Abstract
The mammalian supraoptic nucleus (SON) is a neuroendocrine center in the brain regulating a variety of physiological functions. Within the SON, peptidergic magnocellular neurons that project to the neurohypophysis (posterior pituitary) are involved in controlling osmotic balance, lactation, and parturition, partly through secretion of signaling peptides such as oxytocin and vasopressin into the blood. An improved understanding of SON activity and function requires identification and characterization of the peptides used by the SON. Here, small-volume sample preparation approaches are optimized for neuropeptidomic studies of isolated SON samples ranging from entire nuclei down to single magnocellular neurons. Unlike most previous mammalian peptidome studies, tissues are not immediately heated or microwaved. SON samples are obtained from ex vivo brain slice preparations via tissue punch and the samples processed through sequential steps of peptide extraction. Analyses of the samples via liquid chromatography mass spectrometry and tandem mass spectrometry result in the identification of 85 peptides, including 20 unique peptides from known prohormones. As the sample size is further reduced, the depth of peptide coverage decreases; however, even from individually isolated magnocellular neuroendocrine cells, vasopressin and several other peptides are detected.
Keywords: supraoptic nuclei, peptidome, neuropeptides, peptide processing, prohormone, single cell, magnocellular neuron, sample preparation, MALDI-MS, ESI-MS/MS
Short abstract
Mammalian peptidome studies are extended to smaller, well-defined brain structures ranging from an individual brain nucleus to individual neurons. Specifically, we characterize the peptidome of the supraoptic nucleus and individual magnocellular neurons within the nucleus. With the use of a coronal brain slice preparation and optimized sample preparation strategies to reduce postmortem degradation, 85 peptides have been characterized, including 20 peptides previously not reported.
Introduction
Neuropeptides are involved in behavioral, cognitive, and homeostatic functions throughout the brain and body. Feeding behavior, circadian rhythms, sleep, learning, and memory are just a few physiological functions regulated by these biologically active peptides.1,2 Neuropeptides are synthesized as larger protein precursors and undergo a series of processing steps to produce bioactive neuropeptides. Processing steps include cleaving the precursor at mono- or dibasic sites, as well as numerous post-translational modifications (PTMs) such as amidation, acetylation, pyroglutamination, and disulfide bond formation.2−4 Modifications such as acetylation can be required for resistance to degradation as well as biological activity.(5)
Before the advent of modern mass spectrometry (MS), neuropeptide characterization was performed by isolating and sequencing one peptide at a time using N-terminal sequencing methods such as Edman degradation. After the peptide has been characterized, appropriate antibodies are created and quantitative peptide measurements performed using radioimmunoassay. Although largely effective, one disadvantage of Edman degradation is that peptides containing N-terminal PTMs (acetylation, cyclic glutamate or glutamine) cannot be directly sequenced. Furthermore, the peptide concentration required for sequencing is typically higher than what is required for MS structural characterization.
Recent developments in sample separation and MS analysis have improved identification and characterization of peptides from biological samples ranging from brain structures6−12 to single cells.13−17 When analyzing complex systems such as the mammalian brain, MS has been restricted to relatively large morphological structures: as examples, an entire hypothalamus, striatum, or hippocampus.6−10 In many cases, obtaining adequate amounts of analyte requires large sample sizes or working with high concentration peptides from neuroendocrine structures such as the pituitary that have significant amounts of peptides.(18) Postmortem degradation also becomes an issue due to proteolytic enzymes present in sample tissues.19,20 To improve sampling efficacy, enzyme deactivation has received recent attention, with studies using a variety of techniques that include using preservatives such as dihydroxybenzoic acid (DHB),(12) focused microwave irradiation before decapitation10,11,21,22 or microwave irradiation after decapitation,7,20 or boiling the tissue immediately after decapitation.(9) The effects of protein degradation on rodent tissue have been studied using MS-based approaches.7,9,20 For instance, Che and colleagues(7) found variations in the levels of some hypothalamic peptides when a 20-s delay was introduced between decapitation and microwaving, and Sköld and co-workers(20) showed that detectable neuropeptide levels greatly decreased when the time between animal sacrifice and microwaving was as brief as 1 min. Moreover, levels continued to decrease during longer delays.
Although data demonstrating the effectiveness of microwave treatment in preventing peptide degradation makes inherent sense and the research is certainly of high quality, it is also surprising when considering the large body of literature on ex vivo brain slice experiments. The brain slice preparation is a well-established technique used in studies investigating mechanisms of circadian rhythmicity, learning, neuronal repair, and synaptic plasticity. Typically, brain slices are kept functional for more than 24 h ex vivo.23−25 Furthermore, it has been shown that altered neurosecretory vesicle pools and other cell organelles recover and stay constant for hours following brain slice preparation.25,26 Advantages of the method include the ability to combine functional tests of peptide release with peptidome measurements,(27) and to clearly delineate small, anatomically defined brain structures such as individual brain nuclei from fresh (non heat-treated or microwaved) samples.
Here, we test the ability to isolate peptides using the established physiological preparation of the fresh brain slice. We combine the aforementioned protocols for peptidomic studies of the brain with a specific focus on the supraoptic nucleus (SON). Although the SON has been intensively studied,28−38 this may be the first mammalian neuropeptidomics examination of this small, functionally and morphologically defined cluster of cells by means of mass spectrometry. In addition to the characteristic products of the SON, oxytocin, and Arg-vasopressin, there are many SON-derived peptides colocalized within this nucleus, presumably at lower levels. In situ hybridization, and immunohistochemical and MS studies have reported several bioactive peptides in the SON coming from different precursors such as secretogranin II,(39) chromogranin A,(40) cocaine- and amphetamine-regulated transcript (CART),41,42 and neuropeptide Y.(43) However, we speculate that these peptides are a subset of the total peptides present in the SON.
Because of its physiological importance, abundance of neuropeptides, and easy isolation, we identify and characterize the SON peptidome. In analyses using liquid chromatography (LC) electrospray ionization (ESI) tandem MS (MS/MS) as well as LC matrix-assisted laser desorption/ionization (MALDI) MS/MS, we identify 85 peptides, a majority of which correspond to known or previously characterized peptides.7−10 Moreover, 20 peptides have been unambiguously identified with MS/MS and appear not to have been reported previously.
In addition, we examined selected sections of the SON. In mammals, the hypothalamic-neurohypophysial system contains populations of well-defined neurosecretory cells producing cell-to-cell signaling peptides that participate in a variety of peripheral regulatory mechanisms. These cells, the magnocellular neurons (MCNs) of the SON, are part of the hypothalamic neuroendocrine system. They are relatively large for mammalian neurons (∼30 μm in diameter) and produce significant amounts of neuropeptides, which are released into the circulatory system. Interestingly, MCNs appear to release their peptides dendritically,44,45 making them an interesting target for cellular, and potentially even subcellular, studies on dense core vesicle targeting and release.
Can we adapt single cell MS to interrogate individual MCNs? Because of its high sensitivity and salt tolerance, MALDI MS allows analysis not only of the peptidome of a neural tissue extract, but also of a single cell.13−17 Investigation of an individual neuron using single-cell MALDI MS enables identification of peptides processed from the same precursor, and also determination of colocalized neuropeptides from different precursors. Jimenez et al.(14) used ∼100-μm diameter molluscan neurons to mass fingerprint the neuropeptides, and several have worked with smaller, ∼20-μm insect neurons.15,46,47 More recently, Rubakhin et al.16,17 successfully performed MALDI MS on individual 8−10-μm diameter mammalian pituitary cells and characterized a number of pro-opiomelanocortin (POMC) and CART-related peptides. We profile a single MCN with MALDI, without introducing exogenous labels or enzymatic treatments, in order to characterize peptide precursors and their products found within this cell population.
Experimental Section
Chemicals
All reagents were obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO), unless otherwise noted. Water was prepared by a Milli-Q filtration system (Millipore, Bedford, MA). The peptides standards used for external calibration were provided by Bruker Daltonics, Inc. (Billerica, MA).
