Abstract
Summary
Background
Atopic eczema is characterized by Th2-dominant immunity with the cytokine interleukin 13 and the transcription factor GATA binding protein 3 playing a critical role.
Objectives
We assessed the association of polymorphisms in the IL13 and GATA3 genes with childhood eczema.
Methods
A birth cohort (n = 1456) was established on the Isle of Wight in 1989 and followed at the ages of 1 (n = 1167), 2 (n = 1174), 4 (n = 1218) and 10 years (n = 1373) to determine the prevalence of allergic disease including eczema. At 4 and 10 years, skin prick testing was performed. Whole blood samples (n = 923) were obtained at the 10-year assessment, stored frozen, and genotyped. Five polymorphisms from IL13 and seven from GATA3 were genotyped for this analysis. Repeated measurement analyses were conducted for the occurrence of eczema at ages 1, 2, 4 and 10 years. All analyses were adjusted for maternal and paternal eczema, low birth weight (< 2500 g), breastfeeding ≥ 3 months and age.
Results
IL13 was not associated with childhood eczema. For GATA3, the single nucleotide polymorphism (SNP) rs2275806 (promoter region) showed an increased odds ratio for atopic eczema independent of whether the comparison group had a positive skin prick test. The SNP rs444762 (intron 3 region) was associated with atopic eczema in comparison with children without eczema. The increased relative risks remained significant after adjustment for multiple testing only for rs2275806 (P < 0Æ05).
Conclusions
A SNP in GATA3 is associated with atopic eczema. This finding highlights the importance of GATA3 as an immune-modulating gene in atopic eczema.
Keywords: atopy, cytokines, eczema, genetics, single nucleotide polymorphisms
Childhood eczema is a chronic inflammatory skin condition closely associated with atopic diathesis. Interleukin (IL)-13 is a key cytokine in atopic diseases as it induces the production of IgE by human B cells, as well as having other proinflammatory effects.1 IL-13 is mainly produced by activated T cells of the Th2 type (CD4+). Skin biopsy specimens of acute and chronic lesions in eczema are characterized by an infiltration with inflammatory cells, including CD4+ T lymphocytes.2 IL-13 activity is increased in eczema, particularly in acute skin lesions, as evidenced by increased expression of IL-13 mRNA.3
GATA binding proteins were originally suggested as regulating gene transcription in inflammatory cells, such as eosinophils and basophils.4 Since then, GATA binding protein 3 (GATA-3) has emerged as having a critical role in the development of Th2-type immune responses mediated by IL-13 and related cytokines.5 Macaubas et al.6 have demonstrated that following allergen stimulation, the processes of differentiation of naive T cells towards Th2 cells in atopics are associated with rapid upregulation of GATA-3, whereas T cells from nonatopics display equally rapid GATA-3 downregulation under similar conditions. Further, in patients with atopic eczema, mRNA for GATA-3 was found to be elevated in peripheral blood mononuclear cells, which normalized following successful therapy.7
Despite its immunological relevance to Th2 immune responses, the role of the GATA3 gene in allergic disorders has not been extensively studied. GATA3 contains six exons and is located on human chromosome 10p14–15. Pykaälaäinen et al.8 examined a Finnish population for the associations between single nucleotide polymorphisms (SNPs) in GATA3, along with STAT4 and STAT6 genes, and asthma patients with high IgE or eosinophils. While they found no significant allele associations, they did identify three 9-SNP haplotypes of GATA3 associated with asthmatics with high IgE.
The IL13 gene is located at chromosome 5q31–33, a region previously identified as having susceptibility loci for eczema.9 SNPs in the IL13 gene have been suggested as possibly mediating genetic susceptibly to eczema.10-12 However, not all studies have found an association between IL13 polymorphisms and eczema. Chang et al.13 were unable to find an association of a number of relevant cytokine genes, including IL13, with eczema in Chinese patients.
We investigated the association of common SNPs in GATA3 and IL13 genes and examined gene–gene interaction. To our knowledge, this is the first study investigating the association of GATA3 in eczema. This work also extends previous work on eczema and IL13, as this analysis is based on a birth cohort with a comprehensive assessment of eczema from infancy to 10 years of age. In addition, this work will differentiate eczema according to the children's atopic status.
Materials and methods
A whole-population birth cohort (n = 1456) was established on the Isle of Wight in 1989 to study the natural history of asthma and allergic disorders and to identify genetic and environmental risk factors important in their development. These children have been followed at the ages of 1 (n = 1167), 2 (n = 1174), 4 (n = 1218) and 10 years (n = 1373). The local Research Ethics Committee approved the study and parental consent was obtained at recruitment and subsequently at each follow-up. The island is close to the British mainland, semi-rural, with no heavy industry. The population is 99% white. At birth, information was collected on the family history of atopy, including maternal and paternal eczema, and potential environmental risk factors such as weight, breastfeeding and exposure to smoking, and was updated at each follow-up.
Detailed questionnaires were completed with the parents for each child regarding allergy prevalence at each follow-up. Eczema was defined as chronic or chronically relapsing, itchy dermatitis lasting more than 6 weeks with characteristic morphology and distribution.
At 4 and 10 years, skin prick testing was performed in most children attending the Research Centre to a standard battery of common allergens (ALK, Horsholm, Denmark). Inhalant allergens tested were house dust mite, cat, dog, Alternaria alternata, Cladosporium herbarium, grass pollen mix and tree pollen mix. Food allergens tested were cows' milk, soya, hens' egg, peanut and cod. Positive and negative controls were included. A positive skin prick test (SPT) was defined as having at least one allergen test response with mean weal diameter 3 mm greater than the negative control. Atopy was defined as a positive SPT at either 4 or 10 years of age. Children with eczema and positive SPT were regarded as having atopic eczema.
