Abstract
This study addresses the properties of a newly identified internal ribosome entry site (IRES) contained within the mRNA of the homeodomain protein Gtx. Sequential deletions of the 5′ untranslated region (UTR) from either end did not define distinct IRES boundaries; when five nonoverlapping UTR fragments were tested, four had IRES activity. These observations are consistent with other cellular IRES analyses suggesting that some cellular IRESes are composed of segments (IRES modules) that independently and combinatorially contribute to overall IRES activity. We characterize a 9-nt IRES module from the Gtx 5′ UTR that is 100% complementary to the 18S rRNA at nucleotides 1132–1124. In previous work, we demonstrated that this mRNA segment could be crosslinked to its complement within intact 40S subunits. Here we show that increasing the number of copies of this IRES module in the intercistronic region of a dicistronic mRNA strongly enhances IRES activity in various cell lines. Ten linked copies increased IRES activity up to 570-fold in Neuro 2a cells. This level of IRES activity is up to 63-fold greater than that obtained by using the well characterized encephalomyocarditis virus IRES when tested in the same assay system. When the number of nucleotides between two of the 9-nt Gtx IRES modules was increased, the synergy between them decreased. In light of these findings, we discuss possible mechanisms of ribosome recruitment by cellular mRNAs, address the proposed role of higher order RNA structures on cellular IRES activity, and suggest parallels between IRES modules and transcriptional enhancer elements.
In eukaryotes, the initiation of mRNA translation is generally thought to occur by a cap-binding/-scanning mechanism, although some mRNAs are translated efficiently despite the lack of a cap structure or a free 5′ end (for reviews, see refs. 1–5). In some of these cases, sequences contained within the mRNA directly recruit the translation machinery. These internal ribosome entry sites (IRESes) are defined functionally by using dicistronic mRNAs. A nucleotide sequence functions as an IRES if, when present in the intercistronic region of a dicistronic mRNA, it can direct translation of the second cistron in a manner that is independent of the first cistron. IRESes have been identified in the 5′ UTRs of both cellular and viral mRNAs, but they have been most extensively characterized in picornaviral mRNAs as well defined segments of approximately 450 nt (e.g., see ref. 6) that have been categorized on the basis of sequence and structural similarities (reviewed in ref. 7). In contrast, little is known about the IRESes of cellular mRNAs. For most cellular IRESes, distinct boundaries have been difficult to determine by deletion analysis (8–12), and fragments as short as 55 nt have been shown to have IRES activity (13).
Inasmuch as cellular IRESes contain no obvious sequence similarity to each other or to picornaviral IRESes, it has been suggested that they resemble each other in secondary structure (14). RNA-folding analyses have indicated that some cellular IRESes may contain a Y-type stem–loop structure followed by a stem loop immediately upstream of the initiation codon. This Y-shaped conformation was predicted to occur within IRESes from the mRNAs that encode the Ig heavy chain-binding protein (Bip), fibroblast growth factor-2, and the Antennapaedia gene product, and this conformation is reportedly similar to one contained within the IRESes of picornavirus, pestivirus, and hepatitis C virus (15). RNA conformations, including Y-shaped secondary structures, have subsequently been predicted to occur within several other cellular IRESes (e.g., refs. 9, 10, 16, and 17). In the case of the c-sis mRNA, three independent fragments have IRES activity, and each of these fragments is proposed to contain a Y-type stem–loop structure (16).
There is no independent physical evidence demonstrating that these Y-shaped RNA conformations actually occur, however, and for many cellular IRESes, the ability to internally initiate translation is not correlated with the presence of the proposed secondary structure. For example, 5′ or 3′ deletions of the Bip (8) IRES still have IRES activity even though these fragments do not contain the predicted Y-type stem–loop structure. The lack of correlation between proposed RNA conformations and IRES activity has also been observed in other studies. For example, the vascular endothelial growth factor and c-myc IRESes both contain two nonoverlapping fragments with IRES activity (9, 10). In addition, the Bip IRES has three nonoverlapping fragments with IRES activity (8), none of which contain the proposed Y-type stem–loop structure or at best contain only part of it.