Animal Sacrifice and Brain Slice Treatments
The animals and sacrifice procedures used are delineated in greater detail elsewhere.(48) Briefly, 6−12 week old Long-Evans/BluGill male and female rats (University of Illinois at Urbana−Champaign) were sacrificed by decapitation in accordance with protocols established by the University of Illinois Institutional Animal Care and Use Committee and in accordance with all state and federal regulations. The brain was quickly removed and the hypothalamic coronal brain slices (500 μm) containing the paired SON were prepared as previously reported.(23) Slices were maintained in Earle’s balanced salt solution, pH 7.2, at 37 °C supplemented with 24.6 mM glucose, 26.2 mM sodium bicarbonate, and 5 mg/L gentamicin and saturated with 95% O2−5% CO2. Brain slices were transferred to physiological phosphate buffered saline (PBS) and the SON (∼1 mm3 of brain tissue) was removed from the ex vivo coronal brain slice by punching a 1-mm plug or cutting out the nucleus on the surface of a plastic Petri dish placed on a bed of ice. Because the SON is situated directly adjacent to the optic tracts, these structures served as easily identifiable landmarks for SON collection. These samples may include small amounts of tissue from the nearby optic tract. After the SON punches were collected, the PBS was removed and the tissue processed for peptide extraction.
Multistage Peptide Extraction
SONs from both hemispheres from 12–17 animals were pooled and collected in 1.5-mL plastic vials. To increase the number of peptides extracted and eventually detected with MS, tissue extraction was performed in multiple stages with each experiment repeated at least three times. During the first stage of peptide extraction, 300 μL of sterile deionized (DI) water (MilliQ), preheated to 90 °C, was added to the SON tissue samples, capped, and placed into a boiling water bath for 10 min. Next, the samples were centrifuged (Mini Centrifuge, model C1301, Denville, Woodbridge, NJ), the supernatant was transferred to a new vial, and the resulting pellets were frozen at −80 °C until the second stage of extraction. The second stage extraction was processed using 300 μL of acidified acetone (40:6:1 acetone/water/HCl) for 1 h at 4 °C. The sample was vortexed for 1 min and centrifuged at 14 000 rpm for 5 min, and the supernatant was collected. The third extraction stage used 300 μL of 0.25% acetic acid and followed the same sample processing as stage two. All extracts were dried using a SpeedVac concentrator (Thermo Scientific, San Jose, CA) and reconstituted with 10 μL of solvent A (aqueous 5% acetonitrile (ACN) containing 0.1% formic acid (FA) and 0.01% trifluoroacetic acid (TFA). After reconstitution, the three samples were pooled and filtered through a 10 kDa molecular weight cutoff tube (Millipore) (see Supporting Information, Figure S1).
Liquid Chromatography−Mass Spectrometry (LC-MS)
Chromatographic separation of the extracted peptides and mass spectrometric analyses were performed using a capLC (Micromass, U.K.) LC system coupled with an HCT Ultra-PTM Discovery system ion-trap mass spectrometer (Bruker Daltonics) equipped with an electrospray ionization source. For LC-MALDI-time-of-flight (TOF/TOF)-MS analysis, the capLC was coupled with a robotic fraction spotter (ProteineerFC, Bruker Daltonics). The samples were loaded onto a trap column (PepMap, C18, 5 μm, 100 Å, Dionex, Sunnyvale, CA) using a manual injector (Valco Instruments Co., Inc., Houston, TX) and washed for 5 min. The trapped peptides were then eluted in reverse direction onto a reverse phase capillary column (LC Packings, 300-μm inner diameter × 15 cm, C18 PepMap100, 100 Å) using a solvent gradient at 2 μL/min flow rate. The solvent gradient was generated using solvent A and solvent B (aqueous 95% ACN containing 0.1% FA and 0.01%TFA). The 70-min gradient run for LC separation included three steps: 5−80% solvent B in 15−55 min (linear); 80% solvent B for 55−60 min (isocratic); 80−5% solvent B in 60−65 min (linear).
The MS data acquisition and the subsequent MS/MS of selected analytes were performed in a data-dependent manner using the Esquire software (Bruker Daltonics). For each MS scan, three peptides were selected to be fragmented, for 300−500 ms, based on their charge (preferably +2) and intensity. Dynamic exclusion of previously fragmented precursor ions was set to 2 spectra for a period of 60 s. The MS and MS/MS scans were performed in the range of m/z 300−1500 and 50−2000, respectively.
For LC-MALDI-TOF/TOF-MS analysis, chromatographic separation was performed using a 45-min gradient run comprising the following parameters: 5−80% solvent B in 10−30 min (linear); 80% solvent B for 30−35 min (isocratic); 80−5% solvent B in 35−45 min (linear). The chromatographic elutant was monitored using a UV detector (Waters, Milford, MA) and subsequently spotted onto a ground steel MALDI sample plate at the rate of 60 s/spot using ProteineerFC. Immediately after spotting, 1 μL of α-cyano-4-hydroxy cinnamic acid matrix was applied to each spot.
For LC-Fourier transform (FT) MS analysis, separations were accomplished using a ProteoPep II C18 column from New Objective (Woburn, MA), with an inner diameter of 75 μm and length of 10 cm. The chromatographic gradient was performed using a 1D NanoLC pump from Eksigent Technologies (Dublin, CA) operating at 300 nL/min with the following gradient parameters: 10% solvent B for 0−10 min (isocratic); 10−30% solvent B for 10−20 min (linear); 30−100% solvent B for 20−80 min (linear); 80−5% solvent B for 80−100 min (linear). Peptides were detected on a 7 T linear trap quadrupole (LTQ)-FT instrument from Thermo Scientific, and a setting of 15 V was used for mild ion activation in the ESI source. The LTQ-FTMS method consisted of three scan events, all in profile mode with four microscans used per full scan. An initial, full scan event was performed at a high resolution (100 000) in FT mode. The second scan event was a data-dependent collision-activated dissociation (CAD) MS/MS scan of the most abundant peak in the previous full scan, again with FT mode detection (50 000 resolution). MS/MS settings were as follows: isolation width = 4 m/z; minimum signal threshold = 50 000 counts; normalized collision energy (NCE) = 40%; duration = 200 ms, and q-factor = 0.5. Dynamic exclusion was used for a duration of 6 min per precursor to avoid redundant fragmentation data sets. The third scan was a full scan detected in the ion trap (low resolution). All neuropeptide identifications for this system were accomplished using ProSightPC software (Thermo Scientific) as described below. The intact and CAD fragmentation data were used to search a database of predicted neuropeptides.
Matrix Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI TOF/TOF MS)
Each sample spot was analyzed by a Bruker Ultraflex II TOF/TOF instrument with a delayed ion extraction and a frequency tripled Nd:YAG laser operating at 1064 nm and 50 Hz (Bruker Daltonics). Mass spectra were accumulated over 2 to 3 individual acquisitions of 300 laser shots each. Intense peaks from the MS scan of samples were manually analyzed and selected for further analysis using the MS/MS feature of the Ultraflex II instrument. Several peaks were assigned based on accurate mass matches without MS/MS data, normally with the confidence of these assignments enhanced by the detection of other related peptides from the same prohormone.
Single Cell Analysis
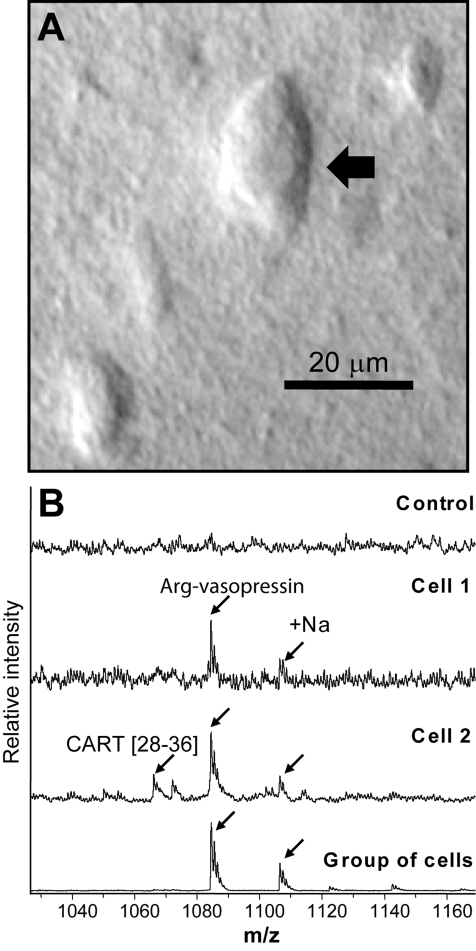
Individual SON neurons and small groups of cells were isolated in accordance with the detailed protocol described previously.(17) Briefly, brain sections were immersed in solution containing 33% glycerol and 67% of modified Gey’s balanced salt solution (mGBSS, in mM, 1.5 CaCl2, 4.9 KCl, 0.2 KH2PO4, 11 MgCl2, 0.3 MgSO4, 138 NaCl, 27.7 NaHCO3, 0.8 Na2HPO4, 25 HEPES and 10 glucose, pH 7.2) and kept in this solution during all isolation steps. Addition of glycerol stabilizes cellular structures during tissue and cell manipulation as well as prevents dehydration of the specimen. Small, several-hundred micrometers in diameter, regions of the SON were removed from the slices and placed on a glass slide positioned on an inverted light microscope (see Figure 3A). Under visual control, fine-tipped glass needles were used to isolate, pick up, and transfer individual neurons and small groups of cells onto a metal MALDI sample plate. Pipettes pulled from borosilicate glass were used to deposit MALDI matrix-saturated DHB in water onto specimens.
