Abstract
Histone acetyltransferase (HAT) is an enzyme required for chromatin remodeling and transcriptional activation. Sarcoendoplasmic reticulum Ca2+ ATPase (SERCA) is an ATP coupled Ca2+ ion pump involved in metabolic arrest. Both these enzymes are present in Plasmodia and have been selected as molecular targets for in silico studies of some new non-resistant antimalarial drugs like artemisinin, curcumin and diarylheptanoids along with some other inhibitors reported in literature. Ten top inhibitors have also been generated based on common pharmacophore from ZINC database. The HAT enzyme was modeled with the help of the Modeller software and the SERCA enzyme pdb file was obtained from the protein data bank. Ligbuilder was used for structure based drug designing, which generated a common pharmacophore of the ligands. Molegro was used to perform virtual screening of the hits from the pharmacophore based Zinc database search and known inhibitors of the enzymes from the literature survey. Curcumin shows good and optimal binding to both HAT and SERCA enzymes; therefore it might be a good inhibitor of these key enzymes in Plasmodium. Curcumin is reported to act synergistically with artemisinin which forms covalent adducts with the transmembrane proteins (SERCA enzyme) and inactivates them, thus inhibiting the activity of Plasmodium parasite. This combination has already been reported to be effective in malaria treatment. Some other diarylheptanoids besides curcumin showed better binding to both the enzymes. Therefore, a combination of artemisinin and diarylheptanoids can prove to be better combination for antimalarial therapy. Different formulations involving curcumin, artimisinin and diarylheptanoids may result in a more potent antimalarial drug.
Keywords: molecular target, malaria, Plasmodium, histone acetyltransferase, anti-malarial drugs
Background
Malaria is fast becoming a serious threat to public health throughout the world especially in endemic regions like India and Africa. The malarial parasite Plasmodium falciparum is responsible for around one million deaths each year. Its life cycle involves many morphologically distinct stages, which alternate between a vertebrate and an invertebrate host. Like other eukaryotes, the malaria parasite DNA is organized into nucleosomes and the genome encodes components of chromatin-remodeling complexes. Falciparum species is more common in Africa while in India vivax is more common. Plasmodium has developed resistance to most of the existing drugs e.g. Mefloquin and Chloroquin, as well as the combination of drugs. The reason has been attributed to the decreased susceptibility of resistance transporters (PfCRT) for chloroquin and (PfMDR1) for multidrug resistance [1]. Therefore, there is a growing need to develop newer drugs and drug combinations to inhibit the growth of protozoa. This requires work on new molecular targets in Plasmodium and designing inhibitors with good parasiticidal activity and also which are cost effective to the underprivileged population of the developing world and impose less financial burden on their governments. In the present study we have selected two enzymes present in Plasmodium as molecular targets viz. Histone acetyltransferase and Sarcoendoplasmic reticulum ATPase.
Histone acetyltransferases (HAT) are enzymes that acetylate conserved lysine residues at N-terminal tails of core histone proteins by transferring an acetyl group from acetyl CoA to lysine to form τ-N-acetyl lysine, neutralizing their positive charges. Thus presumably the affinity of the histones is reduced for the DNA (which is negatively charged), which lead to nucleosomes being unfolded and increase access to transcriptional factors. Thus, histone acetylation is generally linked to transcriptional activation generally associated with euchromatin. Lysine acetylation and other post translational modifications of histones generate binding sites for specific protein-protein interaction domains, such as the acetyl-lysine binding bromodomain [2]. Its inhibition will lead to transcriptional deactivation and therefore the enzyme can be used as a novel drug target in Plasmodium. Ion pumping is one of the most energetically taxing physiological processes in cells, and ion motive ATPases are likely loci to be differentially regulated in models of metabolic arrest. We propose that the deactivation of sarcoendoplasmic reticulum (SER) calcium-ATPase(SERCA) would potentially contribute to the overall suppression of metabolism [3]. In case of inhibition of activity of these two enzymes the Plasmodium would not be able to complete its life cycle. In the present work we have docked some latest antimalarials in use like artimisinin and curcumin. In addition we have selected ten top inhibitors from ZINC database. The synergistic effect of formulations of these herbal products is also expected to yield excellent results.
Methodology
The following software's were used, MODELLER for modeling the Histone acetyltransferase protein, SPDB Viewer for loop modeling, LIGBUILDER for structure based drug designing, MOLEGRO for docking and virtual screening.