DNA isolation and genotyping
Anticoagulated whole blood samples (n = 923) were obtained at the 10-year interview and stored frozen. Genomic DNA was isolated from these samples using QIAamp DNA Blood Kits (Qiagen, Valencia, CA, U.S.A.) or the ABI PRISM™ 6100 Nucleic Acid PrepStation (Applied Biosystems, Foster City, CA, U.S.A.). Polymorphisms were examined using SNPper,14 HapMap15 (http://www.hapmap.org/) and Applied Biosystems (https://products.appliedbiosystems.com/) databases. Genotyping was conducted by biotin–streptavidin-based pyro-sequencing performed on PSQ-96 instrumentation (Biotage AB, Uppsala, Sweden). Genotypes were accepted when pyrograms were graded as ‘pass’ by the Pyrosequencing software. Three SNPs were genotyped by fluorogenic 5′ nuclease chemistry polymerase chain reaction (PCR) using Assays on Demands kits cycled on a 7900HT Sequence Detection System (SDS; Applied Biosystems), in which case the genotypes were based on clustering as determined by SDS 2.1 software. Approximately 20% of all genotypes were replicated as a quality control measure.
The rationale for selecting markers in IL13 and GATA3 was to capture the genetic variation across each gene in an efficient manner. SNP selection was based on information gathered from SNPper, Applied Biosystems and HapMap databases regarding SNP validation, allele frequencies, potential function and linkage disequilibrium (LD) in relevant test populations (HapMap CEU). We also prioritized IL13 SNPs previously reported to be associated with asthma-related pheno-types.12,16-21 We subsequently selected five IL13 and 10 GATA3 SNPs to test in our population, genotyping these first in approximately 100 subjects to determine informativeness and LD in our population, and then in all available subjects for the IL13 SNPs and a subset of seven GATA3 SNPs (Table 1) as three GATA3 SNPs were uninformative (rs11567901) or were in LD (rs570613 and rs2280015) with other SNPs in our population. These fully genotyped SNPs provide information on the entire IL13 and GATA3 genes. Two SNPs located in exons were examined in this study. While the rs2229359 SNP in GATA3 exon 3 is a synonymous variant, the rs20541 SNP is a coding variant in IL13 exon 4 with the common allele (G) coding for arginine and the minor allele (A) encoding gluta-mine at amino acid 144. The other validated GATA3 exonic SNP identified in our database search, rs11567901, was not polymorphic in our population. While no specific functional consequences have been prescribed to the several promoter and 3′ untranslated region variants we examined, they may have as of yet unidentified functional significance. The intronic SNPs examined are not located at known splice sites.
Table 1.
Genotype for IL13 and GATA3 single nucleotide polymorphism (SNPs)
| SNP | Locationa | Type | Genotypes | Genotype frequencies | |
|---|---|---|---|---|---|
| ILI3 | Chromosome 5 | ||||
| rs1800925 | 132020708 | 5′ promoter | CC/CT/TT | 577/295/35 (907) | |
| rs2066960 | 132022334 | Intron 1 | CC/CA/AA | 729/ 15 7/8 (894) | |
| rs1295686 | 132023742 | Intron 3 | CC/CT/TT | 483/240/25 (748) | |
| rs20541 | 132023863 | Exon 4b | GG/GA/AA | 583/291/32 (906) | |
| rs1295685 | 132024344 | 3′ UTR | GG/GA/AA | 584/280/41 (905) | |
| GATA3 | Chromosome 10 | ||||
| rs4143094 | 8129142 | Intergenic | GG/GT/TT | 497/334/46 (877) | |
| rs2275806 | 8135346 | 5′ promoter | AA/AG/GG | 25 8/482/177 (917) | |
| rs2229359 | 8140653 | Exon 3c | GG/GA/AA | 804/114/4 (922) | |
| rs444762 | 8143266 | Intron 3 | CC/CA/AA | 410/414/99 (923) | |
| rs406103 | 8151627 | Intron 5 | GG/GA/AA | 549/315/58 (922) | |
| rs1058240 | 8156604 | 3′ UTR | TT/TC/CC | 595/298/27 (920) | |
| rs379568 | 8165825 | 3′ UTR | GG/GA/AA | 644/188/10 (842) | |
SNP location data from Applied Biosystems database.
A nonsynonymous (amino acid change, arginine > glutamine) polymorphism.
A synonymous (no amino acid change) polymorphism. UTR, untranslated region.
Statistical analysis
Each SNP was tested for Hardy–Weinberg equilibrium using Haploview 3.2 software22 (http://www.broad.mit.edu/mpg/haploview/index.php). Estimates of LD between SNPs in each gene were calculated using D′ and r2.23
Using SAS/STAT® version 9.1 (SAS Institute, Cary, NC, U.S.A.), statistical analysis was performed on the data only from children who had complete information on IL13/GATA3 genotypes, and eczema phenotype. χ2 tests were used to compare the sample used in the analysis with that which was followed up at age 10 years. Repeated measurement analyses were conducted using Generalized Estimating Equations24 (PROC GENMOD). In order to provide unbiased parameter estimates and standard errors, this method takes into account correlation of observations collected on the same subject across successive points in time. To avoid collinearity of different SNPs, we tested models in a SNP-by-SNP approach controlling for confounders.