Figure 3.
(A) Photomicrograph of a neuron isolated manually from a glycerol-treated brain slice. (B) Mass spectra acquired from individual MCN neurons using saturated DHB matrix: top trace, mass spectrum from a neuron isolated from a brain slice treated with glycerol; middle traces, individual neuron spectra; bottom trace, a group of SON cells.
Data Analysis
Tandem MS data obtained from the ESI-MS/MS and MALDI-TOF/TOF systems were selected and exported to the Biotools software program (Bruker Daltonics) for further analysis. The data were converted to a Mascot (Matrix Science, London, U.K.) generic file format and searched against mouse and rat IPI databases using Peaks software (Bioinformatics Solutions, Inc., Waterloo, CA). Search parameters for the Peaks software included unsuspected cleavage site, N-terminal pyro-glutamic acid, and C-terminal amidation. Subsequently, the search results were confirmed by manual de novo analysis using the Data Analysis software (Bruker Daltonics). Complimentary searches were also performed with Mascot using the MSDB database and an in-house database of neuropeptide prohormones. Parameters for the Mascot search included enzyme specified as “none,” and variable modifications were considered, such as acetylation of N-terminal, amidation of C-terminal, pyro-Glu of Q and pyro-Glu of E. The peptide mass tolerance for MS/MS data was set at 0.5 Da. The MS/MS data were manually verified to contain at least three consecutive amino acid fragment ions for confirming positive identification of a peptide. In addition, MALDI MS data were analyzed by direct comparison of measured molecular weights of the putative neuropeptides to the NeuroPred prediction engine (http://neuroproteomics.scs.uiuc.edu/neuropred.html)49,50 using an in-house-generated database of known rat prohormones. Mass matches within 150 ppm were required for positive identifications.
Analysis of FTMS Data
Data files (*.raw) were reduced to a ProSightPC Upload Format file (*.puf) using an in-house program (cRAWler), which groups MS/MS data from all precursors of the same protein into one experiment. An experiment consisted of one precursor mass and all fragment ion masses derived from it. Within this program, molecular weights of precursor and product ions were also determined using a highly modified THRASH algorithm.(51) Fragment ions for all MS/MS scans were required to have an S/N ratio of 5:1 or greater to be considered for database searching. Each *.puf file typically contained hundreds of experiments from a single LC-FTMS run. The experiments within these files were then searched against two types of databases using ProSightPC52,53 (Thermo Scientific) and/or neuroProsight (http://neuroprosight.scs.uiuc.edu/), a highly annotated mouse proteome database of intact proteins, and a database of predicted mouse neuropeptides generated by the NeuroPred tool described above. Two types of searches were performed against these databases. First, an Absolute Mass search was done to find any proteins/peptides that were identified as they appeared in the database. Second, a Biomarker search was done to retrieve any endogenously cleaved pieces of proteins/peptides in the databases. In all cases, the mass tolerance for fragment ions was ±5 ppm. All retrieved proteins and peptides needed to meet two requirements to be considered a positive identification: (1) an expect score of less than 10−5 and, (2) no unexplained mass shifts between the observed and theoretical intact masses.(54) Identifications that could not be assigned unambiguously were validated using the Sequence Gazer tool to account for errors in determination of the monoisotopic mass of the protein.
Results
Peptidomics studies provide detail on the peptides present in entire brains to selected brain regions. This study represents a downscaling of sample sizes for mammalian neuropeptidomics analysis to the level of an identified nucleus, here the hypothalamic SON. In comparison with previous neuropeptidomics methods devised for larger brain regions, our analyses show a comparable number of peptide identifications. Because the ability to work with select, anatomically well-defined nuclei requires precise registration of the samples, it is easier to work with intact brain slices than heat-treated or microwaved tissue, especially as the anatomical features required for identification of the region may be altered by such treatments.7,20
One question of particular interest was whether or not neuropeptides would be detected in such samples, given that prior work showed a large reduction in neuropeptide levels and an increase in peptide fragments from proteins shortly after decapitation.11,21 Our results demonstrate that, even if peptides are released and degraded immediately after death, the neurosecretory vesicles and peptide levels recover. After sampling the SON by tissue punch from a brain slice, the sample is denatured using heat. Increasing sample temperature by heating with 90 °C DI water reduces protease activity and serves as an extraction solution. We see almost no difference between extracting peptides using DI water extraction and an acidified acetone extraction (Supporting Information, Figure S1); however, fewer peaks were seen in the acetic acid extraction. Although there is variation in the data between extraction solutions, each approach produced reproducible mass spectra. Because some peptides are extracted more efficiently using different conditions, our final method involves treating the SON tissue with a sequential series of solvent extractions. By increasing the tissue temperature in water, followed by two-stage extraction using acidified acetone and acetic acid, we achieve an increased recovery of the SON peptidome.
Using this optimized approach, we detect 20 peptides characterized using MS/MS that were not reported in prior hypothalamus peptidome studies (Table 1). The quality of mass spectra (Supporting Information, Figure S1) and the number of products detected (Supporting Information, Table S1) indicate that the peptides are not affected by the lack of immediate microwaving and the delay between slice preparation and sample extraction.
Table 1. Peptides from Known Neuropeptide Precursorsa.