The published results seem more consistent with the notion that some cellular IRESes may be composed of numerous short segments that either have independent IRES activity or that together affect the overall IRES activity. In the present study, we provide evidence to support this hypothesis. We show that the 196-nt 5′ UTR of the Gtx homeodomain mRNA contains an IRES, that multiple nonoverlapping fragments of this 5′ UTR have IRES activity, and that one of these fragments contains a 9-nt segment that can function independently as an IRES. Multiple copies of this 9-nt sequence increase IRES activity synergistically. This 9-nt segment is 100% complementary to the 18S rRNA at nucleotides 1132–1124. In an earlier study, we used photochemical crosslinking to show that this same mRNA sequence could bind to 40S ribosomal subunits by crosslinking to its complement within the 18S rRNA (18). We discuss the possibility that cellular IRESes are modular assemblies consisting of subsequences that can function independently and also cooperatively. To facilitate discussion, we refer to these minimal functional units as IRES modules.
Materials and Methods
Dicistronic mRNA Analyses.
Dicistronic constructs used for this study are based on the pGL3-R2 (RP) and pGL3-R2h (RPh) dicistronic reporter vectors [kindly provided by Anne E. Willis, University of Leicester, Leicester, United Kingdom (9)]. These vectors encode a dicistronic mRNA with the Renilla (sea pansy) luciferase gene upstream of the Photinus (firefly) luciferase gene. An SV40 promoter and enhancer drive expression. The RPh vector contains a 60-bp inverted repeat located 5′ of the first cistron. The 5′ UTR of the Gtx mRNA (19) was synthesized as two overlapping oligonucleotides, amplified by PCR with pfu DNA polymerase (Stratagene), and inserted into the intercistronic region as an EcoRI to NcoI fragment immediately upstream of the Photinus luciferase initiation codon in both the RP and RPh vectors. Other inserts were similarly cloned into the intercistronic region by using the EcoRI and NcoI restriction sites or by using the SpeI and EcoRI restriction sites as indicated in supplemental Table 1 (see the PNAS web site, www.pnas.org). The sequence of all constructs was verified. Plasmids containing the chloramphenicol acetyl transferase (CAT) gene instead of the Photinus luciferase gene were derived by deleting the luciferase gene and replacing it with the CAT gene by using the NcoI and BamHI restriction sites (RCh). The CAT gene was from the pCAT3 control vector (Promega).
Cell lines were obtained from American Type Culture Collection and maintained according to their instructions. Dicistronic reporter constructs were transfected into cells by using fugene 6 (Roche Molecular Chemicals). Transfection efficiencies were normalized by cotransfection with the pCMVβ vector (CLONTECH). Cells were harvested after 24 h, and luciferase activity was determined by using the dual reporter assay system (Promega). β-Galactosidase activity was assayed by using the Fluoreporter lacZ kit (Molecular Probes), and fluorescence was measured by using the Millipore Cytofluor 2450 system. CAT enzyme activity was measured by using N-butyryl CoA according to technical bulletin no. 84 (Promega).
RNA Analysis.
Northern blot analysis was performed as previously described (20) by using RNA prepared from the neuronal cell line Neuro 2a (N2a), extracted by the guanidium thiocyanate method (21). poly(A)+ RNA was selected by using Oligotex oligo(dT) beads (Qiagen, Chatsworth, CA). Northern blot hybridizations were performed by using Rapid hyb buffer (Amersham) with a probe containing the entire Photinus luciferase coding sequence. This probe was obtained by digesting the dicistronic construct with the NcoI and SalI restriction sites. Hybridizations were performed at 65°C; wash stringency was 65°C in 0.1× saline sodium citrate.
Results
The 5′ UTR of Gtx mRNA Contains an IRES.
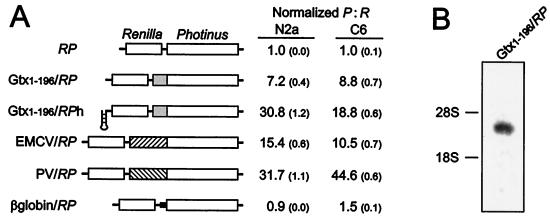
The 5′ UTR of the Gtx mRNA is 196 nt long (19) and contains several complementary sequence matches to 18S rRNA, one of which was the focus of a previous study (18). We postulated that mRNA sequences such as these might directly interact with complementary sequences within the 18S rRNA and lead to translation initiation events. To test this notion, the Gtx 5′ UTR was tested in the intercistronic region of a dicistronic mRNA (Gtx1–196/RP, Fig. 1A) that contains the Renilla luciferase coding sequence as the first cistron and the Photinus luciferase coding sequence as the second cistron (9). The insertion of the 196-nt Gtx 5′ UTR in the intercistronic region of this dicistronic construct enhanced expression of the downstream cistron 7.2-fold over background in the rat neuronal cell line N2a and 8.8-fold in the rat glioma cell line C6. This IRES activity was approximately 47% of that of the encephalomyocarditis virus (EMCV) IRES in N2a cells and 84% of that of EMCV in C6 cells, when tested in the same vector system (EMCV/RP). Translation of the β-globin mRNA is very cap dependent (e.g., refs. 22 and 23), and insertion of the 5′ UTR of this mRNA between cistrons served as a control with no IRES activity (β-globin/RP). To show that the enhanced expression of the second cistron did not depend on the translation of the first cistron, the translation of the first cistron was inhibited. This was accomplished by using an equivalent construct that contained an inverted repeat, with the potential to form a stable hairpin structure, in the 5′ UTR (Gtx1–196/RPh). When normalized to the activity of the RP vector, the hairpin structure increased IRES activity up to 31-fold over background. A Northern blot analysis using poly(A)+ RNA isolated from N2a cells transfected with the Gtx1–196/RP vector and probed with a Photinus luciferase fragment showed that the cells expressed only one detectable mRNA of the correct expected size (Fig. 1B). This argues against the possibility that enhanced expression of the downstream Photinus luciferase gene resulted from functionally monocistronic mRNAs that were generated by unusual splicing events or RNA fragmentation.
