Abstract
Flaviviruses are a major cause of infectious disease in humans. Dengue virus causes an estimated 50 million cases of febrile illness each year, including an increasing number of cases of hemorrhagic fever. West Nile virus, which recently spread from the Mediterranean basin to the Western Hemisphere, now causes thousands of sporadic cases of encephalitis annually. Despite the existence of licensed vaccines, yellow fever, Japanese encephalitis and tick-borne encephalitis also claim many thousands of victims each year across their vast endemic areas. Antiviral therapy could potentially reduce morbidity and mortality from flavivirus infections, but no effective drugs are currently available. This article introduces a collection of papers in Antiviral Research on molecular targets for flavivirus antiviral drug design and murine models of dengue virus disease that aims to encourage drug development efforts. After reviewing the flavivirus replication cycle, we discuss the envelope glycoprotein, NS3 protease, NS3 helicase, NS5 methyltransferase and NS5 RNA-dependent RNA polymerase as potential drug targets, with special attention being given to the viral protease. The other viral proteins are the subject of individual articles in the journal. Together, these papers highlight current status of drug discovery efforts for flavivirus diseases and suggest promising areas for further research.
Keywords: Flavivirus, dengue virus, West Nile virus, yellow fever virus, Japanese encephalitis virus, envelope glycoprotein, RNA-dependent RNA polymerase, helicase, protease, methyltransferase, antiviral therapy
Introduction
This article introduces a collection of papers in Antiviral Research focusing on targets for flavivirus drug discovery, which recognizes the importance of flaviviruses as agents of human disease and the urgent need to develop new vaccines and effective therapies. The genus Flavivirus contains more than 53 members, including yellow fever (YF), dengue (DEN), West Nile (WN), Japanese encephalitis (JE) and tick-borne encephalitis (TBE) (Gubler, 2007). Although licensed vaccines are available for YFV, JEV and TBE (Mackenzie et al., 2004), none have been developed for other flaviviral diseases. Efforts for vaccine development for dengue have been a continuous challenge for decades, the main issue being the inability of vaccines to protect simultaneously against all four antigenically distinct serotypes. A further barrier to vaccine development is the sporadic nature of infections caused by agents such as WNV, JEV and TBEV, which could only be completely prevented by carrying out universal immunization across huge geographic regions. In the absence of vaccines, drugs for specific therapy are needed, but no antiviral medications are approved for use against the flaviviruses. Ribavirin suppresses the replication of some agents in vitro, but demonstrations of in vivo activity have been limited to a few rodent models (Leyssen et al., 2008). There is thus a need for new antivirals that can reduce viremia during early stages of infection, block viral replication in the brain in cases of encephalitis, or modulate host responses to prevent or treat disease (Bray, 2008).
This article begins by describing the flaviviral replication cycle, then, briefly summarizes the role of each viral protein in entry, replication, assembly and maturation and its potential as a drug target. Five other papers examine the same proteins in much greater detail. One describes individual steps in the processing and maturation of the envelope (E) glycoprotein and shows how they could potentially be blocked by small-molecule drugs (Perera et al., 2008), while another describes novel approaches to developing anti-helicase compounds (Lescar et al., 2008). Two different activities of the NS5 protein are then considered as therapeutic targets: the methyltransferase (Dong et al., 2008) and the RNA-dependent RNA polymerase (Malet et al., 2008). The fifth paper reviews the current status of mouse model development for dengue fever and dengue hemorrhagic fever/shock syndrome (Yauch and Shestra, 2008).
II. Flavivirus virion and genome structure
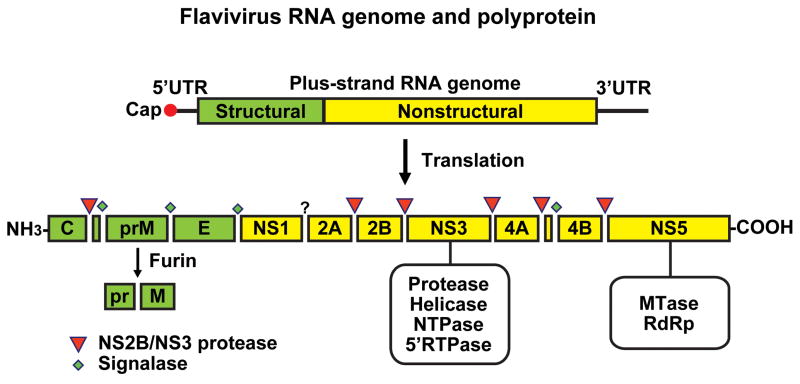
The mature flavivirus virion is smooth and spherical, with a diameter of 500 Å. The genome is packaged by the viral capsid protein (C) in a host-derived lipid bilayer in which 180 copies of the envelope protein (E) are embedded (Mukhopadhyay et al., 2005). The E protein is initially complexed with the precursor membrane protein (prM) during assembly of the virions in the endoplasmic reticulum forming immature particles. The immature particles are transported to trans-Golgi compartment where they undergo maturation by the cellular serine protease, furin, which mediates cleavage of prM to M resulting in homodimerization of E protein to form fusion-competent mature particles before release into circulation (Stiasny and Heinz, 2006). The single-stranded positive-sense RNA genome contains a single long open reading frame flanked by 5′-and 3′ untranslated regions, which have secondary structures that are essential for the initiation of translation and for replication (Figure 1) (Lindenbach et al., 2007). The 5′ end of the genome has a type 1 cap, but the 3′ end lacks a poly-A tail. Translation of the genome by the host cell machinery produces a polyprotein comprising the viral structural and non-structural proteins that are required for replication and assembly of new virions.
Figure 1.
Schematic representation of flavivirus genome organization and polyprotein processing. The 11 kb positive-sense, single-stranded RNA genome contains a single open reading frame which encodes 3 structural proteins (capsid (C), precursor membrane (prM) and envelope (E)) and 7 non-structural proteins (NS1-NS2A, NS2B, NS3, NS4A, NS4B, NS5). The open reading frame is flanked by untranslated regions. Sites of polyprotein cleavage mediated by the viral NS2B-NS3 and by host signalase and furin are shown, and the enzymatic activities of NS3 and NS5 are also indicated. See text for further information. (Courtesy of Pei-Yong Shi).
III. Viral replication cycle
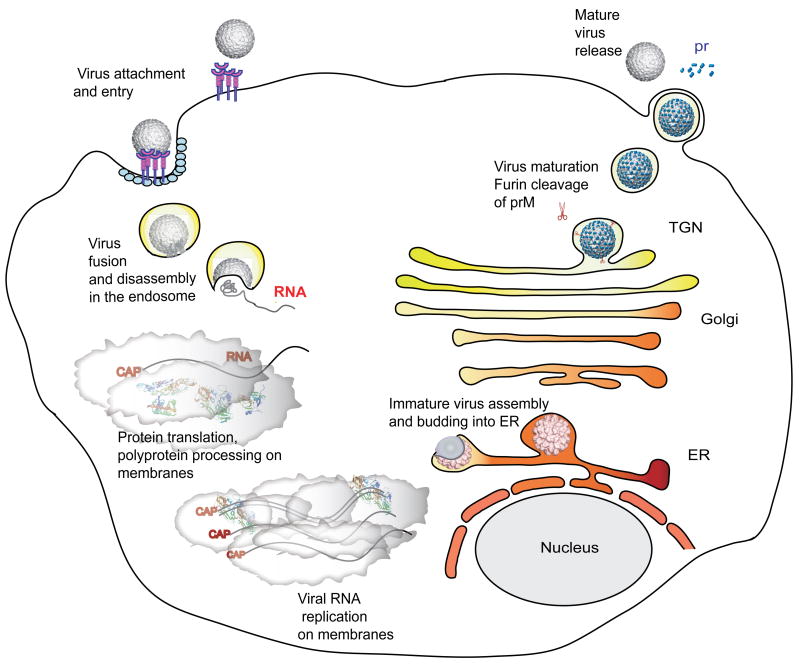
Host cells for flaviviral infection include monocytes, macrophages and dendritic cells (Marianneau et al., 1999, Tassaneetrithep et al., 2003, Barba-Spaeth et al., 2005, Lozach et al., 2005, Krishnan et al., 2007). The virus attaches to the cell surface, mediated by the E protein, and enters the cell by receptor-mediated endocytosis (Figure 2). Low pH in the endosomal compartment triggers fusion of the viral and host cell membrane mediated by structural reorganization of E, which leads to the release of the nucleocapsid and viral RNA into the cytoplasm. Translation of the RNA generates a polyprotein that is co-translationally and post-translationally processed by the virus-encoded serine protease, NS2B/NS3, and by host-encoded proteases, including signalase and furin, to produce the 3 structural proteins and 7 nonstructural proteins in the order C-prM-E -NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5 (Rice et al., 1985).
Figure 2.
The flavivirus replication cycle. Virions bind to cell-surface attachment molecules and receptors and are internalized through endocytosis. In the low pH of the endosome, viral glycoproteins mediate fusion of the viral and cellular membranes, allowing disassembly of the virion and release of its RNA into the cytoplasm. The viral RNA is translated into a polyprotein that is processed by viral and cellular proteases. Viral non-structural proteins then replicate the genomic RNA. Virion assembly occurs at the ER membrane. Capsid protein and viral RNA are enveloped by the membrane and its embedded glycoproteins to form immature virus particles, which are then transported through the secretory pathway. In the low pH of the trans-Golgi network (TGN), prM is cleaved by furin. Mature virions are then released into the cytoplasm. See text for further information. (Courtesy of Rushika Perera and Richard Kuhn.)