To determine whether genetic polymorphisms in the IL13 and GATA3 genes are associated with eczema dependent on atopy status, we stratified our analyses. After analysing all cases of eczema (eczema vs. no eczema), we investigated genotype association in children with atopic eczema compared with those without eczema. This step provides the effect of genetic polymorphisms on the combination of eczema and atopy. In a second step, we compared the occurrence of atopic eczema with children who have no eczema and no positive SPT at age 4 or 10 years. This analysis determines whether genetic polymorphisms also constitute a risk for eczema in atopic children. In all models, we included three IL13 SNPs and five GATA3 SNPs. Thus, the association of one SNP is mutually adjusted for all others. In addition, we adjusted for maternal and paternal eczema, low birth weight (< 2500 g), breastfeeding ≥ 3 months, and age. To control for false-positive (type I) error rate, when multiple tests are conducted, we adjusted the statistical significance of the SNPs for false discovery.25 We also tested whether IL13 and GATA3 SNPs showed an interaction on a multiplicative scale.
Results
Data on IL13 and GATA3 genotypes and eczema phenotypes were available for 894 children. Table 2 describes demographic characteristics for children who participated in the largest follow-up at 10 years (n = 1373) and those used in the analysis (n = 894). There were no significant differences between the samples.
Table 2.
Characteristics of children seen at age 10 years and those used in the analysis
| Characteristics | Seen at 10 years, n/N (%) |
Used in the analysis, n/N (%) |
P-value | ||
|---|---|---|---|---|---|
| Boy | 697/1373 (50·8) | 447/894 (50·0) | 0·72 | ||
| Maternal eczema | 219/1318 (16·6) | 149/852 (17·5) | 0·59 | ||
| Paternal eczema | 103/1316 (7·8) | 71/850 (8·4) | 0·66 | ||
| Low birth weight (< 2500 g) | 49/1336 (3·7) | 30/868 (3·5) | 0·79 | ||
| Breast-fed (≥ 3 months) | 563/1231 (45·8) | 394/822 (47·9) | 0·35 | ||
| Smoke exposure | |||||
| Maternal smoking during pregnancy | 319/1366 (23·4) | 185/891 (20·8) | 0·27 | ||
| Not during pregnancy, but ETS during childhood |
430/1366 (31·5) | 286/891 (32·1) | 0·86 | ||
| Any skin prick test positive (age 4 or 10 years) |
261/824 (31·7) | 235/716 (32·8) | 0·63 | ||
ETS, environmental tobacco smoke.
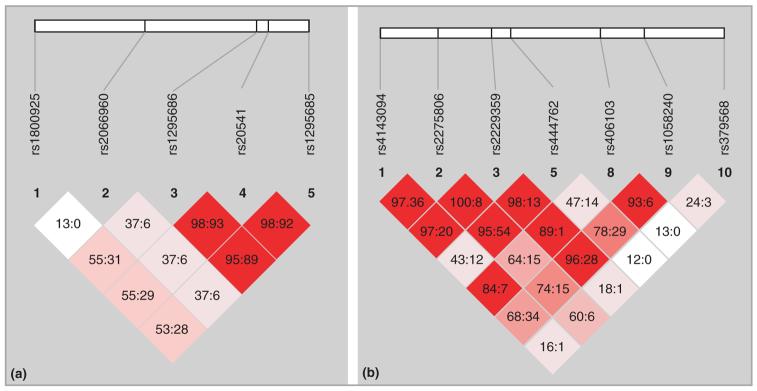
All the SNPs were in Hardy-Weinberg equilibrium. Three of the SNPs (rs1295686, rs20541 and rs1295685) in IL13 were in a block based on LD measures (D′ ≥ 0·95 and r2 ≥ 0·89; Fig. 1). For further analysis, we therefore used only the coding SNP (rs20541). Also, SNPs rs4143094, rs2275806 and rs2229359 in GATA3 were in a block based on D′ (D′ ≥ 0·97; Fig. 1) and, therefore, further analysis was done using only SNP rs2275806. SNP rs2275806 was selected as the representative SNP for this block as it was located in the 5′ promoter and had the highest minor allele frequency.
Fig 1.
Using the Haploview program, three single nucleotide polymorphisms of the IL13 gene and of the GATA3 gene appeared to be in a block. D′ and r2 are pairwise linkage disequilibrium (LD) determinants. (a) IL13 LD plot, D′ : r2 values displayed; (b) GATA3 LD plot, D′ : r2 values displayed.
The period prevalence of eczema increased steadily from infancy (9·6%) to age 10 years (13·1%) (Table 3). However, the prevalence of atopic eczema (eczema with positive SPT) remained at around 6% throughout the first decade of life. This indicates that new-onset eczema in later childhood is largely nonatopic.
Table 3.
