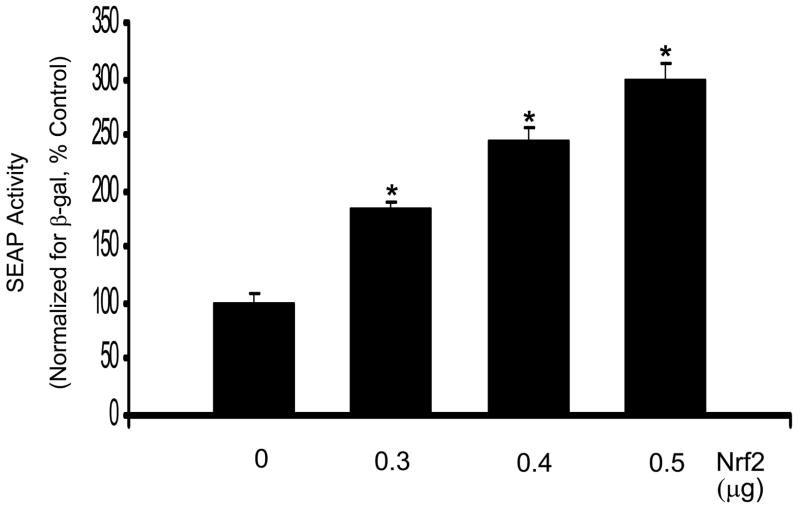
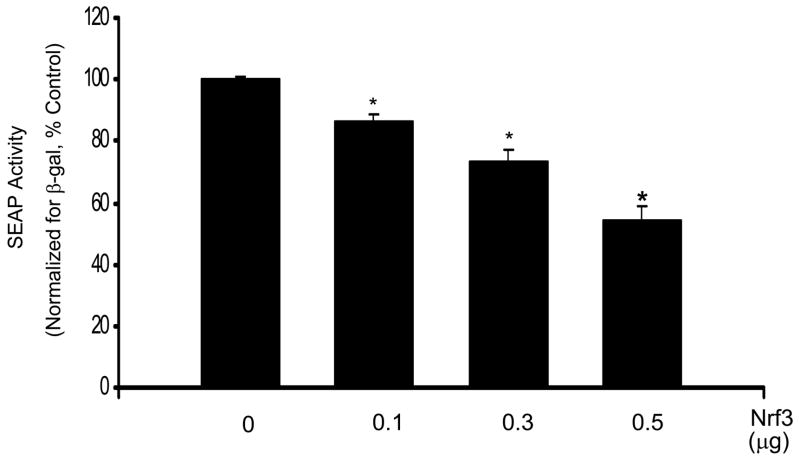
Nrf2 is thought to bind to AREs and up-regulate transcription [30]. To verify whether Nrf2 regulates Prdx6 gene expression, an Nrf2 expression plasmid pNrf2 and the reporter plasmid-SEAP were co-transfected into A549 cells. Increasing expression of Nrf2 in this manner significantly increased the Prdx6 promoter activity in a dose dependent fashion (Fig. 5A). These results suggest that Nrf2 is involved in the regulation of Prdx6 gene expression. On the other hand, Nrf3 is another member of the Nrf family that binds to the ARE [31]. Increasing expression of Nrf3 in a co-transfection experiment led to concentration-dependent repression of Prdx6 ARE-mediated SEAP gene expression (Fig. 5B). The transfection of A549 cells with 0.5μg of pcDNA-Nrf3 resulted in a 60% repression of SEAP gene expression.
Fig. 5.
