Abstract
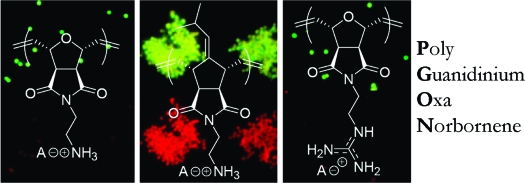
Polyguanidinium oxanorbornene (PGON) was synthesized from norbornene monomers via ring-opening metathesis polymerization. This polymer was observed to be strongly antibacterial against Gram-negative and Gram-positive bacteria as well as nonhemolytic against human red blood cells. Time-kill studies indicated that this polymer is lethal and not just bacteriostatic. In sharp contrast to previously reported SMAMPs (synthetic mimics of antimicrobial peptides), PGON did not disrupt membranes in vesicle-dye leakage assays and microscopy experiments. The unique biological properties of PGON, in same ways similar to cell-penetrating peptides, strongly encourage the examination of other novel guanidino containing macromolecules as powerful and selective antimicrobial agents.
Introduction
We report that poly guanidinium oxanorbornene (PGON) possesses a remarkable combination of antimicrobial activity against both Gram-negative and Gram-positive bacteria as well as low hemolytic activity against human red blood cells (RBCs). Standard antibacterial, bacteria cell-viability, and hemolysis assays all give data that is encouraging for the development of powerful, yet nontoxic, antibacterial materials.(1) Additionally, the properties of PGON surpass that of other cationic polymers, in particular Poly-1 and Poly-3 from our previous study.(2) Vesicle investigations show that PGON is not membrane-disruptive, supporting a previous study indicating it also has properties similar to poly arginine and other cell-penetrating peptides (CPPs) like HIV-TAT.(3) As a result, PGON represents an exciting and fundamentally novel entry in the expanding field of synthetic mimics of antimicrobial peptides (SMAMPs).1,2,4,5
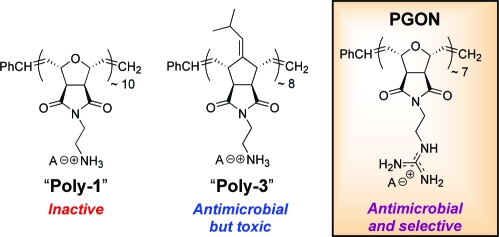
While several antimicrobial peptides (AMPs) contain the amino acid arginine along with other basic residues, SMAMPs have rarely used the guanidine functionality. Instead, the focus has been almost exclusively on amines.2,6 In this work, a new Boc-protected guanidinium oxanorbornene monomer was polymerized via ring-opening metathesis polymerization (ROMP) using the third generation Grubbs’ catalyst to afford synthetic poly arginine, PGON (PDI = 1.07, MW of the deprotected polymer-TFA salt = 2.5 kDa,). This strategy is in contrast to a postpolymerization functionalization technique recently published that afforded polynorbornenes that efficiently promote cellular internalization.(7) Lastly, polyamine norbornenes, Poly-1 and Poly-3, with similar MWs were prepared for comparison.
Experimental Section
Materials
Solvents and reagents for the monomer synthesis were purchased from Aldrich and used without further purification. For the polymer synthesis the second generation Grubbs’ catalyst, tricyclohexylphosphine, (1,3-dimesitylimidazolidine-2-ylidine)benzylideneruthenium dichloride was purchased from Aldrich. This compound was converted to a Grubbs’ third generation catalyst by reaction with 3-bromopyridine from Aldrich. Polymerizations were performed in dry CH2Cl2 from Acros (sealed under nitrogen with molecular sieves). Chloroform solutions of lipids were obtained from Avanti Polar Lipids. Calcein dye was purchased from Aldrich. The BacLight Staining Kit L-7012 was obtained from Invitrogen Corp.
Synthesis of PGON
The synthesis of PGON starts with oxanorbornene 1,(1) and to this salt (1.4 g, 4.34 mmol) N,N′-di-Boc-1H-pyrazole-1-carboxamidine (4.0 g, 13.02 mmol) and triethylamine (4.1 g, 40 mmol) were added with 60 mL of solvent (90% CH3CN, 10% H2O). This solution was stirred at room temperature for 12 h. The solution was then diluted with ethyl acetate and washed twice each with H2O and brine. The organic layer was evaporated and purified by silica column chromatography using a MeOH in CH2Cl2 gradient to afford the Boc-protected guanidinium oxanorbornene monomer, 2.
