Abstract
A major challenge in microarray design is the selection of highly specific oligonucleotide probes for all targeted genes of interest, while maintaining thermodynamic uniformity at the hybridization temperature. We introduce a novel microarray design framework (Thermodynamic Model-based Oligo Design Optimizer, TherMODO) that for the first time incorporates a number of advanced modelling features: (i) A model of position-dependent labelling effects that is quantitatively derived from experiment. (ii) Multi-state thermodynamic hybridization models of probe binding behaviour, including potential cross-hybridization reactions. (iii) A fast calibrated sequence-similarity-based heuristic for cross-hybridization prediction supporting large-scale designs. (iv) A novel compound score formulation for the integrated assessment of multiple probe design objectives. In contrast to a greedy search for probes meeting parameter thresholds, this approach permits an optimization at the probe set level and facilitates the selection of highly specific probe candidates while maintaining probe set uniformity. (v) Lastly, a flexible target grouping structure allows easy adaptation of the pipeline to a variety of microarray application scenarios. The algorithm and features are discussed and demonstrated on actual design runs. Source code is available on request.
INTRODUCTION
DNA microarray technology has developed into a well-established, powerful high-throughput method in biological research. The flexibility of the technology allows a variety of applications, including the study of genome-scale gene expression patterns, genotyping and genetic mapping, DNA–protein interactions (ChIP-chip), and comparative genomic hybridizations (CGH) (1–6). The predominant use of microarrays is still in the domain of gene expression profiling. One strength of the technology is its high dynamic range, with modern platforms yielding 5–6 orders of magnitude (7,8). Also, microarrays can directly probe low-copy-number targets, including regulatory transcripts and transcription factors. Despite the widespread success of microarrays, however, the interpretation of gene expression measurements has remained a challenge (9). Many modern methods for microarray data analysis attempt to detect biologically meaningful patterns or signatures in the data, and thus particularly rely on accurate measurements (10,11). Highly specific probes with uniform hybridization behaviour are, therefore, crucial for accurate quantitative modelling and further advancement of inference methods in microarray analysis (12,13).
While a large number of tools for rapid array design are currently available, high-quality probe design requires the prediction of microarray probe hybridization behaviour (14) in a complex mixture, which is an intrinsically hard problem (15). The computational complexity of predicting individual probe behaviour is considerably reduced through ad hoc heuristics, such as replacing thermodynamic modelling with local sequence similarity criteria. These have been motivated by experimental exploratory studies of microarray probe specificity (16). Sequence similarity can be tested very efficiently with suffix trees (17–20) or the popular BLAST tool (21–32).
As calculations for one probe can already be quite elaborate, most of the current research in microarray design has focused on dealing with the inherent difficulty of probe selection in a high-dimensional search space (33). Typically, greedy approaches select the first probe candidates matching design criteria, and thus narrowing the pool of fully considered candidates early. For example, restricting guanine–cytosine (GC) content excludes probes with extreme probe–target melting temperatures. Filters for low sequence complexity regions avoid areas intrinsically prone to non-specific hybridization. Concerns about probe secondary structure have motivated sequence palindrome-based filtering heuristics (21,26,27), while more recent advances have introduced a new class of thermodynamic models (34–39) for the prediction of probe structure and also probe–target binding behaviour (22,23,25,30).
Several design challenges, however, remain that are central to achieving high-performance microarrays. While very efficient, the conventional use of greedy searches and fixed parameter thresholds often leads to suboptimal probe choices, compared with approaches that determine optimal parameters and probes by non-greedy optimization (18,40,41). Ideally, microarray design would draw from a thorough comparison of all possible probes and their properties. Essentially, for each probe candidate, one thus wants the best possible prediction of probe characteristics, such as the sensitivity and specificity of probe signal response to the concentration of its target transcript (12,15).
For example, during sample labelling, the primer type and the limited processivity of the reverse transcriptase affect the likelihood that a certain sequence region is present in the pool of labelled transcripts. As a consequence, the location of the probe binding site along the transcript can affect probe performance. The difficulty of determining the actual parameters for a model of this process has precluded a quantitative consideration of these effects. Most probe design tools, therefore, try to handle these biases using a greedy preference for probe binding sites in the terminal regions which are enriched in labelled products. This, however, comes at the expense of forfeiting potentially better probes in different locations.