Genotypes for IL13 single nucleotide polymorphisms (SNPs) and prevalence of eczema at different ages
|
IL13 polymorphisms |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1800925 |
rs2066960 |
rs20541 |
|||||||||
| CC | CT | TT | CC | AC | AA | GG | AG | AA | Total % (n/N) | ||
| All eczema cases and controls | n = 577 | n = 295 | n = 35 | n = 729 | n = 157 | n = 8 | n = 583 | n = 291 | n = 32 | ||
| Eczema at 1 year | 10·8 | 9·4 | 14·0 | 10·9 | 10·7 | 25·0 | 10·5 | 12·8 | 9·7 | 9·6 (132/1374) | |
| Eczema at 2 years | 10·2 | 12·3 | 13·0 | 11·2 | 9·6 | 25·0 | 9·2 | 14·0 | 17·9 | 10·3 (127/1231) | |
| Eczema at 4 years | 10·9 | 12·8 | 15·9 | 11·6 | 12·3 | 14·3 | 11·1 | 13·9 | 3·7 | 11·9 (145/1214) | |
| Eczema at 10 years | 17·0 | 16·6 | 32·0 | 16·2 | 14·7 | 37·5 | 16·0 | 16·5 | 18·8 | 13·1 (178/1358) | |
| Atopic eczema cases and controls | n = 520 | n = 267 | n = 28 | n = 655 | n = 144 | n = 6 | n = 522 | n = 264 | n = 29 | ||
| Atopic eczema at 1 year | 6·8 | 4·5 | 12·9 | 6·1 | 6·4 | 25·0 | 6·0 | 6·8 | 3·5 | 5·7 (61/1074) | |
| Atopic eczema at 2 years | 6·2 | 5·4 | 9·4 | 5·8 | 6·1 | 2·0 | 4·8 | 7·9 | 8·0 | 5·4 (54/991) | |
| Atopic eczema at 4 years | 6·7 | 8·1 | 11·5 | 6·8 | 9·0 | 14·3 | 6·5 | 8·9 | 3·7 | 6·4 (66/1026) | |
| Atopic eczema at 10 years | 6·9 | 7·1 | 10·7 | 6·9 | 7·6 | 16·7 | 6·3 | 8·3 | 10·3 | 6·5 (68/1050) | |
Unless indicated, all values are percentages.
There was no clear association of IL13 genotype and eczema assessed from birth to age 10 years (Table 3). A trend was seen for children with the minor allele (AA) of SNP rs2066960 and (TT) of SNP rs1800925 to have higher prevalence of eczema, but the difference did not reach statistical significance. No association was observed for GATA3 and eczema, without stratification. However, significant differences were observed with χ2 tests in the occurrence of atopic eczema at 2 and 10 years with rs2275806 and at 1 year with rs444762 (Table 4).
Table 4.
Genotypes for GATA3 single nucleotide polymorphisms (SNPs) and prevalence of eczema at different ages
|
GATA3 polymorphisms |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1058240 |
rs2275806 |
rs379568 |
rs406103 |
rs444762 |
||||||||||||
| TT | CT | CC | AA | AC | CC | GG | AG | AA | GG | AG | AA | CC | AC | AA | ||
| All eczema cases and controls |
n = 595 | n = 297 | n = 27 | n = 258 | n = 482 | n = 176 | n = 644 | n = 187 | n = 10 | n = 547 | n = 315 | n = 58 | n = 409 | n = 414 | n = 98 | |
| Eczema at 1 year | 10·9 | 12·3 | 4·0 | 12·3 | 11·9 | 7·7 | 11·3 | 11·9 | 10·0 | 11·7 | 10·8 | 7·4 | 11·2 | 12·2 | 6·5 | |
| Eczema at 2 years | 12·4 | 8·5 | 16·0 | 8·6 | 13·4 | 9·5 | 11·9 | 11·1 | 0·0 | 11·3 | 11·5 | 8·2 | 10·5 | 12·4 | 9·1 | |
| Eczema at 4 years | 11·0 | 12·9 | 12·5 | 10·7 | 13·3 | 10·0 | 12·6 | 9·2 | 0·0 | 11·6 | 12·9 | 9·3 | 10·3 | 13·8 | 10·9 | |
| Eczema at 10 years | 16·1 | 16·5 | 11·1 | 14·7 | 17·6 | 14·2 | 17·1 | 14·4 | 20·0 | 16·6 | 15·6 | 15·5 | 15·2 | 17·2 | 16·3 | |
| Atopic eczema cases and controls |
n = 533 | n = 269 | n = 26 | n = 226 | n = 437 | n = 163 | n = 573 | n = 172 | n = 9 | n = 488 | n = 290 | n = 51 | n = 365 | n = 377 | n = 88 | |
| Atopic eczema at 1 year | 5·7 | 7·4 | 4·0 | 4·5 | 7·6 | 4·9 | 5·6 | 8·3 | 10·0 | 6·3 | 6·1 | 5·7 | 4·2a | 8·7 | 3·3 | |
| Atopic eczema at 2 years | 6·7 | 4·4 | 8·7 | 2·3 | 8·1 | 5·9a | 6·5 | 5·3 | 0·0 | 5·3 | 7·5 | 4·3 | 4·1 | 7·8 | 5·9 | |
| Atopic eczema at 4 years | 6·7 | 7·8 | 8·7 | 5·0 | 8·5 | 7·1 | 6·8 | 8·1 | 0·0 | 6·3 | 9·5 | 3·9 | 5·1 | 9·1 | 7·9 | |
| Atopic eczema at 10 years | 6·6 | 7·8 | 7·7 | 3·1 | 9·2 | 7·4a | 7·0 | 7·0 | 11·1 | 6·8 | 8·3 | 3·9 | 5·2 | 9·0 | 6·8 | |
Unless indicated, all values are percentages.
P ≤ 0·05 in %2 χ2 tests comparing prevalence for the three genotypes of one SNP.
To determine the associations of IL13 and GATA3 with childhood eczema, we conducted repeated measurement regression analysis adjusting for confounding variables. Due to the limited number of children homozygous for minor alleles at all SNPs (< 5%), minor allele homozygous and heterozygous genotypes were grouped together. IL13 was neither associated with eczema nor with atopic eczema in childhood (Table 5).