This monomer was polymerized by ring-opening metathesis polymerization (ROMP) using a derivative of the second generation Grubbs’ catalyst, [(H2Imes)(3-Br-py)2-(Cl)2Ru=CHPh].(2) The polymerization for PGON entailed adding to a test tube monomer (100 mg) plus catalyst (27.4 mg). The test tube was capped with a septum and purged with N2 for 5 min, then 1 mL dry CH2Cl2 was injected. The N2 line was removed and the clear, brown solution was stirred at 28 °C for 30 min after which 0.4 mL of ethyl vinyl ether was injected to terminate the polymer. After stirring for 15 min, the solution was added dropwise to 300 mL of stirring pentane to precipitate the polymer. The pentane solution was stirred an additional 30 min and left standing undisturbed for an hour. The precipitate was then collected by a fine sinter funnel. The polymer was then redissolved in 1 mL of CH2Cl2, reprecipitated, collected, and then dried by vacuum for 8 h.
The Boc-protected polymers were deprotected by stirring 100 mg in 8 mL of 1:1 TFA/CH2Cl2 for 2 h. The solution was concentrated to an oil by rotary evaporator set at 40 °C and residual TFA was removed by sonicating the oil in more CH2Cl2 and evaporating the solvent again by rotary evaporator. The resulting solid was placed under vacuum for 2 h. Finally, the solid was fully dissolved in 4 mL of H2O and filtered through a polyethersulphone (PES) syringe filter (Whatman, 25 mm diameter, 0.45 μm pore) and freeze-dried for 48 h to give an eggshell colored soft solid. Final deprotected polymers were stored at −20 °C.
Boc-Protected Guanidinium Oxanorbornene Monomer, 2
1H NMR (300 MHz, DMSO-d6): δ 11.44 (1H, s), 8.35 (1H, t, J = 6.0 Hz), 6.56 (2H, s), 5.10 (2H, s), 3.52 (2H, m), 3.43 (2H, m), 2.85 (2H, s), 1.47 (9H, s), 1.36 (9H, s). 13C NMR (75 MHz, DMSO-d6): δ 176.3, 156.6, 153.0, 136.5, 83.3, 80.9, 47.6, 39.0, 38.4, 28.3, 28.1. HR-MS (FAB+) calcd, 451.49; found, 451.22.
Boc-Protected PGON
1H NMR (300 MHz, DMSO-d6): δ 11.51 (1H, br), 8.47 (1H, br), 5.90 (trans) and 5.70 (cis; 2H total, br), 4.89 (cis) and 4.39 (trans; 2H total, br), 1.45 (9H, s), 1.38 (9H, br), cis/trans ratio = 46:54.
GPC of Boc-protected PGON: Mn = 3.2 kDa, PDI = 1.07 (After deprotection the TFA-salt PGON has an approximate weight of 2.5 kDa).
PGON
1H NMR (300 MHz, DMSO-d6): δ 7.88 (1H, br), 7.36 (4H, br), 5.96 (1H, br), 5.74 (1H, br), 4.92 (1H, br), 4.43 (1H, br) 3.47 (4H, br).
Microbial and Hemolysis Assays
Bacteria suspensions of Ec, Sm, Sa, and Bs, (Escherichia coli D31, Serratia marcescens ATCC 43861, Staphylococcus aureus ATCC 25923, and Bacillus subtilis ATCC 8037) were grown in Mueller−Hinton Broth (MHB) overnight at 37 °C, diluted with fresh MHB to an optical density of 0.001 at 600 nm (OD600). This OD600 gave an initial bacterial concentration of ∼105 cells/mL. This suspension was mixed with different concentrations of freshly prepared polymer solutions dissolved in DMSO (40 mg/mL stock solution) by serial dilutions in a 96-well plate, and incubated for 6 h at 37 °C. The OD600 was measured for bacteria suspensions that were incubated in the presence of polymer solution or only MHB. Antibacterial activity was expressed as minimal inhibitory concentration (MIC), the concentration at which more than 90% inhibition of growth was observed after 6 h. All experiments were run in quadruplicate.