NS3 (70 kDa) and NS5 (104 kDa) are the best characterized nonstructural proteins, with multiple enzyme activities that are required for viral replication. NS3 has three distinct activities: serine protease together with the cofactor NS2B, required for polyprotein processing; helicase/NTPase activity, required for unwinding the double-stranded replicative form of RNA; RNA triphosphatase, required for capping nascent viral RNA (Falgout et al., 1991; Zhang, L. et al., 1992; Arias et al., 1993, Li, H. et al., 1999; Benarroch et al., 2004). Mutations that affect each activity impair viral replication (Matusan et al., 2001a,b). NS5 is the largest and most highly conserved flaviviral protein, with greater than 75% sequence identity across all DEN serotypes. It contains two distinct enzymatic activities, separated by an interdomain region: an S-adenosyl methyltransferase (SAM) (Egloff et al., 2002, Ray et al., 2006) and an RNA-dependent RNA polymerase (RdRp) (Grun and Brinton, 1986, Chu and Westaway, 1987, Tan et al., 1996, Ackermann and Padmanabhan, 2001, Guyatt et al., 2001). NS1 (46 kDa) is required for flavivirus replication and presumably involved in negative-strand synthesis by an unknown mechanism. A large deletion in YFV NS1 abolished viral replication but can be complemented in trans by functional expression from Sindbis virus vector (Lindenbach and Rice, 1997). Furthermore, a temperature-sensitive mutation (Arg-299) is defective in viral replication at 39°C and fails to accumulate negative strand RNA but is functional at 32° C, suggesting that NS1 is required for negative strand RNA synthesis (Muylaert et al., 1997). NS2A (22 kDa) is a small hydrophobic transmembrane protein that is involved in generation of virus-induced membranes during virus assembly (Leung, 2008). NS4A (16 kDa) is an integral membrane protein which may induce membrane rearrangements to form the viral replication complex (Miller et al., 2007; Roosendaal et al., 2006). NS4B (27 kDa) inhibits the type I interferon response of host cells (Munoz-Jordan et al., 2005), and may modulate viral replication via its interaction with NS3 (Umareddy et al., 2006).
Viral RNA replication occurs in the rough endoplasmic reticulum (ER) and in Golgi-derived membranes called vesicle packets (VP) (Chu and Westaway, 1992; Mackenzie, 2005). The nonstructural proteins and dsRNA are concentrated in the VP, constituting the site of viral RNA synthesis (Mackenzie et al., 1998; Westaway et al., 1997, 1999). The newly synthesized viral RNA is extruded in the intermembrane space of the double-membrane VPs, from which it exits into the cytoplasm by an unknown mechanism (Uchil and Satchidanandam, 2003). Assembly of virus particles occurs in the lumen of the rough ER. The first step in this process is the coating of the newly synthesized viral RNA with the C protein (Khromykh and Westaway, 1996; see also Perera et al. in this volume). Next, E and PrM hetero-dimerize and envelope the nucleocapsid, forming an immature virus particle that buds from the RER lumen into the Golgi (MacKenzie and Westway, 2001). However, the mechanism of interaction of the C protein within the nucleocapsid is still not clear. Maturation of virus particles occurs in the trans-Golgi network, where prM is cleaved to M by furin, along with conformational rearrangements of E (Li, L., et al., 2008; Mukhopadhyay et al., 2005; Yu, I.M., et al., 2008). This is an essential step for the virus in the transition from fusion-incompetent and non-infectious virus particles to mature, fusion-competent, and infectious virions. The mature particles eventually exit from the host cell by exocytosis.
IV. Targeting critical functions of individual flaviviral proteins
Identification of small molecules that specifically inhibit critical steps in the viral life cycle requires detailed biochemical and structural characterization of the essential viral proteins. Development of high through-put screening assays that include both biochemical assays and cellular assays is also an essential step in the lead-finding process. Both types of assays have their pros and cons. Although in vitro assays can be more target-specific and can utilize available structural information for optimization of inhibitors before testing in cells, there is always a risk that active compounds will be unable to penetrate the cellular membrane. Cellular assays obviate this issue, but target de-convolution can be a laborious process. Once interesting lead compounds have been identified, the next stage is to test them in a physiologically relevant small animal model. The review by Yauch and Shreshta (2008) discusses the current status of the development of mouse models of dengue and dengue HF. Rodent models are also available for WN, YF and JE viral infection (Charlier et al., 2004; Holbrook and Gowen, 2008).
A. Structural proteins as drug targets
As described above, the viral C, prM and E proteins undergo a number of conformational changes during the entry, assembly and exit of viral particles during the course of their maturation. These changes potentially offer targets for inhibition by antiviral drugs.
1. C protein
The highly basic 11 kDa C protein interacts with viral genomic RNA, forming the nucleocapsid (NC). The minimal functional region required for dimerization for virus assembly has been characterized (Jones et al., 2003; Patkar et al., 2007). The capsid protein comprises an internal hydrophobic sequence that mediates membrane association (Ma et al., 2004). The capsid folds into a dimer, in which each monomer contains four alpha helices. The N and C termini contain charged residues, of which the C-terminal region may be involved with RNA association (Ma et al., 2004; Wang et al., 2002). Dimerization of C is induced by interaction with DNA or RNA (Kiermayr et al., 2004). The development of an in vitro assembly system would be very useful for identifying compounds that block capsid dimerization or capsid-RNA interaction which could lead to identification of inhibitors that block either of these steps.
2. M and E proteins
The 26-kDa glycosylated precursor of M protein, prM, is processed from a polyprotein in the ER by the host signalase by cleavages at capsid-prM site at its N-terminus and prM-E site at its carboxy terminus. The association of prM with E produces non-infectious, immature virus particles. The arrangement of prM in the prM-E heterodimers in the immature particles protects the fusion loop of E protein from premature fusion. The “immature” particles transit through a low pH environment of the Golgi compartment, and a reversible conformational/morphological change occurs in E protein prior to processing of prM. Cleavage of prM to M by cellular serine protease, furin, in the trans-Golgi network results in an irreversible conformational change in E. The peptide cleaved off from prM (pr) is retained on the virion, and is released only after the virion has been secreted out and exposed to neutral pH, thus protecting the E protein from premature fusion (Li, L., et al., 2008; Yu, I.M., et al., 2008). The 53 kDa E protein in its mature dimeric form is the major surface component of the virion. In this form, the E protein is competent for cell surface attachment, fusion and virus entry into host cells as described by Perera et al. (2008).
Virus entry followed by endosomal acidification induces structural changes, in which E rearranges from 90 homodimers in neutral pH to 60 homotrimers in acidic pH (Allison et al., 1995; Stiasny et al., 2004; Stiasny and Heinz, 2006). The fusion loop on the DII domain that was buried in the DI/DIII pocket is now exposed in the fusogenic state of E trimer prior to its insertion into the host cell membrane,
In the drug discovery point of view, development of a robust high-throughput assay based on protein-protein interactions would be very useful to screen conformational transitions of prM and E. Perera et al. (2008) review recent advances in understanding the dynamics of the flavivirus E protein and suggest three regions within the protein that could be targeted by antivirals: the β-OG ligand binding pocket, E-protein rafts in the mature virus and E homotrimers. The availability of structural information from immature and mature particles is thus paving the way for rational drug design.
B. Non-structural proteins as drug targets
1. Protease
Within the N-terminal 180 amino acid residues of NS3 protein, was identified to include a trypsin-like serine protease domain (the catalytic triad residues being His51-Asp75-Ser135) by sequence comparison (Bazan and Fletterick, 1989, Gorbalenya et al., 1989). Using in vitro transcription and translation of precursor protein containing the NS2B-NS3 protease domain as well as polyprotein precursors expressed from recombinant vaccinia virus vectors established that for cleavages of protease sensitive sites, in cis and trans, NS2A-NS2B, NS2B-NS3 (a cis-cleavage), NS3-NS4A, NS4B-NS5, are performed by a heterodimeric complex of NS2B and NS3 (Chambers et al., 1990; 1991; 1993; Preugschat et al., 1990; Wengler et., 1991; Falgout et al., 1991; 1993; Zhang, L., et al., 1992; Clum et al., 1997). The viral protease has a preference for two basic amino acid residues (Arg-Arg, Arg-Lys, Lys Arg or occasionally Gln-Arg) at the P2 and P1 positions preceding the cleavage sites, followed by Gly, Ala or Ser at the P1′ position (Chambers et al., 1990). Since this polyprotein processing is a prerequisite for assembly of viral replicase complex, the viral protease represents an attractive therapeutic target.
NS2B is an integral membrane protein (Clum et al., 1997). NS2B and NS3 were colocalized within distinct paracrystalline or convoluted membranes structures suggesting that these may be the sites of polyprotein processing (Westaway et al., 1997). The hydrophobic regions of NS2B are likely required for membrane association of the polyprotein precursor rendering the protease sensitive sites in the optimal context for cis- and trans-cleavages. An in vitro protease assay using a DENV2 NS2B-NS3 protease precursor demonstrated that cotranslational insertion of NS2B into exogenously added canine pancreatic microsomal membranes is required for efficient cis cleavage of 2B-3 site in vitro. NS2B contains 3 hydrophobic domains flanking a conserved hydrophilic region of ~45 amino acid residues. Deletion of the three hydrophobic regions abrogated the membrane requirement for the NS2B-NS3 cleavage in vitro suggesting that for the cis cleavage of the NS2B-NS3 site in vitro, the hydrophobic regions are dispensable (Clum et al., 1997). A precursor devoid of the hydrophobic regions but containing the conserved hydrophilic domain linked to the NS3 protease domain through a carboxy terminal region of NS2B containing the NS2B-NS3 cleavage site was expressed in E. coli. The precursor, expressed as insoluble inclusion bodies, was purified by denaturation and refolding. The purified protein was active in cleaving the [35S]methionine-labeled NS4B-NS5N-ter precursor as well as a fluorogenic peptide substrate, t-butyl-oxycarbonyl(Boc)-Gly-Arg-Arg-7-amino-4-methyl coumarin, AMC (Yusof et al., 2000).