| precursor | sequence | ESI MS (m/z) | obs. mass (Da) | error (ppm) | LC-ESI-FTMS | LC-MALDI-TOF/TOF | LC-ESI ion trap | LC-MALDI-TOF MS |
|---|---|---|---|---|---|---|---|---|
| Arg-vasopressin[24−32] | A.C*YFQNC*PRGamide.G | 1083.49 | −42.5 | X | ||||
| Provasospressin[24−35] | A.CYFQNCPRGGKR.A | 1427.67 | −7.0 | X | ||||
| Provasopressin[157−168] | T.QESVDSAKPRVY | 1377.59 | 65.3 | X | ||||
| Provasopressin[156−168] | G.TQESVDSAKPRVY | 1478.69 | 33.8 | X | ||||
| Provasopressin[155−168] | A.GTQESVDSAKPRVY | 1535.69 | 39.1 | X | ||||
| Provasopressin[158−168] | Q.ESVDSAKPRVY | 1249.59 | 32.0 | X | ||||
| Provasopressin[154−168] | L.AGTQESVDSAKPRVY | 1606.69 | 62.2 | X | ||||
| Provasopressin[151−168] | L.VQLAGTQESVDSAKPRVY | 1947.09 | −41.1 | X | ||||
| Provasopressin[160−168] | S.VDSAKPRVY | 1033.56 | −9.7 | X | ||||
| Provasopressin[36−43] | R.ATSDMELR.Q | 921.47 | −54.3 | X | ||||
| CART[28−36] | A.pQEDAELQPR.A | 534.68 | 1067.52 | −26.2 | X | X | ||
| CART[82−86] | R.IPIYE.K | 633.34 | 633.34 | 4.7 | X | X | ||
| TRH[178−199] | R.FIDPELQRSWEEKEGEGVLMPE.K | 873.40 | 2617.24 | 0.0 | X | X | X | |
| Proenkephalin A[100−104] | R.YGGFM.K | 573.23 | −10.5 | X | ||||
| Proenkephalin A[198−209] | R.SPQLEDEAKELQ.K | 693.80 | 1385.67 | −7.2 | X | X | ||
| Proenkephalin A[198−209] | R.(Ac)SPQLEDEAKELQ.K | 1427.67 | 7.0 | X | ||||
| Proenkephalin A[219−229] | R.VGRPEWWMDYQ.K | 1465.65 | −6.1 | X | X | |||
| Proenkephalin A[114−133] | K.MDELYPVEPEEEANGGEILA.K | 735.10 | 2203.56 | 190.6 | X | X | ||
| Proenkephalin A[219−238] | R.VGRPEWWMDYQKRYGGFLKR.F | 2571.30 | 0.0 | X | ||||
| Protachykinin 1[58−68] | R.RPKPQQFFGLMGamide.K | 1347.70 | 5.2 | X | ||||
| Protachykinin 1[98−110] | R.HKTDSFVGLMGKR.A | 1474.74 | 20.3 | X | ||||
| VIP[111−124] | R.ISSSISEDPVPVKR.Q | 1512.73 | 52.9 | X | ||||
| VIP[156−169] | R.SSEGDSPDFLEELE.K | 1552.75 | −70.8 | X | ||||
| POMC[103−120] | R.AEEETAGGDGRPEPSPREamide.G | 941.97 | 1881.90 | −31.9 | X | X | ||
| POMCI[124−136] | R.SYSMEHFRWGKPVamide.G | 811.80 | 1621.76 | 12.3 | X | |||
| POMC[124−137] | R.(Ac)SYSMEHFRWGKPVG.K | 1721.91 | −63.9 | X | ||||
| Somatostatin[89−100] | R.SANSNPAMAPRE.R | 622.70 | 1243.50 | 47.4 | X | X | X | |
| Somatostatin[91−100] | A.NSNPAMAPRE.R | 543.80 | 1085.60 | −101.3 | X | |||
| Somatostatin[103−116] | K.AGCKNFFWKTFTSC | 1638.80 | −42.7 | X | ||||
| Secretogranin 1[585−594] | R.SFAKAPHLDL.K | 549.73 | 1097.44 | 132.1 | X | X | ||
| Secretogranin 1[597−611] | R.pQYDDGVAELDQLLHY.R | 879.00 | 1760.87 | −45.4 | X | X | ||
| Secretogranin 1[435−451] | R.LLDEGHDPVHESPVDTA.K | 915.90 | 1829.82 | 10.9 | X | X | X | |
| Secretogranin 1[49−61] | S.SAPTITPECRQVL.R | 707.80 | 1413.70 | 21.2 | X | |||
| Secretogranin 1[47−56] | S.KSSAPTITPE.C | 514.25 | 1028.45 | 97.2 | X | |||
| Secretogranin 1[21−34] | S.APVDNRDHNEEMVT.R | 1625.74 | −18.5 | X | ||||
| Secretogranin 1[383−395] | R.NHPDSELESTANR.H | 1468.73 | −54.5 | X | ||||
| Secretogranin 1[383−394] | R.NHPDSELESTAN.R | 1312.48 | 53.3 | X | ||||
| Secretogranin 1[572−584] | R.PFSEDVNWGYEKR.S | 1625.74 | 6.2 | X | ||||
| Secretogranin 2[571−583] | R.IPAGSLKNEDTPN.R | 678.22 | 1354.67 | 0.0 | X | |||
| Secretogranin 2[529−568] | R.VPSPGSSEDDLQEEEQLEQAIKEHLGQGSSQEMEKLAKVS.K | 4366.08 | −2.3 | X | ||||
| Secretogranin 2[287−297] | R.(Ac)SGHLGLPDEGN.R | 1136.49 | 17.6 | X | ||||
| Secretogranin 3[23−36] | A.FPKPEGSQDKSLHN.R | 792.30 | 1582.78 | −6.3 | X | |||
| Secretogranin 5[198−210] | K.SVPHFSEEEKEPE | 1542.79 | −71.3 | X | ||||
| Secretogranin 5[198−210] | K.(Ac)SVPHFSEEEKEPE | 1584.84 | −94.7 | X | ||||
| Secretogranin 5[180−195] | R.SVNPYLQGKRLDNVVA.K | 1771.87 | 50.8 | X | ||||
| Secretogranin 5[180−195] | R.(Ac)SVNPYLQGKRLDNVVAKK.S | 2070.23 | −33.8 | X | ||||
| Secretogranin 5[177−189] | R.KRRSVNPYLQGKR.L | 1600.83 | 62.5 | X | ||||
| Proenkephalin B[235−247] | R.SQENPNTYSEDLDV | 806.09 | 1609.61 | 37.3 | X | |||
| Proenkephalin B[211−218] | R.PKLKWDNQ.K | 514.79 | 1027.56 | −9.7 | X | |||
| Proenkephalin B[47−59] | C.SLECQDLVPPSEE.W | 722.21 | 722.21 | 13.8 | X | |||
| Proenkephalin B[166−174] | R.YGGFLRKYP.K | 1099.60 | 1099.60 | −18.2 | X | |||
| Proenkephalin B[221−234] | R.YGGFLRRQFKVVTR.S | 1725.98 | 1725.98 | −46.4 | X | |||
| Proenkephalin B[228−248] | R.pQFKVVTRSQENPNTYSEDLDV | 2451.61 | 2451.61 | −24.5 | X | |||
| Cortistatin[20−33] | L.LLLSGIAASALPLE.S | 1366.71 | 1366.71 | 65.8 | X | X | ||
| Somatotropin[208−216] | R.RFAESSCAF | 1016.49 | −49.2 | X | ||||
| ProSAAS[245−260] | R.LENSSPQAPARRLLPP. | 1744.37 | 136.4 | X | ||||
| ProSAAS[245−260] | R.LENSSPQAPARRLLPP.(+Na) | 1766.37 | 132.2 | X | ||||
| ProSAAS[42−59] | R.SLSAASAPLAETSTPLRL.R | 892.89 | 1783.97 | 0.6 | X | X | X | |
| ProSAAS[42−58] | R.SLSAASAPLAETSTPLR.L | 1670.80 | 47.9 | X | ||||
| ProSAAS[42−57] | R.SLSAASAPLAETSTPL.R | 757.29 | 1514.58 | 132.0 | X | |||
| ProSAAS[221−242] | R.AVDQDLGPEVPPENVLGALLRV.K | 2300.20 | 13.5 | X | ||||
| ProSAAS[206−218] | G.SSEPEAAPAPRRL.R | 1379.53 | 132.6 | X | ||||
| ProSAAS[62 −75] | R.AVPRGEAAGAVQEL.A | 1366.54 | 131.7 | X | ||||
| ProSAAS[45- 59] | S.AASAPLAETSTPLRL.R | 1496.68 | 93.5 | X | ||||
| ProSAAS[44- 59] | L.SAASAPLAETSTPLRL.R | 1583.71 | 88.4 | X | ||||
| Neurotensin[150−164] | R.pQLYENKPRRPYILKR.A | 1955.99 | 56.2 | X | ||||
| Neurotensin[150−162] | R.pQLYENKPRRPYIL.A | 1671.91 | 0.0 | X | ||||
| PACAP[51−68] | R.GMGENLAAAA VDDRAPLT.K | 1770.90 | −22.6 | X | ||||
| Progonadoliberin I[24−33] | S.pQHWSYGLRPGamide.G | 1181.31 | 111.6 | X | ||||
| Progonadoliberin I[24−33]+Na | S.pQHWSYGL RPGamide.G(+Na) | 1204.52 | 33.2 | X | ||||
| ProMCH[131−143] | R.EIGDEENSAKFPIamide.G | 724.73 | 1446.45 | 160.4 | X | X | ||
| Interleukin-3[54−62] | K.LPVSGLNNS.D | 899.39 | 33.4 | X |
Period (.) indicates cleavage sites. Bold type indicates putative novel peptides. “X” indicates type of MS instrument used to identify peptides. Tandem MS data was obtained in all cases except for the last column, indicating MALDI TOF data. Asterik (*) indicates the Cys−Cys bonds.
Here, we characterized and identified 85 peptides, many of which include peptides derived from known prohormones (see Tables 1 and 2). Che et al.(7) reported 41 neuropeptides from three brain regions (hypothalamus, hippocampus, and striatum) using a quadrupole time-of-flight mass spectrometer equipped with a capillary/nano-LC system. Dowell and co-workers(9) detected 56 neuropeptides from the rat hypothalamus and striatum using a different extraction protocol and 1D LC-MS/MS, 2D LC-MS/MS, and MALDI-FTMS. Our approach uses a smaller brain region from ex vivo brain slices and multistage extraction protocols along with LC-MALDI MS, LC-ESI MS, and LC-ESI FTMS, resulting in the identification of several unreported peptides. The results are consistent with previous studies that characterized peptides in larger brain regions discussed above.
