Abstract
This study was designed to provide a genome-wide analysis of the effects of luteinizing hormone (LH) versus steroid ablation/replacement on gene expression in the developed corpus luteum (CL) in primates during the menstrual cycle. On Days 9–11 of the luteal phase, female rhesus monkeys were left untreated (control) or received a GnRH antagonist Antide (A), A + LH, A + LH + the 3β-hydroxysteroid dehydrogenase inhibitor Trilostane (TRL) or A + LH + TRL + a progestin R5020. On Day 12 of the luteal phase, CL were removed and samples of RNA from individual CL were hybridized to Affymetrix™ rhesus macaque total genome microarrays. The greatest number of altered transcripts was associated with the ablation/replacement of LH, while steroid ablation/progestin replacement affected fewer transcripts. Replacement of LH during Antide treatment restored the expression of most transcripts to control levels. Validation of a subset of transcripts revealed that the expression patterns were similar between microarray and real-time PCR. Analyses of protein levels were subsequently determined for two transcripts. This is the first genome-wide analysis of LH and steroid regulation of gene transcription in the developed primate CL. Further analysis of novel transcripts identified in this data set can clarify the relative role for LH and steroids in CL maintenance and luteolysis.
Keywords: corpus luteum, luteinizing hormone, microarray, steroids, progesterone
Introduction
Corpora lutea (CL) appear very similar, in terms of structure, function and regulation, between old-world monkeys (rhesus macaques) and humans. Humans and macaques have similar length menstrual cycles (luteal phases of the cycles are approximately the same length), neither relies on a uterine signal to induce luteolysis in non-gravid cycles and both exhibit an absolute requirement for luteinizing hormone (LH) from the anterior pituitary to maintain luteal structure–function (Stouffer, 2006).
Treatment of rhesus macaques with an antagonist of gonadotrophin-releasing hormone (GnRH; Antide, to inhibit LH production) results in rapid destruction of developed CL [as evidenced by a dramatic drop in progesterone (P) secretion], but the functional lifespan of these CL can be rescued by pulsatile administration of exogenous LH (Duffy et al., 1999b). Primate CL are remarkably sensitive to local LH levels as macaque luteinized granulosa/luteal cells respond to acute LH stimulation by rapidly secreting P in vitro, and LH pulses are entrained to P pulses by mid-luteal phase in vivo (Molskness et al., 1991; Stouffer, 2003). It is generally accepted that LH exerts its effects on targets, i.e. luteal cells, via the LH receptor (LHR), a seven transmembrane domain G-protein-coupled receptor capable of activating adenylate cyclase in the ovary (Benyo and Zeleznik, 1997; Zeleznik and Somers, 1999; Priyanka and Medhamurthy, 2007), and in other cells/tissues (Ascoli et al., 2002). However, the exact mechanism(s) by which LH exerts its luteotropic effects on the primate CL have yet to be elucidated.
Local autocrine/paracrine factors produced in response to LH may also be important in controlling CL structure–function. For example, it is hypothesized that locally produced P is the primary luteotropin in the primate CL, and alone may be sufficient to sustain luteal function, i.e. the so-called ‘Rothchild hypothesis’ (Rothchild, 1981). Treatment of rhesus monkeys during the mid-luteal phase with Trilostane (TRL), an inhibitor of 3β-hydroxysteroid dehydrogenase (3β-HSD), results in onset of premature luteolysis within 4 days of treatment (Duffy et al., 1994). Luteal phase length is reduced by approximately half, whereas endogenous LH levels remain relatively unchanged following TRL administration. The so-called ‘genomic’ progesterone receptors (PGRs; ligand-activated transcription factors) are present in macaque luteal cells and co-localize with 3β-HSD, supporting the premise of the Rothchild hypothesis (Stouffer and Duffy, 1995). But, receptors for other sex steroids [e.g. estrogen receptor β (ESR2) and androgen receptor (AR)] as well as mRNAs for putative membrane-associated steroid receptors were also detected in primate CL (Duffy et al., 1999a, 2000; Engmann et al., 2006). Genomic steroid hormone receptors are ligand-activated transcription factors that bind to regulatory elements on DNA with the assistance of several co-factors to modify gene transcription (Lonard and O’Malley, 2005). Thus, the actions of LH on CL structure–function are likely a composite of direct LH receptor-mediated effects and indirect effects resulting from LH-regulated local factors, e.g. steroids including P.
To investigate both the direct and indirect (via local steroids including P) effects of LH on the primate CL during the menstrual cycle, an in vivo model was developed in which ablation and replacement of LH (Duffy et al., 1999b) and steroid ablation/progestin replacement could be performed in rhesus monkeys during the same experimental protocol (Young and Stouffer, 2004). Previous ablation/replacement experiments on the primate CL focused primarily on the effects of LH versus P on individual gene families, including proteases and caspases involved in tissue reorganization (Young et al., 2002, 2004; Young and Stouffer 2004; Peluffo et al., 2005).
Recently, attempts were made to broadly evaluate LH regulation of genes in the macaque CL (Yadav et al., 2004; Xu et al., 2005). Xu et al. (2005) either left rhesus monkeys untreated or administered a single bolus of Antide in the mid-luteal phase, the CL were removed and total cDNA hybridized to a limited (11 600 transcripts) human spotted array. The resulting data successfully identified a novel group of genes, including the corticotropin-releasing hormone (CRH)/urocortin (UCN)-receptor-binding protein system, that appear to be involved in the maintenance of CL structure/function (Xu et al., 2007). Yadav et al. (2004) used the GnRH antagonist Cetrorelix©, administered to bonnet monkeys in the mid-luteal phase, to identify changes in mRNA transcripts by differential display. Seven transcripts, including mRNA for low-density lipoprotein (LDL) receptor, were identified as changing in expression after LH withdrawal (Yadav et al., 2004).
While these approaches were successful in identifying novel gene regulation by LH in the CL, further opportunities now exist for global (genome-wide) evaluation of gene activities in macaque tissues (Noriega et al., 2007; Parent et al., 2008) including the rhesus CL (Bogan et al., 2008b) using the Affymetrix™ Rhesus Macaque Genome Expression Array. Building on these validated studies, we utilized the rhesus array to further identify changes in gene expression in the macaque CL in response to LH and steroid ablation/replacement as described earlier. It was expected that LH-dependent changes in gene expression (directly regulated by LH) could be contrasted and compared with changes in the expression of genes that were steroid (especially P) dependent (indirectly regulated by LH).
Materials and Methods
The studies were performed at the Oregon National Primate Research Center (ONPRC) on the West Campus of Oregon Health & Science University (OHSU). All procedures and protocols were approved by the ONPRC/OHSU Animal Care and Use Committee, in accordance with the National Institute of Health (NIH) Guidelines for Care and Use of Laboratory Animals. Adult, female rhesus macaques (Macaca mulatta) exhibiting regular menstrual cycles were selected for the study and were under the direct care of the ONPRC Division of Animal Resources (DAR).
Monkeys (n = 22) received one of the following treatments for 3 days beginning on Day 9 of the luteal phase (mid-luteal phase, the time of peak CL function). Some females were left untreated to establish gene/protein expression in normal CL (control group; n = 4). Alternatively, ablation of LH was accomplished by treating females with a GnRH antagonist [Antide obtained from the Contraceptive Development Branch, Center for Population Research, National Institute of Child Health and Human Development (NICHD), 3 mg/kg once per day, n = 5]. This treatment significantly lowers LH and P levels within 1 day, with baseline levels present by the end of the experimental period (Duffy et al., 1999b). Recombinant human LH (40 IU, three times daily) was also administered in a pulsatile manner (A + LH; n = 4), which returns systemic LH and P to control levels (Duffy et al., 1999b). In addition, steroid production by the CL in response to LH replacement was blocked by oral administration of the 3β-HSD inhibitor TRL (A + LH + TRL; 600 mg b.i.d, n = 4), which suppresses systemic P levels and induces premature luteolysis at this dosage and duration of treatment in rhesus monkeys (Duffy et al. 1994). Actions of P were restored in a subset of TRL-treated animals by treatment with a synthetic progestin, R5020 (A + LH + TRL + R5020; 2.5 ng/ml once per day, n = 5), at a dosage and duration that prevents the indices of luteolysis brought on by TRL treatment in macaque monkeys (Young and Stouffer, 2004). Following the 3-day treatment interval, CL were surgically removed and half of each tissue was individually processed for total RNA, and the other half for total protein as described previously (Peluffo et al., 2005). Anesthesia of animals and lutectomy were performed by the surgical veterinary staff (DAR, ONPRC) as described previously (Duffy et al., 2000).