Co-transfection with Nrf2 or Nrf3 influences Prdx6 promoter activity. A, Co-transfection of the Prdx6 promoter-SEAP construct (0.9 μg) with an Nrf2 expression plasmid increases Prdx6 promoter activity in a dose-dependent manner. A549 cells were co-transfected with increasing amounts of Prdx6-pSEAP2 and pNrf2 (an Nrf2 expression plasmid). The total amount of DNA was kept constant by addition of an appropriate amount of the empty plasmid vector pcDNA3. SEAP activity was measured in the medium after 30 h and normalized to cellular β-galactosidase activity. B, Nrf3 negatively regulates Prdx6 gene expression in co-transfection experiments, in a dose-dependent manner. The Prdx6 promoter-SEAP construct (0.9 μg) was co-transfected with increasing amounts of an Nrf3 expression plasmid (pNrf3) into A549 cells. The total amount of DNA was adjusted by addition of an appropriate amount of the empty plasmid vector, pcDNA3. SEAP activity was measured in the medium after 30 h and normalized to β-galactosidase activity from the cells. Values shown are means ± SE (n=3). *, P<0.05 vs. the preceding concentration.
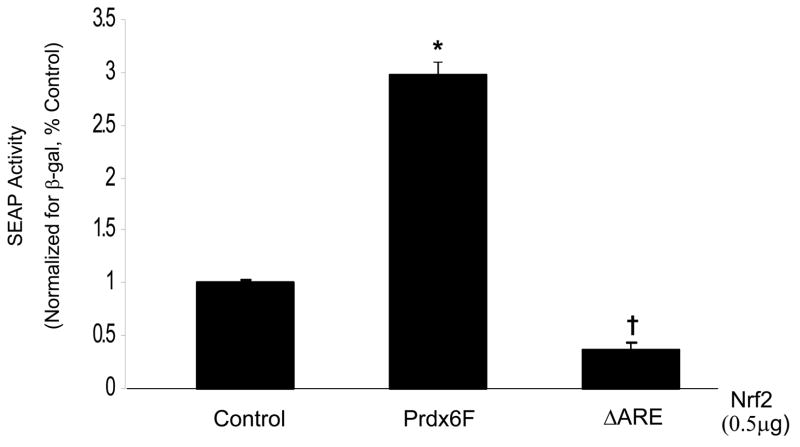
Deletion of the ARE abolishes Prdx6 promoter activity in response to Nrf2
In order to confirm that the region responsible for the Nrf2-mediated activation was the ARE, reporter constructs in which the ARE was deleted were tested in co-transfection experiments with the Nrf2 expression plasmid. SEAP activity from the ARE deletion construct was not induced by Nrf2 cotransfection indicating that the ARE is indispensable for Nrf2-mediated Prdx6 promoter activation (Fig. 6). In addition, the basal activity of the deletion was lower than control indicating that the ARE affects basal promoter activity.
Fig. 6.
Deletion of ARE1 abolishes the effect of Nrf2 on Prdx6 promoter activity. The Prdx6 promoter-SEAP construct with the ARE deletion (0.9μg) was co-transfected independently with Nrf2 expression plasmid (0.5 μg) along with the β-galactosidase expressing plasmid (0.2μg). SEAP activity in the medium was measured after 30 h and normalized to β-galactosidase activity in the cell extract. Control cells not transfected with Nrf2. Prdx6F, full length Prdx6 promoter; ΔARE, Prdx6 promoter with deleted ARE. Values are means ± SE (n=3).*P<0.05 compared with Control; †P<0.01, compared with full length.