Table 2. Other Peptides and Proteins Identifieda.
| precursor | sequence | ESI MS (m/z) | obs. mass (Da) | error (ppm) | LC-ESI-FTMS | LC-MALDI TOF/TOF | LC-ESI ion trap | LC-MALDI-TOF MS |
|---|---|---|---|---|---|---|---|---|
| MBP S[2−18] | M.(Ac)ASQKRPSQR HGSKYLATA.S | 2026.99 | 34.0 | X | ||||
| MBP S[2−16] | M.(Ac)ASQKRPSQRHGSKYLA.S | 1854.90 | 45.8 | X | ||||
| MBP S[2−11] | M.(Ac)ASQKRPSQRHG.S | 1292.59 | 61.9 | X | ||||
| MBP S[183−195] | G.GRDSRSGSPMARR | 1431.69 | 14.0 | X | ||||
| DBI[2−14] | M.(Ac)SQADFDKAAEEVK.R | 1478.70 | −6.8 | X | ||||
| PEBP[9−25] | W.AGPLSLQEVDEPPQHAL.R | 1800.03 | −72.2 | X | ||||
| Cholecalcin[33−44] | K.LLIQSEFPSLL.K | 1258.74 | 1258.74 | −15.1 | X | X | ||
| T-kininogen I[378−385] | S.RPPGFSPF.R | 903.44 | 903.44 | 18.8 | X | X | ||
| Thymosin beta-10[2−44] | M.ADKPDMGEIASFDKAKLKK TETQEKNTLPTKETIEQEKRSEIS | 4932.52 | −2.0 | X | ||||
| Thymosin beta-4[2−44] | M.SDKPDMAEIEKFDKSKLKKTETQEKNPLPSKETIEQEKQAGES | 4960.49 | −2.9 | X | ||||
| Ubiquitin[1−76] | MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLSDYNIQKESTLHLVLRLRGG | 8559.62 | −1.2 | X | ||||
| Tubulin beta-5 chain[1−7] | MREIVHI.Q | 896.03 | 896.03 | 110.9 | X | X | ||
| Cullin 5-AVP-activated calcium-mobilizing receptorVACM-1 | D.KVPGGIEP.M | 795.45 | 5.0 | X |
Period (.) indicates cleavage sites. Bold type indicates putative novel peptides. “X” indicates type of MS instrument used to identify peptides. Tandem MS data was obtained in all cases except for the last column, indicating MALDI TOF data.
The use of multiple MS-based platforms, LC-MALDI MS, LC-ESI MS, and LC-ESI FTMS, enhances peptide and protein coverage as each platform excels at specific types of samples and analytes. Therefore, their combination facilitates detection of larger numbers of peptides from the SON samples. A majority of detected peptides have been previously observed and characterized in different parts of the mammalian brain such as the hypothalamus, striatum, pituitary, and hippocampus.
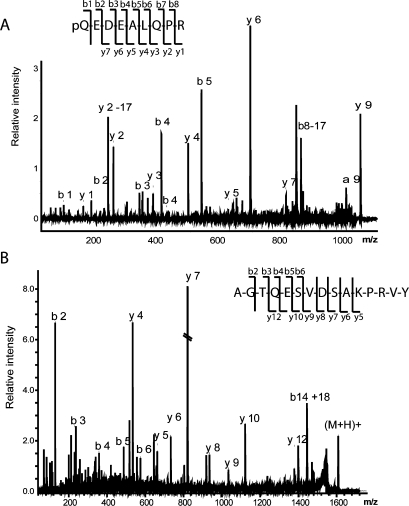
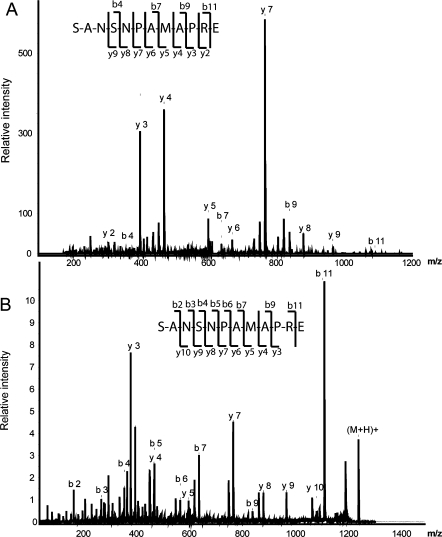
Our data enhances prior peptidome studies on the hypothalamus and striatum.(9) As previously reported peptides from these regions, we also observe the provasopressin[24−32] (AVP), the pyro-Glu form of CART (Figure 1A),(10) POMC[124−136] (melanotropin alpha),7,9 somatostatin[103−116] (somatostatin 14), somatostatin[89−100],7,9 proenkephalin A[100−104] (met-enkephalin), proenkephalin A[219−229], proenkephalin A[198−209],7,9 secretogranin 1[585−594],(9) proSAAS[245−260] (big LEN), proSAAS[42−59] (little SAAS)7,9 and proMCH[131−143].(9) Figure 1 shows the MS/MS fragmentation spectra of the known peptides CART[28−36] (Figure 1A) and provasopressin[154−168] (Figure 1B) identified in the SON, with Figure 2 showing MS/MS results from two MS platforms. In addition to products from known prohormones, several different protein fragments have been identified in this analysis, most of which correspond to pieces of myelin basic protein (MBP), the second most abundant protein in the brain (Table 2).(55)
Figure 1.
MALDI-TOF/TOF (MS/MS) data allows the identification of several peptides in the SON sample: (A) CART[28−36], A.pQEDAELQPR.A, m/z 1067.52 (which was previously characterized in a mouse brain sample); (B) a novel peptide from the vasopressin prohormone: provasopressin[154−168] L.AGTQESVDSAKPRVY, m/z 1606.69.
Figure 2.
Sequencing of known peptides from somatostatin; somatostatin[89−100] R.SANSNPAMAPRE.R, m/z 1243.56, using two tandem MS approaches: (A) MALDI-TOF/TOF and (B) LC-ESI ion trap.
Twenty novel peptides (Table 1) are identified using MS/MS from various precursors: provasopressin, somatostatin, secretogranin 1, secretogranin 3, proenkephalin B, cortistatin, somatotropin, proSAAS, and neurotensin. Figure 1B shows an example of the tandem MS (MS/MS) fragmentation spectra for a novel peptide derived from provasopressin. This peptide presents an unconventional cleavage site at Leu−Ala, a potential product of Leu-X specific protease.(7) A majority of sequenced peptides in the SON are generated by enzymatic cleavage of mono- or dibasic sites of the corresponding prohormones. However, in several cases of the newly discovered peptides, the prohormone cleavage occurs at uncommon processing sites: C-terminus of provasopressin, somatostatin, N-terminus of secretogranin I, N-terminus of secretogranin III, N-terminus signal peptide of cortistatin, N-terminus of proenkephalin B, proSAAS (Table 1). The possibility that these cleavages occur due to extracellular peptide degradation in situ cannot be ruled out.
The peptidome of the SON is as complex as the previously reported peptidome of the entire hypothalamus. At what stage of sample reduction does the peptide complement simplify? To investigate the peptidome of even smaller samples, individual MCN were manually isolated—without enzymatic treatment or in vivo or in vitro labeling of the SON—using coronal brain slices stabilized with glycerol (Figure 3A). Here, MALDI MS of a small group of MCNs provides a cleaner spectrum as compared with the mass profile of single cells, which is not surprising considering the significant increase in the amount of peptides available for analysis. Using MS measurements, AVP (m/z 1083.45) can be unambiguously identified in a low mass range (Figure 3) and neurophysin II (m/z 9842.29) was identified in the higher mass range (Supporting Information, Figure S2), corroborating previous reports of neurophysin prohormones being present in MCN neurons. Different peaks are visible in the higher range mass spectra (Supporting Information). Because the amount of material in an individual cell is too small for MS/MS approaches, we assign these peaks based on sequenced peaks from the SON study and the mass match within the individual MCN. Interestingly, using such mass matching, we identified a CART-related peptide, CART[28−36] (Figure 3B). Many peaks were unidentifiable due to the low amount found in a single cell. Reduction of sample size to the single cell level reduces the depth of peptide coverage but provides a more streamlined linkage of functional, morphological and biochemical information.