The kinetic parameters and substrate specificity of DENV2 protease were reported (Khumthong et al., 2002, 2003; Chanprapaph et al., 2005). Recently, it was shown that shortening the linker to five amino acid residues from the C-terminal region of the NS2B hydrphilic domain yielded a soluble, noncovalently associated heterodimeric WNV protease which was active in cleaving a fluorogenic peptide substrate, Boc-Gly-Lys-Arg-AMC (Mueller et al., 2007). Leung et al. showed that the linker between the cofactor, NS2B hydrophilic region, and the NS3 protease domain (NS3-pro) could be substituted with G4-S-G4 linker and the precursor could be expressed in E. coli as a very active protease in a soluble, non-cleavable form, thus obviating the denaturation and refolding steps in the purification of the protease (Leung et al., 2001). Using this active non-cleavable form of DENV2 and WNV proteases, a number of groups reported the substrate specificity, kinetic parameters, and profiles of peptide-based viral protease inhibitors. A suitable enzymatic substrate was identified by functional profiling using tetra peptide and octapeptide libraries comprising ~13,000 substrates (Li, J., et al., 2005). Detailed specificity studies have led to the design of robust screening assays in high-throughput formats, employing both colorimetric and fluorescent readouts (Nall et al., 2004; Li, J., et al., 2005; Chappell et al., 2005; 2006; 2007; Gouvea et al., 2007; Knox et al., 2006; Lohr et al., 2007; Radichev et al., 2008; Shiryaev et al., 2006; Shiryaev et al., 2007a, b, c; Yin et al., 2006a,b)
While all these studies utilized a truncated NS3 containing NS3-pro domain alone, Bera et al. (2007) recently reported characterization of the biochemical properties of WNV protease in which the NS2B hydrophilic domain and the full length NS3 were connected by the non-cleavable Gly-rich linker, described above. These authors identified two autolytic and intramolecular cleavage sites; the one at the NS2B/NS3 site was rapid and occurred during protein purification, whereas the cleavage at the second site, Arg459↓Gly460, located within the C-terminal RNA helicase region, was slower. These studies, taken together, revealed that the protease activity of NS2B-NS3 does not require cleavage of NS2B/NS3 site or free N-terminus of NS3 protease domain (Leung et al., 2001; Bera et al., 2007).
The crystal structure of the non-cleavable form of the WNV protease with a Gly-rich linker between the ~47 amino acid residue NS2B hydrophilic domain and the WNV NS3 protease domain was solved (Erbel et al., 2006); subsequently, a second crystal structure of WNV protease was reported in which the bovine pancreatic trypsin inhibitor replaced the tetrapeptide substrate-based inhibitor used in solving the first structure (Aleshin et al., 2007). In the presence of the cofactor, the WNV protease domain is complexed with a peptide inhibitor, revealing a direct interaction of NS2B with the active site of NS3pro. The C-terminus of the WNV NS2B wraps around NS3pro, and is involved in the formation of the S2 and S3 pockets (Erbel et al., 2006). The interaction of the protease with the inhibitor results in an induced fit conformation of the active site. In contrast, the DEN2 NS2B does not interact with the substrate binding site in the absence of the inhibitor as suggested from the crystal structure of the noncleavable form of DENV2 NS2B cofactor peptide linked to the NS3pro through the Gly-rich linker (Erbel et al., 2006). One of the challenges foreseen in developing an active-site serine-protease inhibitors is to identify compounds that block the viral enzyme, but not host serine proteases.
In a recent study, the previously described in vitro protease assays (Yusof et al., 2000, Mueller et al., 2007) were adapted to a high-throughput format using WNV (EG101 strain) protease expressed and purified from E. coli. From a screen of ~32,000 compounds against WNV protease, 212 compounds were identified to inhibit the protease activity using the substrate Boc-Gly-Lys-Arg-AMC in the 50% to 86% range. Other selection criteria were applied such as (i) being active at ≤ 50 μM, (ii) molecular weight < 500 daltons, (iii) calculated value of the logarithm of octanol-water partition coefficient (C lop P) < 5, (iv) the sum of nitrogen and oxygen atoms (H-bond acceptors) < 10, (v) < 5 H-bond donor atoms, (vi) < 10 rotatable bonds and finally those containing peptide-like (e.g. N-H-C=O) bonds that would mimic protease substrate. After applying these criteria, 98 compounds were selected which were clustered into three core structures and five groups based on their relatedness of chemical structures. Among the limited number of compounds selected for further characterization, the core structure 1 containing the 8-OH quinoline (R1) derivatives with different R2 and R3 substitutions were found to be the most active. The Ki values of compounds A and B for the WNV protease were 3.2 ± 0.3 μM and 3.4 ± 0.6 μM, respectively, whereas they were 10-fold less effective in the assays performed with the DENV2 protease and its preferred substrate, Boc-Gly-Arg-Arg-AMC (28.6 ± 5.1 μM and 30.2 ± 8.6 μM, respectively). Compound A was cytotoxic to Vero cells over a wider range of drug concentrations tested, whereas compound B had a moderate cytotoxicity (CC50 =140 ± 1.98 ± μM
Compound B was assayed for its inhibition of WNV replication in Vero cells infected with virus-like particles containing WNV replicon that encodes Renilla luciferase reporter (Pierson et al., 2006). In this system, the WNV replicon RNA encoding the reporter is delivered into the cytoplasm by infection of cells with virus-like particles, which then initiates a cascade of events such as translation, polyprotein processing, assembly of viral replicase complex and viral RNA replication. This system can be used to assay the effects of inhibitors of any of these steps. Since an inhibitor of the viral protease is expected to interfere with polyprotein processing, an early step affecting all subsequent steps, its potency could be assessed precisely by monitoring the reporter gene expression as a function of inhibitor concentration. Using this system, compound B was found to inhibit WNV replication with an EC50 value of 1.4 ± 0.3 μM and the selectivity index of ~ 100. The kinetic analysis and molecular docking studies indicated that compound B binds near the substrate binding site and thus inhibits the activity of the enzyme. Further work such as crystal structure of the protease in a complex with the inhibitor is necessary to better understand the mode of inhibition by this class of compounds, optimize and identify more potent inhibitors (Mueller et al., 2008 Mueller et al., in press). Recently, using a HTS approach, a different class of inhibitor compounds was identified from the National Institutes of Health’s compound library (Johnston et al., 2007). The kinetic analysis of these “hits” showed that these are uncompetitive inhibitors of WNV NS2B-NS3pro. These authors concluded that the identified compounds seem to interfere with the functional interaction between the cofactor NS2B and NS3pro domains (Johnston et al., 2007).
2. NS3 helicase
As described by Lescar et al. (2008), the flaviviral RNA helicase domain is located following the NS3pro domain within the C-terminal three-fouth of NS3. It contains 7 conserved motifs associated with the Super-Family 2 (SF2) class of NTPases and RNA helicases. NS3 helicase activity is thought to be required for melting secondary structures prior to initiation of RNA synthesis or for resolving RNA duplexes, either to separate dsRNA intermediates formed during viral RNA synthesis or as a translocase that can remove proteins bound to viral RNA. Strand separation is an energy-dependent reaction driven by ATP hydrolysis. Therefore, all RNA helicases have ATPase activity; this activity of flaviviral RNA helicases can hydrolyze non-specifically any nucleoside triphosphate (hence known as “NTPase”) and purines are preferred over pyrimidine nucleoside triphosphates. This activity is stimulated by the addition of single-stranded polyribonucleotides (Li et al., 1999; Suzich et al., 1993; Tamura et al., 1993; Warrener et al., 1993).
A conserved, positively charged motif (RKRK in DENV2) modulates the RNA-stimulated NTPase, RNA helicase and 5′ RNA triphosphatase activities of NS3. The mutagenesis of the four basic residues abolished the RNA-stimulated NTPase activity although the basal NTPase activity of NS3 was still retained. Binding of NS3 to single-stranded RNA was also abolished as well as other RNA binding dependent activities such as 5′ RNA triphosphatase and RNA helicase are affected by the mutagenesis of basic amino acid residues (Li, H., et. al. 1999; Yon et al., 2005). Mutations of critical residues that abolish helicase activity prevent viral replication (Matusan et al., 2001b). Drugs that target unwinding activity could act in 3 ways: inhibiting ATPase activity by interfering with ATP binding or hydrolysis; by preventing nucleic acid binding; or blocking unwinding by sterically hindering the translocation of helicase.
The flaviviral helicase has been a more challenging target for drug development than other nonstructural proteins, mainly because its mechanisms of action are not well understood. There is also a problem with selectivity for compounds that inhibit via the ATP binding site and these compounds are likely to be cytotoxic to the host. At the technical level, traditional assays for screening helicases are very time consuming. New assays using DNA substrates have recently been developed in a high-throughput format (Frick, 2003) but few options are available for assays using RNA as a substrate. The crystal structure of the helicase domain has been reported for DEN, YF and JE viruses (Wu, 2005 et al;Xu, 2006 et al; Yamashita, 2008). The RNA binding tunnel is located at the center, surrounded by residues emanating from the three domains. More recently, the structures of the full-length NS3 protein with the protease and helicase domains have been reported for Kunjin based on SAX analysis (Mastrangelo et al., 2007) and DEN4 virus (Luo et al., 2008), the latter being the first reported 3D structure for a flavivirus full length NS3 protein.
Several compounds have been identified as RNA helicase of inhibitors of flavi- and other RNA viruses (Borowski et al., 2007; Carta et al., 2006; Frick, 2007; Johansson et al., 2003; Maga et al., 2005; Wu et al., 2006; Xi, 2007). Borowski et al., (2002) reported a WNV helicase inhibitor which also was active in cellular assays (IC50 = 25–30 μM). Halogenated benztrioles have also been shown to inhibit WNV helicase (Borowski et al., 2003). More recently, ring-expanded nucleosides (RENs) have also been reported to inhibit JEV and WNV helicases (Zhang, N., et al., 2003a,b). While most reported inhibitors target the NTPase activity of NS3, mutation studies by Sampath et al. (2006) showed residual helicase activity for ATPase mutants, leading to the question whether targeting allosteric sites, such as the pockets lining the RNA binding tunnel, would be a better alternative.