Microarray analysis and real-time PCR validation
All rhesus macaque genome arrays were processed by the OHSU Affymetrix Microarray Core (AMC), OHSU. The AMC performed the RNA expression profiling, amplification and hybridization of samples (http://www.ohsu.edu/gmsr/amc/amc_technology.html#assay). Following data normalization using the robust multichip average algorithm, the web-based GeneSifter© software (http://www.genesifter.net/web/) was used to identify transcripts from log2-transformed data that significantly changed expression between treatment groups by pairwise comparison analysis (t-test, P < 0.05), and for the entire experimental group by multiple comparison analysis procedures (one-way ANOVA, P < 0.05; see http://www.genesifter.net/web/webinars.php for further explanation of analytical methods by GeneSifter©). The Affymetrix™ Rhesus Genome Array contains 52 024 rhesus probe sets for approximately 47 000 rhesus transcripts, plus several control probe sets; array design is based on the Baylor School of Medicine’s rhesus macaque whole-genome shotgun assembly and GenBank® sequence-tagged sites (STSs), expressed sequence tag and mRNA sequences (http://www.affymetrix.com/support/technical/datasheets/rhesus_datasheet.pdf). There are multiple probes sets (transcripts) for individual gene products on this array (Spindel et al., 2005). Therefore, microarray data will be referred to as ‘transcripts’, whereas real-time PCR data will be referred to as ‘mRNA’ values.
Data files were submitted to the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo: series GSE12 281). To validate the microarray results, selected genes were analyzed by real-time PCR to quantify changes in mRNA levels that occur following LH and/or P ablation/replacement. The real-time PCR probes were designed as described previously (Xu et al., 2006) and labeled with the 5′ reporter dye FAM and the 3′ quencher dye MGBNFQ. Sequences for real-time probes and primers are listed in Table I, along with gene identifiers. All real-time studies were performed and analyzed as reported previously (Xu et al., 2006). The data were subjected to one-way ANOVA by the use of SigmaStat© software (Version 2.0). When significant (P < 0.05), differences between treatment groups were analyzed by pairwise multiple comparison procedures (Tukey test or Student–Newman–Keuls method as appropriate).
Table I.
Real-time PCR primer and probe sequences used to validate microarray expression
| Genea | NCBI Accession Number | Forward Primer (5′–3′) | Probe (5′–3′) 6FAM-Sequence-MGBNFQ | Reverse Primer (5′–3′) |
|---|---|---|---|---|
| MHC1 | U84789 | CAGATAGATAAGGCGGGAGCTACT | CAGTGACAGTTCCC | GCCGTGAGAGACACATCAGAAC |
| SFRP4 | NM_001032962 | CTGCGAGCCCCTCATGAA | CACAGCTGGCCCGAA | GTCATAGACGGGCAGCTCATC |
| CCL3 | BV209119 | GGCCTGCTGTAGGCAGTCAT | CAGACTGACCAATGTGTATC | ACAGCCCTGACCAAAAGCAT |
| CAT | NM_001752 | TCCATTCGATCTCACCAAGGT | CAA GGACTACCCTCTCAT | CCGGTTTAAGACCAGTTTACCAA |
| StAR | BV208869 | GTGCGTGTGTACAAGGCAGAA | CGGCAGCCCTCTG | GGTGTTGCTGTGCAGTGAATG |
| ADAMTS-5 | XM_001103439 | TTCATGTTTCCCTCAGATAGTGATG | ACACATGTCATAGAAACAA | TGGTTTATTTGCTCTAAGCCCTCTA |
| MMP-9 | XM_001104871 | AGGCGCTCATGTACCCTATGTAC | CCCCCCTTGCATAAG | TGCCGGATGCCATTCAC |
| MMP-19 | XM_001111542 | TGGCCCCCAACTCCATT | TGTCTTAGACAGCCCTTC | ACCATTGACCTCCAGAAAAGAGAT |
| CASP2 | NM_032982 | GCACTGAGGGAGACCAAGCA | TCACCACCCTTTCCGGGCTTCAG | CACAGCTCAACGGTGGGAGTA |
| IL1RN | U65590 | TGGCACTTGGAGACGTGTATG | CCTCGGCCTGTCTC | CGAGTCTTTTTTCCTGCTCTGAA |
| AHRIP | NM_003977 | TCCAAAAACGTGTGATACAGGAA | CTCCCGGACTTTC | TAGTGGAACGTGGCCTTGGT |
| ARNT2 | BV208380 | CCAGTCCCAGGGAATGAATG | CTCCCGGCAGCACT | CACGTTCTCGCATAGCTTATGG |
| VEGF | M27281 X15997 | ACGAAGTGGTGAAGTTCATGGA | TCAGCGCAGCTACT | CTCATCAGGGTACTCCTGGAAGA |
| PRLR | NM_000949 | AGCTGAGTGGGAGACCCATTT | AAACAGAGTTTAAGATTCTC | GCGAACCTGGACAAGGTATTTC |
| CRHBP | NM_001882 | GACAGACCCCAACCTCTTTCC | CCCTGGTCGTTCCAC | TGGAGAAGCTGCAGTTTCGA |
| INHBA | BV166389 | AGAGCACCTGGCACGTCTTC | CAGCATCCAGCGGTT | ACGTCCAGGGAGCTTTTTCC |
| SC4MOL | AB169840 | TGGAGTGTAGCGGCACAATC | CGGCTCACTGCAAC | GGAGGCTGAGGCAGGAGAAT |
| LOC693394 | XM_001081982 | CACGTCATCGACTACATCTTGGA | CCTGGACTCGCATCC | TCTGGTGATGGAGGCTGACA |
| ESR2 | AB006589 | TCAGTTGAGGTCAGGAGTTTGAAA | CAACATGGCGAAACC | ACCACCATGCCTGGCTAATT |
| AR | NM_001032911 | GCAGGCAAGAGCACTGAAGATA | TGCTGAGTATTCCCC | CCTTTGGTGTAACCTCCCTTGA |
| PGR | XM_001095317 | CATGTCAGTGGGCAGATGCT | TATTTTGCACCTGATCTAA | ATTCTTTCATCCGCTGTTCATTC |
| PGRMC1 | XM_001100639 | CCAGGACCCCCGCATAC | CATGGCCATCAACG | TTTGGTCACGTCGAACACCTT |
aBased on NCBI Entrez Gene Terminology.
Western blot analysis
Immunoblot (western blot) analysis was performed to permit comparison of the patterns of protein levels of two selected gene products versus their mRNA levels in individual CL. Electrophoresis was performed using gradient gels (10–20% liner with 4% stacking gel; BioRad Laboratories, Hercules, CA, USA) with 20 µg of protein in denaturing SDS loading buffer (3% SDS, 7 mM EDTA, 7 mM EGTA, 7% glycerol, 0.36 M Tris, 3.6 mM DTT and 3.6% bromophenol blue) per individual sample.
After electrophoresis, proteins were transferred to Immun-Blot™ PVDF membranes (0.2 µM; BioRad) overnight at 4°C. Membranes were blocked with a neutral protein (3% NFM) at 4°C overnight and then incubated with the following primary antibodies according to the manufacturer’s protocols: mouse anti-human secreted frizzled related protein-4 (SFRP4) whole sera antibody (final dilution 1:1000; Abnova/Affinity Bioreagents H00006424-A01) or rabbit anti-human steroidogenic acute regulatory (StAR) protein (final concentration 1 µg/ml; Fisher/Affinity Bioreagents PA1-560). All horseradish peroxidase-labeled secondary antibodies were from the same source (Zymed Laboratories, Invitrogen Corp., Carlsbad, CA, USA), specific to the host animal of the primary antibody, and used at a final dilution of 1:2000.
For the quantification of protein levels from individual CL, a luminescent signal was generated using western blotting luminol reagent (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and detected with Kodak X-OMAT film (Eastman Kodak Co., Rochester, NY, USA). Densitometry analysis was performed using a gel documentation system and Quantity One software (Bio-Rad). To quantify protein levels, the blots were stripped by a gentle method (successive PBS and TBST washes, Abcam Inc., Cambridge, MA, USA) to remove all antibodies. This method was confirmed for each antibody by incubation of stripped blots with only the secondary antibody and then developed for any luminescent signal as described earlier. Blots were then blocked again overnight and re-probed for β-actin levels (Tesone et al., 2005) by incubation with the primary antibody rabbit anti-human β-actin (0.8 µg/ml; Abcam ab8227), and secondary antibody as before. All data were subject to one-way ANOVA (similar to real-time PCR data) and are presented as the intensity of protein of interest: β-actin intensity. Because of high background to signal ratio for SFRP4 blots containing +TRL and +R5020 samples, for both β-actin and SFRP4 samples, background intensity was subtracted from average intensity for all bands (regardless of treatment group) before normalization for SFRP4 expression.
