Abstract
Genome-wide DNA rearrangements occur in many eukaryotes but are most exaggerated in ciliates, making them ideal model systems for epigenetic phenomena. During development of the somatic macronucleus, Oxytricha trifallax destroys 95% of its germ line, severely fragmenting its chromosomes, and then unscrambles hundreds of thousands of remaining fragments by permutation or inversion. Here we demonstrate that DNA or RNA templates can orchestrate these genome rearrangements in Oxytricha, supporting an epigenetic model for sequence-dependent comparison between germline and somatic genomes. A complete RNA cache of the maternal somatic genome may be available at a specific stage during development to provide a template for correct and precise DNA rearrangement. We show the existence of maternal RNA templates that could guide DNA assembly, and that disruption of specific RNA molecules disables rearrangement of the corresponding gene. Injection of artificial templates reprogrammes the DNA rearrangement pathway, suggesting that RNA molecules guide genome rearrangement.
Parental RNA transcripts and microRNAs are critical for programming development in metazoa1–4, raising the possibility that altered RNA molecules can reprogramme patterning on a developmental or evolutionary timescale5. Despite the suggestion of template-directed events involving “an ancestral RNA-sequence cache”6 there has been limited evidence for a direct role of RNA as a template of information across generations7,8. Information transfer from RNA to DNA usually involves polymerization9. Here we show that RNA molecules can also organize DNA rearrangements, expanding the epigenetic influence of RNA beyond gene expression and priming or directing DNA and RNA synthesis, editing, modification or repair9–11.
O. trifallax is a unicellular eukaryote harbouring two kinds of nuclei: germline micronuclei and somatic macronuclei. Diploid micronuclei are transcriptionally inert during vegetative growth but they transmit the germline genome through subsequent generations. Effectively polyploid macronuclei provide all vegetative gene expression, but degrade after fertilization, when new micronuclei and macronuclei develop. DNA differentiation in ciliates such as Oxytricha (also called Sterkiella) involves massive chromosome fragmentation and deletion of transposons and internally eliminated sequences (IESs), accomplishing 95% genome reduction, compressing a 1 Gb germline into one-twentieth of the space. Rearrangement of the remaining short segments (macronuclear destined segments; MDSs) by permutation or inversion, followed by telomere addition and amplification, produces mature macronuclear ‘nanochromosomes’, typically ~2 kb with just one gene12.
Genome-wide rearrangements discard nearly all nongenic DNA in Oxytricha, packing a streamlined gene-rich (~30,000 genes) eukaryotic genome in only 50 Mb. Furthermore, hundreds of thousands of MDSs require unscrambling (reordering or inversion) to interpret the sequence information in their DNA. The remarkable degree of specificity and reproducibility suggests a highly accurate mechanism to programme rearrangements. The parental cell must provide sufficient information to assemble a fully functional macronucleus. The mechanisms that allow perfect recognition of hundreds of thousands of DNA sequences for elimination or reordering remain largely unknown. The discovery of homology-dependent maternal effects that modify DNA deletions may provide a clue13,14.
Homology-dependent trans-nuclear comparison of germline and somatic genomes may regulate these DNA rearrangements. In Tetrahymena, germ-line-specific small RNAs called scan RNAs derive from the micronuclear-limited sequence and target DNA for elimination15. However, this model cannot explain fusion of unlinked or disordered DNA segments in Oxytricha, which can join segments smaller than the length of typical scan RNAs. DNA rearrangements in Tetrahymena are imprecise, involving no permutations.
In other species including Paramecium, DNA rearrangements occur at short direct repeats called pointers. Homologous recombination between identical repeats at MDS–IES boundaries can both remove the IESs between them and reorder MDSs, leaving one copy of the pointer in the final product. Pointers of 2–20 bp in Oxytricha (average length of 4 bp between nonscrambled junctions and 9 bp between scrambled junctions16) generally occur at the 3′ end of every MDS segment n and the 5′ end of segment n + 1, providing a simple linked list of final MDS order. Although necessary for DNA deletion in Paramecium17, 2–20 bp pointers could not unambiguously guide rearrangement18.