Figure 1.
Dicistronic analysis of Gtx IRES activity. (A) A schematic representation of the dicistronic constructs used in this analysis is indicated. Constructs are based on the RP (Renilla-Photinus) and RPh (Renilla-Photinus hairpin) vectors. Inserts include the full Gtx 5′ UTR (Gtx1–196), the EMCV and poliovirus IRESes, and the β-globin 5′ UTR. IRES activities are represented as ratios of Photinus luciferase activity to Renilla luciferase activity after transfection into N2a and C6 cells. Values represent activities that have been normalized to those of the RP vector. Numbers in parentheses represent SEM. (B) Northern blot of poly(A)+ RNA purified from N2a cells transfected with the Gtx1–196/RP dicistronic construct and probed with a fragment of the Photinus luciferase gene. The positions of the 28S and 18S rRNAs are indicated.
Gtx 5′ UTR Contains Shorter Segments with IRES Activity.
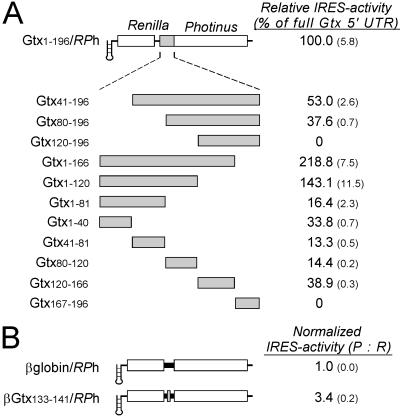
The Gtx 5′ UTR was sequentially deleted from both the 5′ and the 3′ end and was fragmented into five nonoverlapping segments of approximately 40 nt, each of which was tested for IRES activity (Fig. 2A). To minimize the contribution of the first cistron, the RPh vector was used for these constructs. The results of these studies in the N2a cell line indicate that all deletions and fragments have IRES activity, with the exception of constructs Gtx120–196/RPh and Gtx167–196/RPh. Deletion of nucleotides 167–196 increased IRES activity to a level greater than that of the full 5′ UTR (see constructs Gtx1–166/RPh and Gtx1–120/RPh), which may indicate that this sequence inhibits IRES activity or that the IRES activity of nucleotides 1–166 is affected by its location relative to the initiation codon. One of the fragments, contained within construct Gtx120–166/RPh, is of particular interest, because we have shown previously that this fragment contains a segment at nucleotides 133–141 of the Gtx 5′ UTR that is 100% complementary to the 18S rRNA at nucleotides 1132–1124. This Gtx sequence was the focus of an earlier study in which we showed that it could be crosslinked to its complement within intact 40S subunits (18). To determine whether IRES activity is associated with the complementary nucleotides, this sequence was directly tested for IRES activity. The spacing of these nine nucleotides relative to the initiation codon was kept the same as in the Gtx120–166/RPh construct by using a polynucleotide sequence identical to the whole β-globin 5′ UTR (which itself has no detectable IRES activity) as a filler sequence (Fig. 2B). This precaution was taken to avoid possible effects on IRES activity that might result by changing the location of the 9-nt segment relative to the initiation codon. The IRES activity associated with this 9-nt fragment was 3.4-fold over the background.
Figure 2.