Table 5.
Single nucleotide polymorphism of the GATA3 and IL13 genes and repeated assessment of eczema at age 1, 2, 4 and 10 years in all children and in children with atopic eczemaa
| Children with eczema vs. no eczema, n = 748–923; number of observations, n = 2405–3009; number of observations with eczema, n = 318–396 |
Children with atopic eczema vs. no eczema, n = 744–915; number of observations, n = 1809–2303; number of observations with atopic eczema, n = 129–182 |
Atopic eczema vs. nonatopic references, n = 669–730; number of observations, n = 1929–2105; number of observations with atopic eczema, n = 177–195 |
||
|---|---|---|---|---|
| ORb (95% CI) | ORb (95% CI) | ORb (95% CI) | ||
| IL13 | ||||
| rs1800925 | ||||
| TT | 1·67 (0·92–3·03) | 1·41 (0·55–3·61) | 1·34 (0·51–3·55) | |
| CT | 0·92 (0·65–1·31) | 0·81 (0·46–1·41) | 0·80 (0·47–1·37) | |
| CC | 1 | 1 | 1 | |
| rs2066960 | ||||
| AA | 2·39 (0·5 7–10·0) | 2·756 (0·38–20·4) | 1 -81 (0·27–12·14) | |
| AC | 1·03 (0·67–1·58) | 1·2831 (0·96–2·38) | 1·26 (0·68–2·31) | |
| CC | 1 | 1 | 1 | |
| rs20541 | ||||
| AA | 0·86 (0·34–2·19) | 0·80 (0·16–4·02) | 0·61 (0·13–2·96) | |
| GA | 1·29 (0·93–1·80) | 1 -38 (0·83–2·29) | 1 -25 (0·77–2·05) | |
| GG | 1 | 1 | 1 | |
| GATA3 | ||||
| rs1058240 | ||||
| CC | 0·88 (0·30–2·52) | 1·28 (0·31–5·20) | 1·01 (0·24–4·30) | |
| CT | 0·97 (0·70–1·35) | 1·10 (0·66–1·81) | 1·11 (0·68–1·83) | |
| TT | 1 | 1 | 1 | |
| rs2275806 | ||||
| GG | 0·84 (0·52–1·36) | 1·88 (0·88–4·03) | 1·67 (0·80–3·51) | |
| AG | 1·31 (0·92–1·86) | 2·55* (1·39–4·68) | 1·91** (1·05–3·46) | |
| AA | 1 | 1 | 1 | |
| rs379568 | ||||
| AA | 0·63 (0·18–2·25) | 0·88 (0·17–4·42) | 2·03 (0·34–12·09) | |
| GA | 0·92 (0·61 – 1·37) | 1·29 (0·72–2·30) | 1·33 (0·76–2·34) | |
| GG | 1 | 1 | 1 | |
| rs406103 | ||||
| AA | 0·68 (0·36–1·29) | 0·63 (0·21–1·88) | 0·54 (0·19–1·55) | |
| AG | 0·95 (0·67–1·34) | 1·34 (0·81–2·23) | 1·08 (0·66–1·76) | |
| GG | 1 | 1 | 1 | |
| rs444762 | ||||
| AA | 0·87 (0·50–1·51) | 1·29 (0·53–3·16) | 0·96 (0·40–2·28) | |
| AC | 1·1 1 (0·80–1·53) | 1·85* (1·12–3·06) | 1·56 (0·96–2·54) | |
| CC | 1 | 1 | 1 | |
The number of repeated measurements of eczema varies within each analysis depending on completed samples of the individual SNPs. Of the 3375 repeated measurements, 2115 were from children with no eczema and no positive skin prick test (SPT) at ages 4 and 10 years; eczema and positive SPT were found in 212 observations; 216 observations were from children with eczema but not allergic sensitization; and 832 were from children without eczema but with allergic sensitization (atopic eczema was defined as having eczema and at least one positive SPT at age 4 or 10 years of age).
Statistically controlled for age, sex, maternal and paternal history of eczema, low birth weight, and breastfeeding for 3 months and more. OR, odds ratio; CI, confidence interval
P <0·05 prior to adjustment for multiple testing
P < 0·05 after adjustment for multiple testing.
For GATA3, SNP rs2275806 (promoter region) and rs444762 (intron 3 region) showed an increased odds ratio for atopic eczema compared with all children without eczema (Table 5). However, after adjustment for multiple testing (false discovery rate, FDR), the association was no longer statistically significant. SNP rs2275806 (promoter region) also showed an increased odds ratio for atopic eczema compared with all nonatopic and noneczema children (Table 5), which remained statistically significant after adjustment for multiple testing (FDR: P = 0·04). We did not find statistically significant interactions between IL13 and GATA3 SNPs using repeated measurements in a logistic model.
Discussion
We investigated the association of IL13 and GATA3 with childhood eczema from birth up to the age of 10 years. There was no association of IL13 or GATA3 with childhood eczema. A trend was observed in crude analysis (Table 3) with IL13 SNPs; however, when adjusted for confounding variables in logistic regression analysis, no independent effect of IL13 with eczema was seen. Following stratification for atopy, SNP rs2275806 in the promoter region of GATA3 was found to be associated with atopic eczema with the AG genotype predisposing to a twofold increased risk (Table 5).