Protein modeling
MODELLER software was used for modelling the Histone acetyltransferase protein. Doing loop modelling on the SPDBViewer so that 88% of the amino acid residues fall within the acceptable region of the Ramachandran plot did further refinement of the protein structure.
Ligand designing
LIGBUILDER was used for structure based drug designing on the Histone acetyltransferase protein. Ligand was developed by both LINK and GROW methods, 200 ligands by GROW and around 1000 ligands by LINK techniques were generated along with a common parmacophore of the generated ligands. Pharmacophore based search was performed on the ZINC database and around 78 hits were found. Also some known inhibitors of the Histone actyltransferase enzyme were found on Literature survey. The known inhibitors are Isothiazolones, Garcinol, Quinolones, Anacardic Acid and Curcumin. Other diarylheptanoid compounds were also selected. Their 3-D coordinates were generated after drawing their 2-D structures on CORINA software
Virtual screening
Virtual screening of the above compounds on the Histone acetyltransferase enzyme was performed on the MOLEGRO software and the top 10 compounds with the best docking energies were obtained. These compounds were also docked on Sarcoendoplasmic reticulum Ca (2+) ATPase enzyme protein. Sarcoendoplasmic reticulum ATPase enzyme pdb file was obtained from the protein data bank. Curcumin was found to have good binding to both HAT and SERCA enzymes. Diarylheptanoids have shown better binding to both these enzymes therefore suggesting it might be a better inhibitor of these key enzymes in Plasmodium. Curcumin was docked with SERCA enzyme and the curcumin bound SERCA enzyme was saved in the pdb format. This pdb file was docked with the different diarylheptanoids and their docking energies (of curcumin bound SERCA enzyme) were compared to those where diarylheptanoids were docked with the SERCA enzyme (without curcumin) as such (see Table 3, supplementary material).
Discussion
Curcumin (See Table 1 under supplementary material) shows good and optimal binding to both Histone acetyltransferase (HAT) and Sarcoendoplasmic Reticulum Ca (2+) ATPase (SERCA) enzymes, therefore it might be a good inhibitor of these key enzymes in Plasmodium. Curcumin is reported to act synergistically with artemisinin (Figure 1) which forms covalent adduct with the transmembrane proteins (Sarcoendoplasmic Reticulum Ca2+ ATPase enzyme) and inactivates them, thus inhibiting the activity of Plasmodium parasite. This combination has already been reported to be effective in malaria treatment [4]. Some other diarylheptanoids (See Table 1, under supplementary material and Figure 5 and 6) besides curcumin showed better binding than curcumin to both Histone acetyltransferase and Sarcoendoplasmic Reticulum Ca (2+) ATPase enzymes. The table (see Table 1 under supplementary material) shows the binding affinity; inter total Energy, intra steric and total energy of the interaction of Curcumin and Diarylheptanoids with the HAT and the SERCA enzymes. Therefore, a combination of artemisinin and diarylheptanoids can prove to be a better combination for antimalarial therapy. Diarylheptanoids show better binding to curcumin bound SERCA enzyme than to SERCA enzyme directly (see Table 3, under supplementary material and Figure 7) implying that curcumin might be causing some conformational change in the SERCA enzyme leading to better binding of the diarylheptanoids to the enzymes. Curcumin might be having a facilitating effect on the action of diarylheptanoids on the SERCA enzyme. Therefore different formulations involving curcumin, artimisinin and diarylheptanoids may result in a more potent antimalarial drug.
Figure 1.
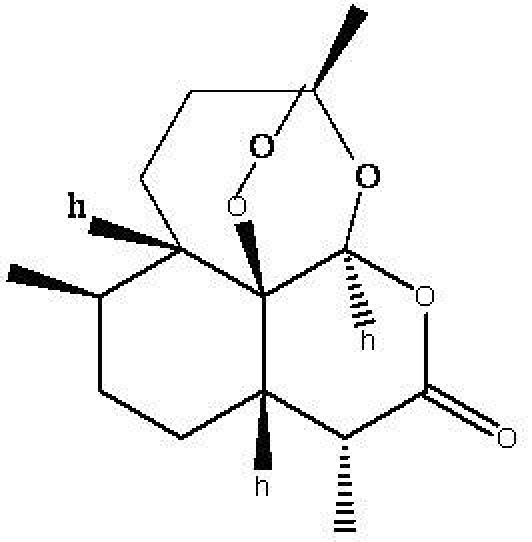
Structure of Artemisinin
Figure 5.