It is noteworthy that transcript secondary structure can make particular target regions inaccessible and hence impede probe hybridization. In a recent study of a genome-scale probe design, up to a third of all probe binding sites were affected (42). Nevertheless, target structure is typically not considered during probe design, perhaps due to the additional computational expense of its prediction. This, however, results in probes with unexpectedly reduced sensitivity.
Identification of potentially cross-hybridizing probes is crucial for controlling probe specificity. The commonly employed sequence-similarity heuristics are fast because they exploit short contiguous sequence match ‘words’ to seed their alignments. As a consequence, however, they may miss less similar, yet thermodynamically relevant cross-matches (15,20). This needs to be considered in a conservative assessment of probe specificity. In addition, sequence comparisons are usually limited to known transcripts, although our catalogue of all actively transcribed genome regions is still far from exhaustive (43–45). Hence, the complete mixture of transcripts that need to be discriminated in a biological sample is not known a priori. This motivates the inclusion of genomic sequence in the detection of potential cross-hybridization.
Finally, groups of very similar targets, such as protein families, alternative splice variants, as well as paralogues and orthologues can severely constrain the selection of specific probes, sometimes even making it impossible to discern targets with a single probe. Explicit support for these scenarios is already beneficial in the design of whole-genome arrays and becomes even more relevant for arrays targeting splice variants or multiple organism strains.
There thus remain many opportunities for improving array design for high-performance microarrays (12,15). We here introduce a Thermodynamic Model-based Oligo Design Optimizer (TherMODO) that showcases a number of ways in which we can advance the state of the art in probe design. In particular, we combine:
An experimentally derived model of position-dependent labelling effects.
Thermodynamic multi-state models of probe hybridization behaviour that also consider target structure.
A calibrated BLAST heuristic for the efficient conservative detection of cross-hybridizations.
Compound scores integrating individual models for a joint assessment of probe specificity and signal uniformity.
Global optimization of probe sets in contrast to a greedy search and user supplied design parameters.
An extensible, flexible design framework that allows for groups of highly similar targets.
The algorithm and its features are discussed and demonstrated with the analyses of an actual design run on the Escherichia coli K12 genome, with complementary results from human.
MATERIALS AND METHODS
Algorithm structure
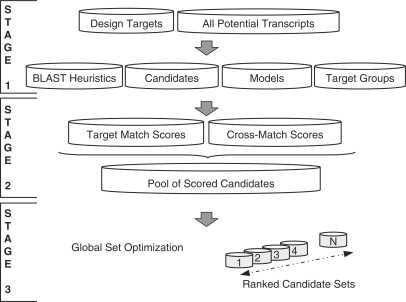
For computational efficiency, TherMODO probe design proceeds in a tiered structure. Figure 1 gives an overview of the algorithm. First, any sequence similarities between the Design Targets and all transcripts are identified in a screen for regions of interest for quantitative modelling (BLAST Heuristics). Probe Candidates are characterized each by Models for the Gibbs free energies of all potential binding reactions, that the probe is accessible (rather than self-folding), and that probe binding sites have been labelled and are accessible (rather than part of transcript secondary structure). Relationships between very similar design targets such as homologues, splice variants or related genomic loci are represented by Target Groups. Combining all the information from the first pipeline stage the algorithm then computes integrated hybridization intensity scores, allowing direct comparisons between intended Target Matches and unwanted cross-hybridization (Cross-Match). All probe candidates are then considered together in a final Global Set Optimization step to choose a probe set with maximal probe specificity and uniformity. Simple file-based data storage is employed to allow easy coarse-grained parallelization that scales linearly on modern multi-core workstations, or even compute clusters (tested for 4–64 threads, data not shown). Due to the multi-tiered architecture, however, allowing a reuse of Stage 1 results for multiple subsequent passes, more complex designs with multiple probe lengths run proportionally faster and, even for a large transcriptome like human, can be run on a modern workstation in less than two days (Table 1).
Figure 1.
Tiered framework structure, overview. In the first stage of the pipeline, sequence similarities between design targets and other transcripts are identified to screen for regions of interest for modelling (BLAST Heuristics). Probe Candidates are characterized each by Models for the Gibbs free energies of all potential binding reactions, that the probe is accessible (rather than self-folding), and that probe binding sites have been labelled and are accessible (rather than part of transcript secondary structure). Relationships between very similar design targets, such as homologues, splice variants, or related genomic loci are represented by Target Groups. In the second stage, all the information from the first phase is combined to compute integrated hybridization intensity scores allowing direct comparisons between intended Target Matches and unwanted cross-hybridization (Cross-Matches). All probe candidates are then considered together in the third and final stage of Global Set Optimization, ranking alternative probe sets by their mean integrated compound score.