Deletion and fragment analysis of the Gtx 5′ UTR after transfection into N2a cells. (A) A schematic representation of the dicistronic constructs used in this analysis is indicated. Constructs are based on the RPh vector. Inserts include the full Gtx 5′ UTR (Gtx1–196) and deletions and fragments of this sequence. The Gtx construct designations indicate the exact nucleotide sequence present in each construct. IRES activities are represented as a percentage of the activity of the full Gtx 5′ UTR. Numbers in parentheses represent SEM. (B) Analysis of a 9-nt segment of the Gtx 5′ UTR. Construct β-globin/RPh contains the full 5′ UTR of the mouse β-globin mRNA. The activity of construct βGtx133–141/RPh is normalized to that of the β-globin control construct βglobin/RPh.
Linked Copies of the 9-nt IRES Module Appear to Function Synergistically.
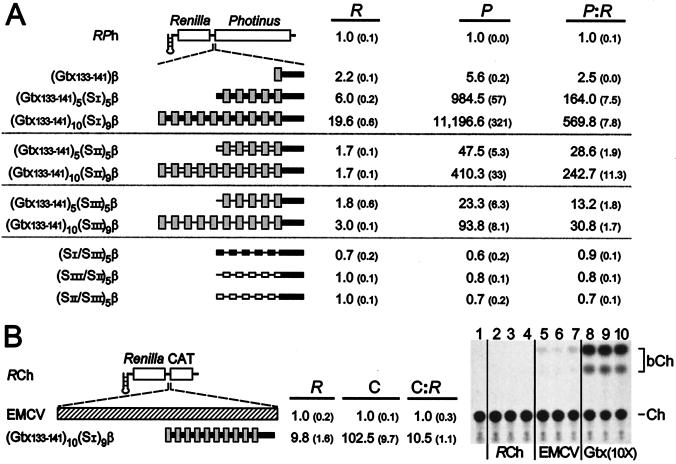
Dicistronic constructs were generated containing 1, 5, or 10 copies of the 9-nt IRES module, spaced 25 nt upstream of the initiation codon by using β-globin 5′ UTR sequences. Constructs with 5 or 10 copies of the IRES module were arbitrarily spaced 9 nt apart by using a repeated segment of the β-globin 5′ UTR (designated SI). The results of transfection experiments in N2a cells indicated that the IRES activity of one copy [(Gtx133–141)β/RPh] was 2.5-fold over background. The activity of five linked copies [(Gtx133–141)5(SI)5β/RPh] was 164-fold over background, and the activity of 10 copies [(Gtx133–141)10(SI)9β/RPh] was 570-fold over background (Fig. 3A). Note that these IRES activities are normalized to that of the parent RPh vector. If normalized to the RP vector, the activity of (Gtx133–141)10(SI)9β/RPh is approximately 6,000-fold over background. The P:R ratio reflects the absolute increase in Photinus luciferase activity for all constructs except those that contain 5 or 10 copies of (Gtx133–141) with spacer SI, in which the expression of both cistrons increased. The level of Renilla expression obtained with construct (Gtx133–141)10(SI)9β/RPh is approximately 20-fold higher than that of construct RPh. A Northern blot analysis of mRNAs from N2a cells indicated that the mRNA levels were not significantly different. It is therefore unlikely that increased mRNA levels caused increased expression of the Renilla gene. A similar enhancement of both cistrons was observed when the Tobacco necrosis virus enhancer was placed in the 3′ UTR of a dicistronic mRNA (24).
Figure 3.
Dicistronic mRNAs containing synthetic IRESes based on multiple linked copies of nucleotides 133–141 of the Gtx 5′ UTR. (A) Schematic representation of the dicistronic constructs used in this analysis is indicated. Constructs are based on the RPh vector and contain 25 nt of the mouse β-globin 5′ UTR sequence (β) immediately upstream of the initiation codon, represented as a thick black line. (Inset) One, five, or ten copies of the Gtx IRES module (Gtx133–141) are indicated as gray bars; constructs with multiple copies are spaced 9 nt apart by one of three spacer sequences, SI, SII, or SIII, shown as a thick black bar, an open bar, and a thin line, respectively. Constructs were transfected into N2a cells, and luciferase activities were normalized for transfection efficiency (see Materials and Methods). Numbers in parentheses represent SEM. (B) Analysis of dicistronic constructs containing the CAT gene as the second cistron. The constructs used in this assay are based on the RCh vector. Inserts include the EMCV IRES and (Gtx133–141)10(SI)9β. Constructs were transfected into N2a cells and assayed for Renilla (R) and CAT (C) activities. CAT activities in the table were determined by liquid scintillation assay and, because CAT activity of the RCh vector was indistinguishable from a negative control, numbers were normalized to the activity of EMCV/RCh. An autoradiogram of a TLC assay of CAT activity is also shown. Lane 1, negative control; lanes 2–4, transfections with RCh; lanes 5–7, transfections with EMCV/RCh (EMCV); lanes 8–10, transfections with [Gtx133–141)10(SI)9β/RCh (Gtx(10X)]. Each lane represents an independent transfection. Ch represents 14C chloramphenicol; bCh represents butyrylated 14C chloramphenicol and is the product of CAT enzyme activity.