Hemolytic activity measurements were performed on all polymers. Freshly drawn human red blood cells (RBC, 30 μL), were suspended in 10 mL of Tris saline (10 mM Tris, 150 mM NaCl, pH 7.2, filtered through PES membrane with 0.22 μm pore size) and rinsed three times by centrifugation (5 min at 1500 rpm) and resuspended in Tris saline. Polymer solutions were prepared in DMSO at 40 mg/mL and further diluted as necessary. Freshly prepared polymer solutions with different concentrations were added to 100 μL of the above-prepared RBC suspension to reach a final volume of 200 μL on a 96-well plate. The resulting mixture was kept at 37 °C for 30 min on a stirring plate. Then the plate was centrifuged (IEC Centra-4B, 10 min at 1500 rpm), and the supernatant in each well was transferred to a new plate. Hemolysis was monitored by measuring the absorbance of the released hemoglobin at 414 nm. Hemolysis (100%) was obtained by adding 10 μL of Triton-X (polyoxyethylene isooctylphenyl ether from Sigma-Aldrich) solution (20 vol % in DMSO), a strong surfactant, to the above-prepared HRBC suspension. The upper limit of polymer concentration that was required to cause 50% hemolysis is reported as HC50, and the absorbance from Tris saline containing no polymer was used as 0% hemolysis. All experiments were run in quadruplicate.
Bactericidal Kinetics Studies
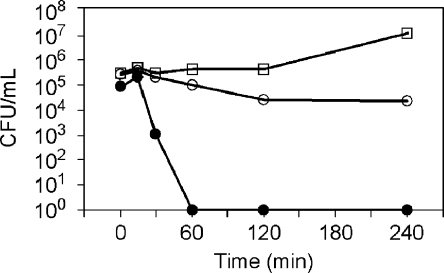
Bactericidal kinetics studies (also known as time-kill assays) were performed on Sa to differentiate growth inhibition from cell death. The protocol was adapted from literature.(3) Cells were grown in MHB to midexponential phase with an OD600 ∼0.3, and then adjusted to an OD600 of 0.001. This OD600 gave an initial bacterial concentration of ∼105 cells/mL, the same as in the MIC studies. Cells were either untreated or treated to PGON at 1× MIC or four times the MIC90 concentration and then placed under rotary agitation at 37 °C. Aliquots were removed at different time points (0, 15, 30, 60, 120, and 240 min) and placed back at 37 °C with agitation. Cells were diluted immediately to remove any effect of PGON. Serial 10-fold dilutions were spread on MHB agar plates and incubated at 37 °C for 18 h to determine viable colony counts. Viable cell counts were plotted on a log-scale graph (CFU/mL vs time) to determine log-reduction of bacterial growth compared to the untreated control.
Dye-Leakage from Large Unilamellar Vesicles
The following is a general procedure to make large unilamellar vesicles (LUV) having a hydrodynamic radius, Rh ∼ 120 nm (by dynamic light scattering). Chloroform solutions of POPE (3.23 mg, 0.0045 mmol) and POPG (1.15 mg, 0.0015 mmol; Avanti Polar Lipids, Inc.) were mixed in a 10 mL round-bottom flask, and the chloroform was removed at room temperature by rotary evaporator to form a uniform film. The flask was placed under vacuum for an additional 6 h. The dried film was hydrated with 1 mL of a 40 mM aqueous calcein (Sigma-Aldrich) solution in Tris buffer without added NaCl. The calcein dye solution was adjusted to pH = 7 prior to adding it to the film. The solution was subjected through five freeze/thaw cycles using liquid nitrogen and warm water. The entire volume of 1 mL of solution was subjected to extrusion (a total of 15 passes) through two stacked 400 nm pore PC membranes (Avanti Polar Lipids, Inc.) at room temperature. Finally, the solution was pressurized through a small column packed with Sephadex-25 (Sigma-Aldrich), eluting with Tris-saline buffer to remove nontrapped calcein dye. Fractions (∼5 drops each) were collected. The vesicle solution can be stored in a vial at 4 °C and diluted as needed for up to 4 days.