Discussion
For the current neuropeptidome study of the SON, hypothalamic coronal brain slices kept in an oxygenated media are used and a complement of peptides detected. How can this be reconciled with prior work showing that tissue denaturation immediately after animal death is required, or large-scale protein and peptide degradation is observed?7,20 The prior peptidome comparisons have been between tissue that is heat- or microwave-treated with various time delays, and not with living brain slices. In fact, peptide-containing dense core vesicles of neurosecretory cells in brain slices lose a fraction of their content initially; however, the contents recover and stay constant for at least 9 h, as shown by Hatton et al.(25) Furthermore, it has been well-established that living brain slices can be used to successfully study the suprachiasmatic nucleus (SCN), the master biological clock. In this case, the experiments demand the maintenance of active tissue for longer than 24 h.23,48 Similarly, Chen et al.(56) found that pituitary adenylate cyclase activating peptide (PACAP) remained active in SCN brain slices longer than 19 h in continuously perfused chambers. On the basis of these studies, and with the information in prior peptidome studies,6−9 we infer that the releasable pool of peptides is rapidly depleted but recovers before the SON is isolated.
Once the SON punch is made and the tissue is removed from the chamber, degradation will occur; we stop this process by using a hot water treatment that also serves to facilitate the peptide extraction processes. After protease deactivation, we employ acidic extraction to complete the peptide extraction. Comparison of the number of characterized SON peptides with the number of identified peptides reported for larger brain structures6−9 supports the efficiency of our strategy.
We used a combination of mass spectrometric approaches (LC-MALDI, LC-ESI, and LC-ESI-FTMS) to increase the coverage in this proteomics investigation and unambiguously identify the peptides. Each approach has its advantages, excelling at characterizing different peptides and modifications. For example, LC-FTMS/MS is a great tool for unambiguously identifying peptides through high-resolution measurements of precursor and product ions. The major drawback to this technique is the lower sensitivity inherent to FT-ICR mass spectrometry. Thus, it is a great tool for making peptide assignments when the ions signals are sufficient, but it trails other techniques in terms of the number of peptides identified.
The detected peptides are the result of processing of known prohormones, with these processing steps either being intracellular (before peptide release) or extracellular (after peptide release), the latter possibly being the result of degradation during sample preparation. A number of detected peptides originate from unconventional cleavage sites, which may be due to extracellular enzymes. In the case of interleukin-3[54−62], the removal of the C-terminal Asp residue may be due to the acidic extraction.7,9 Peptides originating from provasopressin precursor indicate that multiple unconventional, nonspecific cleavages may occur either during the acidic extraction or through enzymatic processing. Because it is difficult to directly relate peak heights of structurally distinct peptides to the amount of peptide present (at least without using calibration standards), it is difficult to know whether some of these unusually processed peptides are present at trace levels compared to the known biologically active peptides.
Surprisingly, one peptide that we did not detect is oxytocin, even though it is in an ideal mass range (m/z 1006.44) for characterization. It should be noted that, when the entire hypothalamus was extracted, oxytocin was not reported in at least one prior study.(9) Nevertheless, there are reports of oxytocin in the hypothalamus identified by mass(6) as well as following N-terminal modification.(8) Considering that 90% of the content in the SON peptides is released dendritically and not somatically or axonally, oxytocin is expected in the SON. Two novel peptides, cortistatin [20−33], and provasopressin[154−168], and a previously characterized peptide, provasopressin[151−168], have Leu-X or Leu−Leu cleavage sites. Che et al.(7) assumed the presence of a Leu-specific enzyme in the processing of corticotropin-like intermediary peptide of POMC prohormone, and Hummon et al.(57) reported Leu−Leu cleavage sites in Aplysia californica prohormones. It is known that in vertebrates, the enzyme renin specifically cleaves Leu−Leu sites.(58) Another enzyme, chymotrypsin, previously characterized in the human, rat, and pig, cleaves Leu−Ala sites.(59) On the basis of these studies, we assume that cortistatin[20−33] and provasopressin[154−168] are cleaved by either rennin-like or chymotrypsin-like enzymes. We further speculate that provasopressin[151−168] and provasopressin[154−168] are processed in intracellular secretory vesicles since they also were observed in prior microwaved samples (data not shown). According to Che et al.,(7) rapid microwave irradiation before decapitation prevents vesicle fusion and neuropeptide release; hence, the peptides remain in secretory vesicles and are available for extraction.
MS/MS is capable of characterizing the PTMs present in neuropeptides, which is important as these PTMs often directly modify the activity or lifetime of the peptide. The most common PTMs in mammalian tissues are N-terminal acetylation and C-terminal amidation.(2) In 1987, Fischer and Spiess(60) identified a glutaminyl cyclase converting enzyme in mammalian brain responsible for the formation of N-terminal pyro-Glu. Additionally, in situ and immunocytochemical studies showed that the SON contains a glutaminyl cyclase-converting enzyme required to create pyro-Glu forms of several peptides.60,61 Here, we detect pyro-Glu forms of CART[28−36],(10) neurotensin precursor[150−164], and progonadoliberin I precursor[24−33]. We also detect amidation of AVP, substance P,(62) POMC[103−120],(7) and POMC[124−136],(62) as shown in Table 1. We also identify novel N-terminal acetylated peptides such as proenkephalin[198−209], POMC[124−137], secretogranin 2[287−297], secretogranin 5[198−210], secretogranin 5[180−195], the pyro-Glu forms of secretogranin 1[597−611], proenkephalin B[228−248], and neurotensin precursor[150−162] (Table 1). Although our processing steps reduce protein degradation, we did detect several peptides from proteins that are not prohormones (Table 2). MBP is the second most abundant protein in the central nervous system and is present in the peripheral nervous system as well.(55) MBP can have different PTMs such as phosphorylation of serine and threonine residues, deamidation of Asn or Gln, citrullination and methylation of arginine residues, and N-terminal acetylation of Thr residues.63−66 MBP plays an important function in axon myelination, but other possible roles of MBP are hinted, such as significant changes in mRNA levels on exposure to cocaine.67,68 Three peptides derived from MBP are detected in the current study of the SON peptidome, one of which exhibits N-terminal acetylation of Thr residues (Table 2).
Our neuropeptidomics study of this small nucleus extends to the single cell level using MALDI MS. Although in SON extracts we are able to identify several fragments derived from provasopressin, our single cell experiments detect the characteristic peptides processed from this hormone. The major drawback of single cell MS is the low amount of material available for analysis; some peptides are often present below the detection level of the employed MS approach. Because of our careful isolation approach, extracellular degradation products from peptides and proteins are not normally detected. Thus, our detection of CART-related peptide in a portion of the studied MCN cells is exciting and suggests that we are observing cell-to-cell differences in the peptide complement in a “homogenous” population of MCNs.
With the ability to perform peptidome studies in well-defined, smaller anatomical regions from the brain slice, coupled with novel methods of characterizing stimulation-dependent peptide release recently reported,(27) the ability to link peptide complement to function has become possible.
Acknowledgments
We thank Jennifer Mitchell and Sufang Huang from the Gillette Laboratory for tissue preparation and useful discussions. This work was funded in part from the W. M. Keck Foundation, the National Institute of Drug Abuse under Award No. DA017940 and Award No. DA018310 to the UIUC Neuroproteomics Center on Cell-Cell Signaling.