Two other spacer sequences and a number of controls were then tested to distinguish between the effects of the 9-nt IRES module and the spacer sequence. Other spacer sequences include a different 9-nt segment of the β-globin 5′ UTR (designated SII) and a 9-nt stretch of poly(A) (designated SIII). IRES activity increased up to 243-fold over background for constructs with SII and up to 31-fold for constructs with SIII. In all cases, increasing the number of copies of the IRES module resulted in increased IRES activity, but the level of activity was affected differentially by the different spacer sequences. Different spacer sequences may affect the higher order structure of the mRNA and alter the presentation of the IRES modules to the translation machinery. Alternatively, some spacer sequences may introduce nucleotides that contribute to the IRES module or that may themselves synergize with the IRES modules. Controls indicate that three different combinations of the spacer sequences themselves, (SI/SIII)5, (SIII/SII)5, and (SII/SIII)5, alternating in position as they do in the experimental constructs, did not themselves have any detectable IRES activity.
To determine whether the IRES activity obtained with the (Gtx133–141)10(SI)9β synthetic IRES is independent of the reporter gene used, the Photinus luciferase gene was replaced with the CAT gene in construct (Gtx133–141)10(SI)9β/RCh. After transfection into N2a cells, thin-layer chromatography (Fig. 3B) and liquid scintillation counting were performed to determine CAT activity. The results show that CAT activity in cell lysates transfected with this synthetic IRES is approximately 103-fold greater than that of the EMCV IRES in a construct containing the same reporter genes (EMCV/RCh), whereas the activity of a control construct (RCh) was indistinguishable from background. As in the RPh vector, the (Gtx133–141)10(SI)9β synthetic IRES also increased Renilla expression significantly (6.1-fold) compared with the control RCh construct. The overall ratio of CAT expression to Renilla luciferase activity for the synthetic IRES [(Gtx133–141)10(SI)9β] was 10.5-fold greater than that of the EMCV IRES. This is a high level of IRES activity, but it is lower (approximately 6-fold) than that obtained in constructs containing the Photinus luciferase gene as the second cistron. This may highlight the influence of the reporter cistron on IRES activity, a feature observed with the EMCV IRES (25).
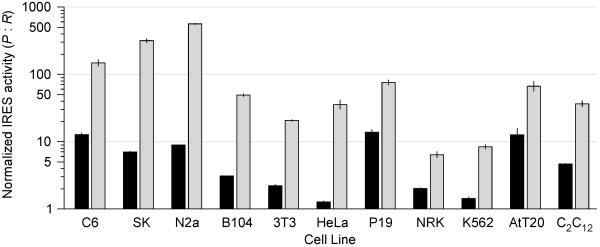
The activity of some viral IRESes depends on cell type (26). Accordingly, to evaluate the activity of the synthetic IRES (Gtx133–141)10(SI)9β, it was tested in a total of 11 cell lines, representative of several different cell types and compared with that of the EMCV IRES. Results indicated that in all cell lines tested, the synthetic IRES was 3.3- (NRK cells) to 63-fold (N2a cells) more active than that of the EMCV IRES, which was very weak in some cell lines (Fig. 4). Note that the data are shown on a logarithmic scale.
Figure 4.
IRES activities in various cell lines. EMCV/RPh and (Gtx133–141)10(SI)9β/RPh were transfected into the cell lines indicated and IRES activity calculated as the ratio of Photinus to Renilla luciferase activities. These activities were normalized to those obtained with the RPh vector in these same cell lines. Activity of the EMCV/RPh construct is indicated as a black bar, and that of the (Gtx133–141)10(SI)9β/RPh construct is indicated as a gray bar. Vertical lines indicate SEM. Note that IRES activities are represented on a logarithmic scale. The following cell lines were used in this study: rat glial tumor line C6; human neuroblastoma SK-N-SH (SK); mouse neuroblastoma N2a; mouse fibroblast B104–1-1 (B104); mouse fibroblast NIH 3T3 (3T3); human epitheloid carcinoma HeLa; mouse embryonal carcinoma P19; rat normal kidney NRK; human chronic myelogenous leukemia K562; mouse pituitary tumor AtT-20 (AtT20); mouse muscle myoblast C2C12.