In refs 19 and 20, the authors proposed an epigenetic model in which an RNA or DNA template derived from the maternal macronucleus guides assembly of the new macronuclear chromosomes. Macronuclear templates could provide a scaffold to organize the layout of segment order and DNA deletion, using strand displacement and branch migration to align pointer pairs for recombination. Here we demonstrate that RNA templates can orchestrate this cascade of DNA rearrangements in Oxytricha. Injection of synthetic templates replaces the cellular programme, producing alternative epigenetically wired DNA rearrangements.
RNAi against putative templates disrupts rearrangement
To test the hypothesis that putative maternal RNA templates influence rearrangement, we induced RNA interference (RNAi) to target homologous RNA degradation. Oxytricha cells, before and during conjugation, were fed Escherichia coli producing double stranded RNA fragments of two macronuclear genes: either telomere-end-binding protein subunit α (TEBPα or α-TBP) or DNA polymerase α (pol-α). The germline versions in O. trifallix are broken into 17 and 48 scrambled segments, respectively21,22.
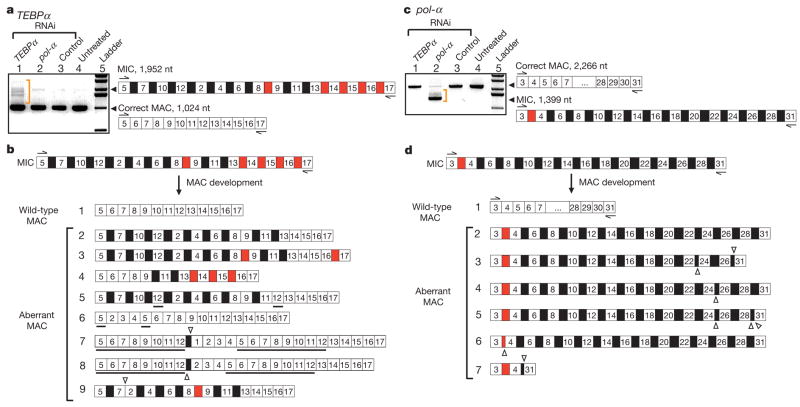
RNA interference against putative RNA templates leads to aberrant (incorrect or completely blocked) gene unscrambling in the resulting progeny (Fig. 1). PCR screening of TEBPα segments 5–17, after treatment with the RNAi vector spanning segments 2–17 in order, reveals DNA molecules longer than the typical macronuclear product, but shorter than the micronuclear precursor (Fig. 1a, lane 1). Sequencing revealed these were partially and/or incorrectly rearranged. Some aberrant forms still contain IESs (Fig. 1b, examples 2–5 and 9), with a bias for retention between scrambled segments (black boxes in Fig. 1). IESs between nonscrambled segments (red in Fig. 1) are usually absent. This suggests different susceptibility between these two kinds of IESs to RNAi treatment, and possibly a mechanistic decoupling, consistent with observations that simple IES removal precedes unscrambling in the actin I gene in the related genus Stylonychia23. Some permutations (partial descrambling) occurred (Fig. 1b, examples 4–6; molecule 4 could be an unscrambling intermediate), and even large duplications (Fig. 1b, examples 7 and 8) or deletions (Fig. 1b, examples 8–9). All deletions occur between short direct repeats that provide alternative (cryptic) pointers for recombination—the type of error that occurs early during rearrangement23.
Figure 1. RNAi against putative RNA templates leads to disruption of DNA rearrangement, with accumulation of aberrant products.
MDS segments (white boxes) and IES regions (black boxes if located between nonconsecutive segments, red if consecutive) are drawn schematically, not to scale. The ladders are 1 kb (Invitrogen). a, PCR amplification of the TEBPα region between segments 5 and 17 from total DNA extracted from the sources treated with: TEBPα RNAi, pol-α RNAi and control double-stranded (ds)RNA (184 nucleotide (nt) dsRNA from feeding vector polylinker), as well as untreated cells. Only cells treated with TEBPα RNAi contain partial or incorrect rearrangements, on the basis of size (orange bracket). Cells treated with TEBPα RNAi were fed with dsRNA covering the region between segments 1 and 16. MAC, macronucleus; MIC, micronucleus. b, The sequence of several TEBPα PCR products between segments 5 and 17 in cells treated with TEBPα RNAi. IESs between both scrambled (black) and nonscrambled (red) MDSs are deleted from some molecules at both correct and incorrect (cryptic) repeats. Open triangles show the locations of cryptic junctions between neighbouring segments (if pointing up) and non-neighbouring segments (pointing down) on the basis of the precursor micronuclear order. Underlined segments are duplications. c, PCR amplification of pol-α between segments 3 and 31 from total DNA extracted from sources treated with: TEBPα RNAi, pol-α RNAi and control dsRNA (as above), as well as untreated cells. Only cells treated with pol-α RNAi show aberrantly rearranged products, on the basis of size (orange bracket). Cells treated with pol-α RNAi were fed with dsRNA covering the region between segments 16 and 29. d, Sequence of several pol-α PCR products between segments 3 and 31 in cells treated with pol-α RNAi.
