Abstract
Purpose
This study evaluated the [-2]proPSA serum marker using a blinded reference specimen set from three NCI Early Detection Research Network centers from men with an indication for prostate biopsy.
Materials and Methods
Serum was collected pre-biopsy from 123 men with no prior biopsy or prostate cancer history. Specimens (cancer cases: 51%; non-cancer controls: 49%) were selected equally from the three sites and analyzed for PSA, free PSA, [-2]proPSA, BPSA, and testosterone (Beckman Coulter ACCESS analyzer).
Results
There was no difference in total PSA concentrations (non-cancer: 6.80±5.20 ng/mL, cancer: 6.94±5.12 ng/mL) between the groups. Overall, %[-2]proPSA had the greatest area under the curve (AUC=0.69) followed by %fPSA (AUC=0.61). For %[-2]proPSA, maximal sensitivity was 60% and specificity was 70%. A logistic regression model combining PSA, BPSA, %fPSA, %[-2]proPSA, [-2]proPSA/BPSA, and testosterone had an AUC of 0.73. In the 2-10 ng/mL PSA range, %[-2]proPSA and the model had the largest AUC (0.73). The AUC for %fPSA was 0.53. Specificities for %[-2]proPSA, the logistic regression model, and %fPSA at 90% sensitivity were 41%, 32%, and 18% and at 95% sensitivity were 31%, 26%, and 16%, respectively.
Conclusions
%[-2]proPSA was the best predictor of prostate cancer detection compared to %fPSA, particularly in the 2-10 ng/mL total PSA range. These findings provide rationale for broader validation studies to determine whether %[-2]proPSA alone can replace other molecular PSA assays (such as %fPSA) for improving the accuracy of prostate cancer early detection and support the utility of well-characterized, carefully collected reference sets to evaluate new biomarkers.
Keywords: prostate cancer, PSA, proPSA, detection, markers
Introduction
It has been recognized that disease-specific isoforms of free prostate specific antigen (PSA) exist in prostate tissue and in serum and may improve some of the current limitations of PSA in the early detection of prostate cancer.1 These enzymatically inactive forms include proenzyme PSA (proPSA), associated with prostate cancer and the peripheral zone of the prostate and BPSA, a degraded form with internal peptide bond cleavages associated with benign prostatic hyperplasia and the transition zone of the prostate.2-4 An additional intact, yet denatured form, termed iPSA, is also found in serum.5
proPSA is the precursor form of PSA and contains a 7 amino acid pro leader peptide. Additional truncated forms of proPSA exist in serum, primarily those with leader sequences of 5, 4, and 2 amino acids. Cleavage of the leader sequences by human kallikrein 2 (hk2) and trypsin to activate PSA decreases with decreasing size of the propeptide leader sequence with [-2]proPSA resistant to activation. [-2]proPSA is the most prevalent form in tumor extracts and immunohisotchemically stains cancer cells more strongly than benign cells.6 The [-2]proPSA form and the other proPSA forms have been studied individually and in combination and suggest a role for these molecular forms of PSA in the early detection of prostate cancer 3, 7-10 as well as in identifying aggressive cancers.11, 12 Recently, automated, chemiluminescent immunoassays for [-2]proPSA as well as for BPSA have been developed.13, 14
Based on preliminary studies, several potential prostate cancer biomarkers, including [-2]proPSA, were identified by the NCI's Early Detection Research Network (EDRN) for further evaluation using a common, retrospective, archival, blinded reference set of serum samples collected at three EDRN sites in men with an indication for prostate biopsy. Here we report on the results of the analysis of the molecular forms of free PSA using newly developed automated immunoassays.
Materials and Methods
The 123 serum specimens analyzed in this study were part of retrospective collections from the three NCI EDRN Prostate Clinical Epidemiology and Validation Centers at Beth Israel Deaconess Medical Center (BIDMC), Boston MA; Johns Hopkins University (JHU), Baltimore, MD; and the University of Texas Health Science Center at San Antonio (UTHSCSA), San Antonio, TX. Specimens were from men with an indication for prostate biopsy and subjects had no prior prostate cancer history. All men had a ≥ 10 core biopsy. Blood was collected prior to biopsy and sera stored frozen at -70 °C for up to six years prior to analysis. Informed consent was obtained under IRB-approved and HIPAA compliant protocols.