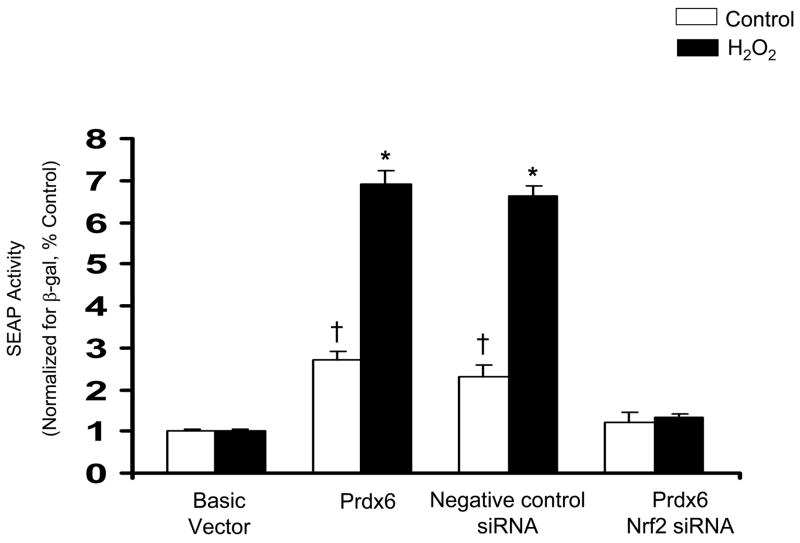
Nrf2 Activation Induced Prdx6 Expression in A549 cells
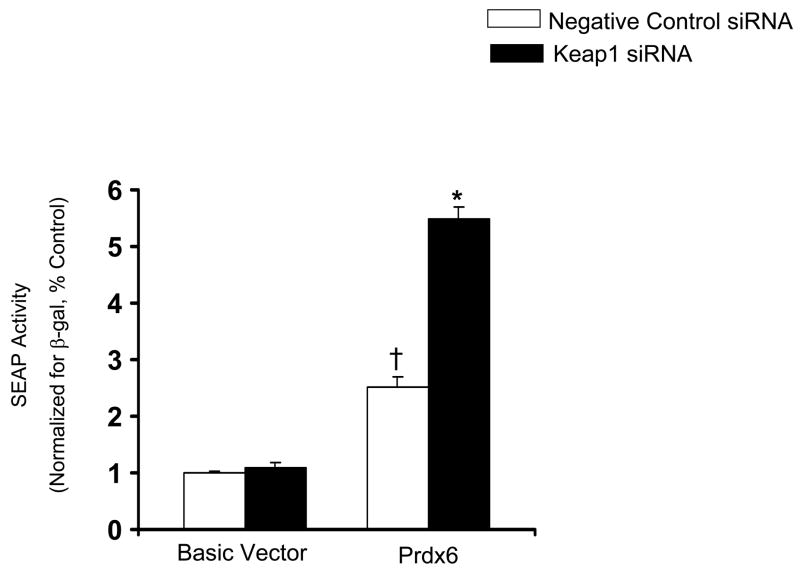
To further confirm that Nrf2 regulates Prdx6 gene expression, the effect of siRNA directed against Nrf2 and its cytoplasmic repressor, Keap1, on Prdx6 promoter-mediated expression were tested. The siRNAs were each transfected independently into A549 cells. A nontargeting siRNA was used as a negative control. The cells transfected with non-targeting siRNA showed no evidence of silencing and contained Nrf2 levels comparable to the vehicle control (data not shown). However, cells transfected with Nrf2 siRNA displayed reduced expression compared with the cells transfected with negative control siRNA in both basal and H2O2 treatment (Fig. 7A). On the other hand, Keap1 siRNA increased reporter gene expression compared with the cells transfected with SS siRNA (Fig. 7B). This increase occurred in the absence of H2O2 treatment. Thus, Keap 1 inhibition of Nrf2 is present in unstressed cells and is released by oxidative stress, allowing Nrf2 to translocate into the nucleus and transcribe Prdx6.
Fig. 7.
Nrf2 participates in induction of Prdx6 expression in A549 cells. A, Induction of the Prdx6 promoter by H2O2 is inhibited by Nrf2 siRNA. A549 cells were transfected with the full-length Prdx6 promoter in the pSEAP2 plasmid and co-transfected with Nrf2 si RNA or nontargeting siRNA as a negative control. The basic vector (no Prdx6) is also shown. After 72 h, cells were incubated for 8h with 400 μM H2O2 and then SEAP activity was measured and normalized to cell β-galactosidase activity. B, A549 cells were transfected with the basic vector or with the full length Prdx6 promoter (Prdx6). Keap1 siRNA or a negative control siRNA were co-transfected. Results were normalized to cotransfected β-galactosidase. Means ± SE (n=3). *P<0.05 compared with corresponding no H2O2, vehicle control, †P<0.05 compared with Basic vector.