RNAi against pol-α leads to almost complete abrogation of DNA elimination or rearrangement (Fig. 1c, lane 2) with no permutations at all, but with several examples of aberrant deletion at cryptic pointers (Fig. 1d, examples 3–7), including one large deletion between segments 4 and 31 (Fig. 1d, example 7). Approximately 85% of pol-α molecules surveyed between segments 3 and 31 (the RNAi vector spanned segments 16–29) 80 h post conjugation were identical in size to the germline precursor (Fig. 1c, lane 2, and Fig. 1d, example 2), indicating no rearrangement (which we confirmed by sequencing).
These results provide strong indirect support for an RNA-template model involving intracellular genome comparisons by means of RNA transcripts from the maternal macronucleus. In principle, RNAi would degrade or negatively influence any homologous RNAs (Supplementary Figs 3 and 4), including templates, yielding aberrant rearrangements of corresponding genes. In our experiments, all aberrant rearrangements are restricted to the targeted gene (that is, pol-α feeding does not influence TEBPα rearrangement and vice versa), suggesting that each macronuclear molecule may have its own RNA template, which acts in a homology-dependent manner.
Early presence of long, bidirectional RNA
The RNAi experiments are consistent with the presence during conjugation of double- or single-stranded maternal RNA transcripts from the parental macronucleus. Single-stranded RNA is a reasonable target for RNAi-induced post-transcriptional gene silencing in eukaryotes. If the maternal templates are RNA, then the question arises whether transcription is uni- or bi-directional, and what are the promoters? Normal gene promoters sometimes initiate transcription downstream of the first MDS; however, both 5′- and 3′-untranscribed regions may be scrambled or interrupted by several IESs. Therefore, messenger RNA templates could not guide complete rearrangement. Another possibility is that telomeres themselves could promote transcription. This may allow unbiased transcription of all nanochromosomes, rather than gene-specific promoters. Complete transcription could also contribute to Oxytricha’s bias for tiny chromosomes with little extraneous DNA.
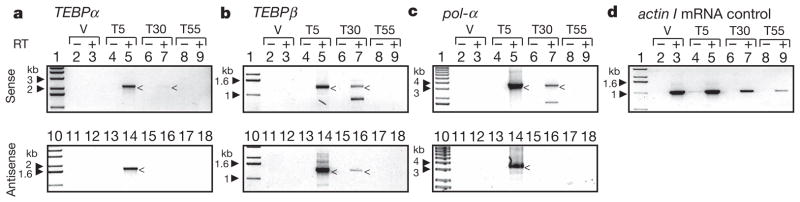
PCR with reverse transcription (RT–PCR) demonstrates the presence of antisense and sense transcripts of macronuclear chromosomes 5–30 h post conjugation (Fig. 2). These RNA transcripts are longer than messenger RNAs, contain telomeres (complementary DNA synthesis began at the telomere), are absent from vegetative cells, and disappear 55 h post conjugation. The presence of such RNA transcripts during early macronuclear development (estimated between meiosis and the peak of polytene chromosome formation) suggests they are available during DNA rearrangement and could therefore have a role. Figure 2 shows RT–PCR products for three different genes: two scrambled (TEBPα, pol-α) and one nonscrambled (TEBPβ or β-TBP); similar results were obtained for two other independent genes (not shown). TEBPα and TEBPβ transcripts both retain introns (pol-α has no introns in the surveyed region).
Figure 2. Long sense and antisense transcripts are present during early development.