Equal numbers of cancer cases (51%) and non-cancer controls (49%) were randomly selected from specimens which were frozen at -70 °C within 12 hours of collection and were distributed equally among the three sites (BIDMC: n=40; JHU: n=43; UTHSCSA: n=40). Specimens were analyzed in a blinded fashion at Beckman Coulter, Inc. on the Beckman Coulter ACCESS 2 immunoassay system (Beckman Coulter, Inc., Chaska, MN) for PSA, free PSA (fPSA), [-2]proPSA, BPSA, and testosterone. The PSA assays are all dual monoclonal sandwich assays using Hybritech antibodies and a chemiluminescent detection system. The assays for PSA, free PSA, and testosterone are commercially available while the assays for [-2]proPSA 14 and BPSA 13 are for research use only. The [-2]proPSA assay is calibrated using [-2]proPSA purified from the AVA12-PSA mammalian cell line while BPSA assay is calibrated using BPSA purified from human seminal plasma. These assays have linear ranges of <1-5,000 pg/mL for [-2]proPSA and <5-5,000 pg/mL for BPSA with inter-assay precision of 4.8-12.5% (20-279 pg/mL) 3.7-7.2% (31-1,816 pg/mL) for [-2]proPSA and BPSA, respectively. Cross-reactivity of other PSA isoforms in the two assays is minimal.
The following PSA derivatives were calculated: %fPSA (fPSA/PSA), %[-2]proPSA ([-2]proPSA/fPSA), %BPSA (BPSA/fPSA) and [−2]proPSA/BPSA. Differences in the cancer and non-cancer groups were assessed using student's t-test and the Mann-Whitney U Test. Results for these parametric and non-parametric statistical tests were similar and therefore only student's t-test results are presented. Diagnostic utility was assessed using ROC analysis and areas under the curve determined in addition to clinical sensitivity and specificity. Statistics were performed with Analyse-it (version 1.73). A logistic regression model combining PSA and its derivatives and testosterone was also generated using R (version 2.3.1) software. The resulting linear predictor score was used to evaluate the combined markers by ROC analysis.
Results
The mean age (±SD) of the 123 subjects in this study was 62.2±8.2 years (41-83 years) with a racial distribution as follows: Caucasian: 90.2%; African American: 6.5%; and Other: 3.2%. In the 120 men in whom information was available, 79% of the non-cancer group had a DRE non-suspicious for cancer, while in the cancer group 71% of men had a DRE non-suspicious for cancer. Within the cancer group, 52% of men had a biopsy Gleason score of 6, 38% had a score of 7, and 10% had a score of 8 or 9.
The utility of the isoforms of free PSA, particularly [-2]proPSA was examined in the entire dataset (PSA range 0.48-33.18 ng/mL) and in the clinically important ranges incorporating 4-10 ng/mL total PSA and 2-10 ng/mL total PSA. Overall, the cancer and non-cancer groups were equivalent with respect to age (non-cancer: 61.7±8.6 years, cancer: 62.6±7.8 years) as well as total PSA concentrations (non-cancer: 6.80±5.20 ng/mL, cancer: 6.94±5.1 ng/mL). A comparison between the two groups for the measured PSA derivatives and calculated values as well as testosterone is shown in Table 1. %fPSA was significantly lower (p<0.05) in the cancer group while [-2]proPSA and %[-2]proPSA were significantly higher in the cancer group.
Table 1.