Nrf2 binds to the Prdx6 promoter in A549 cells
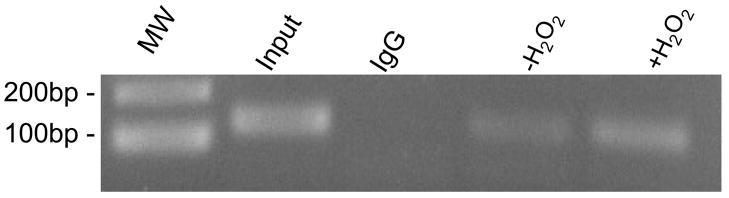
The ChIP assay was used to confirm the recruitment of Nrf2 to Prdx6 in the intact cell. Binding of Nrf2 to the Prdx6 ARE as indicated by PCR of the immunoprecipitated DNA fragments from A549 cells was at a low level under unstimulated conditions (-H2O2) and increased significantly after treatment with H2O2 (Fig. 8) No binding was observed with the immunoglobin G control.
Fig. 8.
A549 cells treated with 400 μM H2O2 for 8 h were processed for ChIP assay using the primer pairs described in Materials and Methods. MW, molecular weight marker; Input, PCR product amplified from chromatin extracts before immunopreciptation; IgG, PCR reaction using chromatin immunoprecipitated with rabbit immunoglobulin G;-H2O2, PCR product obtained after immunoprecipitation of untreated cells with antibody against Nrf2; +H2O2, H2O2 treated cells immunoprecipitated with antibody against Nrf2. A repeat experiment gave similar results.
Discussion
In contrast to the other peroxiredoxins, Prdx6 contains a single conserved cysteine and utilizes GSH as the redox cofactor to reduce lipid hydroperoxides [9]. Unlike the GSH peroxidases, Prdx6 is not a seleno-protein but uses cysteine as its catalytic center. In view of its antioxidant properties, it is not surprising that oxidative stress induced Prdx6 expression [22]. However, the mechanism of such up-regulation has not been addressed previously.
In this study, we examined the human Prdx6 promoter in A549 cells, a human lung epithelial cell line, and rat primary alveolar type II epithelial cells. Our studies documented that A549 cells express Prdx6 and the transcription factor Nrf2, and that Prdx6 mRNA level increased in a dose dependent manner in response to H2O2. Limited studies with primary lung epithelial cells confirmed results obtained with the A549 cell line. Our previous study in a cell line (L2) derived from rat lungs [22] showed that 500μM H2O2 had no effect on LDH release during 24 h of incubation, indicating that the cells remained intact. The standard exposure regimen for A549 cells in the present study was 400 μM H2O2 for 8 h which resulted in minimal loss of cell viability.
The sequence of the Prdx6 promoter was obtained from a BAC clone and a putative enhancer region was examined. In order to map the promoter, we prepared a series of 5′ deletions to assay their effect on activity. Initially we attempted to use firefly luciferase as a reporter gene in these experiments with Renilla luciferase as the control for transfection efficiency. We found that both the firefly and the Renilla luciferase activities decreased dramatically in response to H2O2 (data not shown). Reduction of luciferase activity by NO has been shown to occur through an effect on the stability of its mRNA [32], and we postulated that H2O2 treatment might have a similar effect. Therefore, we switched to the use of secreted alkaline phosphatase (SEAP) as a reporter gene and β-galactosidase as the transfection control. SEAP has the added advantage of being secreted into the medium allowing us to assay multiple time points per dish. Our deletion studies pointed to the region between 410 and 338 nucleotides upstream of the transcriptional start as necessary for both the basal and the H2O2 inducible promoter activity. Within this region, a consensus ARE region is present. The ARE has been described previously as key in the regulation of basal and inducible activity for a variety of genes [33]. Transfection of the Prdx6 ARE deletion constructs into either A549 cells or rat primary type II cells resulted in loss of basal promoter activity and of induction by H2O2.