For presentation of western blot data, samples from individual CL were pooled by treatment group, and then pooled samples underwent electrophoresis and transfer as stated earlier for the detection of each protein of interest (StAR, SFRP4 and β-actin). Following electrophoresis and protein transfer to PVDF membranes, StAR and β-actin blots were developed via the WesternDot™ 625 Western Blot Kit (Invitrogen, Molecular Probes) with the aforementioned primary antibodies according to the manufacturer’s protocol; SFRP4 was developed as described earlier.
Results
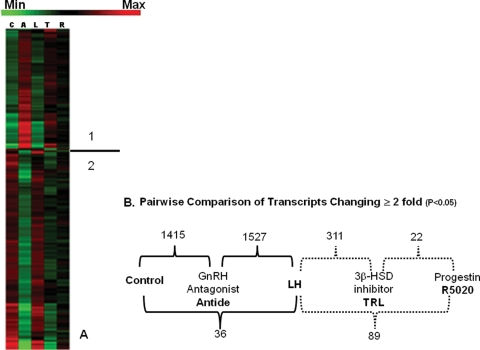
Normalized data files for individual arrays have been uploaded to the National Center for Biotechnology Information’s Gene Expression Omnibus gene expression repository (http://www.ncbi.nlm.nih.gov/geo/ Series GSE12281). Of the more than 47 000 transcripts represented on the rhesus genome arrays, the multiple comparison analysis of GeneSifter© identified ∼2% of all transcripts on the array as changing in expression ≥2-fold between various treatments (one-way ANOVA, P < 0.05); these were then analyzed by the cluster analysis algorithm of GeneSifter© (for further description of cluster analysis, see http://www.genesifter.net/web/resources_cluster.php, and http://www.statsoft.com/textbook/stcluan.html for a description of the cluster analyses performed by the GeneSifter© software). The results are presented as a heat map showing relative levels of expression in Fig. 1A. The heat map is divided into two regions: region 1 (above the line) shows transcripts whose levels increased (red) above controls following treatment with Antide, and region 2 depicts transcripts whose levels decreased (green) with Antide treatment. The majority of transcripts were in the latter region, i.e. levels decreased in expression following Antide treatment. Most transcripts in both regions were restored to near-control levels by LH replacement (see Table II for examples of transcripts not restored by LH replacement, which compares the transcripts that differ significantly between the control and the Antide+LH groups). Many, but not all, of the Antide-sensitive transcripts appear to be affected similarly (either decreased or increased) by steroid withdrawal (TRL), but several of these changes were not as pronounced. Treatment with R5020 reversed many of the effects of TRL, but not necessarily to the level observed with LH replacement.
Figure 1.
(A) Cluster analysis by multiple comparison procedure of GeneSifter© of all transcripts whose levels significantly (P < 0.05) changed in expression ≥2-fold when compared with control CL. The heat map indicates the relative levels of the expression of each transcript, with red indicating increased expression and green indicating decreased expression. The line separates the two regions of different transcript expression: 1: increasing expression with Antide treatment; 2: decreasing expression with Antide treatment. C, control; A, Antide; L, A + LH; T, A + LH + TRL; R, A + LH + TRL + R5020. (B) Summary of pairwise comparison analysis of the number of transcripts identified by GeneSifter© as significantly changing expression ≥2-fold (Student’s t-test, P < 0.05) between indicated treatment groups. Comparisons between complimentary ablation/replacement groups are indicated on the top, whereas pairwise comparisons between control and A + LH, or A + LH and A + LH + TRL + R5020 treatments are depicted below. For example, 1415 transcripts differ significantly between the control and Antide treatment groups, whereas only 36 differ significantly between the control and Antide + LH treatment groups. The number of transcripts whose expression differs between control, Antide and A + LH groups (black brackets) are greater than those between A + LH, A + LH + TRL and A + LH + TRL + R5020 groups (dashed brackets).
Table II.
Transcripts not restored to control levels by LH replacement of Antide-treated CL (≥2-fold differences P < 0.05)
| Affymetrix Gene Transcript | NCBI Accession No. | Antide Effecta | Ratiob |
|---|---|---|---|
| Mamu-DRB mRNA for major histocompatibility complex class II, partial cds, clone:R00205DR09 | AB112035 | Down | 4.7 |
| Pre-B-cell colony enhancing factor 1 | AA873350 | Down | 4.3 |
| Nuclear receptor subfamily 4, group A, member 1 | NM_002135 | Down | 4.3 |
| Prostaglandin F receptor (FP) | NM_000959 | Down | 4.3 |
| gDNA.Hs2.89418.2.S1 FEA = U133PSR GEN = PTGFR DEF = Orthologous to 1555097_a_at prostaglandin F receptor (FP) | BC035694 | Down | 4.2 |
| Pre-B-cell colony enhancing factor 1 | BF575514 | Down | 3.9 |
| Low density lipoprotein receptor (familial hypercholesterolemia) | AI861942 | Down | 3.6 |
| V-fos FBJ murine osteosarcoma viral oncogene homolog | BC004490 | Down | 3.4 |
| gi:47777222 DEF = TUBA1 _1214 Rhesus macaque genomic DNA Macaca mulatta STS genomic clone MMA1214, sequence tagged site. GEN = TUBA1 PROD=tubulin, alpha 1 | Down | 3.3 | |
| Low density lipoprotein receptor (familial hypercholesterolemia) | NM_000527 | Down | 3.3 |
| Hypothetical protein LOC129293 (LOC129293), mRNA | AA005361 | Down | 3.2 |
| gDNA.Hs.106635.0.S2 FEA = U133PSR GEN = PIPPIN DEF = Orthologous to 209981_at RNA-binding protein pippin | AL023553 | Down | 3.0 |
| Transcribed locus | AI248760 | Down | 2.9 |
| Pre-B-cell colony enhancing factor 1 | BC020691 | Down | 2.7 |
| Pre-B-cell colony enhancing factor 1 | NM_005746 | Down | 2.4 |
| Pre-B-cell colony enhancing factor 1 | BF575514 | Down | 2.4 |
| Endothelial cell-specific molecule 1 | NM_007036 | Down | 2.4 |
| Similar to nicotinamide phosphoribosyltransferase (NAmPRTase) (Nampt) (Pre-B-cell colony-enhancing factor 1 homolog) (PBEF) | CB229989 | Down | 2.4 |
| DIS3 mitotic control homolog (S. cerevisiae)-like 2 | AI935717 | Down | 2.4 |
| gi:47776579 DEF = EHD3_613 Rhesus macaque genomic DNA Macaca mulatta STS genomic clone MMA613, sequence tagged site. GEN = EHD3 PROD = EH-domain containing 3 | Down | 2.3 | |
| zinc finger, DHHC-type containing 19 | NM_144637 | Down | 2.2 |
| complement component 4B (Childo blood group) | NM_000592 | Down | 2.2 |
| Transcribed locus | AI873273 | Down | 2.2 |
| Aminolevulinate, delta-, synthase 1 | NM_000688 | Down | 2.2 |
| Transcribed locus, strongly similar to NP_057696.2 mitochondrial solute carrier protein [Homo sapiens] | H69701 | Down | 2.0 |
| Dual specificity phosphatase 1 | AA530892 | Down | 2.0 |
| Amine oxidase, copper containing 2 (retina-specific) | NM_009590 | Down | 2.0 |
| Similar to HLA class I histocompatibility antigen, A-74 alpha chain precursor (MHC class I antigen A*74) (Aw-74) (Aw-19) | AF157401 | Up | 8.4 |
| Neuronal pentraxin I | NM_002522 | Up | 4.1 |
| Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | NM_000602 | Up | 3.5 |
| gi:47776712 DEF = HP_143 Rhesus macaque genomic DNA Macaca mulatta STS genomic clone MMA143, sequence tagged site. GEN = HP PROD = haptoglobin | Up | 3.0 | |
| gi:47777186 DEF = THBS1_279 Rhesus macaque genomic DNA Macaca mulatta STS genomic clone MMA279, sequence tagged site. GEN = THBS1 PROD = thrombospondin 1 | Up | 2.6 | |
| Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | AL574210 | Up | 2.5 |
| ADP-ribosylation factor-like 4C | AW450363 | Up | 2.1 |
| ARTC1 mRNA, complete sequence | BF508813 | Up | 2.0 |
| Serine/threonine kinase 17a | AW194730 | Up | 2.0 |
aEffect of Antide, transcript not restored to near control levels by LH replacement.
bControl: Antide + LH.