Comparison of mean serum values for the non-cancer and cancer groups for all subjects (n=123)
| Non-Cancer | Cancer | p-value | |||||
|---|---|---|---|---|---|---|---|
| Mean±SD | Median | N | Mean±SD | Median | N | ns | |
| PSA, ng/mL | 6.80±5.20 | 5.52 | 60 | 6.94±5.12 | 5.74 | 63 | ns |
| Free PSA, ng/mL | 1.14±1.35 | 0.84 | 60 | 0.94±0.66 | 0.84 | 63 | ns |
| [-2]proPSA, pg/mL | 10.00±5.93 | 7.74 | 60 | 12.70±7.52 | 10.72 | 63 | 0.03 |
| BPSA, pg/mL | 363.5±652.2 | 197.0 | 59 | 299.6±377.9 | 213.3 | 62 | ns |
| %fPSA | 19.1±10.0 | 16.5 | 60 | 15.0±6.72 | 14.8 | 63 | 0.008 |
| %[-2]proPSA | 1.18±0.61 | 1.07 | 60 | 1.53±0.55 | 1.47 | 63 | 0.001 |
| %BPSA | 27.2±18.3 | 22.0 | 59 | 28.9±14.1 | 27.2 | 62 | ns |
| [-2]proPSA/BPSA | 0.078±0.136 | 0.046 | 59 | 0.066±0.041 | 0.060 | 62 | ns |
| Testosterone, ng/mL | 3.77±1.14 | 3.61 | 54 | 3.93±1.51 | 3.85 | 59 | ns |
ns: not significant
Results for the 4-10 ng/mL and 2-10 ng/mL PSA truncated ranges were similar and therefore only the 2-10 ng/mL data are presented here. In that range, similar to the overall range of data (Table 2), %[-2]proPSA was significantly higher (p<0.05) in the cancer group, as was [-2]proPSA alone and the ratio of [-2]proPSA/BPSA. %fPSA was equivalent between the cancer and non-cancer groups.
Table 2.
Comparison of mean serum values for the non-cancer and cancer groups for subjects in the 2-10 ng/mL PSA range (n=89)
| Non-Cancer | Cancer | p-value | |||||
|---|---|---|---|---|---|---|---|
| Mean±SD | Median | N | Mean±SD | Median | N | ns | |
| PSA, ng/mL | 5.82±2.02 | 5.49 | 39 | 5.60±2.01 | 5.41 | 50 | ns |
| Free PSA, ng/mL | 0.92±0.460 | 0.84 | 39 | 0.82±0.42 | 0.80 | 50 | ns |
| [-2]proPSA, pg/mL | 9.31±4.52 | 7.40 | 39 | 11.97±6.38 | 10.09 | 50 | 0.03 |
| BPSA, pg/mL | 289.1±273.6 | 216.5 | 38 | 224.0±149.2 | 197.9 | 49 | ns |
| %fPSA | 16.4±6.82 | 15.4 | 39 | 15.6±6.89 | 15.2 | 50 | ns |
| %[-2]proPSA | 1.10±0.43 | 1.08 | 39 | 1.53±0.53 | 1.47 | 50 | 0.0001 |
| %BPSA | 30.3±20.7 | 23.3 | 39 | 27.4±13.49 | 25.1 | 49 | ns |
| [-2]proPSA/BPSA | 0.050±0.03 | 0.045 | 39 | 0.069±0.040 | 0.063 | 49 | 0.02 |
| Testosterone, ng/mL | 3.92±1.21 | 3.72 | 34 | 3.83±1.38 | 3.70 | 47 | ns |
ns: not significant
ROC analysis of the non-cancer controls and cancer cases in all subjects is shown in Table 3 and Figure 1. Of the PSA derivatives, %[-2]proPSA had the greatest area under the curve (AUC) of 0.69 followed by %fPSA with an AUC of 0.61. At a sensitivity of 90 percent, corresponding specificity was 32% for %fPSA and 37% for %[-2]proPSA while at 95% sensitivity, specificity was 30% for %fPSA and 15% for %[-2]proPSA, respectively. At the optimal cutoff point for %[-2]proPSA (1.4%) corresponding to the maximal sum of sensitivity and specificity, sensitivity was 60% with a specificity of 70%. A logistic regression model was constructed combining PSA, BPSA, %fPSA, %[-2]proPSA, [-2]proPSA/BPSA, and testosterone, although only %[-2]proPSA was significant in the final model. The AUC was 0.73 and sensitivity at 90% and 95% specificity was 39% and 28%, respectively (Figure 1).
Table 3.
ROC analysis for all subjects (n=123)
| N | ROC AUC | 95% Confidence Interval | |
|---|---|---|---|
| PSA | 123 | 0.52 | 0.42-0.63 |
| Free PSA | 123 | 0.53 | 0.42-0.63 |
| [-2]proPSA | 123 | 0.63 | 0.53-0.73 |
| BPSA | 121 | 0.50 | 0.40-0.61 |
| %fPSA | 123 | 0.61 | 0.51-0.71 |
| %[-2]proPSA | 123 | 0.69 | 0.60-0.79 |
| %BPSA | 121 | 0.58 | 0.47-0.68 |
| [-2]proPSA/BPSA | 121 | 0.58 | 0.48-0.69 |
| Testosterone | 113 | 0.52 | 0.42-0.63 |
| Logistic regression model (PSA, BPSA, %fPSA, %[-2]proPSA, [-2]proPSA/BPSA, testosterone) | 113 | 0.73 | 0.64-0.83 |
Figure 1.