Table 2.
Probe design target coverage and probe specificity
| Program | Designed probes (unique) | Perfect specificity/improved |
|---|---|---|
| TherMODO | 4471 (4381) | 4296 (96.1%)/– |
| OligoRankPick | 4471 (4357) | 3915 (87.5%)/550 (12.3%) |
| OligoArray | 4222 (4157) | 3924 (87.7%)/293 (6.6%) |
| YODA | 4110 (4110) | 3825 (85.5%)/282 (6.3%) |
The number of transcript targets covered by each design is shown, with the number of unique probes reflecting the number of distinct transcripts that can be discriminated. Probes with ΔI > Δ I′ = 1012 were considered as having a perfect specificity under realistic laboratory conditions (see Discussion section). The number of probes for difficult targets for which probe specificity could be improved through the TherMODO design process is the last number shown.
Algorithm prerequisites
TherMODO takes advantage of the ViennaRNA package (34,46,47) for the thermodynamic calculations of the intermolecular probe–target duplexes (RNAduplex) as well as the intra-molecular structures of both targets and probes (RNAplfold). The Gibbs free energy from ViennaRNA is adjusted by an empirical correction for substrate bound probes [(48); J. SantaLucia, Jr, personal communication],
that have been determined for microarray platforms where probes are attached to a gel-like slide surface (48). Surface effects of other platforms are still an active field of research (13).
WU-BLAST is used for sequence similarity searches in a fast heuristic approach to identify potential cross-hybridization (W. Gish, personal communication).
BLAST heuristic for cross-match prediction
Target sequences were compared with both strands of all other transcripts and the genome. Sequence similarities could then be used in a heuristic to identify potential binding partners of probe candidates. We included genomic sequences because substantial non-coding regions are known to be actively transcribed (43–45).
The following WU-BLAST (BLASTN) options were employed: a seed alignment word size W = 7, a match score M = 1, a mismatch score N =−1, a gap penalty Q = 3 and a gap extension penalty R = 1. Exploratory studies of probe binding behaviour suggest that a stretch of 13–15 matching nucleotides can already give rise to detectable cross-hybridization (16). For WU-BLAST, the corresponding minimum score threshold S = 13 can be specified directly, thus avoiding the need to calculate the corresponding expect value (E) thresholds that would be necessary for other implementations of the BLAST algorithm (23). The sequence alignments of all matches along the target transcript are stored in the BLAST heuristics data structure, allowing efficient reuse when considering different probe candidates for the same target.
Probe candidates
The user may specify a minimum and maximum oligonucleotide length for probes candidates considered. The E. coli and human probe design runs discussed in this article permitted a range of 65–69 nt as probe lengths. Probes of this length show good sensitivity (49) and consideration of a range of lengths allows more uniform designs. For each target, stretches of mono-, di- and tri-nucleotide repeats of 10 nt or longer are masked as these have been reported to possibly interfere with synthesis chemistry (23).
Models
Quantitative models allow an integrated assessment of a number of probe properties. Results are stored in the Models data structure allowing efficient reuse for different probe candidates.
Labelling probability
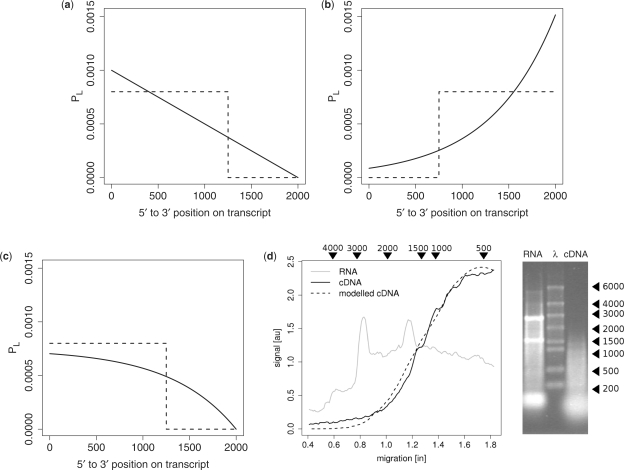
With the processivity of the labelling enzyme represented by the characteristic length λ, the probability of obtaining a labelled transcript of length l follows an exponential distribution (Figure 2b),
For random primed labelling and a target of length x0, the probability of obtaining a labelled transcript at position x relative to the 3′-terminal becomes
We fit this model to data from both gel images and Bioanalyzer data (Figure 2d) to obtain the characteristic length λ. We provide a full derivation of equations and a detailed description of the fitting process and fit results in the Supplementary Material.