RT–PCR of both strands from cells in a vegetative state (V) as well as 5 (T5), 30 (T30) and 55 (T55) hours post conjugation. Filled arrowheads indicate size of relevant markers (1 kb ladder, Invitrogen). Long, intron-containing maternal transcripts appear 5 h post conjugation and disappear 50 h later. The peak of polytene chromosome formation and DNA rearrangements is estimated to occur in this time window. Arrowheads indicate specific products. All non-specific amplification products were confirmed by sequencing to be unrelated. a, b, c, RT–PCR detection of both sense (+) and antisense (−) strands of TEBPα, TEBPβ and pol-α RNA templates, respectively. d, RT–PCR of actin I mRNA as a control for RNA in each sample.
Injection of alternative DNA templates
Together, the detection of sense/antisense transcripts during development plus the RNAi experiments provide support for an RNA-template-based model. To test this model directly, we microinjected synthetic DNA or RNA versions of alternatively rearranged chromosomes into conjugating cells to ask whether this would specify the rearrangement pattern in the offspring.
We designed artificial macronuclear chromosomes to permute the natural order of two DNA segments in both TEBPα (Fig. 3a) and TEBPβ (Fig. 3c). TEBPα is scrambled in the germ line, requiring several DNA permutations to assemble a functional gene during development. TEBPβ normally requires only DNA deletion and no permutation (with identical MDS order in both nuclei). We chose these genes to test whether template-directed rearrangements act genome-wide or are specific to scrambled genes.
Figure 3. Microinjection of alternative DNA templates produces alternatively rearranged chromosomes.
a, Top, wild-type TEBPα macronuclear chromosome (labelled TEBPα wild type) with segments 1–17 colinear; middle, microinjected TEBPα template designed to switch (sw) the order of segments 7 and 8 (TEBPα sw78 template); bottom, map of resulting macronuclear product (TEBPα sw78), to scale. Asterisks, point mutations in synthetic templates; black rectangles, telomeres; open triangles, cryptic pointers to switch segment order; filled triangle, an unspliced 5 bp IES (see Supplementary Fig. 1). b, Left, PCR and restriction analysis of DNA microinjection products (ethidium bromide staining). Inj., injected. Right, single-sided arrows, PCR primers (in bp); scissors, restriction sites (in bp). Lanes 1–4 show presence of wild-type (WT) macronuclear product as well as a product from microinjected cells with segments 7 and 8 switched (TEBPα sw78; lanes 2, 4). Lanes 5–8 distinguish the microinjected template (lanes 7, 8) from the macronuclear product (lanes 5, 6). c, Top, wild-type TEBPβ macronuclear chromosome (TEBPβ wild type) with segments 1–7 colinear; middle, microinjected template designed to switch the order of segments 4 and 5 (TEBPβ sw45 template); and bottom, map of expected macronuclear product (TEBPβ sw45). d, PCR and restriction analysis of DNA microinjection products. Lanes 1–8 use a simple PCR length assay for the presence of a smaller product when the order of segments 4 and 5 has been reversed (lower band): one week (F1 sw-1w) and two weeks (F1 sw-2w) after microinjection, as well as the putative F2 generation (epigenetic inheritance was also observed for putative F3, see Supplementary Fig. 2; 1 kb DNA ladder (Invitrogen)). F2 sw-A and F2 sw-B are the asexual progeny of two independent conjugating pairs in the F1; sw indicates injected cells or their progeny and WT indicates wild-type non-injected controls. Lanes 9–15 confirm the presence of the macronuclear product in which segments 4 and 5 are switched (TEBPβ sw45) in F1 (one week post injection). Lanes 16–22 distinguish the microinjected template from the F1 (one week) macronuclear product; arrowheads in lanes 19 and 20 point to aberrantly rearranged molecules lacking segment 5. e, Lanes 23–26, HindIII and BsrGl Southern analysis of total DNA extracted from the putative F2 generation.