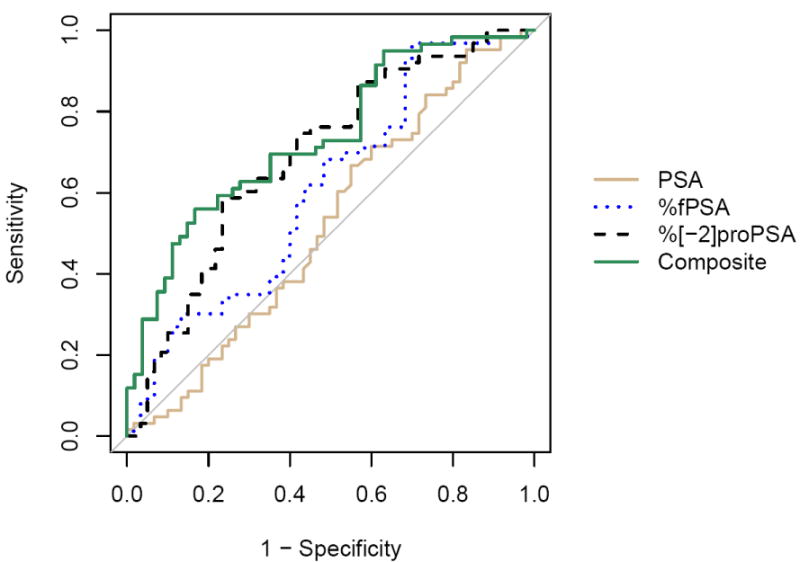
ROC analysis for all subjects (n=123) comparing PSA (AUC=0.52), %fPSA (AUC=0.61), %[-2]proPSA (AUC=0.69), and a composite logistic regression model combining PSA, BPSA, %fPSA, %[-2]proPSA, %[-2]proPSA/BPSA, and testosterone (AUC=0.73)
In the 2-10 ng/mL PSA range, individually, %[-2]proPSA again had the largest AUC (0.73) while the AUC for %fPSA was 0.53 (Table 4, Figure 2). When PSA, BPSA, %fPSA, %[-2]proPSA, [-2]proPSA/BPSA, and testosterone were evaluated together using logistic regression (Figure 2), only %[-2]proPSA was a significant discriminator for prostate cancer (AUC=0.73). The specificities for %fPSA, %[-2]proPSA, and the logistic regression model at 90% sensitivity were 18%, 41%, and 32%, respectively and at 95% sensitivity were 15%, 31%, and 26% respectively.
Table 4.
ROC analysis for subjects in the 2-10 ng/mL PSA range (n=89)
| N | ROC AUC | 95% Confidence Interval | |
|---|---|---|---|
| PSA | 89 | 0.52 | 0.40-0.64 |
| Free PSA | 89 | 0.58 | 0.46-0.69 |
| [-2]proPSA | 89 | 0.65 | 0.53-0.76 |
| BPSA | 87 | 0.55 | 0.43-0.68 |
| %fPSA | 89 | 0.53 | 0.41-0.65 |
| %[-2]proPSA | 89 | 0.73 | 0.63-0.84 |
| %BPSA | 87 | 0.50 | 0.38-0.63 |
| [-2]proPSA/BPSA | 87 | 0.65 | 0.54-0.77 |
| Testosterone | 81 | 0.51 | 0.38-0.64 |
| Logistic regression model (PSA, BPSA, %fPSA, %[-2]proPSA, [-2]proPSA/BPSA, testosterone) | 81 | 0.73 | 0.62-0.84 |
Figure 2.