We did not observe a net clearance of eczema in the population from infancy to age 10 years, with a period prevalence remaining at around 10% up to age 4 years and slightly higher (13%) at the age of 10 years. This is at odds with the presumption that eczema is a disease of early childhood and improves in the vast majority of cases.26 However, a large population-based study concluded that the long-term prognosis of eczema may be worse than some previous studies have suggested.27 In this study 39% of children had age of onset of eczema before 1 year, 29% between 1 and 7 years, and 32% after the age of 7 years. Thus, new-onset eczema is not uncommon in later childhood and recurrence of infantile eczema may further increase the period prevalence in later childhood.
Incomplete examination of the gene and inappropriate or misclassification of the disease may result in either spurious or false-negative associations. We examined both IL13 and GATA3 genes comprehensively and examined many of the SNPs that have been covered in previous studies.8,10-13,16-21 Eczema is a disease of relapse and remission and children can be affected transiently. Thus, misclassification of eczema is possible in cross-sectional studies where the child may be in remission or may have grown out of eczema when the assessment was conducted. However, eczema was determined in this birth cohort with repeated longitudinal assessments from birth to 10 years of age. Thus, we are confident that both our phenotype and genotype data are valid.
Table 2 shows that no selection bias occurred for the subgroup of children used in this analysis (n = 894). This subgroup comes from the population recruited at birth (n = 1456) who chose to provide a blood sample for genetic analysis and where phenotype assessment was available from birth to 10 years. A limitation of our study was that the numbers were relatively small, which may explain the lack of association observed (false-negative or type II statistical error). This shortcoming was overcome by repeated measurement analysis, which not only makes an overall assessment of childhood eczema but, in statistical terms, the number of observations was increased with augmentation in the power of the study. Repeated measurement analysis also has the advantage of giving appropriate weighting to children with persistent disease.
IL-13 plays a pivotal role in the development of allergic responses.28 The IL13 gene is located on chromosome 5q31–33 in the cluster of genes encoding Th2 cytokines. It is, therefore, reasonable to hypothesize that polymorphisms in the IL13 gene may influence the risk of the development of allergic diseases, including eczema. Previous studies have shown that polymorphisms in the IL13 gene are associated with high total serum IgE,21 cord serum IgE29 and atopy.12 With regard to organ-specific allergic disease, an association with asthma has been suggested by some,19,30 but others have failed to confirm this.31,32 Liu et al.33 first reported an association of SNP G4257A (now termed rs20541) with serum IgE and eczema in German children. Subsequently, in Japanese adults, Tsunemi et al.10 reported that the ‘A’ allele of SNP G4257A was increased in patients with eczema compared with controls. A Canadian study further supported this notion by finding an association of atopy and eczema at age 2 years with the same polymorphism.11
Hummelshoj et al.,12 in a primarily caucasian population, found an association with both eczema as well as inhalation allergy with a different SNP (rs1800925). Our findings of a lack of association of IL13 with eczema are confirmatory to those of Chang et al.,13 who studied IL13 SNPs in the Chinese population. They used the PCR primers (genotype by sequencing) that would cover IL13 SNP at position −1070 corresponding to rs1800925. In addition to these SNPs, we investigated three further SNPs but found no association of IL13 with childhood eczema, with or without stratification for atopy. The reason for this lack of association is not clear. False-negative findings of clinically important risks due to a small sample are not likely. Comparing IL13 and GATA3, we see that at least 6·3% of the children in various IL13 geno-types, used as references, had eczema at age 10 years and at least 6·8% in the reference genotypes for GATA3 (Tables 3, 4). When estimating the statistical power for this setting, this study was able to detect a 1·6-fold odds ratio for SNPs in the IL13 and GATA3 genes. However, it is possible that associations are observed only in children with exposure to certain environmental influences in early life. Taking advantage of the birth cohort design and available information on some environmental exposures (such as smoking, pets and infant feeding practices), we re-analysed the data (not shown), but did not find an association.
We did find an association of GATA3 (SNP rs2275806) with atopic but not with nonatopic eczema, which remained significant following adjustment for confounding variables and for FDR. This is consistent with the prominent role of GATA-3 in facilitating Th2 immune responses5,6 and the role this transcription factor plays in Th2/Th1 bias in atopic eczema.7
Interestingly, a recent study34 suggests that GATA-3 may be required to establish epidermal barrier, which is essential in preventing water loss and entry of infections and allergens into the skin. Dysfunction of skin barrier function is critically important in the development of atopic eczema.35 It is possible that the association of GATA3 with atopic eczema in our study may be related both to its role in atopic immune responses, as well as in the development of epidermal barrier function. Animal models illustrate that the two functions are intimately linked. For example, impairment of the skin barrier by two genetically distinct mechanisms leads to innate immune responses, as observed in atopic dermatitis.34
Filaggrin is a key protein that facilitates terminal differentiation of the epidermis and formation of the skin barrier. It has been shown that two independent loss-of-function genetic variants in the gene encoding filaggrin predispose to atopic eczema.36 A defective epidermal barrier may facilitate allergen entry. This is supported by the finding of increased extrinsic (atopic) eczema and allergic sensitization in those with filaggrin gene mutation.37 Conversely, patients with atopic eczema (not carrying filaggrin gene defect) have an acquired defect in filaggrin expression in the skin, modulated by Th2-type immune response. Howell et al. have shown that IL-4 and IL-13 inhibit the expression of filaggrin gene in keratinocytes from patients with atopic eczema.38 The interaction between genes encoding proteins important for epidermal barrier function, such as filaggrin, and those modulating immune responses, such as GATA3, IL4 and IL13, may be further investigated by gene–gene interaction in large populations. In addition, functional genomics studies, by looking at the expression of these genes in those with extrinsic (atopic) and intrinsic (nonatopic) eczema, will be extremely helpful.