Figure 2.
Quantitative models of position-specific labelling effects. (a–c) The probability PL of finding a target region in the labelled transcript pool is on the y-axis. The x-axis indicates the position along the target, here shown for a 2-kb sequence. The dashed lines represent typical thresholds used by traditional design tools [e.g. (23,25)] instead of a quantitative model. (a) An idealized schema demonstrating the effects of random primer placement, shown for an infinite labelling enzyme read-through. (b) The effects of labelling enzyme drop-off, shown for a 3′-terminal anchored primer, assuming a uniform random drop-off rate. (c) A model taking into account both random primer placement and labelling enzyme drop-off. The model shown uses actual parameters obtained from measured gel data (next panel). (d) Gel data for unlabelled RNA (grey line) and cDNA from random-primed labelling (solid line). Fluorescence intensity is plotted as a function of the run-length, with the corresponding DNA ladder size markers also shown at the top of the panel. Peaks due to structural RNAs were removed for the model fit. The dashed line traces the model prediction. The gel itself is pictured on the right with the three lanes, respectively, loaded with unlabelled RNA, a ladder (λ), and labelled cDNA.
The labelling probabilities for both targets and potential binding partners are calculated with this model.
Binding probability
We compute a score reflecting the predicted probe–target binding probability PB at the effective hybridization temperature Thyb. This is one of the few design parameters that need to be set by the user. Past experience has shown that subtracting 20°C from the typical probe–target melting temperature has worked well for probes of length 40–70 nt. For the designs reported here, we have chosen Thyb =69°C. We note that for optimally efficient hybridizations, the physical hybridization temperature that corresponds to Thyb needs to be experimentally calibrated. The binding probability
is proportional to the Boltzmann factor with the Gibbs free energy ΔGtarget. Here, R = 1.9872kcal mol−1K−1 is the universal molar gas constant. For the observed binding energies of oligonucleotide probes and practical reaction temperatures, binding is far from the saturation point and the above equation is a good approximation of the steady-state binding probability. Analogous calculations are performed for other potential binding partners.
Probe accessibility
We use RNAplfold to obtain the probability PP of a probe being accessible, i.e. not forming a stable secondary structure at Thyb.
Binding site accessibility
For reasons of computational efficiency when considering many probes of different sizes for longer transcripts, we calculate the probability PA of a probe binding site being accessible in two steps. First, RNAplfold is used to obtain the accessibility of short seed-like regions in the transcript. These are then combined to calculate the probability that the entire probe binding site is accessible, considering possible stable secondary structures in a 100-base window (see Supplementary Material for details).
Target groups
The Target Groups data structure describes the associations between design target sequences and other similar sequences, such as protein family members, orthologues, paralogues or splice variants. These associations can, for example, be assigned manually, from database information, or by sequence-based method like BLASTN searches or sequence clustering approaches CD-HIT (50). Target Groups allow two types of relationship between a probe and potential binding partners. For each target, a list of ‘primary’ targets states which binding partners a probe must bind, whereas another list of ‘secondary’ targets shows which binding partners a probe may bind (is ‘allowed’ to bind). One can specify entire sequences or sequence regions as binding partners, which allows for a simple integration of genomic regions as binding partners. For each candidate probe, TherMODO will compare the predicted binding partners to those listed in the respective Target Group. The reported Cross-Match score is the strongest cross-match that is not permitted according to the targets ‘may bind’ list.
Design runs
We applied TherMODO to the transcripts reported in the NCBI annotation of E. coli K12 (June 2007). The 4488 annotated transcripts included 102 pseudogenes and 168 structured non-coding RNAs (ncRNAs). Seventeen annotations were less than 65 bp in length and were removed from the design set, leaving 4471 eligible transcripts. Using CD-HIT (50) to cluster the E. coli transcriptome at the default 90% sequence identity threshold, we grouped 179 genes with high sequence similarity into 48 target groups. A total of 11 214 969 probes were evaluated as part of this run.
For comparison, several other, popular probe design tools (20,23,41) were applied to the same data set (see Supplementary Material for details).
We also examined 25 125 human genes from the NCBI RefSeq database (January 2008). Computations were performed on a standard PC (dual-CPU dual-core 2 GHz PC with 8 GB of RAM), running four threads in parallel. Test runs on a modern workstation with eight cores demonstrated that the course grained parallelization easily afforded by the pipeline architecture scaled linearly (Table 1). Additional tests on a small compute cluster (64 threads) further confirmed almost linear scaling of execution times (data not shown).