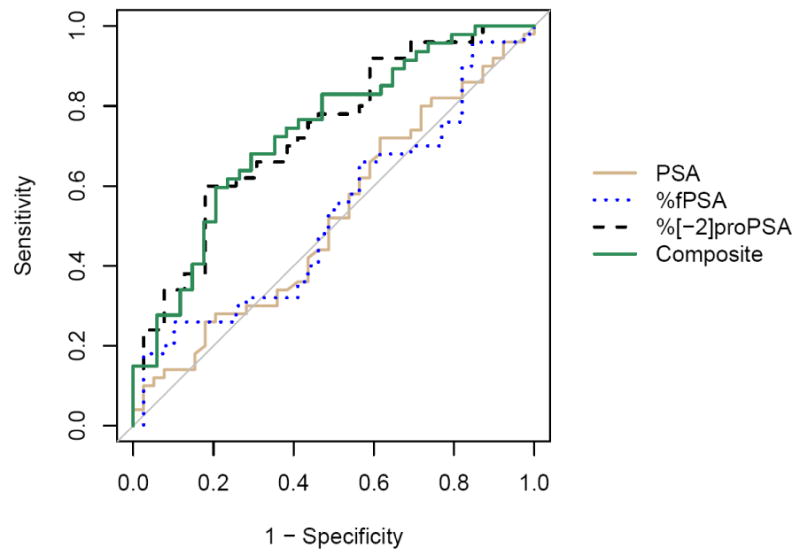
ROC analysis for all subjects with PSA between 2 and 10 ng/mL (n=89) comparing PSA (AUC=0.52), %fPSA (AUC=0.53), %[-2]proPSA (AUC=0.72), and a composite logistic regression model combining PSA, BPSA, %fPSA, %[-2]proPSA, [-2]proPSA/BPSA and testosterone (AUC=0.73, n=81).
Discussion
In the early 1990s, the discovery that PSA is present in serum in both a “free” form and “bound” to protease inhibitors, particularly α1-antichymotrypsin, and that differing proportions of these forms are present in men with and without prostate cancer led to the development of commercial assays for free and complexed PSA that were able to improve the specificity of PSA within defined total PSA ranges.15, 16 The knowledge that free PSA is in fact comprised of cancer (proPSA) and benign (BPSA) prostate specific-forms may explain the limitations of %free PSA in the mid-range of values (another diagnostic gray zone) and further serve to improve the clinical utility of the PSA marker.1
The various truncated and full-length forms of proPSA [-2], [-4], [-5, -7] and the sum of these forms have been variously examined for prostate cancer detection with in general the [-2]proPSA form and the sum showing the greatest value.3, 7, 10 Mixed results with the [-5, -7] propSA form may reflect population or assay differences.17, 18 In this study, [-2]proPSA and BPSA, in addition to total and free PSA, were analyzed using robust assays on an automated immunoassay analyzer. The samples from three different sites, although modest in number, were well-defined clinically, and collected and stored in a uniform manner as well as analyzed in a blinded fashion. In this study [-2]proPSA was best presented as a ratio with free PSA (%[-2]proPSA) and was more useful diagnostically in the truncated PSA ranges of 4-10 ng/mL and 2-10 ng/mL total PSA than overall PSA range. In the 2-10 ng/mL PSA range (data similar to 4-10 ng/mL range, not shown), the area under the ROC curve was 0.73 compared to 0.53 for %fPSA while the sensitivity at 95% specificity was 31% compared to 16% for %fPSA. In a screening study of men with PSA concentrations between 4 and 10 ng/mL, %proPSA (sum) had an AUC of 0.69 and specificity of 19% at 95% sensitivity, which was greater than all other PSA forms.3 Similarly, in a two site study, %proPSA spared 21% of unnecessary biopsies compared to %fPSA (13%) in the 2-10 ng/mL range.7 %proPSA has also shown improvement over %fPSA in the 2.5-4 ng/mL 10 and 2-4 ng/mL 7 PSA ranges, although sample size limited evaluating this range in the current study.
Combining PSA markers has become a standard in the clinical interpretation of free PSA which is best utilized as a ratio with total PSA. Similarly, [-2]proPSA appears best presented as a percentage (or ratio) of free PSA. The multiplicity of PSA forms provides potential opportunities to combine markers with various algorithms and models. In a study in the 4-10 ng/mL PSA range, a multivariate logistic regression model containing total PSA, %fPSA, and the sum of proPSA forms gave the best performance compared to individual markers with an AUC of 0.77 and a specificity of 44% at 90% sensitivity.8 In another preliminary study, the ratio of [-2]proPSA/(fPSA-sum proPSA) was associated with cancer detection.9 In a larger study, [-5, -7]proPSA did not improve specificity for cancer detection alone, but had utility combined with other markers using artificial neural networks.19 In this study, a logistic regression model appeared promising although only the %[-2]proPSA term in the model was a significant predictor.