3. NS5 methyltransferase (MTase)
As described by Dong et al. (2008), the flaviviral NS5 MTase is a very recently studied enzyme that appears to be an attractive drug target. Four enzyme activities are required for 5′-capping of nascent RNA. The NS5 MTase functional domain located in the N-terminal region of the protein (~33-kDa) methylates the N-7 position of the 5′ guanine cap as well as at the ribose 2′-OH position of the first transcribed nucleotide, an adenine (m7GpppAm) (Ray et al., 2006; Egloff et al., 2002). Mutation of critical residues in both methylation activities impair viral replication, indicating that the enzyme plays an essential role in the viral replicative cycle (Ray et al., 2006; Zhou et al., 2007).
Interestingly, the N-7 methylation of guanine cap by WNV NS5 is specific for viral RNA sequence, recognizing distinct elements within the 5′-stem loop of the viral RNA (Dong et al., 2007; Ray et al., 2006). The active site for 2′-OH methylation has been mapped to a tetrad K61-D146-K182-E218 (Ray et al., 2006) but has not been determined for N-7 methylation, although D146 was shown to be essential for this activity. Zhang et al. recently analyzed four variants of D146 mutation, D146L, D146P, D146R, and D146S, for viability in mutant RNA-transfected cells. Genome sequencing of recovered virion RNAs revealed that two classes of adaptive mutations, the first within the 5′ SL (G35U or U38 insertion) and the second within MTase (K61Q) and the polymerase (W751R) domains of NS5 (Zhang, B. et. al., 2008). These results supported the conclusion of these authors that there is a genetic interaction among the MTase, the polymerase and the 5′ SL during replication and that these adaptive mutations conferred survival fitness to D146S mutation which alone confers a lethal phenotype. The crystal structures of WNV and DEN2 MTases have been reported (Egloff et al., 2002, 2007; Zhou et al., 2007). Based on that structural information, Luzhkov et al (2007) reported a high-throughput structure-based virtual screening for 2′-O- methylation inhibitors which resulted in identification of a novel inhibitor. This is the first step in the development of MTase inhibitors, but much work remains to be done before these compounds can be developed as drugs.
4. NS5 RNA-dependent RNA polymerase (RdRp)
Because human cells lack RNA-dependent DNA or RNA polymerases such as the HIV-1 reverse transcriptase or RdRp of flaviviruses, this class of enzymes appears to be one of the most promising targets for antivirals against viruses that utilize polymerases for replication. Therefore, it is not surprising that of the 30 compounds that are currently marketed in the U.S. for treatment of viral infections, 15 of them are polymerase inhibitors, especially nucleoside analogs. the viral polymerase is one of the two favorite targets for HCV drug therapy (Jensen and Ascione, 2008; Liu-Young and Kozal, 2008).
Viral polymerase activity can be targeted using either nucleoside analogs or non-nucleoside compounds, the latter targeting allosteric sites in the protein. Nucleoside analogs must be phosphorylated to 5′-triphosphate where the pro-drug is converted to the active form in order to inhibit the enzyme at the active site. One argument that works in favor of active site inhibitors is that during therapy, chances of development of resistant mutants are lower as compared to an allosteric inhibitor.
Inhibitors of flaviviral RNA replication have been characterized using subgenomic RNA replicon systems (Barklis et al., 2007; Ng et al., 2007; Ray and Shi, 2006) and infectivity assays (Kajaste-Rudnitski et al., 2006; Leyssen et al., 2006; Michaelis et al., 2007; Migliaccio et al., 2003; Paeshuyse et al., 2006; Sidwell et al., 2007; Takhampunya et al., 2006). A potentially useful approach to determine the effect of antiviral compounds is the in vitro RdRP assay which employ lysates from flavivirus-infected and inhibitor-treated cells containing endogenous replicative viral RNAs associated with viral replicase components (Chu and Westaway., 1987; Bartholomeusz and Wright, 1993). This system has not been fully exploited although the feasibility has been demonstrated (Takhampunya et al., 2006). A second in vitro RdRP assay system relies on exogenously added subgenomic RNA templates containing the 5′- and 3′-terminal regions including the cyclization sequences (5′- and 3′-CS1 RNA elements) and infected cell lysates (You and Padmanabhan, 1999; You et. al., 2001) or purified NS5 protein (Tan et al.,1996; Ackermann and Padmanabhan, 2001; Guyatt et al., 2001; Nomaguchi et. al., 2003; 2004; Yu, L. et. al. 2008). The flavivirus NS5 RdRp is capable of de novo RNA synthesis (Ackermann and Padmanabhan, 2001; Selisko et al., 2006). Yap et al., (2007) recently reported a robust miniaturized assay for high-throughput compound screening. These in vitro RdRP assay systems are likely to be useful in studying the mode of action of a nucleoside analog in the triphosphorylated form in de novo initiation, elongation, processivity, or chain termination.
The review by Malet et al. (2008) discusses the drug discovery efforts focused on RdRp with an emphasis on how the structural information can be used for effective drug design. The crystal structures of NS5 of both WNV and DENV3 have recently been reported (Malet et al., 2007; Yap et al., 2007), including a model for the initiation complex and 3′ dGTP binding site. The RdRp of both viruses shows a typical right-hand structure with finger, palm and thumb domains. The region of NS5 that contains a nuclear localization sequence for transporting DENV2 NS5 into the nucleus was also shown to be involved in interaction with NS3 (Johannson et al., 2001). Targeting this interaction between NS3 and NS5 in the context of viral RNA is an option that could be explored. An in vitro assay that incorporates both of these proteins with viral polymerase activity as the readout would be very useful in identifying key replisome inhibitors.
VI. Host proteins as targets of antiviral therapeutics
In any antiviral drug discovery program, it is prudent to consider host cell components as potential targets because virus-host interactions are not only important for viral life cycle but also for pathogenesis. Flaviviruses like other viral pathogens gain entry into the host through specific interaction between a viral protein and host cell receptor(s) and strategy to usurp this interaction could lead to discovery of candidate antiviral drugs. This topic is discussed elegantly by Perera et al. (2008) (in this volume).
There are a number of other cellular proteins reported to bind to the viral RNA and in some cases, the disruption of their binding seem to affect viral replication (for a review, see (Brinton, 2001)). For example, EF-1α, eukaryotic translation elongation factor, binds to the 3′ SL of WNV RNA and DENV4 RNA (Blackwell and Brinton, 1997; De Nova-Ocampo et al., 2002). Mutations that interfere with this binding affect minus strand RNA synthesis without having any effect on translation of viral RNA ((Davis et al., 2007) and the references therein). In addition to eEF-1a, the autoantigen, La protein, and polypyrimidine tract binding protein, PTB, also have been reported to bind to the 3′SL of DENV4 RNA (De Nova-Ocampo et al., 2002; Garcia-Montalvo et al., 2004; Yocupicio-Monroy et al., 2007). Moreover, two members of RNA recognition motif family of RNA binding proteins, T cell intracellular antigen-1 (TIA-1) and TIA-related (TIAR) protein, were also shown to bind to WNV 3′ SL of minus strand RNA with high affinity (Emara and Brinton, 2007; Li et al., 2002). However, the function of these cellular proteins binding to viral RNA in virus life cycle has not yet been elucidated. Thus, the identification and functional characterization of cellular protein in flavivirus life cycle are at their infancy.
VII. Goals for the flavivirus drug discovery effort
This review and the accompanying papers have demonstrated that there are many potential molecular targets for antiviral therapeutics against flaviviruses. However, based on past experience with the development of drugs that are now in clinical use, the RdRP may prove to be the target with the best chance of success. The enzyme forms part of a multimeric complex that plays a crucial role in viral genome replication, but because it is encoded by the viral genome, and has no cellar counterpart, there are no toxicity issues (Malet et al., 2008). Nucleoside analogues which mimic natural NTP substrates could be potent inhibitors, especially if they are mistakenly incorporated into newly synthesized progeny RNAs and if they could potentially be involved in alternate base-pair interactions during replication, yielding lethal mutations into the genome RNA. This strategy has been amply described by others in the literature and is an attractive approach for drug development against RNA viruses that employ RdRP for their replication (Cameron and Castro, 2001; Crotty and Andino, 2002; Crotty et al., 2002; Crotty et al., 2001; Graci et al., 2007; Pariente et al., 2003)
The second attractive target for drug development is the viral protease. The anti-HIV protease inhibitors are examples of this class of drugs that are already in clinical use as a component of highly active antiretroviral therapy (HAART). The flavivirus protease might also prove to be a useful target for drug therapy, although it remains to be seen whether these viral serine proteases are sufficiently different from cellular serine proteases such as furin (which also recognizes two basic amino acid residues at P1 and P2 positions) that the anti-flaviviral protease inhibitors would not pose toxicity issues.
Virus entry inhibitors also offer an attractive choice for antiviral therapeutics. In fact, this class of inhibitors overcomes some of the problems associated with classical inhibitors of proteases and DNA or RNA polymerases such as cellular toxicity and emergence of drug resistance. Virus entry inhibitors have been developed and approved as antivirals for treatment of HIV-1 infections (Bridges, 2003; Cammack, 2001; Citterio and Rusconi, 2007; Daelemans et al., 2007; DeMarco et al., 2006; Kazmierski et al., 2006; Munch et al., 2007; Rusconi et al., 2007).