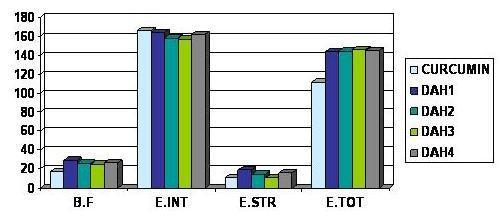
Comparative docking energies of Curcumin and diarylheptanoids on HAT Enzyme
Figure 6.
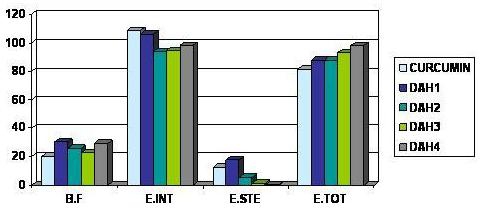
Comparative docking energies of curcumin and diarylheptanoids on SERCA Enzyme
Figure 7.
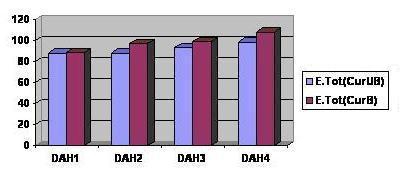
Graph showing comparative docking energies of diarylheptanoids when they are docked to SERCA enzyme and to curcumin bound SERCA enzyme
As artemisinin (Figure 1) is active during the intraerythrocyte stage of infection and contains an endoperoxide function that is essential for its activity, a reasonable hypothesis for the mechanism of action of artemisinin is the reductive activation of its endoperoxide bridge by Fe (II) heme resulting from the digestion of hemoglobin by Plasmodium species. This reaction might lead to dioxygen-derived radicals responsible for oxidative stress within infected erythrocytes or to C-centered radicals OR carbocations that alkylate heme and membrane associated parasite proteins e.g. PfTCTP-Plasmodium Falciparum translationally controlled tumor protein [5,6]. An important target for artemisinin is Pf ATP6-A SERCA.
Curcumin belongs to the class of diarylheptanoids i.e. class of compounds with two aromatic rings separated by 7 aliphatic carbons. Other compounds of this class have substitution of different functional groups at different positions. They were drawn on CORINA server and their 3D coordinates were obtained. They were docked to both Sarcoendoplasmic reticulum Ca (2+) ATPase enzyme and Histone acetyltransferase enzyme and showed better docking energies than Curcumin. Hydrogen bond interactions of Diarylheptanoids and Curcumin to the SERCA enzyme have been shown in Figure 2 and Figure 3 respectively.
Figure 2.
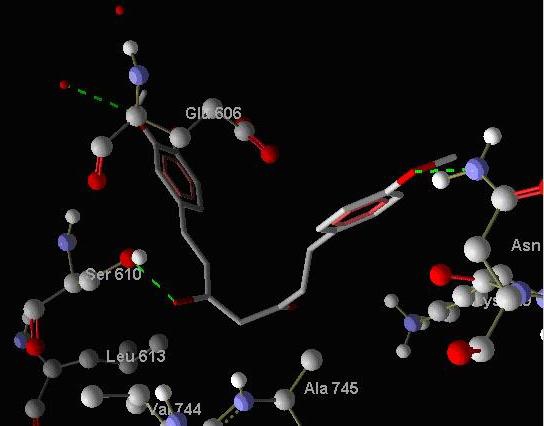
Showing hydrogen bond interactions of binding of DAH1 to SERCA enzyme
Figure 3.
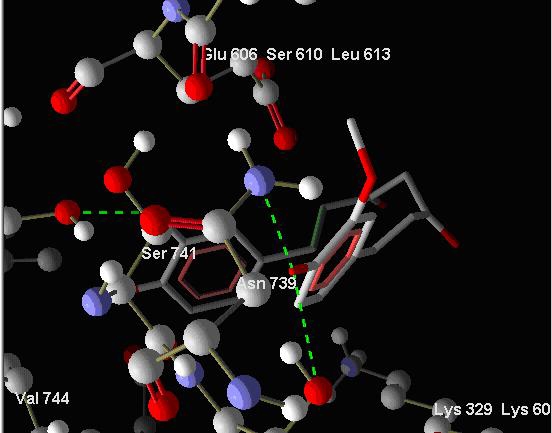
Showing hydrogen bond interactions of binding of Curcumin to SERCA enzyme
The following compounds which were obtained from the pharmacophore based search on Zinc database are the best binding compounds on Histone acetyltransferase enzyme therefore can be effective inhibitors of the Plasmodium parasite. In addition curcumin bound pdb of SERCA enzyme was docked with diarylheptanoids. Three of them showed increase in the docking energy (see Table 3, supplementary material and Figure 7) suggesting that on binding curcumin might be causing some conformational change in the enzyme leading to better binding of diarylheptanoids. Residues involved in hydrogen bond interactions were visualized and compared to those of diarylheptanoid bound directly to the SERCA enzyme (see supplementary material Table 4; Figure 2 and Figure 4).