Although the ARE in Prdx6 has not been studied previously, the Prdx1 gene promoter contains two AREs and is up-regulated by oxidative stress associated with hypoxia/reoxygenation [34]. However, unlike the Prdx6 ARE, neither of the Prdx1 AREs is a perfect match for the ARE consensus sequence. Increases in Prdx6 mRNA were shown in growth-arrested mouse liver cells treated with H2O2, but induction of reporter gene activity could not be demonstrated using a 1.2 Kb fragment of mouse Prdx6 promoter DNA [35]. Interestingly, there are substantial differences between the human and mouse Prdx6 promoters and there is no ARE in the mouse promoter corresponding to the ARE that we identified in the human promoter. Despite this, Prdx6 was induced in mouse lungs by oxidative stress associated with hyperoxia [22]. Possible mechanisms for this paradoxical effect include an ARE in an as yet unidentified region of the mouse promoter or ARE-independent induction of the mouse Prdx6 gene.
A large number of ARE-responsive genes are regulated by Nrf2, a member of the bZIP family of transcription factors [33]. The consensus binding sequence of the ARE, 5′-TGACNNNGC-3′, has been found in the promoter region of drug metabolizing enzymes such as glutathione S-transferase (GST) and NAD(P)H: quinone oxidoreductase (QR) [36]. Interestingly, we have shown that heterodimerization of Prdx6 and GST is required for peroxidase activity of the peroxiredoxin [37] so the presence of an ARE in each gene promoter suggests the possibility of coordinate regulation. Although several other protein transcription factors are known to bind the ARE, DNA binding assays and transfection experiments have indicated that Nrf2 is the most important one. In the present study, deletion of the ARE abolished Prdx6 promoter activity in response to co-transfection of Nrf2 and down-regulation of Nrf2 by siRNA caused decreased expression of the Prdx6 reporter gene in response to H2O2. Further, t-BHQ, a known inducer of Nrf2, increased Prdx6 promoter activity. Finally, ChIP assay indicated increased Nrf2 binding to the ARE sequence in the Prdx6 promoter after H2O2 treatment. These results indicate that Prdx6 expression in response oxidants is regulated by the interaction of Nrf2 with the ARE in the Prdx6 gene promoter region. The Nrf3 transcription factor has been shown in different studies with different genes to be either a positive [38] or a negative [31] regulator. Our studies confirm that for ARE-mediated gene expression, the regulation by Nrf3 is negative.
Nrf2 is present normally in the cytoplasm bound to Keap1 (also known as INrf2). Reactive oxygen species interact with Keap1 to release Nrf2 which, after phosphorylation, translocates to the nucleus where it can activate transcription by heterodimerizing with Small Maf Protein (Smaf) and binding to the ARE [29, 36, 39, 40]. The A549 cell line has been found in one study to possess a G-T transversion in the fourth exon of Keap1 that prevents it from efficiently binding Nrf2 [41]. If A549 cells do contain this mutation, it did not seem to affect the function of Keap1 in suppressing the activation of Nrf2, as the induction of nuclear localization of Nrf2 in response to hypoxia/reoxygenation was similar in these cells to other cell lines [34]. In our experiments, the G-T transversion of Keap 1, if present, would be expected to increase the basal expression and limit the inducibility of the Prdx6 gene. However, the level of induction that we found (three-fold) was similar in A549 and Type II cells and is consistent with our previously published results in L2 cells [22]. Thus, Keap1 function in the A549 cells used in our study appeared not to be compromised.
The term “oxidative stress” may refer to a broad spectrum of conditions that change the cellular redox status, such as increased production of free-radical species within the cell or treatment with pro-oxidant compounds that are thiol reactive and mimic an oxidative insult [33]. The ARE may thus be of importance in regulating the expression of the Prdx6 gene in tissues where H2O2 concentrations are elevated such as demonstrated in some tumors. Future research will be needed to determine whether Nrf2-Prdx6 activation can be suppressed by genetic and or pharmacologic approaches. Prdx6 is elevated in some forms of cancer including bronchogenic and mesothelioma lung cancers [13] and it will be interesting to determine whether suppressing the Nrf2-Prdx6 axis in preclinical models will inhibit their malignant progression or resistance to treatment. Thus, Prdx6 dysregulation in lung cancer and other malignancies may correlate with a poor clinical outcome. A recent publication demonstrated that Prdx6 transfected breast cancer cells grew much faster and metastasized more readily to lungs than control cells [42]. The information provided in the current study raises the possibility that the Nrf2-Prdx6 axis may serve as a potential target for cancer prognosis and therapy.