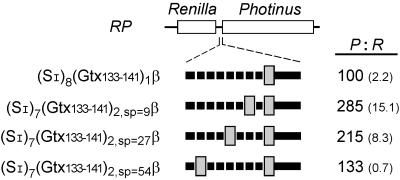
To investigate the potential effects of distance between IRES modules, a series of constructs was generated that contained one or two copies of the 9-nt IRES module spaced various distances apart by using multiple copies of the SI spacer sequence in such a fashion that all constructs were identical in size (Fig. 5). Of those constructs with two copies of the 9-nt IRES module, spacing between the modules was varied from 9 nt to 54 nt (Fig. 5). The IRES activity obtained with one copy of the IRES module increased 2.8-fold when a second copy was introduced 9 nt upstream of the first. This activity decreased to 2.1-fold and to 1.3-fold as the distance between the IRES modules was increased to 27 and 54 nt, respectively. These results indicate that two linked 9-nt IRES modules can interact cooperatively when spaced less than 27 nt apart.
Figure 5.
Effect on IRES activity of varying the spacing between two copies of the 9-nt Gtx IRES module (Gtx133–141). Constructs containing one or two copies of the 9-nt IRES module are indicated schematically. Constructs contain 25 nt of the mouse β-globin 5′ UTR sequence (β) immediately upstream of the initiation codon. Spacing between and upstream of the 9-nt IRES modules was achieved by using a 9-nt spacer sequence (SI). Spacing between two of the 9-nt IRES modules was 9, 27, or 54 nt. IRES activity is the ratio of Photinus to Renilla luciferase activity and is normalized to 100 for the activity of construct (SI)8(Gtx133–141)1β/RP. Numbers in parentheses represent SEM.
Discussion
Some viruses and cellular mRNAs use IRESes to initiate translation in a cap-independent manner, but accumulated evidence indicates that viral and cellular IRESes may be fundamentally distinct. In the present study, we dissected a cellular IRES contained within the 5′ UTR of the Gtx mRNA and identified a 9-nt sequence that can function independently as an IRES module. This result and others in the literature (e.g., refs. 8–10) are not consistent with the notion that Y-shaped RNA conformations are necessarily required for IRES activity in cellular mRNAs (14) and point to the potential importance of short nucleotide motifs. Although RNA conformation may be critical for the activity of some cellular IRESes, as it is in picornavirus, our results raise the possibility that, in some instances, RNA secondary structures may simply alter the accessibility or presentation of individual IRES modules to the translation machinery. The observation that multiple copies of nucleotides 133–141 of the Gtx 5′ UTR increase IRES activity cooperatively suggests that the activity of some cellular IRESes may depend on the strength, number, and spacing of various individual IRES modules. In addition to the Gtx IRES module, we have identified IRES modules within the 5′ UTR of the mRNA that encodes Rbm3, a stress-induced RNA-binding protein (unpublished data). It is expected that further analysis of other cellular IRESes will reveal that many of them have a similar modular composition.
In earlier studies, we showed that a segment of the Gtx 5′ UTR containing nucleotides 133–144 could directly bind to 40S ribosomal subunits by base pairing to the 18S rRNA (18). When tested in the 5′ UTR of a monocistronic reporter construct, this sequence inhibited translation. Mutations of this nucleotide sequence that increased the degree of complementarity to the rRNA decreased the translation of a monocistronic mRNA, whereas those that decreased complementarity increased translation (27). In the present study, nucleotides 133–144 of the Gtx mRNA sequence are shown to function as an IRES module when tested in the intercistronic region of a dicistronic mRNA. This same sequence thus appears to have opposite effects on translation depending on whether it is present in a monocistronic or in a dicistronic mRNA, which may be explained by differences in what these assays measure. Very active IRES modules are likely to be those that recruit the 40S subunit efficiently but still allow it to detach for protein synthesis to occur. Less active IRES modules might include those that recruit poorly because of weak initial interactions with the 40S subunit as well as those that form very stable interactions that effectively sequester the mRNA.