Figure 4.
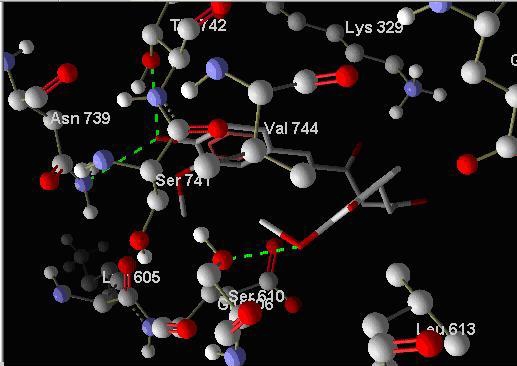
Showing hydrogen bond interactions of binding of DAH1 to Curcumin bound SERCA enzyme
On docking Curcumin is showing good binding to both Histone acetyltransferase and Sarcoendoplasmic reticulum Ca (2+) ATPase enzymes suggesting that its mechanism of action might be involving more than one molecular target in the Plasmodium parasite. Curcumin might be acting synergistically with artemisinin (which forms covalent adducts with the transmembrane proteins of the malarial parasite) to inhibit the SERCA enzyme, hence potentiating its action. Certain other diarylheptanoids show even better binding to the HAT and SERCA enzymes (see Figure 5 and Figure 6) and hence might make a more effective combination with artemisinin than curcumin in inhibiting the malarial parasite. Artemisinin is known to be acting by a pathway, which is independent to curcumin, therefore diarylheptanoids may be better substitutes for curcumin. However, our results have shown that curcumin may potentiate the action of diarylheptanoids in enhancing the activity of artimisinin. Also there are a group of compounds from the Zinc Database search which show good binding affinities with the Histone Acetyltransferase enzyme (see Table 2, shown under supplementary material) and hence can be prospective drugs for combination therapy in malaria. Appropriate combinations will have to be worked out for optimum results.
Conclusion
However since the diarylheptanoids are showing better binding to both Histone acetyltransferase enzyme and Sarcoendoplasmic reticulum Ca (2+) ATPase enzyme, hence they might be better inhibitors of both the key enzymes in the Plasmodium parasite, therefore it might be possible that a combination of Artemisinin and diarylheptanoids can be an even better combination for antimalarial therapy. Also, since diarylheptanoids are showing better binding to curcumin bound SERCA enzyme than the curcumin unbound enzyme, curcumin might be causing some conformational change in the SERCA enzyme leading to better binding of the Diarylheptanoids.Therefore different formulations involving curcumin, artemisinin and diarylheptanoids can be a very effective combination for antimalarial therapy.
Author's contribution
Artimisinin and curcumin mixed formulation is known to be an effective inhibitor of plasmodium parasite, however the present study shows that substitution of curcumin with some diarylheptanoids may prove to be a better combination.
Supplementary material
Acknowledgments
I am thankful to the Director of IIITA Allahabad for providing a supportive environment to carry out research activities.
Footnotes
Citation:Singh & Misra, Bioinformation 3(6): 255-262 (2009)
References
- 1.Evans WC, et al. Ibid Journal of Medical Microbiology. 2007;56:1213. [Google Scholar]
- 2.Tang W, Eisenbrand G. Springer-Verlag, Berlin. 1992;160 [Google Scholar]
- 3.Eckstein-Ludwig U, et al. Nature. 2003;424:957. doi: 10.1038/nature01813. [DOI] [PubMed] [Google Scholar]
- 4.Padmanaban G, et al. Antimicrob Agents Chemother. 2006;50:1859. doi: 10.1128/AAC.50.5.1859-1860.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amit V, et al. J Biol Chem. 1999;274:19383. doi: 10.1074/jbc.274.27.19383. [DOI] [PubMed] [Google Scholar]
- 6.Heinrich C, et al. Antimicrobial agents and chemotherapy. 2004 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