In two independent experiments, microinjecting the double-stranded, synthetic TEBPα chromosome (labelled ‘TEBPα sw78’ template in Fig. 3, where SW indicates switched segment order) into the macronucleus of conjugating pairs of Oxytricha switches the order of segments 7 and 8, and injecting the synthetic TEBPβ molecule (‘TEBPβ sw45’ template) switches the order of segments 4 and 5. Each template slightly adjusts segment boundaries (Supplementary Fig. 1) to recruit existing sequence repeats as alternative pointers at new recombination junctions. Both templates contain single nucleotide substitutions (some creating restriction sites) to distinguish microinjected DNA from processed endogenous genes. Non-injected cells were used as controls. DNA was extracted from progeny one week after microinjection, permitting sufficient asexual growth. Injected cells grew slower than non-injected cells. In both cases, the F1 progeny contained alternatively rearranged macronuclear chromosomes, following the reprogrammed order (TEBPα sw78 and TEBPβ sw45; Fig. 3b, d, e). In one case, the switched order was even the major product (Fig. 3d, lane 3).
Restriction mapping ruled out either the presence of microinjected DNA in the harvested cells or extended copying or incorporation of templates during rearrangement (Fig. 3b, lanes 5–8, and Fig. 3d, lanes 17–22). (Sequence analysis revealed a few exceptions; see section entitled ‘Transfer of nucleotide substitutions’.) For TEBPα, the new junction formed between segments 6 and 8 creates a Tsp509I site, which distinguishes wild-type from switched chromosomes (Fig. 3b, lanes 1–4). For TEBPβ, the presence of two restriction sites (BbvCI and BsrGI) in MDS4 produces differently sized products in switched versus wild-type molecules (Fig. 3d, lanes 10–15). In addition, for TEBPβ, a simple PCR assay measuring the length from segment 4 to 6 yields a smaller product for switched versus wild-type chromosomes, because MDS5 is missing in the switched PCR product (Fig. 3d, lanes 2–8). PCR screening was not feasible for TEBPα because segments 7 and 8 are very small, encoding 6 and 10 amino acids, respectively, within the same α helix of the TEBPα protein24.
We also examined DNA from TEBPβ sw45-injected cells two weeks post injection (Fig. 3d, lanes 4–5). These cells have an increase in the ratio of wild-type to switched chromosomes, possibly owing to a fitness decrease in cells burdened with the altered TEBPβ gene. Premature stop codons in the permuted version could produce a severely altered protein or possibly invoke nonsense-mediated mRNA decay (whereas the TEBPα permutation would cause a modest change in helix B of the encoded protein). Thus, we suggest that the use of synthetic templates to reprogramme DNA rearrangement provides a convenient epigenetic tool for reverse genetics in Oxytricha.
To test whether this epigenetic effect transferred to sexual offspring, we induced sexual conjugation by starving TEBPβ sw45-injected cells after two weeks of vegetative growth. Conjugating pairs were isolated and grown separately. Asexual progeny of individual pairs are denoted sw-A and sw-B. We detected the alternative rearrangement pattern in putative F2 progeny (Fig. 3d, lanes 6–8, and Fig. 3e) and even in some putative F3 (Supplementary Fig. 2), suggesting stable epigenetic inheritance of this alternative form. In one case (F2 sw-B) the newly programmed pattern still predominates over wild type.
All PCR and restriction-based conclusions for TEBPα and TEBPβ (after one week) were confirmed by sequencing. Four out of 28 TEBPα clones and 18 out of 37 TEBPβ clones contained the expected alternative rearrangement. No sequenced TEBPβ products were wild type. One TEBPα molecule contains a 19 bp deletion, and one TEBPβ clone contains a 28 bp insertion (Supplementary Information). Nineteen TEBPβ clones contained aberrant deletions, some at cryptic pointers or imperfect repeats near deletion boundaries (Supplementary Information); 6 out of 19 still correctly adopted the programmed junction between segments 4 and 6 (deleting segment 5), often observed on its own (Fig. 3d, arrowhead in lanes 19 and 20, clones 18 and 19). Aberrant rearrangements may derive from competition between synthetic and endogenous templates or from rogue microinjected molecules with polymerase errors—minor products of amplification. Cloning and re-sequencing the microinjected DNA revealed five correct as well as one aberrant product, which had segment 5 deleted (Supplementary Information). Therefore, such rearrangements could derive from a sub-population of injected templates.