Diluted calcein-loaded vesicle fractions that afforded fluorescence intensities <80 au without surfactant and >800 au after addition of surfactant, were used (Ex. = 490 nm used and Em. = 515 nm monitored). A total of 50 μL of 0.2% Triton-X was used as the strong surfactant, which causes complete vesicle disruption and leakage. Typically, chosen vesicle fractions were diluted 6-fold and 25 μL of this stock solution is added to 2 mL of Tris-saline buffer. A fluorescence intensity of <80 au was verified to be stable over a minute before 25 μL of a polymer stock solution was added (2.5 μg/mL final concentration in cuvette), agitated briefly with the pipettor tip, then monitored at 515 nm. After 5 min, Triton-X was added and the corresponding fluorescence was taken as 100% leakage.
Fluorescence Microscopy of Stained Bacteria Cells
An Olympus BX51 Reflected Fluorescence Microscope (Optical Analysis Corp. Nashua, NH) with a 100 W Mercury Lamp (Chin Technical Corp.) was used for fluorescence studies. A BacLight Kit L-7012 (Invitrogen Corp.) was used as the fluorescence dye in a mixture of 1:1 propidium iodide:SYTO9 to examine Sa in the presence of PGON. It is important to mention that an initial bacterial concentration of ∼108 cells/mL was used for microscopy studies (3 orders of magnitude higher than the MIC and bactericidal kinetics studies) for ease of visualization. The dye mixture was incubated with the bacteria at room temperature for 15 min before adding polymer solution (75 μg/mL final concentration). Solution of cells, dye, and polymer were allowed to sit for 30 min before 50 μL was placed on a slide, fitted with a coverslip, and visualized. Bacteria were viewed under a green filter (excitation/emission, 420−480 nm/520−800 nm) or a red filter (480−550 nm/590−800 nm). To confirm that these experiments agreed with the time-kill experiments discussed above and to confirm that Poly-1 is not lethal or static and that Poly-3 is lethal, cell counts were taken directly from the microscopy experiments. They confirmed that Poly-1 is not active with no reduction in cell count compared to the control (at 30 min and two hours), while Poly-3 and PGON showed a reduction of 8 logs in less than two hours. At the 30 min time point, Poly-3 and PGON showed log reductions of 8 and 4, respectively.
Results and Discussion
Results from microbiological assays(8) showed that PGON had an MIC of 6 μg/mL against E. coli (see Table 1 for abbreviations). Activity against an additional Gram-negative bacteria, S. marcescens, which is not typically susceptible to AMPs, and two Gram-positive bacteria, S. aureus and B. subtilis, were also examined attesting to the broad activity of PGON. For comparison, the Magainin derivative, MSI-78, had an MIC of 12 μg/mL against E. coli. PGONʼs close structural analogue, Poly-1, having a primary amino group instead of a guanidino group, had MICs of 200−400 μg/mL and was thus considered to be relatively inactive. In contrast, the MICs of the more hydrophobic, isobutylidene-derivatized Poly-3 were found to be between 12−25 μg/mL for all bacteria, representing good activity. However, and in sharp contrast to Poly-3, broadly active PGON possessed a lack of toxicity toward RBCs (HC50 = 1500 μg/mL). As a result of PGONʼs high HC50 value, its selectivity of 250 is significantly better than other polymeric SMAMPs.1,2,4 Again, as a comparison, MSI-78 had a selectivity of 10 for E. coli over RBCs using the HC50/MIC ratio.
Table 1. Biological Activities.
| MICa (μg/mL) |
||||||
|---|---|---|---|---|---|---|
| polymer (kDa) | Ec | Sm | Sa | Bs | HC50 | Sel.b |
| Poly-1 (3.3) | 400d | 400 | 400 | 200d | 2150 | 5 |
| Poly-3 (2.9) | 25d | 25 | 25 | 12d | 1 | 0.04 |
| PGON (2.5) | 6 | 50 | 12 | 12 | 1500 | 250 |
| AMP (2.5)c | 12 | >256 | 12 | 2 | 120 | 10 |
MIC = minimum inhibitory concentration, HC50 = hemolytic concentration, Ec = Escherichia coli, Sm = Serratia marcescens, Sa = Staphylococcus aureus, and Bs = Bacillus subtilis.
Selectivity for Ec over RBCs shown (Sel. = HC50/MIC).
Previously reported MIC values measured for Ec,(4d)Sm,(9)Sa,(4d) and Bs(5e) used derivatives of antimicrobial peptide Magainin.