If base pairing between segments of mRNAs and 18S rRNA can lead to ribosome recruitment and translation initiation, this may be related to an earlier observation in which we noted that large numbers of eukaryotic mRNAs contain segments complementary to 18S rRNA (20). If particular subsets of these mRNA sequences function as IRES modules, the implication is that large numbers of different mRNAs may use these sequences to recruit ribosomes. Sequences complementary to 18S rRNA have also been noted within other cellular IRESes (e.g., refs. 11, 12), but these sequences have not yet been investigated experimentally. In the case of picornaviral IRESes, they contain an oligopyrimidine stretch that is complementary to the 3′ end of the 18S rRNA (reviewed in ref. 28). Although this sequence has been shown to be important for activity (e.g., refs. 29 and 30), the ability of this sequence to base pair to the 18S rRNA has not been shown. Similar sequences do not occur in the IRESes of hepatitis C, classic swine fever, and bovine diarrhea pestivirus, although indirect evidence for base pairing interactions between these IRESes and 18S rRNA comes from reconstitution studies (31, 32).
Complementarity to 18S rRNA is unlikely to be the only mechanism by which IRES modules can recruit ribosomes. We expect, for example, that some IRES modules may bind to ribosomes through interactions with ribosomal proteins or components of the translation machinery, such as initiation factors or other intermediary proteins. An example of this comes from a recent study (33) in which an RNA translational control sequence, the iron regulatory element (IRE), was able to function as an IRES module in the presence of an engineered intermediary protein. The IRE binds to the iron-regulatory element-binding protein (IRP-1). By fusing IRP-1 to the C-terminal region of initiation factor eIF4G, this fusion protein becomes an intermediary protein that can recruit the preinitiation complex, presumably though an interaction with initiation factor eIF3. This artificial recruitment of the translation machinery is sufficient to obtain weak but detectable IRES activity when the IRE is inserted in the intercistronic region of a dicistronic mRNA.
The ability to internally initiate translation may be used by some mRNAs under conditions that are not favorable for cap-dependent translation, such as occur during mitosis or poliovirus infection (e.g., ref. 34). However, with the exception of certain viral mRNAs, virtually all eukaryotic mRNAs are monocistronic and contain a cap structure (1, 35). Thus the ability to internally initiate translation might just reflect the ability of a sequence to recruit ribosomes. If this recruitment is sufficient to enhance translation, these sequences may give some mRNAs an advantage over others under competitive situations within the cell. Alternatively, IRES modules in eukaryotic mRNAs may be mechanistically analogous to the prokaryotic Shine and Dalgarno sequence (36), albeit combinatorially more complex because they are multiple and the complementary sequences are not fixed in location either within the mRNA or within the rRNA as they are in their prokaryotic counterparts.
The finding that two, five, and ten copies of the 9-nt Gtx(133–141) IRES module have a synergistic effect on IRES activity may have important implications for understanding the evolution of translational enhancers and regulators and may have parallels with cis-acting transcriptional enhancers for which similar effects have been noted: two or more copies of the same or of different cis-acting transcriptional enhancers have more than an additive effect on transcription (37). One explanation is that cooperative interactions between these weak transcription factors may increase the local concentration of these factors. Synergy appears to result from increased contacts between these protein factors and the core transcription machinery (reviewed by refs. 38, 39). Similarly, weak interactions between components of the translational machinery might be sufficient to explain the synergistic effects seen in the present work. Alternatively, increased recruitment may lead to higher local concentrations of some of the initiation factors that might otherwise be limiting for translation. For example, high levels of eIF4A, which is an RNA helicase, might maintain a more open mRNA secondary structure and allow more efficient translation.
In addition to addressing the basic mechanisms underlying translation initiation, synthetic IRESes of the type generated in this study may be of practical use. The most active synthetic IRESes tested in this study are up to 63-fold more active than the EMCV IRES and appear to be much stronger than most or all of the naturally occurring IRESes that have been characterized up to this time (26, 40). Consequently, extremely sensitive reporter assays are now possible, and these IRESes may be useful to obtain very high levels of protein expression. In addition, we have shown that it is possible to fine tune the level of IRES activity by varying the number of linked IRES modules. The activities and properties of these synthetic IRESes may have a number of applications for research, biotechnology, diagnosis, and gene therapy.
Supplementary Material
Acknowledgments
We thank Erin Powrie for excellent technical assistance and Dr. Joseph Gally for critical reading of the manuscript. This work was supported by a grant from the G. Harold and Leila Y. Mathers Charitable Foundation to G.M.E. G.M.E. is a consultant to Becton Dickinson and Company.