Injection of alternative RNA templates
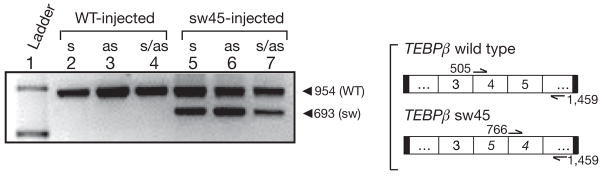
Because we expect that DNA injected in the previous experiments had the opportunity to be transcribed into RNA, we tested whether RNA injection could reprogramme genome rearrangement directly. We injected synthetic TEBPβ RNA templates in both sense and anti-sense directions in which segments 4 and 5 were permuted (TEBPβ sw45 template). Injecting wild-type TEBPβ RNA provided a control. In six independent experiments, we injected sense, antisense and a combination of sense and antisense RNA for both switched and control templates into the cytoplasm of cells during conjugation. The progeny of cells injected with any combination of RNA in the switched orientation produced macronuclear products in which segments 4 and 5 were permuted, in roughly similar proportion to the wild-type product (Fig. 4, lanes 5–7). All results were confirmed by sequencing (Supplementary Information).
Figure 4. Microinjection of alternative RNA templates leads to alternatively rearranged chromosomes.
RNA microinjection of TEBPβ sw45 template. Sense (s), antisense (as), and combined sense and antisense (s/as) RNA templates were microinjected in both wild-type (control) and switched orientations. Lanes 5–7 display the expected macronuclear product if segments 4 and 5 have been switched (lower band). Primers are as in Fig. 3d. Lane 1, 1 kb ladder (Invitrogen).
Although RNA samples were DNase-treated after in vitro transcription and before injection, we tested whether any DNA may have been microinjected and concluded that this was unlikely (Supplementary Fig. 5).
Transfer of nucleotide substitutions
The influence of microinjected templates also extended to different germline alleles of the same gene (Supplementary Information), suggesting that a single sequence can programme the rearrangement pattern of multiple alleles, despite occasional template mismatches. Furthermore, a subset of mutations near pointers occasionally transferred from the synthetic template to rearranged molecules (Supplementary Information). Remarkably, a C-to-T substitution 4 bp from the end of MDS4 in the TEBPβ template transferred to all 40 sequenced molecules (DNA- or RNA-injected) that contained the programmed junction between MDS4 and MDS6 (Supplementary Information). We confirmed the absence of an allele or paralogue containing the ‘T’ nucleotide in O. trifallax strain JRB310 or JRB510 by screening hundreds of JRB310 reads from the Oxytricha genome project, as well as by studying expressed-sequence-tag clones from both strains, sequencing 24 JRB510 PCR clones, and performing BtsCI restriction mapping (not shown). Thus, the source of this T nucleotide is likely to be the template itself, implicating template-directed DNA repair.
Conclusions
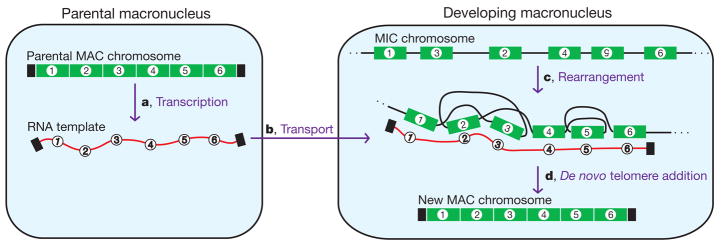
We provide evidence for both the presence of long maternal RNA templates—RNA cached copies of DNA sequences from the previous generation—and the powerful influence of such molecules to guide genome rearrangements, even permuting the order of DNA sequences in vivo (Fig. 5). In particular, we have shown that micro-injection of alternative DNA or RNA templates leads to stable epigenetic inheritance of alternative DNA rearrangement pathways, offering a key informational and regulatory role for RNA in transmitting a genome-rearrangement programme. The templating mechanism seems to be highly accurate, because most alternatively ordered molecules in this study contain precise recombination junctions, instructed by the synthetic template.
Figure 5. Model for RNA guiding of genome rearrangements during macronuclear development in Oxytricha.
a, Bidirectional RNA transcription of all DNA nanochromosomes (including injected DNA) in the old, maternal macronucleus (MAC) before its degradation. b, Transport of these RNA transcripts to the newly developing macronucleus, where they may act as scaffolds to guide rearrangements (deletion, permutation and inversion) of corresponding micronuclear (MIC) DNA sequences (c). This step would be notable and unprecedented, but perhaps possible if there were either local or extensive strand-separation of both the RNA template and the developing DNA (see ref. 20). In this illustration, segments 2 and 3 are switched and segment 5 is inverted (number upside down). d, De novo telomere addition (black rectangles) and amplification completes formation of new macronuclear nanochromosomes.