Previously reported values.(2)
Bactericidal kinetics studies (also known as time-kill assays) were performed on S. aureus to differentiate growth inhibition from cell death and showed that PGON was lethal at 4× the MIC with a 5-log reduction in less than 60 min (Figure 1). Starting with ∼105 cells/mL, aliquots were removed periodically and viable cells counted via plating techniques. The results clearly showed PGONʼs ability to kill bacteria (bactericidal activity) not just inhibit growth (bacteriostatic activity).
Figure 1.
Time kill plots for S. aureus. CFU = colony forming unit; squares = untreated cells; white circles = treated with PGON at 1× MIC; and black circles = at treated with PGON 4× MIC.
Many AMPs and their structural mimics have been shown to be membrane-active with the balance of their hydrophobic/hydrophilic properties critical to their mode of action.(10) Similarly, the biological properties of Poly-1 and Poly-3 were previously explained based on their amphiphilicity, in agreement with the concept that the proper balance of hydrophobic and hydrophilic groups is often essential for the activity of AMPs.(2)Poly-1 was the most hydrophilic and consequently not membrane-active, presumably because it can not sufficiently disrupt the lipid bilayer. In contrast, Poly-3, being significantly more hydrophobic, exhibited good antibacterial activity but unfortunately showed high hemolysis. With its outstanding biological properties, it appeared that PGON had attained a favorable amphiphilicity.
High performance liquid chromatography (HPLC) retention times are routinely exploited in the evaluation of a peptide’s, or peptidomimetic’s relative amphiphilicity.(5b) Retention times for Poly-1, Poly-3, and PGON, were 19.9, 25.5, and 22.4 min, respectively, indicating that the guanidinium groups of PGON resulted in a polymer with intermediate hydrophobic character. Therefore, from the microbiology and HPLC data, it was expected that PGON would exhibit strong membrane-lysis in dye-leakage assays using vesicles composed of phosphatidyl ethanolamine (PE) and phosphatidyl glycerol (PG) phospholipids, which are typical of Gram-negative bacteria.(11)
However, these assays showed that membrane-disruption induced by Poly-1 and Poly-3 did reflect their MICs, while PGON was completely inactive (Figure 2). In other words, Poly-1, which displayed negligible antibacterial activity, caused little dye leakage (16%), while Poly-3, with good antibacterial activity, had significant dye release (87%). On the other hand, vesicles exposed to PGON were seemingly undisturbed (4% leakage). This lack of membrane disruption activity for PGON was unexpected given its high antibacterial efficiency and the fact that its hydrophobicity fell in between the two amine containing polymers.
Figure 2.
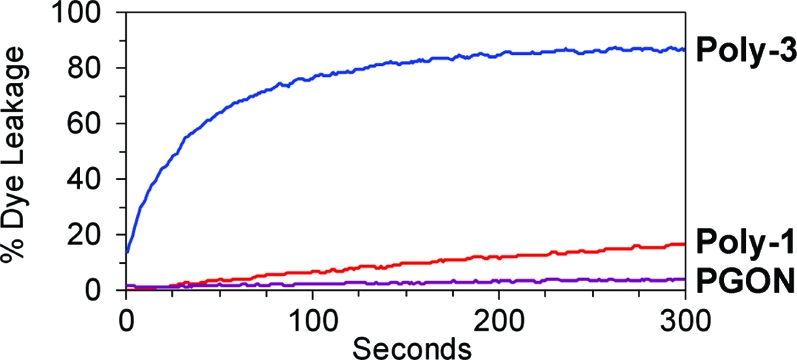
Vesicle dye leakage after polymer addition.
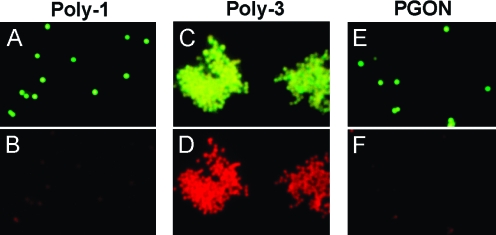
While dye leakage assays are simple models and quite useful in many studies,1b,11 caution should be taken when correlating these results to biological activity. Thus, the effects of these polymers on S. aureus were directly observed using a two-component fluorescence stain (Figure 3). This stain uses a green-emitting SYTO9 dye that aids visualization of all cells and red-emitting propidium iodide, which only enters cells with compromised membranes.
Figure 3.