In summary, %[-2]proPSA was the best predictor of prostate cancer in this blinded standard reference set in the 2-10 ng/mL PSA range. These findings provide rationale for broader validation studies to determine whether %p2PSA alone can supplant other multiple molecular PSA assays for improving accuracy of prostate cancer screening. The study also highlights the utility of the EDRN reference set as a validation tool and the potential value of cancer marker reference sets 20 for evaluating the clinical utility of individual markers, comparing the performance of different markers with a common set of samples, and combining results from different laboratories/assays into a multi-marker panel.
Acknowledgments
The contribution of Beckman Coulter, Inc for the analysis of the samples in this study is gratefully acknowledged.
Grant support: This study was supported by funding from the NCI Early Detection Research Network: (DWC: U24 CA115102, AWP: U01 CA86323, MGS: U01 CA11391, IMT: U01 CA86402, ZF: U01 CA86368)
Abbreviations
- PSA
prostate specific antigen
- fPSA
free PSA
- proPSA
proenzyme PSA
- EDRN
Early Detection Research Network
- IRB
institutional review board
- HIPAA
Health Insurance Portability and Accountability Act
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mikolajczyk SD, Marks LS, Partin AW, Rittenhouse HG. Free prostate-specific antigen in serum is becoming more complex. Urology. 2002;59:797. doi: 10.1016/s0090-4295(01)01605-3. [DOI] [PubMed] [Google Scholar]
- 2.Mikolajczyk SD, Millar LS, Wang TJ, Rittenhouse HG, Marks LS, Song W, et al. A precursor form of prostate-specific antigen is more highly elevated in prostate cancer compared with benign transition zone prostate tissue. Cancer Res. 2000;60:756. [PubMed] [Google Scholar]
- 3.Mikolajczyk SD, Catalona WJ, Evans CL, Linton HJ, Millar LS, Marker KM, et al. Proenzyme forms of prostate-specific antigen in serum improve the detection of prostate cancer. Clin Chem. 2004;50:1017. doi: 10.1373/clinchem.2003.026823. [DOI] [PubMed] [Google Scholar]
- 4.Mikolajczyk SD, Millar LS, Wang TJ, Rittenhouse HG, Wolfert RL, Marks LS, et al. “BPSA,” a specific molecular form of free prostate-specific antigen, is found predominantly in the transition zone of patients with nodular benign prostatic hyperplasia. Urology. 2000;55:41. doi: 10.1016/s0090-4295(99)00372-6. [DOI] [PubMed] [Google Scholar]
- 5.Zhang WM, Leinonen J, Kalkkinen N, Dowell B, Stenman UH. Purification and characterization of different molecular forms of prostate-specific antigen in human seminal fluid. Clin Chem. 1995;41:1567. [PubMed] [Google Scholar]
- 6.Chan TY, Mikolajczyk SD, Lecksell K, Shue M, Rittenhouse HG, Partin AW, et al. Immunohistochemical staining of prostate cancer with monoclonal antibodies to the precursor of prostate-specific antigen. Urology. 2003;62:177. doi: 10.1016/s0090-4295(03)00138-9. [DOI] [PubMed] [Google Scholar]
- 7.Catalona WJ, Bartsch G, Rittenhouse HG, Evans CL, Linton HJ, Amirkhan A, et al. Serum pro prostate specific antigen improves cancer detection compared to free and complexed prostate specific antigen in men with prostate specific antigen 2 to 4 ng/ml. J Urol. 2003;170:2181. doi: 10.1097/01.ju.0000095460.12999.43. [DOI] [PubMed] [Google Scholar]
- 8.Khan MA, Partin AW, Rittenhouse HG, Mikolajczyk SD, Sokoll LJ, Chan DW, et al. Evaluation of proprostate specific antigen for early detection of prostate cancer in men with a total prostate specific antigen range of 4.0 to 10.0 ng/ml. J Urol. 2003;170:723. doi: 10.1097/01.ju.0000086940.10392.93. [DOI] [PubMed] [Google Scholar]