RNA viruses including flaviviruses that replicate in the cytoplasm have evolved their own 5′-capping machinery and are not dependent on the host enzymes which function in the nucleus. Because 5′-capping is required for viral replication (Dong et al., 2008), the enzymes which perform this function are potential targets for drug development. An example of this class are the inhibitors of 5′-capping of respiratory syncytial viral mRNAs by viral RNA dependent RNA polymerase (Liuzzi et al., 2005). Bray et al. (Bray et al., 2000) showed that mice infected with Ebola virus and treated with S-adenosyl-L-homocysteine hydrolase inhibitor were protected from lethality by inhibiting the methylation of 5′-cap. The NS3 RNA helicase of flaviviruses is also a novel target, as this enzyme in a multimeric complex with NS5 is required for viral replication, presumably because it is needed to unwind the double-stranded RNA intermediate during genome replication (described in detail by Lescar et al., 2008).
This collection of papers in Antiviral Research has demonstrated the existence of a range of molecular targets for new antiviral drugs against flaviviruses. The availability of structural information for a number of essential viral proteins and tools for screening the efficacy of inhibitors have provided new opportunities for rational drug design. The development of drugs against multiple and novel targets is a promising approach that will at least partially circumvent the emergence of drug resistance. As new inhibitors are identified, it will be important to assess their physicochemical properties, since these are a determining factor for the pharmacokinetics of any new drug candidate. In the initial stages of lead finding, the choice of targets, the availability of structural information and the availability of assays to screen for activity will strongly influence the success of a lead candidate. As the members of the flavivirus family share similar replication strategies, there is the prospect of identifying broad-spectrum inhibitors with prophylactic or therapeutic activity against a number of different pathogens.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem. 2001;276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16:795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SL, Schalich J, Stiasny K, Mandl CW, Kunz C, Heinz FX. Oligomeric rearrangement of tick-borne encephalitis-virus envelope proteins induced by an acidic pH. J Virol. 1995;69:695–700. doi: 10.1128/jvi.69.2.695-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CF, Preugschat F, Strauss JH. Dengue-2 virus NS2b and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993;193:888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- Barba-Spaeth G, Longman RS, Albert ML, Rice CM. Live attenuated yellow fever 17D infects human DCs and allows for presentation of endogenous and recombinant T cell epitopes. J Expt Med. 2005;202:1179–1184. doi: 10.1084/jem.20051352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barklis E, Still A, Sabri MI, Hirsch AJ, Nikolich-Zugich J, Brien J, Dhenub TC, Scholz I, Alfadhli A. Sultam thiourea inhibition of West Nile virus. Antimicrob Agents Chemother. 2007;51:2642–5. doi: 10.1128/AAC.00007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomeusz AI, Wright PJ. Synthesis of dengue virus RNA in vitro - initiation and the involvement of proteins NS3 and NS5. Arch Virol. 1993;128:111–121. doi: 10.1007/BF01309792. [DOI] [PubMed] [Google Scholar]
- Bazan JF, Fletterick RJ. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989;171:637–639. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- Benarroch D, Selisko B, Locatelli GA, Maga G, Romette JL, Canard B. The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology. 2004;328:208–218. doi: 10.1016/j.virol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Bera AK, Kuhn RJ, Smith JL. Functional characterization of cis and trans activity of the Flavivirus NS2B-NS3 protease. J Biol Chem. 2007;282:12883–12892. doi: 10.1074/jbc.M611318200. [DOI] [PubMed] [Google Scholar]
- Borowski P, Lang M, Haag A, Schmitz H, Choe J, Chen HM, Hosmane RS. Characterization of imidazo 4,5-d pyridazine nucleosides as modulators of unwinding reaction mediated by West Nile virus nucleoside triphosphatase/helicase: Evidence for activity on the level of substrate and/or enzyme. Antimicrob Agents Chemother. 2002;46:1231–1239. doi: 10.1128/AAC.46.5.1231-1239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowski P, Deinert J, Schalinski S, Bretner M, Ginalski K, Kulikowski T, Shugar D. Halogenated benzimidazoles and benzotriazoles as inhibitors of the NTPase/helicase activities of hepatitis C and related viruses. Eur J Biochem. 2003;270:1645–1653. doi: 10.1046/j.1432-1033.2003.03540.x. [DOI] [PubMed] [Google Scholar]
- Borowski P, Lang M, Haag A, Baier A. Tropolone and its derivatives as inhibitors of the helicase activity of hepatitis C virus nucleotide triphosphatase/helicase. Antivir Chem Chemother. 2007;18:103–9. doi: 10.1177/095632020701800206. [DOI] [PubMed] [Google Scholar]
- Bray M, Driscoll J, Huggins JW. Treatment of lethal Ebola virus infection in mice with a single dose of an S-adenosyl-L-homocysteine hydrolase inhibitor. Antiviral Res. 2000;45:135–47. doi: 10.1016/s0166-3542(00)00066-8. [DOI] [PubMed] [Google Scholar]
- Bray M. Highly pathogenic RNA viral infections: challenges for antiviral research. Antiviral Res. 2008;78:1–8. doi: 10.1016/j.antiviral.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Bridges S. Basic science priorities for therapeutics research. Res Initiat Treat Action. 2003;8:25–7. [PubMed] [Google Scholar]
- Cameron CE, Castro C. The mechanism of action of ribavirin: lethal mutagenesis of RNA virus genomes mediated by the viral RNA-dependent RNA polymerase. Curr Opin Infect Dis. 2001;14:757–64. doi: 10.1097/00001432-200112000-00015. [DOI] [PubMed] [Google Scholar]
- Cammack N. The potential for HIV fusion inhibition. Curr Opin Infect Dis. 2001;14:13–6. doi: 10.1097/00001432-200102000-00003. [DOI] [PubMed] [Google Scholar]
- Carta A, Loriga G, Piras S, Paglietti G, Ferrone M, Fermeglia M, Pricl S, La Colla P, Secci B, Collu G, Loddo R. Synthesis and in vitro evaluation of the anti-viral activity of N-[4-(1H(2H)-benzotriazol-1(2)-yl)phenyl]alkylcarboxamides. Med Chem. 2006;2:577–89. doi: 10.2174/1573406410602060577. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, Weir RC, Grakoui A, McCourt DW, Bazan JF, Fletterick RJ, Rice CM. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A. 1990;87:8898–902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Grakoui A, Rice CM. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991;65:6042–50. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Nestorowicz A, Amberg SM, Rice CM. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993;67:6797–807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanprapaph S, Saparpakorn P, Sangma C, Niyomrattanakit P, Hannongbua S, Angsuthanasombat C, Katzenmeier G. Competitive inhibition of the dengue virus NS3 serine protease by synthetic peptides representing polyprotein cleavage sites. Biochem Biophys Res Commun. 2005;330:1237–1246. doi: 10.1016/j.bbrc.2005.03.107. [DOI] [PubMed] [Google Scholar]
- Charlier N, Leyssen P, De Clercq E, Neyts J. Rodent models for the study of therapy against flavivirus infections. Antiviral Res. 2004;63:67–77. doi: 10.1016/j.antiviral.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Chappell KJ, Nall TA, Stoermer MJ, Fang NX, Tyndall JD, Fairlie DP, Young PR. Site-directed mutagenesis and kinetic studies of the West Nile Virus NS3 protease identify key enzyme-substrate interactions. J Biol Chem. 2005;280:2896–903. doi: 10.1074/jbc.M409931200. [DOI] [PubMed] [Google Scholar]
- Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Insights to substrate binding and processing by West Nile Virus NS3 protease through combined modeling, protease mutagenesis, and kinetic studies. J Biol Chem. 2006;281:38448–58. doi: 10.1074/jbc.M607641200. [DOI] [PubMed] [Google Scholar]
- Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Generation and characterization of proteolytically active and highly stable truncated and full-length recombinant West Nile virus NS3. Protein Expr Purif. 2007;53:87–96. doi: 10.1016/j.pep.2006.10.022. [DOI] [PubMed] [Google Scholar]
- Chu PWG, Westaway EG. Characterization of Kunjin virus RNA-dependent RNA-polymerase -- reinitiation of synthesis in vitro. Virology. 1987;157:330–337. doi: 10.1016/0042-6822(87)90275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu PWG, Westaway EG. Molecular and ultrastructural analysis of heavy membrane-fractions associated with the replication of Kunjin virus RNA. Arch Virol. 1992;125:177–191. doi: 10.1007/BF01309636. [DOI] [PubMed] [Google Scholar]
- Citterio P, Rusconi S. Novel inhibitors of the early steps of the HIV-1 life cycle. Expert Opin Investig Drugs. 2007;16:11–23. doi: 10.1517/13543784.16.1.11. [DOI] [PubMed] [Google Scholar]
- Clum S, Ebner KE, Padmanabhan R. Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J Biol Chem. 1997;272:30715–30723. doi: 10.1074/jbc.272.49.30715. [DOI] [PubMed] [Google Scholar]
- Crotty S, Andino R. Implications of high RNA virus mutation rates: lethal mutagenesis and the antiviral drug ribavirin. Microbes Infect. 2002;4:1301–7. doi: 10.1016/s1286-4579(02)00008-4. [DOI] [PubMed] [Google Scholar]
- Crotty S, Cameron C, Andino R. Ribavirin’s antiviral mechanism of action: lethal mutagenesis? J Mol Med. 2002;80:86–95. doi: 10.1007/s00109-001-0308-0. [DOI] [PubMed] [Google Scholar]
- Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci U S A. 2001;98:6895–900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daelemans D, Lu R, De Clercq E, Engelman A. Characterization of a replication-competent, integrase-defective human immunodeficiency virus (HIV)/simian virus 40 chimera as a powerful tool for the discovery and validation of HIV integrase inhibitors. J Virol. 2007;81:4381–5. doi: 10.1128/JVI.02637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco SJ, Henze H, Lederer A, Moehle K, Mukherjee R, Romagnoli B, Robinson JA, Brianza F, Gombert FO, Lociuro S, Ludin C, Vrijbloed JW, Zumbrunn J, Obrecht JP, Obrecht D, Brondani V, Hamy F, Klimkait T. Discovery of novel, highly potent and selective beta-hairpin mimetic CXCR4 inhibitors with excellent anti-HIV activity and pharmacokinetic profiles. Bioorg Med Chem. 2006;14:8396–404. doi: 10.1016/j.bmc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Dong HP, Ray D, Ren SP, Zhang B, Puig-Basagoiti F, Takagi Y, Ho CK, Li HM, Shi PY. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J Virol. 2007;81:4412–4421. doi: 10.1128/JVI.02455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhang B, Shi P-Y. Flavivirus methyltransferase: a novel antiviral target. Antiviral Res. 2008 doi: 10.1016/j.antiviral.2008.05.003. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Decroly E, Malet H, Selisko B, Benarroch D, Ferron F, Canard B. Structural and functional analysis of methylation and 5′-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J Molec Biol. 2007;372:723–736. doi: 10.1016/j.jmb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nature Struct Molec Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- Falgout B, Pethel M, Zhang YM, Lai CJ. Both nonstructural proteins NS2b and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B, Miller RH, Lai CJ. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: Identification of a domain required for NS2B-NS3 protease activity. Journal of Virology. 1993;67:2034–2042. doi: 10.1128/jvi.67.4.2034-2042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN. Helicases as antiviral drug targets. Drug News & Perspectives. 2003;16:355–362. doi: 10.1358/dnp.2003.16.6.829307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN. The hepatitis C virus NS3 protein: a model RNA helicase and potential drug target. Curr Issues Mol Biol. 2007;9:1–20. [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Donchenko AP, Koonin EV, Blinov VM. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989;17:3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun JB, Brinton MA. Characterization of West Nile virus RNA-dependent RNA polymerase and cellular terminal adenylyl and uridylyl transferases in cell-free extracts. J Virol. 1986;60:1113–1124. doi: 10.1128/jvi.60.3.1113-1124.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea IE, Izidoro MA, Judice WA, Cezari MH, Caliendo G, Santagada V, dos Santos CN, Queiroz MH, Juliano MA, Young PR, Fairlie DP, Juliano L. Substrate specificity of recombinant dengue 2 virus NS2B-NS3 protease: influence of natural and unnatural basic amino acids on hydrolysis of synthetic fluorescent substrates. Arch Biochem Biophys. 2007;457:187–96. doi: 10.1016/j.abb.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Graci JD, Harki DA, Korneeva VS, Edathil JP, Too K, Franco D, Smidansky ED, Paul AV, Peterson BR, Brown DM, Loakes D, Cameron CE. Lethal mutagenesis of poliovirus mediated by a mutagenic pyrimidine analogue. J Virol. 2007;81:11256–66. doi: 10.1128/JVI.01028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun JB, Brinton MA. Characterization of West Nile virus RNA-dependent RNA polymerase and cellular terminal adenylyl and uridylyl transferases in cell-free extracts. J Virol. 1986;60:1103–1124. doi: 10.1128/jvi.60.3.1113-1124.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler D, Kuno G, Markoff L. Flaviviruses. In: Knipe DM, Howley PM, editors. Fields virology. 5. Vol. 1. Lippincott William & Wilkins; Philadelphia, PA: 2007. pp. 1153–1253. [Google Scholar]
- Guyatt KJ, Westaway EG, Khromykh AA. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J Virol Methods. 2001;92:37–44. doi: 10.1016/s0166-0934(00)00270-6. [DOI] [PubMed] [Google Scholar]
- Holbrook MR, Gowen BB. Animal models of highly pathogenic RNA viral infections: encephalitis viruses. Antiviral Res. 2008;78:69–78. doi: 10.1016/j.antiviral.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Jensen DM, Ascione A. Future directions in therapy for chronic hepatitis C. Antiviral Ther. 2008;13(Suppl 1):31–6. [PubMed] [Google Scholar]
- Johansson A, Poliakov A, Akerblom E, Wiklund K, Lindeberg G, Winiwarter S, Danielson UH, Samuelsson B, Hallberg A. Acyl sulfonamides as potent protease inhibitors of the hepatitis C virus full-Length NS3 (protease-helicase/NTPase): a comparative study of different C-terminals. Bioorg Med Chem. 2003;11:2551–68. doi: 10.1016/s0968-0896(03)00179-2. [DOI] [PubMed] [Google Scholar]
- Johansson M, Brooks AJ, Jans DA, Vasudevan SG. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-beta and the viral helicase, NS3. J Gen Virol. 2001;82:735–45. doi: 10.1099/0022-1317-82-4-735. [DOI] [PubMed] [Google Scholar]
- Johnston PA, Phillips J, Shun TY, Shinde S, Lazo JS, Huryn DM, Myers MC, Ratnikov BI, Smith JW, Su Y, Dahl R, Cosford NDP, Shiryaev SA, Strongin AY. HTS identifies novel and specific uncompetitive inhibitors of the two-component NS2B-NS3 proteinase of West Nile virus. Assay and Drug Development Technologies. 2007;5:737–750. doi: 10.1089/adt.2007.101. [DOI] [PubMed] [Google Scholar]
- Jones CT, Ma LX, Burgner JW, Groesch TD, Post CB, Kuhn RJ. Flavivirus capsid is a dimeric alpha-helical protein. J Virol. 2003;77:7143–7149. doi: 10.1128/JVI.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajaste-Rudnitski A, Mashimo T, Frenkiel MP, Guenet JL, Lucas M, Despres P. The 2′,5′-oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J Biol Chem. 2006;281:4624–37. doi: 10.1074/jbc.M508649200. [DOI] [PubMed] [Google Scholar]
- Kazmierski WM, Kenakin TP, Gudmundsson KS. Peptide, peptidomimetic and small-molecule drug discovery targeting HIV-1 host-cell attachment and entry through gp120, gp41, CCR5 and CXCR4. Chem Biol Drug Des. 2006;67:13–26. doi: 10.1111/j.1747-0285.2005.00319.x. [DOI] [PubMed] [Google Scholar]
- Khumthong R, Angsuthanasombat C, Panyim S, Katzenmeier G. In vitro determination of dengue virus type 2 NS2B-NS3 protease activity with fluorescent peptide substrates. J Biochem Mol Biol. 2002;35:206–212. doi: 10.5483/bmbrep.2002.35.2.206. [DOI] [PubMed] [Google Scholar]
- Khumthong R, Niyomrattanakit P, Chanprapaph S, Angsuthanasombat C, Panyim S, Katzenmeier G. Steady-state cleavage kinetics for dengue virus type 2 ns2b-ns3(pro) serine protease with synthetic peptides. Protein Pept Lett. 2003;10:19–26. doi: 10.2174/0929866033408228. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Westaway EG. RNA binding properties of core protein of the flavivirus Kunjin. Arch Virol. 1996;141:685–99. doi: 10.1007/BF01718326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiermayr S, Kofler RM, Mandl CW, Messner P, Heinz FX. Isolation of capsid protein dimers from the tick-borne encephalitis flavivirus and in vitro assembly of capsid-like particles. J Virol. 2004;78:8078–8084. doi: 10.1128/JVI.78.15.8078-8084.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JE, Ma NL, Yin Z, Patel SJ, Wang WL, Chan WL, Ranga Rao KR, Wang G, Ngew X, Patel V, Beer D, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of West Nile NS3 protease: SAR study of tetrapeptide aldehyde inhibitors. J Med Chem. 2006;49:6585–90. doi: 10.1021/jm0607606. [DOI] [PubMed] [Google Scholar]
- Krishnan MN, Sukumaran B, Pal U, Agaisse H, Murray JL, Hodge TW, Fikrig E. Rab 5 is required for the cellular entry of dengue and West Nile viruses. J Virol. 2007;81:4881–4885. doi: 10.1128/JVI.02210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescar J, Dahai L, Xu T, Sampath A, Lim SP, Canard B, Vasudevan SG. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from Dengue virus as a target. Antiviral Res. 2008 doi: 10.1016/j.antiviral.2008.07.001. IN PRESS. [DOI] [PubMed] [Google Scholar]
- Leung D, Schroder K, White H, Fang NX, Stoermer MJ, Abbenante G, Martin JL, Young PR, Fairlie DP. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J Biol Chem. 2001;276:45762–45771. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- Leung JY, Pijlman GP, Kondratieva N, Hyde J, Mackenzie JM, Khromykh AA. Role of nonstructural protein NS2A in flavivirus assembly. J Virol. 2008;82:4731–4741. doi: 10.1128/JVI.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyssen P, De Clercq E, Neyts J. The anti-yellow fever virus activity of ribavirin is independent of error-prone replication. Mol Pharmacol. 2006;69:1461–7. doi: 10.1124/mol.105.020057. [DOI] [PubMed] [Google Scholar]
- Leyssen P, De Clercq E, Neyts J. Molecular strategies to inhibit the replication of RNA viruses. Antiviral Res. 2008;78:9–25. doi: 10.1016/j.antiviral.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Clum SR, You S, Ebner KE, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol. 1999;73:3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lim SP, Beer D, Patel V, Wen DY, Tumanut C, Tully DC, Williams JA, Jiricek J, Priestle JP, Harris JL, Vasudevan SG. Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetrapeptide and octapeptide substrate libraries. J Biol Chem. 2005;280:28766–28774. doi: 10.1074/jbc.M500588200. [DOI] [PubMed] [Google Scholar]
- Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: the viruses and their replication. Lippincott William & Wilkins; Philadelphia, USA: 2007. [Google Scholar]