These changes in transcript expression are depicted in Fig. 1B as a pairwise comparison of the number of transcripts differing between treatment groups. The largest number of differentially expressed transcripts was associated with LH ablation/replacement, while steroid ablation/replacement affected fewer transcripts (Fig. 1B). Hierarchical cluster analysis of all treatment groups by GeneSifter© indicated that the Control and A+LH groups were similar in gene expression, whereas the Antide group was most related to the A + LH + TRL and A + LH + TRL + R5020 groups. Because a relatively small number of transcripts differed ≥2-fold between the control and A + LH groups (n = 36, Fig. 1B), the A + LH group can be considered a separate ‘control’ for the steroid ablation/replacement groups for the purpose of further analysis. Pathways significantly [as determined by z-score (Doniger et al., 2003) and ontology abundance] affected by treatments (≥2-fold) are depicted by known gene ontologies in Table III. Many of these significant ontologies are associated with the production of steroids and steroid precursors (e.g. sterol biosynthetic processes and cholesterol biosynthetic processes), and a number are related to the immune system/response (immunoglobulin production, regulation of B cell activation, and others).
Table III.
Ontologies significantly impacted by LH and steroid ablation/replacementa
| Ontology | List | Array | z-score | %b |
|---|---|---|---|---|
| Immune system | ||||
| immunoglobulin production | 5 | 13 | 6.09 | 38 |
| regulation of B cell activation | 5 | 16 | 5.33 | 31 |
| production of molecular mediator of immune response | 5 | 22 | 4.27 | 23 |
| cellular defense response | 10 | 53 | 5.25 | 19 |
| B cell activation | 8 | 46 | 4.40 | 17 |
| response to inorganic substance | 5 | 29 | 3.45 | 17 |
| leukocyte differentiation | 9 | 71 | 3.50 | 13 |
| immune system development | 13 | 135 | 3.08 | 10 |
| negative regulation of immune system process | 6 | 27 | 4.61 | 22 |
| Steroid/cholesterol processes/metabolism | ||||
| sterol biosynthetic process | 11 | 30 | 8.77 | 37 |
| cholesterol biosynthetic process | 8 | 23 | 7.23 | 35 |
| steroid biosynthetic process | 17 | 62 | 9.02 | 27 |
| sterol metabolic process | 16 | 68 | 7.86 | 24 |
| cholesterol metabolic process | 13 | 61 | 6.58 | 21 |
| steroid metabolic process | 26 | 142 | 8.30 | 18 |
| lipid biosynthetic process | 32 | 222 | 7.53 | 14 |
| Cellular morpohology | ||||
| regulation of cell shape | 5 | 31 | 3.26 | 16 |
| regulation of cell morphogenesis | 5 | 32 | 3.17 | 16 |
| cell-substrate adhesion | 10 | 70 | 4.14 | 14 |
| myeloid cell differentiation | 8 | 58 | 3.58 | 14 |
| cell-matrix adhesion | 9 | 66 | 3.76 | 14 |
| chemotaxis | 11 | 100 | 3.33 | 11 |
| taxis | 11 | 100 | 3.33 | 11 |
| Cellular metabolism/biosynthesis | ||||
| isoprenoid metabolic process | 5 | 25 | 3.88 | 20 |
| oxygen and reactive oxygen species metabolic process | 6 | 31 | 4.15 | 19 |
| water-soluble vitamin metabolic process | 7 | 37 | 4.40 | 19 |
| oxidoreduction coenzyme metabolic process | 5 | 31 | 3.26 | 16 |
| vitamin metabolic process | 8 | 55 | 3.76 | 15 |
| cofactor metabolic process | 15 | 122 | 4.39 | 12 |
| membrane lipid biosynthetic process | 10 | 83 | 3.50 | 12 |
| coenzyme metabolic process | 12 | 100 | 3.82 | 12 |
| alcohol metabolic process | 27 | 228 | 5.69 | 12 |
| cellular lipid metabolic process | 52 | 472 | 7.39 | 11 |
| glycoprotein biosynthetic process | 12 | 110 | 3.45 | 11 |
| glycoprotein metabolic process | 14 | 131 | 3.64 | 11 |
| protein amino acid glycosylation | 11 | 103 | 3.22 | 11 |
| biopolymer glycosylation | 11 | 105 | 3.15 | 10 |
| fatty acid metabolic process | 14 | 141 | 3.33 | 10 |
| isoprenoid biosynthetic process | 5 | 15 | 5.56 | 33 |
| Signal transduction | ||||
| activation of phospholipase C activity | 2 | 12 | 2.12 | 17 |
| peptidyl-tyrosine modification | 5 | 36 | 2.85 | 14 |
| positive regulation of I-kappaB kinase/NF-kappaB cascade | 9 | 78 | 3.17 | 12 |
az-score ≥3.0, as determined by GeneSifter© Software Analysis and ≥5 transcripts per ontology, with significant transcripts representing at least 10% of ontology on array.
b% determined by number of transcripts in ontology list divided by total number of transcripts in ontology on array.
Several genes (n = 22), representing transcripts identified from the microarray data set as expressed in the macaque CL, were chosen for validation by real-time PCR. These included novel genes with respect to luteal function, steroid hormone receptors, previously determined LH- and/or steroid-regulated genes, plus ‘negative control’ transcripts that did not change in expression between groups. Table IV lists all validated genes and summarizes the results of microarray expression analysis and one-way ANOVA values for real-time PCR data. The table is divided into examples of gene transcripts whose levels were (i) not altered by treatments (not regulated), (ii) LH-regulated independent of steroid influence and (iii) LH-regulated in a P or (iv) other steroid-dependent manner. Typically, the patterns of mRNA levels determined by real-time PCR resembled those from microarray analysis (Table IV).
Table IV.
Real-time PCR validation of select differentially expressed genes
| Microarray |
Real-time PCR |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Control (C) Microarray relative expression | Antide (A) | A+LH | A+LH+TRL | A+LH+TRL+R5020 | One-Way ANOVA; P | Similar to Microarray Pattern | |
| Not Regulated | AR | 65 ± 5 | ≡C | ≡C | ≡C | ≡C | >0.9 | Yes |
| CASP2 | 16 ± 1 | ≡C | ≡C | ≡C | ≡C | >0.2 | Yes | |
| ESR2 | 23 ± 2 | ≡C | ≡C | ≡C | ≡C | >0.6 | Yes | |
| PGR | 11 ± 1 | ≡C | ≡C | ≡C | ≡C | >0.6 | Yes | |
| LH Regulated | AHRIP | 124 ± 12 | ↓ | ↑ | ≡LH | ≡LH | <0.01 | No |
| IL1RN | 23 ± 4 | ↑ | ↓ | ≡LH | ≡LH | <0.001 | Yes | |
| LH and Progestin Regulated | ARNT2 | 39 ± 5 | ↑ | ↓ | ↑ | ↓ | <0.01 | Yes |
| CRHBP | 75 ± 30 | ↑ | ↓ | ↑ | trend ↓ | <0.001 | Yes | |
| LOC693394 | 26 ± 9 | trend ↑ | ↓ | ↑ | ↓ | <0.03 | Yes | |
| SFRP4 | 30 ± 5 | ↑ | ↓ | ↑ | ↓ | <0.02 | Yes | |
| SC4MOL | 2774 ± 369 | ↓ | ↑ | ↓ | ↑ | <0.02 | Yes | |
| StAR | 8370 ± 569 | ↓ | ↑ | ↓ | ↑ | <0.001 | Yes | |
| LH and Other Steroid Regulated | ADAMTS5 | 24 ± 5 | ↓ | ↑ | ↓ | ≡TRL | <0.01 | Yes |
| CAT | 2803 ± 445 | ↓ | ↑ | ↓ | ≡TRL | <0.05 | Yes | |
| Inhibin BA | 679 ± 138 | ↓ | ↑ | ↓ | ≡TRL | >0.3 | Yes | |
| PRLR | 87 ± 18 | ↓ | ↑ | ↓ | ≡TRL | <0.01 | Yes | |
| VEGF | 282 ± 26 | ↓ | ↑ | ↓ | ≡TRL | <0.05 | Yes | |
| CCL3 | 133 ± 64 | ↑ | ↓ | ↑ | ≡TRL | >0.06 | Yes | |
| MHC1 | 383 ± 232 | ↑ | trend ↓ | ↑ | ≡TRL | <0.01 | No | |
| MMP19 | 132 ± 18 | ↑ | ↓ | ↑ | ≡TRL | <0.05 | Yes | |
| MMP9 | 116 ± 35 | ↑ | ↓ | trend ↑ | ≡TRL | <0.05 | No | |
| PGRMC1 | 2109 ± 82 | ↑ | ↓ | ↑ | ≡TRL | <0.01 | No | |
Control values (±SEM) are presented as a reference. Direction of expression of validated genes by microarray analysis is compared with previous group (e.g. A compared with C), except for genes whose expression did not differ from control. One-way ANOVA values of significance are presented for real-time PCR validation.