Our results are compatible with an earlier result showing that RNA injection in Tetrahymena leads to deletion of homologous sequences during development25. The injected RNA might trigger destruction of homologous RNA templates in Tetrahymena, for example, as in our RNAi experiments.
The transfer of some mutations from the template to the rearranged molecule (Supplementary Information) suggests that RNA could be a template for DNA synthesis or repair very close to the pointer, as in template-guided DNA repair9. A C-to-T mutation in the template that creates a TA dinucleotide close to a new pointer transferred efficiently to all rearranged molecules, whereas substitutions further away from a pointer transferred infrequently, consistent with local polymerase activity. The RNA-guided DNA rearrangements proposed here are distinct from previous indirect roles for RNA in mediating recombination26–30.
The ability of RNA to programme DNA rearrangements suggests new approaches for genome manipulation in vivo, providing a tool for reverse genetics in Oxytricha and possibly in other systems; this also demonstrates an elegant mechanism for RNA-guided recombination that may be widespread, with somatic and evolutionary consequences for genome expression. For example, occasional templating of rearrangements by maternal mRNA could explain the paucity of introns in ciliates12, converting them to IESs31.
METHODS SUMMARY
RNAi
Double-stranded RNA feeding was performed using a Paramecium protocol32, in which ciliates were fed with live bacteria, supplemented with algae. Silencing plasmids contained 1.5 kb of TEBPα or 1.8 kb of the pol-α macronuclear sequence (Supplementary Information).
RT–PCR
First-strand cDNA was synthesized using Superscript III RTS First-Strand cDNA Synthesis Kit (Invitrogen) with either a telomere-anchor primer (Supplementary Information) or oligo(dT)20. Products were amplified for 28–37 cycles with Expand High Fidelity Plus or FastStart High Fidelity PCR Systems (Roche) using a linker primer in combination with gene-specific primers (Supplementary Information).
Template design
For TEBPα, we chose to switch the order of MDS7 and MDS8, which code for a region least likely to affect the telomere-binding domain24. We used MUMmer33 to detect repeats within 14 bp of original pointer boundaries, and chose the longest cryptic repeats that would preserve the reading frame. For TEBPβ, we switched the order of MDS4 and MDS5, which permitted new pointers to be similar to wild-type pointers.
Microinjection of DNA and RNA
Synthetic templates were generated by means of PCR (with point mutations on the primers), followed by restriction digestion, ligating segments in the desired order, and cloning (see Supplementary Information). DNA versions were prepared by telomere-to-telomere PCR, and RNA versions of each strand were prepared by in vitro transcription of PCR products between the vector T7 promoter and the telomere on the opposite side. Approximately 5 pl (picoliters) DNA or 10 pl RNA was injected (Narishige IM 300) into the macronucleus or cytoplasm, respectively, of each cell in a mating pair visualized by phase-contrast inverted microscopy (Zeiss, Axiovert 200). Individual exconjugants were observed under the microscope and each cell displayed morphological features characteristic of sexual reproduction in Oxytricha, such as rounded cell shape or a large macronuclear anlagen. Thus, on the basis of cell morphology, the analysed F2 cells were the progeny of F1, and so on.
Supplementary Material
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Acknowledgments
This work was supported by awards from the NSF and NIH to L.F.L. and the SEAS senior thesis research fund to V.V. We thank J. Wang for technical assistance and all members of the laboratory for discussion.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.N., V.V., Y.Z., T.G.D. and L.F.L. designed experiments; M.N., V.V., Y.Z. and K.S. performed the experiments; T.G.D. provided cells; M.N., V.V., Y.Z. and L.F.L. analysed the data; and M.N., V.V., Y.Z. and L.F.L. wrote the paper.