Abbreviations
- IRES
internal ribosome entry site
- EMCV
encephalomyocarditis virus
- UTR
untranslated region
- N2a
neuronal cell line Neuro 2a
References
- 1.Kozak M. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 2.Gray N K, Wickens M. Annu Rev Cell Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 3.Jackson R J. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 71–112. [Google Scholar]
- 4.van der Velden A W, Thomas A A M. Int J Biochem Cell Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- 5.Gingras A C, Raught B, Sonenberg N. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson R, Pelletier J, Le S-Y, Sonenberg N. J Virol. 1991;65:5886–5894. doi: 10.1128/jvi.65.11.5886-5894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson R J, Kaminski A. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Q, Sarnow P. Nucleic Acids Res. 1997;25:2800–2807. doi: 10.1093/nar/25.14.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoneley M, Paulin F E M, Le Quesne J P C, Chappell S A, Willis A E. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 10.Huez I, Creancier L, Audigier S, Gensac M-C, Prats A-C, Prats H. Mol Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan W, La Celle M, Rhoads R E. J Biol Chem. 1998;273:5006–5012. doi: 10.1074/jbc.273.9.5006. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein J, Sella O, Le S-Y, Elroy-Stein O. J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 13.Oh S-K, Scott M P, Sarnow P. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 14.Le S-Y, Maizel J V., Jr Nucleic Acids Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le S Y, Siddiqui A, Maizel J V., Jr Virus Genes. 1996;12:135–147. doi: 10.1007/BF00572952. [DOI] [PubMed] [Google Scholar]
- 16.Sella O, Gerlitz G, Le S-Y, Elroy-Stein O. Mol Cell Biol. 1999;19:5429–5440. doi: 10.1128/mcb.19.8.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nanbru C, Lafon I, Audigier S, Gensac M-C, Vagner S, Huez G, Prats A-C. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 18.Hu M C-Y, Tranque P, Edelman G M, Mauro V P. Proc Natl Acad Sci USA. 1999;96:1339–1344. doi: 10.1073/pnas.96.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komuro I, Schalling M, Jahn L, Bodmer R, Jenkins N A, Copeland N G, Izumo S. EMBO J. 1993;12:1387–1401. doi: 10.1002/j.1460-2075.1993.tb05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mauro V P, Edelman G M. Proc Natl Acad Sci USA. 1997;94:422–427. doi: 10.1073/pnas.94.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 22.Lockard R E, Lane C L. Nucleic Acids Res. 1978;5:3237–3347. doi: 10.1093/nar/5.9.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keiper B D, Rhoads R E. Nucleic Acids Res. 1997;25:395–402. doi: 10.1093/nar/25.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meulewaeter F, Van Montagu M, Cornelissen M. RNA. 1998;4:1347–1356. doi: 10.1017/s135583829898092x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaminski A, Jackson R J. RNA. 1998;4:626–638. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borman A M, Le Mercier P, Girard M, Kean K M. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tranque P, Hu Michael C-Y, Edelman G M, Mauro V P. Proc Natl Acad Sci USA. 1998;95:12238–12243. doi: 10.1073/pnas.95.21.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheper G C, Voorma H O, Thomas A A M. FEBS Lett. 1994;352:271–275. doi: 10.1016/0014-5793(94)00975-9. [DOI] [PubMed] [Google Scholar]
- 29.Meerovitch K, Nocholson R, Sonenberg N. J Virol. 1991;65:5895–5901. doi: 10.1128/jvi.65.11.5895-5901.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pestova T V, Hellen C U T, Wimmer E. J Virol. 1991;65:6194–6204. doi: 10.1128/jvi.65.11.6194-6204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U T. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pestova T V, Hellen C U T. Virology. 1999;258:249–256. doi: 10.1006/viro.1999.9741. [DOI] [PubMed] [Google Scholar]
- 33.De Gregorio E, Preiss T, Hentze M W. EMBO J. 1999;18:4865–4874. doi: 10.1093/emboj/18.17.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johannes G, Carter M S, Eisen M B, Brown P O, Sarnow P. Proc Natl Acad Sci USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shatkin A J. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 36.Shine J, Dalgarno L. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Busby S, West D, Lawes M, Webster C, Ishihama A, Kolb A. J Mol Biol. 1994;241:341–352. doi: 10.1006/jmbi.1994.1511. [DOI] [PubMed] [Google Scholar]
- 38.Ptashne M, Gann A. Curr Biol. 1998;8:R812–R822. doi: 10.1016/s0960-9822(07)00508-8. [DOI] [PubMed] [Google Scholar]
- 39.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Salas E. Curr Opin Biotechnol. 1999;10:458–464. doi: 10.1016/s0958-1669(99)00010-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.