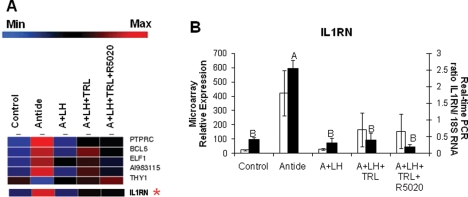
An example of a novel gene identified by microarray as being LH-regulated in the primate CL is interleukin 1 receptor antagonist (IL1RN), depicted in Fig. 2. The IL1RN gene belongs to the ‘negative regulation of immune system process’ ontology based on known Kyoto Encyclopedia of Genes and Genomes—KEGG pathways (similar to Table III, which displays the number of transcripts with known gene ontology pathways); many of the transcripts in this ontology, including IL1RN, were mainly affected by LH ablation/replacement, and to a lesser extent, by steroid ablation/P replacement (Fig. 2A). Results from real-time PCR analysis were comparable with those provided by microarray analysis (Fig. 2B); IL1RN mRNA levels increased following Antide treatment, but this effect was prevented by LH replacement. Treatment with TRL with or without R5020 replacement was without effect.
Figure 2.
A list of regulated transcripts identified by GeneSifter© from the KEGG ontology ‘Negative Regulation of Immune System Process’ (as in Table III) as significantly (one-way ANOVA, P < 0.05) affected by treatments is shown in Panel A (blue indicates minimum expression, whereas red indicates maximum). A comparison of interleukin 1 receptor antagonist (IL1RN; mean ± SEM) results is depicted in Panel B, with white bars representing microarray values (GeneSifter© one-way ANOVA, P < 0.05), and black bars representing real-time PCR data (one-way ANOVA, P < 0.001; different letters signify treatment groups with significantly different values, P < 0.05).
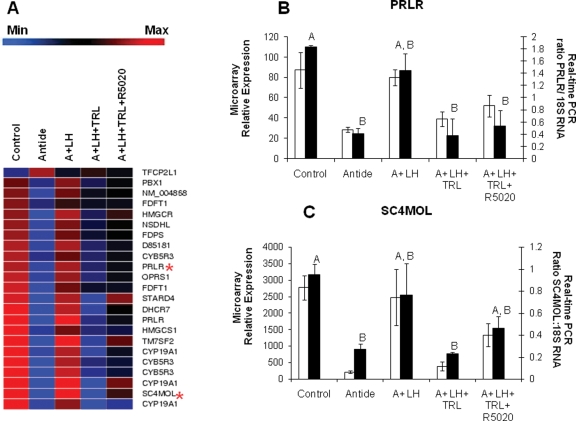
Regulated members of the ‘steroid biosynthetic process’ ontology, based on known KEGG pathways (similar to the results from Table III), and two validated genes from this category are presented in Fig. 3. Most of these transcripts are regulated similarly by LH and steroid ablation (Fig. 3A) in that transcript levels declined with Antide or TRL treatment. While LH replacement typically prevented the decline induced by Antide, progestin replacement with R5020 was less effective in preventing TRL’s effects, except in some instances (e.g. STARD4, CYP19A1). Results from real-time PCR assays of the prolactin receptor (PRLR; Fig. 3B) and sterol-C4-methyl oxidase-like (SC4MOL; Fig. 3C) mRNAs were comparable with the results obtained by microarray. Levels of both mRNAs were decreased by Antide treatment, and LH replacement maintained levels similar to control. Likewise, TRL treatment resulted in mRNA levels similar to Antide, but R5020 replacement either did not affect mRNA levels (PRLR; Fig. 3B), or only partially maintained mRNA at control levels (SC4MOL; Fig. 3C).
Figure 3.
A list of transcripts identified by GeneSifter© from the KEGG ontology ‘Steroid Biosynthetic Process’ (similar to Table III) as significantly (one-way ANOVA, P < 0.05) affected by treatments is shown in Panel A (blue indicates minimum expression, whereas red indicates maximum expression). The red stars indicate the relative expression of transcripts for the PRLR and SC4MOL. Real-time PCR results for PRLR (Panel B; one-way ANOVA, P < 0.01) and SC4MOL (Panel C; one-way ANOVA, P = 0.01) are compared with microarray results (P < 0.05). See the legend of Fig. 2 for more details.
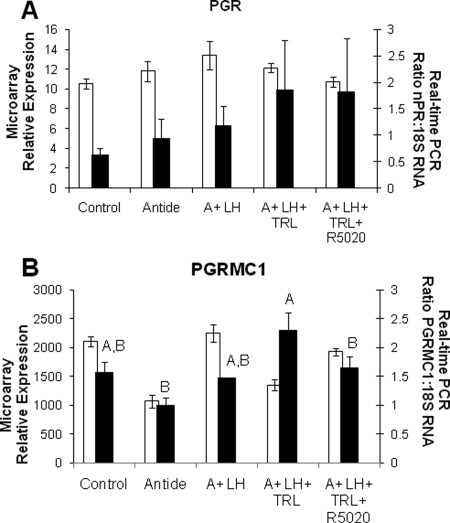
The expression of transcripts for genomic steroid hormone receptors as measured by microarray was low, and the pattern did not change throughout the experimental groups; examples include the PGR (Table IV and Fig. 4A), the AR and estrogen receptor β (AR and ESR2, respectively; Table IV). The only putative steroid hormone receptor whose transcript levels differed significantly with treatment, based on microarray and real-time PCR analyses, was the non-classical progestin receptor: progesterone receptor membrane component 1 (PGRMC1; Fig. 4B). This transcript was highly expressed, but the pattern of mRNA expression differed between real-time PCR data and the microarray for both A + LH and A + LH + TRL treatment groups (Fig. 4B).
Figure 4.
Microarray and real-time PCR expression patterns for the genomic PGR (Panel A; GeneSifter© and real-time PCR one-way ANOVA, P > 0.6) and P receptor membrane component 1 (PGRMC1; Panel B; GeneSifter© one-way ANOVA, P < 0.05; real-time PCR one-way ANOVA, P < 0.01). Statistics for PGR and PGRMC1 are depicted for real-time PCR results (mean ± SEM), with white bars representing microarray values, and black bars representing real-time data. Different letters signify treatment groups with significantly different values (P < 0.05).
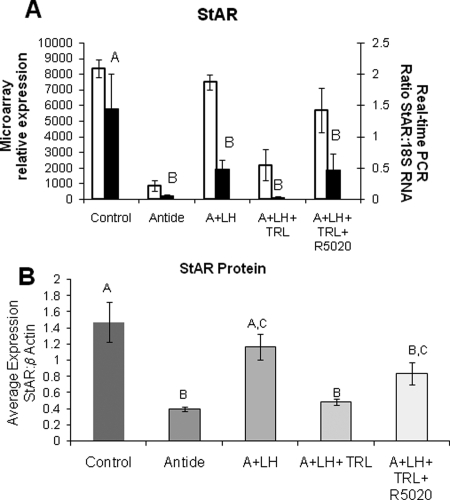
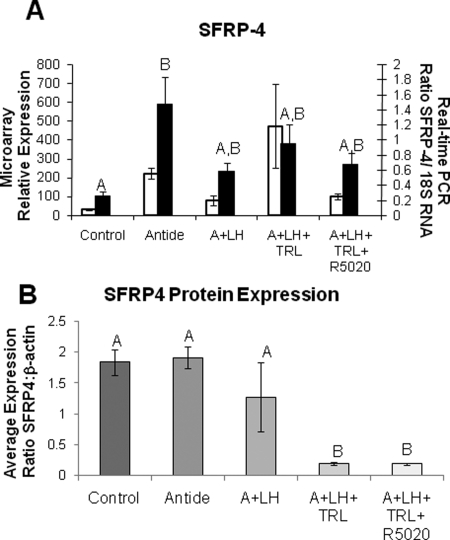
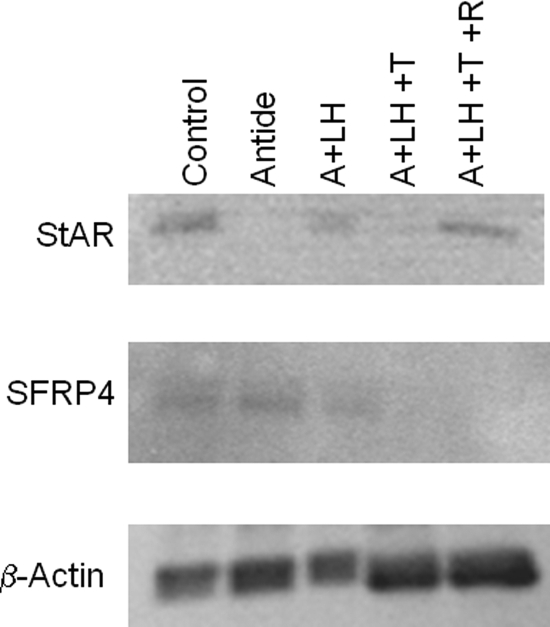
Two gene products, StAR protein and SFRP4, were chosen to determine whether there was any correlation between their mRNA and protein levels. One gene displayed high levels of mRNA expression (StAR; Fig. 5A), whereas expression of mRNA for the other gene was moderate (SFRP4; Fig. 6A). Again, the patterns of expression for mRNA data from real-time PCR matched microarray analysis for both genes. The mRNA levels for StAR were markedly decreased by Antide treatment, but maintained near-control levels by LH replacement, and comparable results were obtained by TRL treatment and R5020 replacement (Fig. 5A). Notably, the pattern of StAR protein levels between treatment groups was similar to that of mRNA levels (Fig. 5B). In contrast, SFRP4 mRNA levels increased 6-fold with Antide treatment, and replacement of LH reduced, but did not completely restore mRNA to control levels; TRL treatment with or without R5020 replacement had a minimal effect when measured by real-time PCR (Fig. 6A). Although the levels of protein for SFRP4 changed significantly across treatment groups (Fig. 6B; P < 0.001), the expression pattern of the protein did not match that of the mRNA. Protein levels were unchanged between LH ablation and replacement groups, whereas steroid ablation by TRL treatment lowered the expression levels, and R5020 replacement had no further effect. The western blots for pooled samples for StAR and SFRP4 are shown in Fig. 7.