Fluorescence microscopy of S. aureus (cells ∼ 1 μm in diameter) incubated with a stain to visualize membrane-disruption and polymer for 30 min. Top row shows emission from S. aureus using a green filter setting, while the bottom row shows the identical field using a red filter.
From these microscopy studies, Poly-1 appeared to be inactive (also confirmed by cell viability assays under the same microscopy conditions),(8) and showed green intact individual S. aureus cells (Figure 3A). In contrast, Poly-3 led to gross aggregation and clear membrane damage inferred from the dramatic increase in red fluorescence intensity (Figure 3D), while under the green filter the cells now appeared more yellow compared to the Poly-1 or PGON trials (compare Figure 3C to 3A and 3E). Cell viability assays confirmed Poly-3 is lethal under these conditions.(8) Interestingly, PGON, which is significantly bactericidal, as demonstrated by cell viability assays, did not aggregate bacteria nor allow propidium iodide to enter the cells. These results agreed completely with the dye leakage experiments in which Poly-1 and PGON caused little to no membrane damage relative to Poly-3. This apparent ability to transverse the membrane without disruption is also consistent with CPP-like activity.(14) At the same time, studies with a dye-labeled version of PGON did support interactions between PGON and bacterial cells under the same conditions but due to the small size of bacteria (nominally 1 μm), it was not possible to see if PGON was inside.(8)
Accordingly, these results suggest that PGONʼs ability to effectively kill bacteria likely occurs via a different mechanism than gross membrane trauma. Likely targets would then be anionic macromolecules such as essential membrane proteins or RNA/DNA. Although no data is yet available on these other targets, CPPs and polyarginine, are well-known to efficiently cross membranes and bind DNA.3a,3d,12,13 Recently, we demonstrated the ability of these PGONʼs, with various chain lengths, to traverse membranes as well as other properties reminiscent of CPPs.(14) Therefore, the proposal of intercellular targets is within reason.
Conclusions
There is no doubt that PGON is potently antimicrobial without gross membrane disruption. This leads to a lack of RBC lysis and thus improved selectivity compared to AMPs and previous primary amine containing SMAMPs. The recent demonstration that PGON-like polymers are effective transduction domains implies intercellular targets are responsible for the antimicrobial properties and lack of mammalian toxicity.7,14 Although no data is yet available on these targets, it is clear that these PGONs represent an interesting and potentially extremely important new class of molecules. At the same time, highly selective antimicrobial polymers, such as PGON, can be extremely valuable in materials designed to prevent Biofilm formation. Targets of opportunity include cardiovascular, orthopedic, and other implants that account for 1 million implant-associated infections and 3 billion U.S. dollars of healthcare costs annually.(1d)
Acknowledgments
The National Institutes of Health and the Office of Naval Research are acknowledged for their generous support.
Supporting Information Available
Synthetic schemes, instrumentation, and dye-labeled polymer synthesis. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- a Madkour A. E.; Tew G. N. Polym. Int. 2008, 57, 6–10. [Google Scholar]; b Gabriel G. J.; Som A.; Madkour A. E.; Eren T.; Tew G. N. Mater. Sci. Eng., R 2007, 57, 28–64. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Klibanov A. M. J. Mater. Chem. 2007, 17, 2479–2482. [Google Scholar]; d Hetrick E. M.; Schoenfisch M. H. Chem. Soc. Rev. 2006, 35, 780–789. [DOI] [PubMed] [Google Scholar]