In summary, the ARE within the Prdx6 promoter is a key regulator of basal transcription of the Prdx6 gene, as well as inducibility under conditions of oxidative stress. Results from this study provide important insights into molecular mechanisms underlying oxidative lung injury. A more complete understanding of the Keap1-Nrf2-ARE pathway in the lung may provide opportunities to prevent oxidant-induced injury and disease pathogenesis.
Acknowledgments
We are grateful to Dr. Anil K. Jaiswal (University of Maryland School of Medicine) for kindly providing the cDNAs encoding Nrf2 and Nrf3, Dr. Shyam Biswal (Johns Hopkins University, Maryland) for gifts of siRNA, and Drs. Tatyana Milovanova and Jonni Moore for assistance with cell viability studies. This work was supported by HL P01-75587. I.C was supported as an NRSA post-doctoral fellow by HL T32-07748. This work has been presented in part at the Experimental Biology Meetings in 2005, 2006 and 2007.
Abbreviations
- Prdx6
Peroxiredoxin 6
- ARE
antioxidant responsive element
- t-BHQ
tert-butylhydroquinone
- Nrf2
NF-E2-related factor 2
- Nrf3
NF-E2-related factor 3
- Keap1
Kelch-like ECH-associated protein1
- siRNA
short interfering RNA
- ChIP
chromatin immunoprecipitation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gilston V, Williams MA, Newland AC, Winyard PG. Hydrogen peroxide and tumour necrosis factor-alpha induce NF-kappaB-DNA binding in primary human T lymphocytes in addition to T cell lines. Free Radic Res. 2001;35:681–691. doi: 10.1080/10715760100301201. [DOI] [PubMed] [Google Scholar]
- 2.Imai H, Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic Biol Med. 2003;34:145–169. doi: 10.1016/s0891-5849(02)01197-8. [DOI] [PubMed] [Google Scholar]
- 3.Clerch LB, Massaro D. Tolerance of rats to hyperoxia. Lung antioxidant enzyme gene expression. J Clin Invest. 1993;91:499–508. doi: 10.1172/JCI116228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahman I, Li XY, Donaldson K, Harrison DJ, MacNee W. Glutathione homeostasis in alveolar epithelial cells in vitro and lung in vivo under oxidative stress. Am J Physiol. 1995;269:L285–292. doi: 10.1152/ajplung.1995.269.3.L285. [DOI] [PubMed] [Google Scholar]
- 5.Chae HZ, Kim IH, Kim K, Rhee SG. Cloning, sequencing, and mutation of thiol-specific antioxidant gene of Saccharomyces cerevisiae. J Biol Chem. 1993;268:16815–16821. [PubMed] [Google Scholar]
- 6.Kang SW, Chae HZ, Seo MS, Kim K, Baines IC, Rhee SG. Mammalian peroxiredoxin isoforms can reduce hydrogen peroxide generated in response to growth factors and tumor necrosis factor-alpha. J Biol Chem. 1998;273:6297–6302. doi: 10.1074/jbc.273.11.6297. [DOI] [PubMed] [Google Scholar]
- 7.Rhee SG, Kang SW, Netto LE, Seo MS, Stadtman ER. A family of novel peroxidases, peroxiredoxins. Biofactors. 1999;10:207–209. doi: 10.1002/biof.5520100218. [DOI] [PubMed] [Google Scholar]
- 8.Kang SW, Baines IC, Rhee SG. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem. 1998;273:6303–6311. doi: 10.1074/jbc.273.11.6303. [DOI] [PubMed] [Google Scholar]
- 9.Fisher AB, Dodia C, Manevich Y, Chen JW, Feinstein SI. Phospholipid hydroperoxides are substrates for non-selenium glutathione peroxidase. J Biol Chem. 1999;274:21326–21334. doi: 10.1074/jbc.274.30.21326. [DOI] [PubMed] [Google Scholar]
- 10.Chen JW, Dodia C, Feinstein SI, Jain MK, Fisher AB. 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J Biol Chem. 2000;275:28421–28427. doi: 10.1074/jbc.M005073200. [DOI] [PubMed] [Google Scholar]