Figure 5.
StAR mRNA expression (Panel A; white bars indicate microarray data, black bars indicate real-time data: see the legend of Fig. 2 for more details on microarray and real-time data; one-way ANOVA, P < 0.001) and protein levels (Panel B; one-way ANOVA, P < 0.001; means ± SEM) in individual CL normalized to β-actin as compared between treatment groups.
Figure 6.
SFRP4 expression in ablation/replacement CL. See the legend of Fig. 5 for details. The pattern of SFRP4 protein expression, although significant (one-way ANOVA, P < 0.001), does not match the expression of SFRP4 mRNA (real-time PCR one-way ANOVA, P < 0.02).
Figure 7.
Representative immunoblots for StAR, SFRP4 and β-actin loading control for pooled samples of different treatment groups. Note that as presented in Fig. 5A and B, StAR protein expression matches mRNA expression patterns, whereas SFRP4 protein expression does not match mRNA expression. β-Actin was used for loading control.
Discussion
This study provides a genome-wide analysis of macaque luteal transcripts that are either directly or indirectly regulated by LH. All normalized data files have been uploaded to the National Center for Biotechnology Information’s Gene Expression Omnibus gene expression repository (http://www.ncbi.nlm.nih.gov/geo/; series GSE12281), which allows for further analyses to be performed by independent investigators. The current microarray analysis confirmed earlier reports of LH-regulated transcripts in macaque luteal tissue. For example, our preliminary analysis of LH-regulated genes in the macaque CL from Antide-treated monkeys identified CRHBP mRNA as markedly up-regulated by LH withdrawal (Xu et al., 2005). This evidence led to the detailed characterization of a novel local system in the macaque CL—the CRH/UCN receptor-binding protein system—that may modulate LH action and hence regulate primate luteal structure–function (Xu et al., 2007). The current results expand our understanding of the control of CRHBP expression and, possibly, action, with the finding that steroid withdrawal and R5020 replacement mimicked the effects of Antide treatment and LH replacement. Thus, CRHBP appears indirectly regulated by LH via steroid hormone production, particularly the local actions of P. Also, the current results confirm and extend the evidence from differential gene display in the bonnet monkey CL that LH withdrawal markedly suppressed the expression of LDL receptor mRNA, a key membrane component for cholesterol uptake into primate luteal cells (Yadav et al., 2004).
However, the current analysis also identified many novel transcripts whose expression is LH-regulated in the macaque CL. For example, mRNA for the IL1RN, a suppressor of the immune system, that was first detected in the ovaries of rats (Kol et al., 1999) and later reported to increase interleukin-1β-stimulated prostaglandin (E, F2α) release from human luteal cells (Miceli et al., 2003), was up-regulated by LH withdrawal. This LH action appears independent of local steroid action, since LH treatment restored IL1RN mRNA to control levels and steroid withdrawal did not mimic the effect of Antide treatment. Unexpectedly, PRLR mRNA expression in the macaque CL was down-regulated by LH withdrawal. Unlike in some non-primate species [e.g. rodents (Herz et al., 1986)], PRL and PRL-like hormones are not considered major luteotropic hormones for the primate CL (Stouffer, 2006). However, a recent microarray analysis (Bogan et al., 2008b) of gene expression in the macaque CL throughout the menstrual cycle discovered appreciable PRLR expression that declined at luteolysis; moreover, the expression pattern for PRLR mRNA and protein were comparable. There are high levels of prolactin (-like) proteins circulating during pregnancy and lactation (Bachelot and Binart, 2007). The possibility proposed by Knobil and colleagues (Richardson et al., 1985) that PRLR ligands could contribute to the maintenance of luteal function in primates, perhaps during pregnancy and lactation, warrants re-evaluation.
Notably, a small but significant number (n = 36, Fig. 1B and Table II) of Antide-sensitive transcripts in the macaque CL were not affected by LH replacement. Of particular interest is the expression of LDL receptor (discussed earlier), a key component in cholesterol entrance into the cell, and prostaglandin F receptor (PTGFR), a potent, local and perhaps physiologic luteolytic factor in primates (Yadav et al., 2004; Bogan et al., 2008a). Expression of both LDL receptor mRNA and PTGFR declined markedly with Antide treatment, and this effect was not significantly altered by LH replacement. Although the LH-replacement regimen restored the expression of many Antide-sensitive gene products, and this regimen prevents the functional–structural regression of the CL in Antide-treated monkeys (Duffy et al., 1999b), it is possible that our LH protocol was sub-optimal for a cohort of LH-regulated genes. Alternatively, there is evidence of GnRH receptor expression (Chakrabarti et al., 2008) and GnRH effects (Siler-Khodr et al., 2004) in primate ovaries and CL. Though it is currently controversial whether GnRH serves as an important local factor in primates, as it does in rodents (Gupta and Flaws, 2005), we cannot rule out that Antide affects luteal GnRH signaling and action. Indeed, the Antide-sensitive, LH-insensitive gene products could be markers of local GnRH action in the primate CL.
Of the approaching 1500 LH-sensitive transcripts in the macaque CL, less than one-third of this cohort was affected similarly by steroid ablation, and even fewer by progestin replacement (Fig. 1B). This evidence implies that many LH-regulated genes are independent of steroid action; their expression may be regulated directly by LH signaling pathways, or indirectly by other LH-regulated local factors (e.g. CRH/UCN, VEGF, PGs) or LH-regulated receptors/binding proteins for other hormones or local factors (PRLR, CRHBP) in the CL. That progestin replacement only restored transcript levels for some genes may again (as for LH replacement) be a limitation of our R5020 regimen. Nevertheless, this regimen prevents endocrine and histological indices of functional and structural luteolysis (Young and Stouffer, 2004), in the macaque CL, and unequivocally restored mRNA levels of select steroid-sensitive genes (see what follows). It is possible that the numbers or types of cells expressing some of these steroid-sensitive transcripts changed after 3 days of treatment. Previous histological analyses of Antide- and A + LH + TRL-treated CL revealed that steroidogenic cells were smaller in volume, and the numbers of immune cells were greater, compared with control, A+LH and A + LH + TRL + R5020 treatments (Young and Stouffer, 2004); but these observations were qualitative. Alternatively, it is possible that by mid-late luteal phase (Day 9–12), the CL is not as responsive to progestin as luteal tissue from the early-to-mid-luteal phase. We reported previously that the percent of PR-positive cells and PR mRNA levels in the macaque CL decline after mid-luteal phase (Hild-Petito et al., 1988; Duffy et al., 1999b). Moreover, there is evidence that gene expression in the CL in response to steroid withdrawal and progestin replacement differs between the early and mid-late luteal phase [e.g. ADAMTS-1, a disintegrin and metalloproteinase with thrombospondin repeats-1 (Young et al., 2004)]. This differential response could also be related to changes in the ratio of PR A:B isoforms as the luteal lifespan progresses (Duffy et al., 1997). The current analysis also detected appreciable mRNA levels for the novel membrane progestin receptor PGRMC1 in macaque luteal tissue. A physiologic role for PGRMC1 in the CL has yet to be defined, but it could play a role in mediating the LH-like, luteotropic effects of P by preventing apoptosis of luteinized cells (Engmann et al., 2006). Finally, since the 3β-HSD inhibitor, TRL, markedly suppresses production of other steroids relying on P as a precursor, those steroid-sensitive transcripts that were not restored by R5020 replacement could be altered by androgen or estrogen withdrawal. The macaque CL expresses AR and ER (particularly ERβ) during its lifespan (Duffy et al., 1999a, 2000). Moreover, Duffy et al. (2000) reported that ERβ mRNA levels decreased around luteolysis and that steroid withdrawal at mid-luteal phase (Day 6) increased ERβ expression, whereas R5020 replacement returned expression to control levels. Further studies are needed at earlier stages of luteal development and function, and while replacing estrogen or androgen, as well as progestin, to clarify the LH- and steroid-regulated gene processes in the primate CL.