GATA-3 regulates the function of Th2 cytokines such as IL-4, IL-5 and IL-13.39 Hence, the increased risk of a GATA3 polymorphism may explain why some studies found an association of the IL13 gene with eczema and others did not. It is possible that studies showing a link of IL13 and eczema studied subjects who also had appropriate GATA3 SNP. There was no clear evidence of gene–gene (GATA3–IL13) or gene–environment (smoking) interactions in our sample. However, this requires further investigation as interactions with GATA3 may be different in various developmental stages. Ouyang et al.39 speculated that GATA-3 may act early during a temporarily restricted window of development.
At present we cannot be certain of the functional consequence of the rs2275806 SNP in the GATA3 gene; it is located upstream of the gene and to date has no known function. However, due to its 5′ location it may be involved in gene promotion or transcription factor binding. Thus, we believe that it merits further investigations, especially in studies that assess the role of interleukins, as GATA-3 has a regulatory role in immune development.
Acknowledgments
The authors gratefully acknowledge the cooperation of the children and parents who participated in this study, and appreciate the hard work of Mrs Sharon Matthews and the Isle of Wight research team in collecting phenotype data. The authors thank Hans Cheng for use of Pyrosequencing equipment and Dennis Shubitowski for technical assistance. This study was funded in part by National Institutes of Health R01 AI061471. The 10-year follow-up of this study was funded with the assistance of the National Asthma Campaign, U.K. (grant no. 364).
Footnotes
Conflicts of interest
None declared.
References
- 1.Punnonen J, Aversa G, Cocks BG, et al. Interleukin 13 induces interleukin 4-independent IgG4 and IgE synthesis and CD23 expression by human B cells. Proc Natl Acad Sci U.S.A. 1993;90:3730–4. doi: 10.1073/pnas.90.8.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung DYM, Bhan AK, Schneeberger EE, Geha RS. Characterization of the mononuclear cell infiltrate in atopic dermatitis using monoclonal antibodies. J Allergy Clin Immunol. 1983;71:47–56. doi: 10.1016/0091-6749(83)90546-8. [DOI] [PubMed] [Google Scholar]
- 3.Hamid Q, Naseer T, Minshall EM, et al. In vivo expression of IL-12 and IL-13 in atopic dermatitis. J Allergy Clin Immunol. 1996;98:225–31. doi: 10.1016/s0091-6749(96)70246-4. [DOI] [PubMed] [Google Scholar]
- 4.Zon LI, Yamaguchi Y, Yee KA, et al. Expression of mRNA for the GATA-binding proteins in human eosinophils and basophils: potential role in gene transcription. Blood. 1993;81:3234–41. [PubMed] [Google Scholar]
- 5.Chakir H, Wang H, Lefebvre DE, et al. T-bet/GATA-3 ratio as a measure of the Th1/Th2 cytokine profile in mixed cell populations: predominant role of GATA-3. J Immunol Methods. 2003;278:157–69. doi: 10.1016/s0022-1759(03)00200-x. [DOI] [PubMed] [Google Scholar]
- 6.Macaubas C, Lee PT, Smallacombe TB, et al. Reciprocal patterns of allergen-induced GATA-3 expression in peripheral blood mononuclear cells from atopics vs. non-atopics. Clin Exp Allergy. 2002;32:97–106. doi: 10.1046/j.0022-0477.2001.01288.x. [DOI] [PubMed] [Google Scholar]
- 7.Arakawa S, Hatano Y, Katagiri K. Differential expression of mRNA for Th1 and Th2 cytokine-associated transcription factors and suppressors of cytokine signalling in peripheral blood mononuclear cells of patients with atopic dermatitis. Clin Exp Immunol. 2004;135:505–10. doi: 10.1111/j.1365-2249.2004.02405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pykäläinen M, Kinos R, Valkonen S, et al. Association analysis of common variants of STAT6, GATA3, and STAT4 to asthma and high serum IgE phenotypes. J Allergy Clin Immunol. 2005;115:80–7. doi: 10.1016/j.jaci.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Beyer K, Nickel R, Freidhoff L, et al. Association and linkage of atopic dermatitis with chromosome 13q12–14 and 5q 31–33 markers. J Invest Dermatol. 2000;115:906–8. doi: 10.1046/j.1523-1747.2000.00096.x. [DOI] [PubMed] [Google Scholar]
- 10.Tsunemi Y, Saeki H, Nakamura K, et al. Interleukin-13 gene polymorphism G4257A is associated with atopic dermatitis in Japanese patients. J Dermatol Sci. 2002;30:100–7. doi: 10.1016/s0923-1811(02)00065-8. [DOI] [PubMed] [Google Scholar]
- 11.He JQ, Chan-Yeung M, Becker AB, et al. Genetic variants of the IL13 and IL4 genes and atopic diseases in at-risk children. Genes Immun. 2003;4:385–9. doi: 10.1038/sj.gene.6363985. [DOI] [PubMed] [Google Scholar]
- 12.Hummelshoj T, Bodtger U, Datta P, et al. Association between an interleukin-13 promoter polymorphism and atopy. Eur J Immunogenet. 2003;30:355–9. doi: 10.1046/j.1365-2370.2003.00416.x. [DOI] [PubMed] [Google Scholar]
- 13.Chang YT, Lee WR, Yu CW, et al. No association of cytokine gene polymorphism in Chinese patients with atopic dermatitis. Clin Exp Dermatol. 2006;31:419–23. doi: 10.1111/j.1365-2230.2006.02124.x. [DOI] [PubMed] [Google Scholar]