- 11.Manevich Y, Reddy KS, Shuvaeva T, Feinstein S, Fisher A. Structure and phospholipase function of peroxiredoxin 6: Identification of the catalytic triad and its role in phospholipid substrate binding. J Lipid Res. 2007 doi: 10.1194/jlr.M700299-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Mo Y, Feinstein SI, Manevich Y, Zhang Q, Lu L, Ho YS, Fisher AB. 1-Cys peroxiredoxin knock-out mice express mRNA but not protein for a highly related intronless gene. FEBS Lett. 2003;555:192–198. doi: 10.1016/s0014-5793(03)01199-2. [DOI] [PubMed] [Google Scholar]
- 13.Kinnula VL, Lehtonen S, Sormunen R, Kaarteenaho-Wiik R, Kang SW, Rhee SG, Soini Y. Overexpression of peroxiredoxins I, II, III, V, and VI in malignant mesothelioma. J Pathol. 2002;196:316–323. doi: 10.1002/path.1042. [DOI] [PubMed] [Google Scholar]
- 14.Lehtonen ST, Svensk AM, Soini Y, Paakko P, Hirvikoski P, Kang SW, Saily M, Kinnula VL. Peroxiredoxins, a novel protein family in lung cancer. Int J Cancer. 2004;111:514–521. doi: 10.1002/ijc.20294. [DOI] [PubMed] [Google Scholar]
- 15.Manevich Y, Sweitzer T, Pak JH, Feinstein SI, Muzykantov V, Fisher AB. 1-Cys peroxiredoxin overexpression protects cells against phospholipid peroxidation-mediated membrane damage. Proc Natl Acad Sci U S A. 2002;99:11599–11604. doi: 10.1073/pnas.182384499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pak JH, Manevich Y, Kim HS, Feinstein SI, Fisher AB. An antisense oligonucleotide to 1-cys peroxiredoxin causes lipid peroxidation and apoptosis in lung epithelial cells. J Biol Chem. 2002;277:49927–49934. doi: 10.1074/jbc.M204222200. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Phelan SA, Forsman-Semb K, Taylor EF, Petros C, Brown A, Lerner CP, Paigen B. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J Biol Chem. 2003;278:25179–25190. doi: 10.1074/jbc.M302706200. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Feinstein SI, Manevich Y, Ho YS, Fisher AB. Lung injury and mortality with hyperoxia are increased in peroxiredoxin 6 gene-targeted mice. Free Radic Biol Med. 2004;37:1736–1743. doi: 10.1016/j.freeradbiomed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Feinstein SI, Manevich Y, Ho YS, Fisher AB. Peroxiredoxin 6 gene-targeted mice show increased lung injury with paraquat-induced oxidative stress. Antioxid Redox Signal. 2006;8:229–237. doi: 10.1089/ars.2006.8.229. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Manevich Y, Feinstein SI, Fisher AB. Adenovirus-mediated transfer of the 1-cys peroxiredoxin gene to mouse lung protects against hyperoxic injury. Am J Physiol Lung Cell Mol Physiol. 2004;286:L1188–1193. doi: 10.1152/ajplung.00288.2003. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Phelan SA, Manevich Y, Feinstein SI, Fisher AB. Transgenic mice overexpressing peroxiredoxin 6 show increased resistance to lung injury in hyperoxia. Am J Respir Cell Mol Biol. 2006;34:481–486. doi: 10.1165/rcmb.2005-0333OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HS, Manevich Y, Feinstein SI, Pak JH, Ho YS, Fisher AB. Induction of 1-cys peroxiredoxin expression by oxidative stress in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2003;285:L363–369. doi: 10.1152/ajplung.00078.2003. [DOI] [PubMed] [Google Scholar]