Nevertheless, the changes in mRNA levels of several genes in response to LH withdrawal and replacement were mimicked by steroid withdrawal and progestin replacement. For example, StAR expression exhibited a similar pattern following either withdrawal/replacement regimen (Fig. 5), strongly suggesting that the changes evoked by LH were indirect and are due to LH-stimulated P production by the tissue. Abundant levels of StAR protein are expressed in the developing and developed human CL, and these levels decline between the mid- and late-luteal phase (Devoto et al., 2001). Likewise, expression of other enzymes in the ontology ‘steroid biosynthetic processes’ (Fig. 3), SC4MOL and CYP19A1, exhibited a similar pattern following either LH or steroid withdrawal/replacement regimen. Thus, P may regulate a select number of key genes that are critical for luteal structure–function around the onset of luteolysis at mid-late luteal phase. As noted from our initial analyses of mRNA and protein levels, it will be important to follow genomic analyses with evaluations of the translation and activity of gene products. The patterns of mRNA and protein expression were similar for StAR, suggesting that transcriptional regulation is a key part of progestin regulation of StAR production. However, the patterns of mRNA and protein levels for secreted frizzled related protein 4 (SFRP4) were not comparable, suggesting that regulation of SFRP4 can occur at the translational level. This concept is consistent with previous evidence (Young and Stouffer 2004; Peluffo et al., 2005) that LH/progestin regulation of proteases (caspases, MMPs) can occur at either/both the mRNA, protein or enzyme activity level. Further analysis of the actions of LH and steroids at the transcriptional, translational and post-translational levels is needed to understand the regulation of luteal structure–function in primates.
In summary, this microarray experiment broadly defines the changes in gene expression in the primate CL that are regulated by LH around the onset of luteolysis in the menstrual cycle. The data suggest that LH regulates the expression of many genes involved in the maintenance of CL structure–function independently of steroid action. However, steroid withdrawal and R5020 replacement indicate that progestin is involved in the regulation of key pathways that regulate the CL lifespan (Stouffer, 2003), and this may involve actions via a non-classical PR. Also, steroid-sensitive genes whose activity was not restored by R5020 replacement could be regulated by androgens or estrogens. Further studies on LH/steroid withdrawal and replacement during CL development in the early-luteal phase and analysis of the patterns of both mRNA and protein expression in macaque CL are needed. During revision of this manuscript, a microarray analysis of a limited sample of primate CL after short-term LH ablation was published online (Priyanka et al., 2008). Comparisons of similar studies will help to elucidate the relative roles of LH and steroids on gene expression in the primate CL during the menstrual cycle.
Funding
This work was supported by several grants from the National Institutes of Health (NIH) [R01 HD20869 to R.L.S., R01 HD42000 to J.D.H., U54 HD18185 (Molecular and Cellular Biology Core), P51 RR00163]; plus a National Research Service Award (NRSA) Institutional Fellowship [T32 HD007133 to CVB].
Acknowledgements
The authors are indebted to Dr Kelly Young for establishing the dosage paradigm, collecting the CL and processing CL for mRNA. We also thank Dr Marina Peluffo for processing CL for protein. We acknowledge the critical expertise of the staff in the Division of Animal Resources, ONPRC for their animal caretaking and assistance with treatment protocols, and especially the surgery staff for their role in the collection of ovarian tissues.
References
- Ascoli M, Fanelli F, Segaloff DL. The lutropin/choriogonadotropin receptor, a 2002 perspective. Endocr Rev. 2002;23:141–174. doi: 10.1210/edrv.23.2.0462. [DOI] [PubMed] [Google Scholar]
- Bachelot A, Binart N. Reproductive role of prolactin. Reproduction. 2007;133:361–369. doi: 10.1530/REP-06-0299. [DOI] [PubMed] [Google Scholar]
- Benyo DF, Zeleznik AJ. Cyclic adenosine monophosphate signaling in the primate corpus luteum: maintenance of protein kinase A activity throughout the luteal phase of the menstrual cycle. Endocrinology. 1997;138:3452–3458. doi: 10.1210/endo.138.8.5346. [DOI] [PubMed] [Google Scholar]
- Bogan RL, Murphy MJ, Stouffer RL, Hennebold JD. Prostaglandin synthesis, metabolism, and signaling potential in the rhesus macaque corpus luteum throughout the luteal phase of the menstrual cycle. Endocrinology. 2008;a 149:5861–5871. doi: 10.1210/en.2008-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan RL, Murphy MJ, Stouffer RL, Hennebold JD. Systematic determination of differential gene expression in the primate corpus luteum during the luteal phase of the menstrual cycle. Mol Endocrinol. 2008;b 22:1260–1273. doi: 10.1210/me.2007-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti N, Subbarao T, Sengupta A, Xu F, Stouffer RL, Sridaran R. Expression of mRNA and proteins for GnRH I and II and their receptors in primate corpus luteum during menstrual cycle. Mol Reprod Dev. 2008;75:1567–1577. doi: 10.1002/mrd.20898. [DOI] [PubMed] [Google Scholar]
- Devoto L, Kohen P, Gonzalez RR, Castro O, Retamales I, Vega M, Carvallo P, Christenson LK, Strauss JF., III Expression of steroidogenic acute regulatory protein in the human corpus luteum throughout the luteal phase. J Clin Endocrinol Metab. 2001;86:5633–5639. doi: 10.1210/jcem.86.11.7982. [DOI] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DM, Hess DL, Stouffer RL. Acute administration of a 3 beta-hydroxysteroid dehydrogenase inhibitor to rhesus monkeys at the midluteal phase of the menstrual cycle: evidence for possible autocrine regulation of the primate corpus luteum by progesterone. J Clin Endocrinol Metab. 1994;79:1587–1594. doi: 10.1210/jcem.79.6.7989460. [DOI] [PubMed] [Google Scholar]
- Duffy DM, Wells TR, Haluska GJ, Stouffer RL. The ratio of progesterone receptor isoforms changes in the monkey corpus luteum during the luteal phase of the menstrual cycle. Biol Reprod. 1997;57:693–699. doi: 10.1095/biolreprod57.4.693. [DOI] [PubMed] [Google Scholar]
- Duffy DM, Abdelgadir SE, Stott KR, Resko JA, Stouffer RL, Zelinski-Wooten MB. Androgen receptor mRNA expression in the rhesus monkey ovary. Endocrine. 1999;a 11:23–30. doi: 10.1385/ENDO:11:1:23. [DOI] [PubMed] [Google Scholar]
- Duffy DM, Stewart DR, Stouffer RL. Titrating luteinizing hormone replacement to sustain the structure and function of the corpus luteum after gonadotropin-releasing hormone antagonist treatment in rhesus monkeys. J Clin Endocrinol Metab. 1999;b 84:342–349. doi: 10.1210/jcem.84.1.5362. [DOI] [PubMed] [Google Scholar]
- Duffy DM, Chaffin CL, Stouffer RL. Expression of estrogen receptor alpha and beta in the rhesus monkey corpus luteum during the menstrual cycle: regulation by luteinizing hormone and progesterone. Endocrinology. 2000;141:1711–1717. doi: 10.1210/endo.141.5.7477. [DOI] [PubMed] [Google Scholar]
- Engmann L, Losel R, Wehling M, Peluso JJ. Progesterone regulation of human granulosa/luteal cell viability by an RU486-independent mechanism. J Clin Endocrinol Metab. 2006;91:4962–4968. doi: 10.1210/jc.2006-1128. [DOI] [PubMed] [Google Scholar]