Supporting Information Available
Table of total number of identified and novel peptides from known prohormones, figures of the different protocols used to extract peptides from the SON and higher molecular weight range from a small group of MCNs. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Hokfelt T.; Broberger C.; Xu Z. Q.; Sergeyev V.; Ubink R.; Diez M. Neuropeptides-an overview. Neuropharmacology 2000, 39 (8), 1337–56. [DOI] [PubMed] [Google Scholar]
- Strand F. L.Neuropeptides: Regulators of Physiological Processes; MIT Press: Cambridge, MA, 1999. [Google Scholar]
- Fricker L. D.; Carboxypeptidase E. Annu. Rev. Physiol. 1988, 50, 309–21. [DOI] [PubMed] [Google Scholar]
- Li L.; Sweedler J. Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu. Rev. Anal. Chem. 2008, (1), 451–483. [DOI] [PubMed] [Google Scholar]
- Polevoda B.; Sherman F. The diversity of acetylated proteins. GenomeBiology 2002, 3, reviews0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che F. Y.; Biswas R.; Fricker L. D. Relative quantitation of peptides in wild-type and Cpe(fat/fat) mouse pituitary using stable isotopic tags and mass spectrometry. J. Mass Spectrom. 2005, 40 (2), 227–37. [DOI] [PubMed] [Google Scholar]
- Che F. Y.; Lim J.; Pan H.; Biswas R.; Fricker L. D. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol. Cell. Proteomics 2005, 4 (9), 1391–405. [DOI] [PubMed] [Google Scholar]
- Che F. Y.; Zhang X.; Berezniuk I.; Callaway M.; Lim J.; Fricker L. D. Optimization of neuropeptide extraction from the mouse hypothalamus. J. Proteome Res. 2007, 6 (12), 4667–76. [DOI] [PubMed] [Google Scholar]
- Dowell J. A.; Heyden W. V.; Li L. Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC. J. Proteome Res. 2006, 5 (12), 3368–75. [DOI] [PubMed] [Google Scholar]
- Svensson M.; Skold K.; Svenningsson P.; Andren P. E. Peptidomics-based discovery of novel neuropeptides. J. Proteome Res. 2003, 2 (2), 213–9. [DOI] [PubMed] [Google Scholar]
- Theodorsson E.; Stenfors C.; Mathe A. A. Microwave irradiation increases recovery of neuropeptides from brain tissues. Peptides 1990, 11 (6), 1191–7. [DOI] [PubMed] [Google Scholar]
- Romanova E. V.; Rubakhin S. S.; Sweedler J. V. One-step sampling, extraction, and storage protocol for peptidomics using dihydroxybenzoic acid. Anal. Chem. 2008, 80 (9), 3379–86. [DOI] [PubMed] [Google Scholar]
- Hummon A. B.; Amare A.; Sweedler J. V. Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom. Rev. 2006, 25 (1), 77–98. [DOI] [PubMed] [Google Scholar]
- Jimenez C. R.; van Veelen P. A.; Li K. W.; Wildering W. C.; Geraerts W. P.; Tjaden U. R.; van der Greef J. Neuropeptide expression and processing as revealed by direct matrix-assisted laser desorption ionization mass spectrometry of single neurons. J. Neurochem. 1994, 62 (1), 404–7. [DOI] [PubMed] [Google Scholar]
- Neupert S.; Johard H. A.; Nassel D. R.; Predel R. Single-cell peptidomics of drosophila melanogaster neurons identified by Gal4-driven fluorescence. Anal. Chem. 2007, 79 (10), 3690–4. [DOI] [PubMed] [Google Scholar]
- Rubakhin S. S.; Churchill J. D.; Greenough W. T.; Sweedler J. V. Profiling signaling peptides in single mammalian cells using mass spectrometry. Anal. Chem. 2006, 78 (20), 7267–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubakhin S. S.; Sweedler J. V. Characterizing peptides in individual mammalian cells using mass spectrometry. Nat. Protoc. 2007, 2 (8), 1987–97. [DOI] [PubMed] [Google Scholar]
- Boonen K.; Landuyt B.; Baggerman G.; Husson S. J.; Huybrechts J.; Schoofs L. Peptidomics: the integrated approach of MS, hyphenated techniques and bioinformatics for neuropeptide analysis. J. Sep. Sci. 2008, 31 (3), 427–45. [DOI] [PubMed] [Google Scholar]
- Minamino N.; Tanaka J.; Kuwahara H.; Kihara T.; Satomi Y.; Matsubae M.; Takao T. Determination of endogenous peptides in the porcine brain: possible construction of peptidome, a fact database for endogenous peptides. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2003, 792 (1), 33–48. [DOI] [PubMed] [Google Scholar]
- Skold K.; Svensson M.; Norrman M.; Sjogren B.; Svenningsson P.; Andren P. E. The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: stathmin 2−20 and peptides as sample quality indicators. Proteomics 2007, 7 (24), 4445–56. [DOI] [PubMed] [Google Scholar]
- Nylander I.; Stenfors C.; Tan-No K.; Mathe A. A.; Terenius L. A comparison between microwave irradiation and decapitation: basal levels of dynorphin and enkephalin and the effect of chronic morphine treatment on dynorphin peptides. Neuropeptides 1997, 31 (4), 357–65. [DOI] [PubMed] [Google Scholar]
- Parkin M. C.; Wei H.; O′Callaghan J. P.; Kennedy R. T. Sample-dependent effects on the neuropeptidome detected in rat brain tissue preparations by capillary liquid chromatography with tandem mass spectrometry. Anal. Chem. 2005, 77 (19), 6331–8. [DOI] [PubMed] [Google Scholar]
- Gillette M. U. The suprachiasmatic nuclei: circadian phase-shifts induced at the time of hypothalamic slice preparation are preserved in vitro. Brain Res. 1986, 379 (1), 176–81. [DOI] [PubMed] [Google Scholar]
- Groos G.; Hendriks J. Circadian rhythms in electrical discharge of rat suprachiasmatic neurones recorded in vitro. Neurosci. Lett. 1982, 34 (3), 283–8. [DOI] [PubMed] [Google Scholar]
- Hatton G. I.; Doran A. D.; Salm A. K.; Tweedle C. D. Brain slice preparation: hypothalamus. Brain Res. Bull. 1980, 5 (4), 405–14. [DOI] [PubMed] [Google Scholar]
- Fiala J. C.; Kirov S. A.; Feinberg M. D.; Petrak L. J.; George P.; Goddard C. A.; Harris K. M. Timing of neuronal and glial ultrastructure disruption during brain slice preparation and recovery in vitro. J. Comp. Neurol. 2003, 465 (1), 90–103. [DOI] [PubMed] [Google Scholar]
- Hatcher N. G.; Atkins N. J.; Annangudi S. P.; Forbes A. J.; Kelleher N. L.; Gillette M. U.; Sweedler J. V. Mass spectrometry-based discovery of circadian peptides. Proc. Natl. Acad. Sci. U.S.A. 2008, 105 (34), 12527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong W. E.; Hatton G. I. Morphological changes in the rat supraoptic and paraventricular nuclei during the diurnal cycle. Brain Res. 1978, 157 (2), 407–13. [DOI] [PubMed] [Google Scholar]
- Brimble M. J.; Dyball R. E.; Forsling M. L. Oxytocin release following osmotic activation of oxytocin neurones in the paraventricular and supraoptic nuclei. J. Physiol. 1978, 278, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajkowska B. Electron microscopic lesions of supraoptic and para-ventricular nuclei of rat thalamus in adrenal insufficiency. Pol. Med. Sci. Hist. Bull. 1975, 15 (5−6), 591–605. [PubMed] [Google Scholar]
- Gajkowska B.; Borowicz J. Electron microscopic observations of the supraoptic and paraventricular nuclei of rat brain in chronic morphine poisoning and after drug withdrawal. Neuropatol. Pol. 1976, 14 (3), 363–70. [PubMed] [Google Scholar]
- Knigge K. M.; Piekut D. T.; Berlove D. J. Immunocytochemistry of magnocellular neurons of supraoptic and paraventricular nuclei of normal and Brattleboro rats with vasopressin anti-idiotype antibody. Cell Tissue Res. 1986, 246 (3), 509–13. [DOI] [PubMed] [Google Scholar]
- Landgraf R.; Ludwig M. Vasopressin release within the supraoptic and paraventricular nuclei of the rat brain: osmotic stimulation via microdialysis. Brain Res. Bull. 1991, 558 (2), 191–6. [DOI] [PubMed] [Google Scholar]
- Morris J. F. Organization of neural inputs to the supraoptic and paraventricular nuclei: anatomical aspects. Prog. Brain Res. 1983, 60, 3–18. [DOI] [PubMed] [Google Scholar]
- Noto T.; Hashimoto H.; Doi Y.; Nakajima T.; Kato N. Biorhythm of arginine- vasopressin in the paraventricular, supraoptic and suprachiasmatic nuclei of rats. Peptides 1983, 4 (6), 875–8. [DOI] [PubMed] [Google Scholar]