References
- 1.King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- 2.Tadros W, Lipshitz HD. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev Dyn. 2005;232:593–608. doi: 10.1002/dvdy.20297. [DOI] [PubMed] [Google Scholar]
- 3.Tang F, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rassoulzadegan M, et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 5.Herbert A, Rich A. RNA processing and the evolution of eukaryotes. Nature Genet. 1999;21:265–269. doi: 10.1038/6780. [DOI] [PubMed] [Google Scholar]
- 6.Lolle SJ, Victor JL, Young JM, Pruitt RE. Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature. 2005;434:505–509. doi: 10.1038/nature03380. [DOI] [PubMed] [Google Scholar]
- 7.Peng P, Chan SW, Shah GA, Jacobsen SE. Plant genetics: increased outcrossing in hothead mutants. Nature. 2006;443:E8. doi: 10.1038/nature05251. [DOI] [PubMed] [Google Scholar]
- 8.Lolle SJ, Pruitt RE, Victor JL, Young JM. Lolle et al. reply. Nature. 2006;443:E8–E9. [Google Scholar]
- 9.Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 11.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 12.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chalker DL, Yao MC. Non-mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol Cell Biol. 1996;16:3658–3667. doi: 10.1128/mcb.16.7.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duharcourt S, Keller AM, Meyer E. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol Cell Biol. 1998;18:7075–7085. doi: 10.1128/mcb.18.12.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 16.Prescott DM, DuBois ML. Internal eliminated segments (IESs) of Oxytrichidae. J Eukaryot Microbiol. 1996;43:432–441. doi: 10.1111/j.1550-7408.1996.tb04502.x. [DOI] [PubMed] [Google Scholar]
- 17.Mayer KM, Forney JD. A mutation in the flanking 5′-TA-3′ dinucleotide prevents excision of an internal eliminated sequence from the Paramecium tetraurelia genome. Genetics. 1999;151:597–604. doi: 10.1093/genetics/151.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landweber LF, Kuo TC, Curtis EA. Evolution and assembly of an extremely scrambled gene. Proc Natl Acad Sci USA. 2000;97:3298–3303. doi: 10.1073/pnas.040574697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prescott DM, Ehrenfeucht A, Rozenberg G. Template-guided recombination for IES elimination and unscrambling of genes in stichotrichous ciliates. J Theor Biol. 2003;222:323–330. doi: 10.1016/s0022-5193(03)00037-7. [DOI] [PubMed] [Google Scholar]
- 20.Angeleska A, Jonoska N, Saito M, Landweber LF. RNA-guided DNA assembly. J Theor Biol. 2007;248:706–720. doi: 10.1016/j.jtbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Prescott JD, DuBois ML, Prescott DM. Evolution of the scrambled germline gene encoding α-telomere binding protein in three hypotrichous ciliates. Chromosoma. 1998;107:293–303. doi: 10.1007/s004120050311. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman DC, Prescott DM. Evolution of internal eliminated segments and scrambling in the micronucleargene encoding DNA polymerase α in two Oxytricha species Oxytricha novo is extremely scrambled. Nucleic Acids Res. 1997;25:1883–1889. doi: 10.1093/nar/25.10.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Möllenbeck M, et al. The pathway to detangle a scrambled gene. PLoS Biol. doi: 10.1371/journal.pone.0002330. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 25.Yao MC, Fuller P, Xi X. Programmed DNA deletion as an RNA-guided system of genome defense. Science. 2003;300:1581–1584. doi: 10.1126/science.1084737. [DOI] [PubMed] [Google Scholar]
- 26.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derr LK, Strathern JN. A role for reverse transcripts in gene conversion. Nature. 1993;361:170–173. doi: 10.1038/361170a0. [DOI] [PubMed] [Google Scholar]
- 28.Moore JK, Haber JE. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 29.Nevo-Caspi Y, Kupiec M. cDNA-mediated Ty recombination can take place in the absence of plus-strand cDNA synthesis, but not in the absence of the integrase protein. Curr Genet. 1997;32:32–40. doi: 10.1007/s002940050245. [DOI] [PubMed] [Google Scholar]
- 30.Teng SC, Kim B, Gabriel A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 31.Chang WJ, et al. Intron evolution and information processing in the DNA polymerase alpha gene in spirotrichous ciliates: a hypothesis for interconversion between DNA and RNA deletion. Biol Direct. 2007;2:6. doi: 10.1186/1745-6150-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galvani A, Sperling L. RNA interference by feeding in Paramecium. Trends Genet. 2002;18:11–12. doi: 10.1016/s0168-9525(01)02548-3. [DOI] [PubMed] [Google Scholar]
- 33.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.