- 23.Papaiahgari S, Kleeberger SR, Cho HY, Kalvakolanu DV, Reddy SP. NADPH oxidase and ERK signaling regulates hyperoxia-induced Nrf2-ARE transcriptional response in pulmonary epithelial cells. J Biol Chem. 2004;279:42302–42312. doi: 10.1074/jbc.M408275200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Liu H, Dickinson DA, Liu R-M, Postlethwait EM, Laperche Y, Forman HJ. [gamma]-Glutamyl transpeptidase is induced by 4-hydroxynonenal via EpRE/Nrf2 signaling in rat epithelial type II cells. Free Radical Biology and Medicine. 2006;40:1281. doi: 10.1016/j.freeradbiomed.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieber M, Smith B, Szakal A, Nelson-Rees W, Todaro G. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer. 1976;17:62–70. doi: 10.1002/ijc.2910170110. [DOI] [PubMed] [Google Scholar]
- 26.Lehtonen ST, Markkanen PM, Peltoniemi M, Kang SW, Kinnula VL. Variable overoxidation of peroxiredoxins in human lung cells in severe oxidative stress. Am J Physiol Lung Cell Mol Physiol. 2005;288:L997–1001. doi: 10.1152/ajplung.00432.2004. [DOI] [PubMed] [Google Scholar]
- 27.Dobbs LG, Gonzalez R, Williams MC. An improved method for isolating type II cells in high yield and purity. Am Rev Respir Dis. 1986;134:141–145. doi: 10.1164/arrd.1986.134.1.141. [DOI] [PubMed] [Google Scholar]
- 28.Kim HS, Pak JH, Gonzales LW, Feinstein SI, Fisher AB. Regulation of 1-cys peroxiredoxin expression in lung epithelial cells. Am J Respir Cell Mol Biol. 2002;27:227–233. doi: 10.1165/ajrcmb.27.2.20010009oc. [DOI] [PubMed] [Google Scholar]
- 29.Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 30.Jain AK, Bloom DA, Jaiswal AK. Nuclear Import and Export Signals in Control of Nrf2. J Biol Chem. 2005;280:29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 31.Sankaranarayanan K, Jaiswal AK. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. J Biol Chem. 2004;279:50810–50817. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- 32.Fan X, Roy E, Zhu L, Murphy TC, Kozlowski M, Nanes MS, Rubin J. Nitric oxide donors inhibit luciferase expression in a promoter-independent fashion. J Biol Chem. 2003;278:10232–10238. doi: 10.1074/jbc.M209911200. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 34.Kim YJ, Ahn JY, Liang P, Ip C, Zhang Y, Park YM. Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 2007;67:546–554. doi: 10.1158/0008-5472.CAN-06-2401. [DOI] [PubMed] [Google Scholar]
- 35.Gallagher BM, Phelan SA. Investigating transcriptional regulation of Prdx6 in mouse liver cells. Free Radic Biol Med. 2007;42:1270–1277. doi: 10.1016/j.freeradbiomed.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 36.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci U S A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manevich Y, Feinstein SI, Fisher AB. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc Natl Acad Sci U S A. 2004;101:3780–3785. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K, Hayashi N, Yamamoto M. Molecular cloning and functional characterization of a new Cap’n’ collar family transcription factor Nrf3. J Biol Chem. 1999;274:6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- 39.Dhakshinamoorthy S, Jaiswal AK. c-Maf negatively regulates ARE-mediated detoxifying enzyme genes expression and anti-oxidant induction. Oncogene. 2002;21:5301–5312. doi: 10.1038/sj.onc.1205642. [DOI] [PubMed] [Google Scholar]
- 40.Motohashi H, Yamamoto M. Carcinogenesis and transcriptional regulation through Maf recognition elements. Cancer Sci. 2007;98:135–139. doi: 10.1111/j.1349-7006.2006.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang XZ, Li DQ, Hou YF, Wu J, Lu JS, Di GH, Jin W, Ou ZL, Shen ZZ, Shao ZM. Identification of the functional role of peroxiredoxin 6 in the progression of breast cancer. Breast Cancer Res. 2007;9:R76. doi: 10.1186/bcr1789. [DOI] [PMC free article] [PubMed] [Google Scholar]