- Rosenwasser A. M.; Trubowitsch G.; Adler N. T. Circadian rhythm in metabolic activity of suprachiasmatic, supraoptic and raphe nuclei. Neurosci. Lett. 1985, 58 (2), 183–7. [DOI] [PubMed] [Google Scholar]
- Sawchenko P. E.; Swanson L. W. The organization and biochemical specificity of afferent projections to the paraventricular and supraoptic nuclei. Prog. Brain Res. 1983, 60, 19–29. [DOI] [PubMed] [Google Scholar]
- Yamashita H. Effect of baro- and chemoreceptor activation on supraoptic nuclei neurons in the hypothalamus. Brain Res. 1977, 126 (3), 551–6. [DOI] [PubMed] [Google Scholar]
- Ang C. W.; Dotman C. H.; Winkler H.; Fischer-Colbrie R.; Sonnemans M. A.; Van Leeuwen F. W. Specific expression of secretogranin II in magnocellular vasopressin neurons of the rat supraoptic and paraventricular nucleus in response to osmotic stimulation. Brain Res. 1997, 765 (1), 13–20. [DOI] [PubMed] [Google Scholar]
- Majdoubi M. E.; Metz-Boutigue M. H.; Garcia-Sablone P.; Theodosis D. T.; Aunis D. Immunocytochemical localization of chromogranin A in the normal and stimulated hypothalamo-neurohypophysial system of the rat. J. Neurocytol. 1996, 25 (7), 405–16. [PubMed] [Google Scholar]
- Koylu E. O.; Couceyro P. R.; Lambert P. D.; Ling N. C.; DeSouza E. B.; Kuhar M. J. Immunohistochemical localization of novel CART peptides in rat hypothalamus, pituitary and adrenal gland. J. Neuroendocrinol. 1997, 9 (11), 823–33. [DOI] [PubMed] [Google Scholar]
- Larsen P. J.; Seier V.; Fink-Jensen A.; Holst J. J.; Warberg J.; Vrang N. Cocaine- and amphetamine-regulated transcript is present in hypothalamic neuroendocrine neurones and is released to the hypothalamic-pituitary portal circuit. J. Neuroendocrinol. 2003, 15 (3), 219–26. [DOI] [PubMed] [Google Scholar]
- Hooi S. C.; Richardson G. S.; McDonald J. K.; Allen J. M.; Martin J. B.; Koenig J. I. Neuropeptide Y (NPY) and vasopressin (AVP) in the hypothalamo-neurohypophysial axis of salt-loaded or Brattleboro rats. Brain Res. Bull. 1989, 486 (2), 214–20. [DOI] [PubMed] [Google Scholar]
- Pow D. V.; Morris J. F. Dendrites of the hypothalamic magnocellular neurons release neurohypophysal peptides by exocytosis. Neuroscience 1989, 32 (2), 435–39. [DOI] [PubMed] [Google Scholar]
- Sabatier N.; Caquineau C.; Douglas A. J.; Leng G. Oxytocin released from magnocellular dendrites: a potential modulator of alpha-melanocyte-stimulating hormone behavioral actions?. Ann. N.Y. Acad. Sci. 2003, 1994, 218–24. [DOI] [PubMed] [Google Scholar]
- Ma P. W.; Garden R. W.; Niermann J. T.; M O. C.; Sweedler J. V.; Roelofs W. L. Characterizing the Hez-PBAN gene products in neuronal clusters with immunocytochemistry and MALDI MS. J. Insect Physiol. 2000, 46 (3), 221–30. [DOI] [PubMed] [Google Scholar]
- Neupert S.; Predel R. Mass spectrometric analysis of single identified neurons of an insect. Biochem. Biophys. Res. Commun. 2005, 327 (3), 640–5. [DOI] [PubMed] [Google Scholar]
- Tischkau S. A.; Mitchell J. W.; Pace L. A.; Barnes J. W.; Barnes J. A.; Gillette M. U. Protein kinase G type II is required for night-to-day progression of the mammalian circadian clock. Neuron 2004, 43 (4), 539–49. [DOI] [PubMed] [Google Scholar]
- Amare A.; Hummon A. B.; Southey B. R.; Zimmerman T. A.; Rodriguez-Zas S. L.; Sweedler J. V. Bridging neuropeptidomics and genomics with bioinformatics: prediction of mammalian neuropeptide prohormone processing. J. Proteome Res. 2006, 5 (5), 1162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southey B. R.; Amare A.; Zimmerman T. A.; Rodriguez-Zas S. L.; Sweedler J. V. NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 2006, 34 (Web Server issue), W267–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D. M.; Zubarev R. A.; McLafferty F. W. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J. Am. Soc. Mass Spectrom. 2000, 11 (4), 320–32. [DOI] [PubMed] [Google Scholar]
- LeDuc R. D.; Taylor G. K.; Kim Y. B.; Januszyk T. E.; Bynum L. H.; Sola J. V.; Garavelli J. S.; Kelleher N. L. ProSight PTM: an integrated environment for protein identification and characterization by top-down mass spectrometry. Nucleic Acids Res. 2004, 32 (Web Server issue), W340–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G. K.; Kim Y. B.; Forbes A. J.; Meng F.; McCarthy R.; Kelleher N. L. Web and database software for identification of intact proteins using ”top down” mass spectrometry. Anal. Chem. 2003, 75 (16), 4081–6. [DOI] [PubMed] [Google Scholar]
- Parks B. A.; Jiang L.; Thomas P. M.; Wenger C. D.; Roth M. J.; Boyne M. T. II; Burke P. V.; Kwast K. E.; Kelleher N. L. Top-down proteomics on a chromatographic time scale using linear ion trap fourier transform hybrid mass spectrometers. Anal. Chem. 2007, 79 (21), 7984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs J. M. Myelin basic protein: a multifunctional protein. Cell. Mol. Life Sci. 2006, 63 (17), 1945–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.; Buchanan G. F.; Ding J. M.; Hannibal J.; Gillette M. U. Pituitary adenylyl cyclase-activating peptide: a pivotal modulator of glutamatergic regulation of the suprachiasmatic circadian clock. Proc. Natl. Acad. Sci. U.S.A. 1999, 96 (23), 13468–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummon A. B.; Kelley W. P.; Sweedler J. V. A novel prohormone processing site in Aplysia californica: the Leu-Leu rule. J. Neurochem. 2002, 82 (6), 1398–405. [DOI] [PubMed] [Google Scholar]
- Skeggs L. T.; Dorer F. E.; Levine M.; Lentz K. E.; Kahn J. R. The biochemistry of the renin-angiotensin system. Adv. Exp. Med. Biol. 1980, 130, 1–27. [DOI] [PubMed] [Google Scholar]
- Bauer C. A.; Thompson R. C.; Blout E. R. The active centers of Streptomyces griseus protease 3, alpha-chymotrypsin, and elastase: enzyme-substrate interactions close to the scissile bond. Biochemistry 1976, 15 (6), 1296–9. [DOI] [PubMed] [Google Scholar]
- Fischer W. H.; Spiess J. Identification of a mammalian glutaminyl cyclase converting glutaminyl into pyroglutamyl peptides. Proc. Natl. Acad. Sci. U.S.A. 1987, 84 (11), 3628–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockers T. M.; Kreutz M. R.; Pohl T. Glutaminyl-cyclase expression in the bovine/porcine hypothalamus and pituitary. J. Neuroendocrinol. 1995, 7 (6), 445–53. [DOI] [PubMed] [Google Scholar]
- Eipper B. A.; Stoffers D. A.; Mains R. E. The biosynthesis of neuropeptides: peptide alpha-amidation. Annu. Rev. Neurosci. 1992, 15, 57–85. [DOI] [PubMed] [Google Scholar]
- Deibler G. E.; Martenson R. E. Chromatographic fractionation of myelin basic protein. Partial characterization and methylarginine contents of the multiple forms. J. Biol. Chem. 1973, 248 (7), 2392–6. [PubMed] [Google Scholar]
- Moscarello M. A.; Pang H.; Pace-Asciak C. R.; Wood D. D. The N terminus of human myelin basic protein consists of C2, C4, C6, and C8 alkyl carboxylic acids. J. Biol. Chem. 1992, 267 (14), 9779–82. [PubMed] [Google Scholar]
- Wood D. D.; Moscarello M. A. The isolation, characterization, and lipid-aggregating properties of a citrulline containing myelin basic protein. J. Biol. Chem. 1989, 264 (9), 5121–7. [PubMed] [Google Scholar]
- Zand R.; Li M. X.; Jin X.; Lubman D. Determination of the sites of posttranslational modifications in the charge isomers of bovine myelin basic protein by capillary electrophoresis-mass spectroscopy. Biochemistry 1998, 37 (8), 2441–9. [DOI] [PubMed] [Google Scholar]
- Albertson D. N.; Pruetz B.; Schmidt C. J.; Kuhn D. M.; Kapatos G.; Bannon M. J. Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. J. Neurochem. 2004, 88 (5), 1211–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon M.; Kapatos G.; Albertson D. Gene expression profiling in the brains of human cocaine abusers. Addict. Biol. 2005, 10 (1), 119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.