- Liu-Young G, Kozal MJ. Review: hepatitis C protease and polymerase inhibitors in development. AIDS Patient Care STDS. 2008;22(6):449–57. doi: 10.1089/apc.2007.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi M, Mason SW, Cartier M, Lawetz C, McCollum RS, Dansereau N, Bolger G, Lapeyre N, Gaudette Y, Lagace L, Massariol MJ, Do F, Whitehead P, Lamarre L, Scouten E, Bordeleau J, Landry S, Rancourt J, Fazal G, Simoneau B. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J Virol. 2005;79:13105–15. doi: 10.1128/JVI.79.20.13105-13115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr K, Knox JE, Phong WY, Ma NL, Yin Z, Sampath A, Patel SJ, Wang WL, Chan WL, Rao KR, Wang G, Vasudevan SG, Keller TH, Lim SP. Yellow fever virus NS3 protease: peptide-inhibition studies. J Gen Virol. 2007;88:2223–7. doi: 10.1099/vir.0.82735-0. [DOI] [PubMed] [Google Scholar]
- Lozach PY, Burleigh L, Staropoli I, Navarro-Sanchez E, Harriague J, Virelizier JL, Rey FA, Despres P, Arenzana-Seisdedos F, Amara A. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN)-mediated enhancement of dengue virus infection is independent of DC-SIGN internalization signals. J Biol Chem. 2005;280:23698–23708. doi: 10.1074/jbc.M504337200. [DOI] [PubMed] [Google Scholar]
- Luo D, Xu T, Hunke C, Gruber G, Vasudevan SG, Lescar J. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzhkov VB, Selisko B, Nordqvist A, Peyrane F, Decroly E, Alvarez K, Karlen A, Canard B, Aqvist J. Virtual screening and bioassay study of novel inhibitors for dengue virus mRNA cap (nucleoside-2′ O)-methyltransferase. Bioorganic & Medicinal Chemistry. 2007;15:7795–7802. doi: 10.1016/j.bmc.2007.08.049. [DOI] [PubMed] [Google Scholar]
- Ma LX, Jones CT, Groesch TD, Kuhn RJ, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proc Nat Acad Sci USA. 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- MacKenzie JM, Westaway EG. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J Virol. 2001;75:10787–10799. doi: 10.1128/JVI.75.22.10787-10799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- Mackenzie J. Wrapping things up about virus RNA replication. Traffic. 2005;6:967–977. doi: 10.1111/j.1600-0854.2005.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G, Gemma S, Fattorusso C, Locatelli GA, Butini S, Persico M, Kukreja G, Romano MP, Chiasserini L, Savini L, Novellino E, Nacci V, Spadari S, Campiani G. Specific targeting of hepatitis C virus NS3 RNA helicase. Discovery of the potent and selective competitive nucleotide-mimicking inhibitor QU663. Biochemistry. 2005;44:9637–44. doi: 10.1021/bi047437u. [DOI] [PubMed] [Google Scholar]
- Malet H, Egloff MP, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, Mackenzie JM, Khromykh AA, Davidson AD, Canard B. Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J Biol Chem. 2007;282:10678–10689. doi: 10.1074/jbc.M607273200. [DOI] [PubMed] [Google Scholar]
- Malet H, Massé N, Selisko B, Romette J-L, Alvarez K, Guillemot JC, Hughes H, Yap TH, Vasudevan S, Lescar J, Canard B. The flavivirus polymerase as a target for drug discovery. Antiviral Res. 2008 doi: 10.1016/j.antiviral.2008.06.007. IN PRESS. [DOI] [PubMed] [Google Scholar]
- Marianneau P, Steffan AM, Royer C, Drouet MT, Jaeck D, Kirn A, Deubel V. Infection of primary cultures of human Kupffer cells by dengue virus: No viral progeny synthesis, but cytokine production is evident. J Virol. 1999;73:5201–5206. doi: 10.1128/jvi.73.6.5201-5206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo E, Milani M, Bollati M, Selisko B, Peyrane F, Pandini V, Sorrentino G, Canard B, Konarev PV, Svergun DI, de Lamballerie X, Coutard B, Khromykh AA, Bolognesi M. Crystal structure and activity of Kunjin virus NS3 helicase; Protease and helicase domain assembly in the full length NS3 protein. J Molec Biol. 2007;372:444–455. doi: 10.1016/j.jmb.2007.06.055. [DOI] [PubMed] [Google Scholar]
- Matusan AE, Kelley PG, Pryor MJ, Whisstock JC, Davidson AD, Wright PJ. Mutagenesis of the dengue virus type 2 NS3 proteinase and the production of growth-restricted virus. J Gen Virol. 2001a;82:1647–1656. doi: 10.1099/0022-1317-82-7-1647. [DOI] [PubMed] [Google Scholar]
- Matusan AE, Pryor MJ, Davidson AD, Wright PJ. Mutagenesis of the Dengue virus type 2 NS3 protein within and outside helicase motifs: Effects on enzyme activity and virus replication. J Virol. 2001b;75:9633–9643. doi: 10.1128/JVI.75.20.9633-9643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis M, Kleinschmidt MC, Doerr HW, Cinatl J., Jr Minocycline inhibits West Nile virus replication and apoptosis in human neuronal cells. J Antimicrob Chemother. 2007;60(5):981–6. doi: 10.1093/jac/dkm307. [DOI] [PubMed] [Google Scholar]
- Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, Bartholomew L, Bosserman MR, Ceccacci A, Colwell LF, Cortese R, De Francesco R, Eldrup AB, Getty KL, Hou XS, LaFemina RL, Ludmerer SW, MacCoss M, McMasters DR, Stahlhut MW, Olsen DB, Hazuda DJ, Flores OA. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem. 2003;278:49164–70. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem. 2007;282:8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- Mueller NH, Yon C, Ganesh VK, Padmanabhan R. Characterization of the West Nile virus protease substrate specificity and inhibitors. Int J Biochem Cell Biol. 2007;39:606–614. doi: 10.1016/j.biocel.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Mueller NH, Pattabiraman N, Ansarah-Sobrinho C, Viswanathan P, Pierson TC, Padmanabhan R. Identification and biochemical characterization of small molecule inhibitors of West Nile Virus serine protease by a high throughput screen. Antimicrobial Agents and Chemotherapy. 2008 doi: 10.1128/AAC.01508-07. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- Munch J, Standker L, Adermann K, Schulz A, Schindler M, Chinnadurai R, Pohlmann S, Chaipan C, Biet T, Peters T, Meyer B, Wilhelm D, Lu H, Jing W, Jiang S, Forssmann WG, Kirchhoff F. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell. 2007;129:263–75. doi: 10.1016/j.cell.2007.02.042. [DOI] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005;79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muylaert IR, Galler R, Rice CM. Genetic analysis of the yellow fever virus NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J Virol. 1997;71:291–298. doi: 10.1128/jvi.71.1.291-298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nall TA, Chappell KJ, Stoermer MJ, Fang NX, Tyndall JDA, Young PR, Fairlie DP. Enzymatic characterization and homology model of a catalytically active recombinant West Nile virus NS3 protease. J Biol Chem. 2004;279:48535–48542. doi: 10.1074/jbc.M406810200. [DOI] [PubMed] [Google Scholar]
- Ng CY, Gu F, Phong WY, Chen YL, Lim SP, Davidson A, Vasudevan SG. Construction and characterization of a stable subgenomic dengue virus type 2 replicon system for antiviral compound and siRNA testing. Antiviral Res. 2007;76:222–31. doi: 10.1016/j.antiviral.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Nomaguchi M, Ackermann M, Yon C, You S, Padmanbhan R. De Novo Synthesis of Negative-Strand RNA by Dengue Virus RNA-Dependent RNA Polymerase In Vitro: Nucleotide, Primer, and Template Parameters. J Virol. 2003;77:8831–42. doi: 10.1128/JVI.77.16.8831-8842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomaguchi M, Teramoto T, Yu L, Markoff L, Padmanabhan R. Requirements for West Nile virus (−)- and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J Biol Chem. 2004;279:12141–51. doi: 10.1074/jbc.M310839200. [DOI] [PubMed] [Google Scholar]
- Paeshuyse J, Leyssen P, Mabery E, Boddeker N, Vrancken R, Froeyen M, Ansari IH, Dutartre H, Rozenski J, Gil LH, Letellier C, Lanford R, Canard B, Koenen F, Kerkhofs P, Donis RO, Herdewijn P, Watson J, De Clercq E, Puerstinger G, Neyts J. A novel, highly selective inhibitor of pestivirus replication that targets the viral RNA-dependent RNA polymerase. J Virol. 2006;80:149–60. doi: 10.1128/JVI.80.1.149-160.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariente N, Airaksinen A, Domingo E. Mutagenesis versus inhibition in the efficiency of extinction of foot-and-mouth disease virus. J Virol. 2003;77:7131–8. doi: 10.1128/JVI.77.12.7131-7138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patkar CG, Jones CT, Chang YH, Warrier R, Kuhn RJ. Functional requirements of the yellow fever virus capsid protein. J Virol. 2007;81:6471–6481. doi: 10.1128/JVI.02120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R, Khaliq M, Kuhn RJ. Closing the door on flaviviruses: Entry as a target for antiviral drug design. Antiviral Res. 2008 doi: 10.1016/j.antiviral.2008.05.004. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Sanchez MD, Puffer BA, Ahmed AA, Geiss BJ, Valentine LE, Altamura LA, Diamond MS, Doms RW. A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virology. 2006;346:53–65. doi: 10.1016/j.virol.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Preugschat F, Yao CW, Strauss JH. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990;64:4364–74. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radichev I, Shiryaev SA, Aleshin AE, Ratnikov BI, Smith JW, Liddington RC, Strongin AY. Structure-based mutagenesis identifies important novel determinants of the NS2B cofactor of the West Nile virus two-component NS2B-NS3 proteinase. J Gen Virol. 2008;89:636–41. doi: 10.1099/vir.0.83359-0. [DOI] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao YW, Dong HP, Deas TS, Zhou YS, Li HM, Shi PY. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Shi PY. Recent advances in flavivirus antiviral drug discovery and vaccine development. Recent Patents Anti-Infect. Drug Disc. 2006;1:45–55. doi: 10.2174/157489106775244055. [DOI] [PubMed] [Google Scholar]
- Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol. 2006;80:4623–4632. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi S, Scozzafava A, Mastrolorenzo A, Supuran CT. An update in the development of HIV entry inhibitors. Curr Top Med Chem. 2007;7:1273–89. doi: 10.2174/156802607781212239. [DOI] [PubMed] [Google Scholar]
- Sampath A, Xu T, Chao A, Luo DH, Lescar J, Vasudevan SG. Structure-based mutational analysis of the NS3 helicase from dengue virus. J Virol. 2006;80:6686–6690. doi: 10.1128/JVI.02215-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selisko B, Dutartre H, Guillemot JC, Debarnot C, Benarroch D, Khromykh A, Despres P, Egloff MP, Canard B. Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology. 2006;351:145–158. doi: 10.1016/j.virol.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Shiryaev SA, Aleshin AE, Ratnikov BI, Smith JW, Liddington RC, Strongin AY. Expression and purification of a two-component flaviviral proteinase resistant to autocleavage at the NS2B-NS3 junction region. Protein Expr Purif. 2007a;52:334–9. doi: 10.1016/j.pep.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev SA, Kozlov IA, Ratnikov BI, Smith JW, Lebl M, Strongin AY. Cleavage preference distinguishes the two-component NS2B-NS3 serine proteinases of Dengue and West Nile viruses. Biochem J. 2007b;401:743–52. doi: 10.1042/BJ20061136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev SA, Ratnikov BI, Aleshin AE, Kozlov IA, Nelson NA, Lebl M, Smith JW, Liddington RC, Strongin AY. Switching the substrate specificity of the two-component NS2B-NS3 flavivirus proteinase by structure-based mutagenesis. J Virol. 2007c;81:4501–9. doi: 10.1128/JVI.02719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev SA, Ratnikov BI, Chekanov AV, Sikora S, Rozanov DV, Godzik A, Wang J, Smith JW, Huang Z, Lindberg I, Samuel MA, Diamond MS, Strongin AY. Cleavage targets and the D-arginine-based inhibitors of the West Nile virus NS3 processing proteinase. Biochem J. 2006;393:503–11. doi: 10.1042/BJ20051374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidwell RW, Barnard DL, Day CW, Smee DF, Bailey KW, Wong MH, Morrey JD, Furuta Y. Efficacy of orally administered T-705 on lethal avian influenza A (H5N1) virus infections in mice. Antimicrob Agents Chemother. 2007;51:845–51. doi: 10.1128/AAC.01051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Bressanelli S, Lepault J, Rey FA, Heinz FX. Characterization of a membrane-associated trimeric low-pH-induced form of the class II viral fusion protein E from tick-borne encephalitis virus and its crystallization. J Virol. 2004;78:3178–3183. doi: 10.1128/JVI.78.6.3178-3183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Heinz FX. Flavivirus membrane fusion. J Gen Virol. 2006;87:2755–66. doi: 10.1099/vir.0.82210-0. [DOI] [PubMed] [Google Scholar]
- Suzich JA, Tamura JK, Palmer Hill F, Warrener P, Grakoui A, Rice CM, Feinstone SM, Collett MS. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–8. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takhampunya R, Ubol S, Houng HS, Cameron CE, Padmanabhan R. Inhibition of dengue virus replication by mycophenolic acid and ribavirin. J Gen Virol. 2006;87:1947–52. doi: 10.1099/vir.0.81655-0. [DOI] [PubMed] [Google Scholar]
- Tamura JK, Warrener P, Collett MS. RNA-stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology. 1993;193:1–10. doi: 10.1006/viro.1993.1097. [DOI] [PubMed] [Google Scholar]
- Tan BH, Fu JL, Sugrue RJ, Yap EH, Chan YC, Tan YH. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996;216:317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfherer C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL, Steinman RM, Schlesinger S, Marovich MA. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. 2003;197:823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil PD, Satchidanandam V. Architecture of the flaviviral replication complex -Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J Biol Chem. 2003;278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- Umareddy I, Chao A, Sampath A, Gu F, Vasudevan SG. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J Gen Virol. 2006;87:2605–2614. doi: 10.1099/vir.0.81844-0. [DOI] [PubMed] [Google Scholar]
- Wang SH, Syu WJ, Huang KJ, Lei HY, Yao CW, King CC, Hu ST. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J Gen Virol. 2002;83:3093–3102. doi: 10.1099/0022-1317-83-12-3093. [DOI] [PubMed] [Google Scholar]
- Warrener P, Tamura JK, Collett MS. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol. 1993;67:989–996. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G, Czaya G, Farber PM, Hegemann JH. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J Gen Virol. 1991;72:851–8. doi: 10.1099/0022-1317-72-4-851. [DOI] [PubMed] [Google Scholar]
- Westaway EG, MacKenzie JM, Kenney MT, Jones MK, Khromykh AA. Ultrastructure of Kunjin virus-infected cells: Colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway EG, Khromykh AA, Mackenzie JM. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology. 1999;258:108–117. doi: 10.1006/viro.1999.9683. [DOI] [PubMed] [Google Scholar]
- Wu JH, Bera AK, Kuhn RJ, Smith JL. Structure of the flavivirus helicase: Implications for catalytic activity, protein interactions, and proteolytic processing. J Virol. 2005;79:10268–10277. doi: 10.1128/JVI.79.16.10268-10277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YS, Lin WH, Hsu JT, Hsieh HP. Antiviral drug discovery against SARS-CoV. Curr Med Chem. 2006;13:2003–20. doi: 10.2174/092986706777584988. [DOI] [PubMed] [Google Scholar]
- Xi XG. Helicases as antiviral and anticancer drug targets. Curr Med Chem. 2007;14:883–915. doi: 10.2174/092986707780362998. [DOI] [PubMed] [Google Scholar]
- Xu T, Sampath A, Chao A, Wen DY, Nanao M, Chene P, Vasudevan SG, Lescar J. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 angstrom. J Virol. 2005;79:10278–10288. doi: 10.1128/JVI.79.16.10278-10288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Unno H, Mori Y, Tani H, Moriishi K, Takamizawa A, Agoh M, Tsukihara T, Matsuura Y. Crystal structure of the catalytic domain of Japanese encephalitis virus NS3 helicase/nucleoside triphosphatase at a resolution of 1.8 angstrom. Virology. 2008;373:426–436. doi: 10.1016/j.virol.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-Angstrom resolution. J Virol. 2007;81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch LE, Shresta S. Mouse models of dengue virus infection and disease. Antiviral Res. 2008 doi: 10.1016/j.antiviral.2008.06.010. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Patel SJ, Wang WL, Chan WL, Ranga Rao KR, Wang G, Ngew X, Patel V, Beer D, Knox JE, Ma NL, Ehrhardt C, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg Med Chem Lett. 2006a;16:40–3. doi: 10.1016/j.bmcl.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Yin Z, Patel SJ, Wang WL, Wang G, Chan WL, Rao KR, Alam J, Jeyaraj DA, Ngew X, Patel V, Beer D, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of Dengue virus NS3 protease. Part 1: Warhead. Bioorg Med Chem Lett. 2006b;16:36–9. doi: 10.1016/j.bmcl.2005.09.062. [DOI] [PubMed] [Google Scholar]
- Yon C, Teramoto T, Mueller N, Phelan J, Ganesh VK, Murthy KH, Padmanabhan R. Modulation of the nucleoside triphosphatase/RNA helicase and 5′-RNA triphosphatase activities of dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J Biol Chem. 2005;280:27412–27419. doi: 10.1074/jbc.M501393200. [DOI] [PubMed] [Google Scholar]
- You S, Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J Biol Chem. 1999;274:33714–22. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- You S, Falgout B, Markoff L, Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J Biol Chem. 2001;276:15581–91. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- Yu L, Nomaguchi M, Padmanabhan R, Markoff L. Specific requirements for elements of the 5′ and 3′ terminal regions in flavivirus RNA synthesis and viral replication. Virology. 2008;374:170–85. doi: 10.1016/j.virol.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem. 2000;275:9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- Zhang LW, Mohan PM, Padmanabhan R. Processing and localization of dengue virus type–2 polyprotein precursor NS3-NS4a-NS4b-NS5. J Virol. 1992;66:7549–7554. doi: 10.1128/jvi.66.12.7549-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Chen HM, Koch V, Schmitz H, Liao CL, Bretner M, Bhadti VS, Fattom AI, Naso RB, Hosmane RS, Borowski P. Ring-expanded (“fat”) nucleoside and nucleotide analogues exhibit potent in vitro activity against Flaviviridae NTPases/helicases, including those of the West Nile virus, hepatitis C virus, and Japanese encephalitis virus. J Med Chem. 2003a;46:4149–4164. doi: 10.1021/jm030842j. [DOI] [PubMed] [Google Scholar]
- Zhang N, Chen HM, Koch V, Schmitz H, Minczuk M, Stepien P, Fattom AI, Naso RB, Kalicharran K, Borowski P, Hosmane RS. Potent inhibition of NTPase/helicase of the West Nile virus by ring-expanded (“Fat”) nucleoside analogues. J Med Chem. 2003b;46:4776–4789. doi: 10.1021/jm030277k. [DOI] [PubMed] [Google Scholar]
- Zhou YS, Ray D, Zhao YW, Dong HP, Ren SP, Li Z, Guo Y, Bernard KA, Shi PY, Li HM. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Dong H, Zhou Y, Shi PY. Genetic interactions among the West Nile virus methyltransferase, the RNA-dependent RNA polymerase, and the 5′ stem-loop of genomic RNA. J Virol. 2008;82:7047–58. doi: 10.1128/JVI.00654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]