- 9.Naya Y, Fritsche HA, Bhadkamkar VA, Mikolajczyk SD, Rittenhouse HG, Babaian RJ. Evaluation of precursor prostate-specific antigen isoform ratios in the detection of prostate cancer. Urol Oncol. 2005;23:16. doi: 10.1016/j.urolonc.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 10.Sokoll LJ, Chan DW, Mikolajczyk SD, Rittenhouse HG, Evans CL, Linton HJ, et al. Proenzyme psa for the early detection of prostate cancer in the 2.5-4.0 ng/ml total psa range: preliminary analysis. Urology. 2003;61:274. doi: 10.1016/s0090-4295(02)02398-1. [DOI] [PubMed] [Google Scholar]
- 11.Catalona WJ, Bartsch G, Rittenhouse HG, Evans CL, Linton HJ, Horninger E, et al. Serum pro-prostate specific antigen preferentially detects aggressive prostate cancers in men with 2 to 4 ng/ml prostate specific antigen. J Urol. 2004;171:2239. doi: 10.1097/01.ju.0000127737.94221.3e. [DOI] [PubMed] [Google Scholar]
- 12.de Vries SH, Raaijmakers R, Blijenberg BG, Mikolajczyk SD, Rittenhouse HG, Schroder FH. Additional use of [-2] precursor prostate-specific antigen and “benign” PSA at diagnosis in screen-detected prostate cancer. Urology. 2005;65:926. doi: 10.1016/j.urology.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 13.Weinzierl CF, Lafranechere SM, Su SX, Arockiasamy DA, Hanson TP, Mizrahi IA. Analytical performance of the Access BPH-A assay from Beckman Coulter. Clin Chem. 2006;52:E48. [Google Scholar]
- 14.Weinzierl CF, Su SX, Pierson TB, Arockiasamy DA, Mizrahi IA, Broyles DA, et al. Measuring [-2]proPSA in serum: analytical performance of the Access p2PSA assay from Beckman Coulter. Clin Chem. 2007;52:A178. [Google Scholar]
- 15.Catalona WJ, Smith DS, Wolfert RL, Wang TJ, Rittenhouse HG, Ratliff TL, et al. Evaluation of percentage of free serum prostate-specific antigen to improve specificity of prostate cancer screening. JAMA. 1995;274:1214. [PubMed] [Google Scholar]
- 16.Partin AW, Brawer MK, Bartsch G, Horninger W, Taneja SS, Lepor H, et al. Complexed prostate specific antigen improves specificity for prostate cancer detection: results of a prospective multicenter clinical trial. J Urol. 2003;170:1787. doi: 10.1097/01.ju.0000092695.55705.dd. [DOI] [PubMed] [Google Scholar]
- 17.Bangma CH, Wildhagen MF, Yurdakul G, Schroder FH, Blijenberg BG. The value of (-7, -5) pro-prostate-specific antigen and human kallikrein-2 as serum markers for grading prostate cancer. BJU Int. 2004;93:720. doi: 10.1111/j.1464-410X.2003.04733.x. [DOI] [PubMed] [Google Scholar]
- 18.Lein M, Semjonow A, Graefen M, Kwiatkowski M, Abramjuk C, Stephan C, et al. A multicenter clinical trial on the use of (-5, -7) pro prostate specific antigen. J Urol. 2005;174:2150. doi: 10.1097/01.ju.0000181221.72017.ca. [DOI] [PubMed] [Google Scholar]
- 19.Stephan C, Meyer HA, Kwiatkowski M, Recker F, Camman H, Loening SA, et al. A (-5, -7) proPSA based artificial neural network to detect prostate cancer. Eur Urol. 2006;50:1014. doi: 10.1016/j.eururo.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Skates SJ, Horick NK, Moy JM, Minihan AM, Seiden MV, Marks JR, et al. Pooling of case specimens to create standard serum sets for screening cancer biomarkers. Cancer Epidemiol Biomarkers Prev. 2007;16:334. doi: 10.1158/1055-9965.EPI-06-0681. [DOI] [PubMed] [Google Scholar]