- 14.SNPper . The Children's Hospital Informatics Program (CHIP) Bioinformatics Tools. Harvard School of Medicine; Boston, MA: 2005. Available at: http://snpper.chip.org (last accessed 31 January 2008) [Google Scholar]
- 15.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vercelli D. Genetics of IL-13 and functional relevance of IL-13 variants. Curr Opin Allergy Clin Immunol. 2002;2:389–93. doi: 10.1097/00130832-200210000-00004. [DOI] [PubMed] [Google Scholar]
- 17.Donfack J, Schneider DH, Tan Z, et al. Variation in conserved noncoding sequences on chromosome 5q and susceptibility to asthma and atopy. Respir Res. 2005;6:145. doi: 10.1186/1465-9921-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffjan S, Ostrovnaja I, Nicolae D, et al. Genetic variation in immunoregulatory pathways and atopic phenotypes in infancy. J Allergy Clin Immunol. 2004;113:511–18. doi: 10.1016/j.jaci.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 19.Howard TD, Whittaker PA, Zaiman AL, et al. Identification and association of polymorphisms in the interleukin-13 gene with asthma and atopy in a Dutch population. Am J Respir Cell Mol Biol. 2001;25:377–84. doi: 10.1165/ajrcmb.25.3.4483. [DOI] [PubMed] [Google Scholar]
- 20.Puthothu B, Krueger M, Forster J, et al. Association between severe respiratory syncytial virus infection and IL13/IL4 haplotypes. J Infect Dis. 2006;193:438–41. doi: 10.1086/499316. [DOI] [PubMed] [Google Scholar]
- 21.Graves PE, Kabesch M, Halonen M, et al. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol. 2000;105:506–13. doi: 10.1067/mai.2000.104940. [DOI] [PubMed] [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 23.Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–94. [PubMed] [Google Scholar]
- 24.Stokes ME, Davis CS, Koch GG. Categorial Data Analysis Using the SAS System. SAS Institute Inc.; Cary, NC: 1995. [Google Scholar]
- 25.Yang Q, Cui J, Chazaro I, Cupples LA. Power and type I error rate of false discovery rate approaches in genome-wide association studies. BMC Genet. 2005;6(Suppl 1):S134. doi: 10.1186/1471-2156-6-S1-S134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stifler WC. A 21 year follow-up of infantile eczema. J Pediatr. 1996;66:166–77. doi: 10.1016/s0022-3476(65)80274-8. [DOI] [PubMed] [Google Scholar]
- 27.Williams HC, Strachan DP. The natural history of childhood eczema. Observations from the British 1958 birth cohort study. Br J Dermatol. 1998;139:834–9. doi: 10.1046/j.1365-2133.1998.02509.x. [DOI] [PubMed] [Google Scholar]
- 28.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Immunol Rev. 2004;202:175–90. doi: 10.1111/j.0105-2896.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 29.Sadeghnejad A, Karmaus W, Arshad SH, Ewart S. IL13 gene polymorphism association with cord serum immunoglobulin E. Pediatr Allergy Immunol. 2007;18:288–92. doi: 10.1111/j.1399-3038.2006.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Pouw Kraan TC, van Veen A, Boeije LC, et al. An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun. 1999;1:61–5. doi: 10.1038/sj.gene.6363630. [DOI] [PubMed] [Google Scholar]
- 31.Leung TF, Tang NL, Chan IH, et al. A polymorphism in the coding region of interleukin-13 gene is associated with atopy but not asthma in Chinese children. Clin Exp Allergy. 2001;31:1515–21. doi: 10.1046/j.1365-2222.2001.01212.x. [DOI] [PubMed] [Google Scholar]
- 32.Hakonarson H, Bjornsdottir US, Ostermann E, et al. Allelic frequencies and patterns of single-nucleotide polymorphisms in candidate genes for asthma and atopy in Iceland. Am J Respir Crit Care Med. 2001;164:2036–44. doi: 10.1164/ajrccm.164.11.2101086. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Nickel R, Beyer K, et al. An IL-13 coding region variant is associated with high total serum IgE level and atopic dermatitis in the German multicenter atopy study (MAS-90) J Allergy Clin Immunol. 2000;106:167–70. doi: 10.1067/mai.2000.107935. [DOI] [PubMed] [Google Scholar]
- 34.Strong CG, Wertz PW, Wang C, et al. Lipid defect underlies selective skin barrier impairment of an epidermal-specific deletion of Gata-3. J Cell Biol. 2006;175:661–70. doi: 10.1083/jcb.200605057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung DY, Boguniewicz M, Howell MD, et al. New insights into atopic dermatitis. J Clin Invest. 2004;113:651–7. doi: 10.1172/JCI21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer CN, Irvine AD, Terron-Kwiatkowski A, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 37.Weidinger S, Illig T, Baurecht H, et al. Loss-of-function variations within the filaggrin gene predispose for atopic dermatitis with allergic sensitizations. J Allergy Clin Immunol. 2006;118:214–19. doi: 10.1016/j.jaci.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Howell MD, Kim BE, Gao P, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol. 2007;120:150–5. doi: 10.1016/j.jaci.2007.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouyang W, Ranganath SH, Weindel K, et al. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–55. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]