- Ilker M. F.; Nüsslein K.; Tew G. N.; Coughlin E. B. J. Am. Chem. Soc. 2004, 126, 15870–15875. [DOI] [PubMed] [Google Scholar]
- a Miyatake T.; Nishihara M.; Matile S. J. Am. Chem. Soc. 2006, 128, 12420–12421. [DOI] [PubMed] [Google Scholar]; b Henriques S. T.; Melo M. N.; Castanho M. A. R. B. Biochem. J. 2006, 399, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Rothbard J. B.; Kreider E.; VanDeusen C. L.; Wright L.; Wylie B. L.; Wender P. A. J. Med. Chem. 2002, 45, 3612–3618. [DOI] [PubMed] [Google Scholar]; d Futaki S.; Suzuki T.; Ohashi W.; Yagami T.; Tanaka S.; Ueda K.; Sugiura Y. J. Biol. Chem. 2001, 276, 5836–5840. [DOI] [PubMed] [Google Scholar]; e Mitchell D. J.; Kim D. T.; Steinman L.; Fathman C. G.; Rothbard J. B. J. Pept. Res. 2000, 56, 318–325. [DOI] [PubMed] [Google Scholar]
- For polymeric examples, see:; a Sambhy V.; Peterson B. R.; Sen A. Angew. Chem., Int. Ed. 2008, 47, 1250–1254. [DOI] [PubMed] [Google Scholar]; b Mowery B. P.; Lee S. E.; Kissounko D. A.; Epand R. F.; Epand R. M.; Weisblum B.; Stahl S. S.; Gellman S. H. J. Am. Chem. Soc. 2007, 129, 15474–15476. [DOI] [PubMed] [Google Scholar]; c Kuroda K.; DeGrado W. F. J. Am. Chem. Soc. 2005, 127, 4128–4129. [DOI] [PubMed] [Google Scholar]; d Arnt L.; Nüsslein K.; Tew G. N. J. Polym. Sci., Part A: Polym. Chem. 2004, 42, 3860–3864. [Google Scholar]; e Tew G. N.; Liu D.; Chen B.; Doerksen R. J.; Kaplan J.; Carroll P. J.; Klein M. L.; DeGrado W. F. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 5110–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For peptidomimetic examples, see; a Gabriel G. J.; Tew G. N. Org. Biomol. Chem. 2008, 6, 417–423. [DOI] [PubMed] [Google Scholar]; b Schmitt M. A.; Weisblum B.; Gellman S. H. J. Am. Chem. Soc. 2007, 129, 417–428. [DOI] [PubMed] [Google Scholar]; c Tew G. N.; Clements D.; Tang H.; Arnt L.; Scott R. W. Biochim. Biophys. Acta 2006, 1758, 1387–1392. [DOI] [PubMed] [Google Scholar]; d Patch J. A.; Barron A. E. J. Am. Chem. Soc. 2003, 125, 12092–12093. [DOI] [PubMed] [Google Scholar]; e Porter E. A.; Wang X.; Lee H.-S.; Weisblum B.; Gellman S. H. Nature 2000, 404, 565–565. [DOI] [PubMed] [Google Scholar]
- Radzishevsky I. S.; Rotem S.; Bourdetsky D.; Navon-Venezia S.; Carmeli Y.; Mor A. Nat. Biotechnol. 2007, 25, 657–659. [DOI] [PubMed] [Google Scholar]
- Kolonko E. M.; Kiessling L. L. J. Am. Chem. Soc. 2008, 130, 5626–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See Supporting Information for details.
- Ge Y.; MacDonald D. L.; Holroyd K. J.; Thornsberry C.; Wexler H.; Zasloff M. Antimicrob. Agents Chemother. 1999, 43, 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden K. A. Nat. Rev. Microbiol. 2005, 3, 238–250. [DOI] [PubMed] [Google Scholar]
- Som A.; Tew G. N. J. Phys. Chem. B 2008, 112, 3495–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai N.; Futaki S.; Matile S. Soft Matter 2006, 2, 636–641. [DOI] [PubMed] [Google Scholar]
- a Pantos A.; Tsogas I.; Paleos C. M. Biochim. Biophys. Acta 2008, 1778, 811–823. [DOI] [PubMed] [Google Scholar]; b Schroeder T.; Niemeier N.; Afonin S.; Ulrich A. S.; Krug H. F.; Braese S. J. Med. Chem. 2008, 51, 376–379. [DOI] [PubMed] [Google Scholar]; c Deglane G.; Abes S.; Michel T.; Prevot P.; Vives E.; Debart F.; Barvik I.; Lebleu B.; Vasseur J.-J. ChemBioChem 2006, 7, 684–692. [DOI] [PubMed] [Google Scholar]; d Fillon Y. A.; erson J. P.; Chmielewski J. J. Am. Chem. Soc. 2005, 127, 11798–11803. [DOI] [PubMed] [Google Scholar]; e Funhoff A. M.; Van Nostrum C. F.; Lok M. C.; Fretz M. M.; Crommelin D. J. A.; Hennink W. E. Bioconjugate Chem. 2004, 15, 1212–1220. [DOI] [PubMed] [Google Scholar]
- Hennig A.; Gabriel G. J.; Tew G. N.; Matile S.. J. Am. Chem. Soc. 2008, 130, 10338−10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.