- Gupta RK, Flaws JA. Gonadotropin-releasing hormone (GnRH) analogues and the ovary: do GnRH antagonists destroy primordial follicles? Fertil Steril. 2005;83:1339–1342. doi: 10.1016/j.fertnstert.2005.01.089. [DOI] [PubMed] [Google Scholar]
- Herz Z, Khan I, Jayatilak PG, Gibori G. Evidence for the secretion of decidual luteotropin: a prolactin-like hormone produced by rat decidual cells. Endocrinology. 1986;118:2203–2209. doi: 10.1210/endo-118-6-2203. [DOI] [PubMed] [Google Scholar]
- Hild-Petito S, Stouffer RL, Brenner RM. Immunocytochemical localization of estradiol and progesterone receptors in the monkey ovary throughout the menstrual cycle. Endocrinology. 1988;123:2896–2905. doi: 10.1210/endo-123-6-2896. [DOI] [PubMed] [Google Scholar]
- Kol S, Donesky BW, Ruutiainen-Altman K, Ben-Shlomo I, Irahara M, Ando M, Rohan RM, Adashi EY. Ovarian interleukin-1 receptor antagonist in rats: gene expression, cellular localization, cyclic variation, and hormonal regulation of a potential determinant of interleukin-1 action. Biol Reprod. 1999;61:274–282. doi: 10.1095/biolreprod61.1.274. [DOI] [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW. Expanding functional diversity of the coactivators. Trends Biochem Sci. 2005;30:126–132. doi: 10.1016/j.tibs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Miceli F, Tropea A, Minici F, Navarra P, Lanzone A, Apa R. Interleukin-1 beta stimulates progesterone production by in vitro human luteal cells: evidence of a mediatory role of prostaglandins. J Clin Endocrinol Metab. 2003;88:2690–2694. doi: 10.1210/jc.2002-020819. [DOI] [PubMed] [Google Scholar]
- Molskness TA, Zelinski-Wooten MB, Hild-Petito SA, Stouffer RL. Comparison of the steroidogenic response of luteinized granulosa cells from rhesus monkeys to luteinizing hormone and chorionic gonadotropin. Biol Reprod. 1991;45:273–281. doi: 10.1095/biolreprod45.2.273. [DOI] [PubMed] [Google Scholar]
- Noriega NC, Garyfallou VT, Kohama SG, Urbanski HF. Glutamate receptor subunit expression in the rhesus macaque locus coeruleus. Brain Res. 2007;1173:53–65. doi: 10.1016/j.brainres.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent AS, Matagne V, Westphal M, Heger S, Ojeda S, Jung H. Gene expression profiling of hypothalamic hamartomas: a search for genes associated with central precocious puberty. Horm Res. 2008;69:114–123. doi: 10.1159/000111815. [DOI] [PubMed] [Google Scholar]
- Peluffo MC, Young KA, Stouffer RL. Dynamic expression of caspase-2, -3, -8, and -9 proteins and enzyme activity, but not messenger ribonucleic acid, in the monkey corpus luteum during the menstrual cycle. J Clin Endocrinol Metab. 2005;90:2327–2335. doi: 10.1210/jc.2004-2214. [DOI] [PubMed] [Google Scholar]
- Priyanka S, Medhamurthy R. Characterization of cAMP/PKA/CREB signaling cascade in the bonnet monkey corpus luteum: expressions of inhibin-alpha and StAR during different functional status. Mol Hum Reprod. 2007;13:381–390. doi: 10.1093/molehr/gam015. [DOI] [PubMed] [Google Scholar]
- Priyanka S, Jayaram P, Sridaran R, Medhamurthy R. Genome-wide gene expression analysis reveals a dynamic interplay between luteotropic and luteolytic factors in the regulation of corpus luteum function in the bonnet monkey (Macaca radiata) Endocrinology. 2008 doi: 10.1210/en.2008-0840. 6 November Epub PMID 18988674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DW, Goldsmith LT, Pohl CR, Schallenberger E, Knobil E. The role of prolactin in the regulation of the primate corpus luteum. J Clin Endocrinol Metab. 1985;60:501–504. doi: 10.1210/jcem-60-3-501. [DOI] [PubMed] [Google Scholar]
- Rothchild I. The regulation of the mammalian corpus luteum. Recent Prog Horm Res. 1981;37:183–298. doi: 10.1016/b978-0-12-571137-1.50009-8. [DOI] [PubMed] [Google Scholar]
- Siler-Khodr TM, Yu FQ, Wei P, Tao SX, Liu YX. Contraceptive action of a gonadotropin-releasing hormone II analog in the rhesus monkey. J Clin Endocrinol Metab. 2004;89:4513–4520. doi: 10.1210/jc.2004-032087. [DOI] [PubMed] [Google Scholar]
- Spindel ER, Pauley MA, Jia Y, Gravett C, Thompson SL, Boyle NF, Ojeda SR, Norgren RB., Jr Leveraging human genomic information to identify nonhuman primate sequences for expression array development. BMC Genomics. 2005;6:160. doi: 10.1186/1471-2164-6-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer RL. Progesterone as a mediator of gonadotrophin action in the corpus luteum: beyond steroidogenesis. Hum Reprod Update. 2003;9:99–117. doi: 10.1093/humupd/dmg016. [DOI] [PubMed] [Google Scholar]
- Stouffer RL. Structure, function, and regulation of the corpus luteum. In: Knobil E, Neill JD, editors. Knobil and Neill’s Physiology of Reproduction. Amsterdam, Boston: Elsevier; 2006. pp. 475–526. [Google Scholar]
- Stouffer RL, Duffy DM. Receptors for sex steroids in the primate corpus luteum. New insight into gonadotropin and steroid action. Trends Endocrinol Metab. 1995;6:83–89. doi: 10.1016/1043-2760(94)00214-o. [DOI] [PubMed] [Google Scholar]
- Tesone M, Stouffer RL, Borman SM, Hennebold JD, Molskness TA. Vascular endothelial growth factor (VEGF) production by the monkey corpus luteum during the menstrual cycle: isoform-selective messenger RNA expression in vivo and hypoxia-regulated protein secretion in vitro. Biol Reprod. 2005;73:927–934. doi: 10.1095/biolreprod.105.039875. [DOI] [PubMed] [Google Scholar]
- Xu J, Stouffer RL, Searles RP, Hennebold JD. Discovery of LH-regulated genes in the primate corpus luteum. Mol Hum Reprod. 2005;11:151–159. doi: 10.1093/molehr/gah157. [DOI] [PubMed] [Google Scholar]
- Xu J, Hennebold JD, Stouffer RL. Dynamic expression and regulation of the corticotropin-releasing hormone/urocortin-receptor-binding protein system in the primate ovary during the menstrual cycle. J Clin Endocrinol Metab. 2006;91:1544–1553. doi: 10.1210/jc.2005-2776. [DOI] [PubMed] [Google Scholar]
- Xu J, Xu F, Hennebold JD, Molskness TA, Stouffer RL. Expression and role of the corticotropin-releasing hormone/urocortin-receptor-binding protein system in the primate corpus luteum during the menstrual cycle. Endocrinology. 2007;148:5385–5395. doi: 10.1210/en.2007-0541. [DOI] [PubMed] [Google Scholar]
- Yadav VK, Muraly P, Medhamurthy R. Identification of novel genes regulated by LH in the primate corpus luteum: insight into their regulation during the late luteal phase. Mol Hum Reprod. 2004;10:629–639. doi: 10.1093/molehr/gah089. [DOI] [PubMed] [Google Scholar]
- Young KA, Stouffer RL. Gonadotropin and steroid regulation of matrix metalloproteinases and their endogenous tissue inhibitors in the developed corpus luteum of the rhesus monkey during the menstrual cycle. Biol Reprod. 2004;70:244–252. doi: 10.1095/biolreprod.103.022053. [DOI] [PubMed] [Google Scholar]
- Young KA, Hennebold JD, Stouffer RL. Dynamic expression of mRNAs and proteins for matrix metalloproteinases and their tissue inhibitors in the primate corpus luteum during the menstrual cycle. Mol Hum Reprod. 2002;8:833–840. doi: 10.1093/molehr/8.9.833. [DOI] [PubMed] [Google Scholar]
- Young KA, Tumlinson B, Stouffer RL. ADAMTS-1/METH-1 and TIMP-3 expression in the primate corpus luteum: divergent patterns and stage-dependent regulation during the natural menstrual cycle. Mol Hum Reprod. 2004;10:559–565. doi: 10.1093/molehr/gah079. [DOI] [PubMed] [Google Scholar]
- Zeleznik AJ, Somers JP. Regulation of the primate corpus luteum: cellular and molecular perspectives. Trends Endocrinol Metab. 1999;10:189–193. doi: 10.1016/s1043-2760(98)00145-3. [DOI] [PubMed] [Google Scholar]