Table 1.
Approximate computation times of microarray probe design forE. coli and human
| Organism | Genes | Transcriptome | Time (1×) | Time (5×) |
|---|---|---|---|---|
| E. coli | ∼4 000 | 4 Mb | 1 h 30 min | 3 h 40 min |
| H. sapiens | ∼25 000 | 76 Mb | 19 h | <2 days |
The table shows the speed up of computation times through reuse of Stage 1 data at subsequent design passes. The first time given (1×) is for the analysis of probes of uniform length, here, 65-mers. The second time given (5×) is for five passes for probes of different lengths, here, 65–69-mers. Tests were run on a modern eight-core workstation (see Methods section).
Labelling assay and analysis
To determine the model parameters for the labelling model introduced above, RNA extracted from E. coli was reverse transcribed using the AffinityScript HC Reverse Transcriptase component of the FairPlay III Microarray Labelling Kit (Stratagene, Cat. No. 252012) and random hexamer primers (MWG Biotech AG) according to the instruction manual provided with the kit. The reverse transcription (RT) was repeated three times and the RNA and resulting cDNA samples were then analysed by both capillary electrophoresis (Agilent 2100 Bioanalyzer, RNA nano LabChip) and on a 1% agarose gel. Gel images were quantified using imageJ (http://rsb.info.nih.gov/ij/). Using size markers, the measured fluorescence signal distributions of the samples where then transformed into molecule length distributions (Figure 2d). See Supplementary Material for further details.
RESULTS AND DISCUSSION
The tiered algorithm structure (Figure 1, see Methods section) allows an exploitation of the power of quantitative models for target labelling and hybridization behaviour, while keeping the overall computational complexity sufficiently low to still permit large-scale array designs. Even designs for a large transcriptome like human can be run in less than two days on a modern workstation (Table 1).
A discussion of the quantitative models employed is followed by an examination of their impact on probe quality assessment. This is illustrated on an actual probe design run for E. coli and with complementary data for human. In particular, parameter estimation through calibration experiments is demonstrated for a popular transcript labelling method and the employed sequence similarity-based heuristic is calibrated on multiple data sets. We then formulate an integrated score for probe assessment allowing global probe set optimization.
Finally, we provide a characterization of sample probe design results in relation to established alternative designs (20,23,41).
Modelling and calibration
Where computationally feasible, modern quantitative models considerably improve probe design, with modelled hybridization behaviour being the best predictor of probe quality (14,42).
Quantitative effect of probe binding position along the target
The choice of labelling/amplification protocol not only determines whether one needs to design probes for the sense or anti-sense strands (51), but also affects the likelihood that a certain target region is present in the pool of labelled transcripts. Traditional probe design tools that try to take this into account typically exhibit a greedy preference for probes located near the 3′-end of the target (23), appropriate for oligo-dT-primed labelling. Similarly, probes near the 5′-end can be preferred for random primer-based labelling (20). Also, many tools impose a fixed threshold on probe position, excluding candidates considered too distant from the terminal (23). In either case, this greedy preference often means forfeiting better probes further away from the terminal. Although the benefits of a quantitative model are increasingly being recognized (28), the problem of determining model parameters has remained a challenge for the principled integration of labelling models in probe design.
While model parameters will strongly depend on the exact protocols employed, they can be obtained from measurements of labelled and unlabelled cDNA length distributions. A model for a simple RT-based labelling protocol, for example, needs to account for primer placement (e.g. oligo-dT anchored or randomly along the target) and RT enzyme drop-off. Figure 2a demonstrates how random primer placement affects the likelihood of finding a target region in the labelled transcript pool, shown on the y-axis. The x-axis indicates the position along the target (shown for a 2 kb sequence). The idealized schema in this figure assumes infinite RT read-through. Conversely, the schema in Figure 2b plots the effect of RT enzyme drop-off, for a 3′-terminal anchored primer. Assuming a uniform random drop-off rate leads to an exponential length distribution ∝ exp (−(x0-x)/λ for a target length x0 and a characteristic drop-off length λ. This means that <37% of labelled transcripts will reach length λ, and <14% will reach 2λ.
Both digital gel images, Figure 2d, and Bioanalyzer data (Supplementary Material) were analysed for a random-primed indirect labelling protocol. The fit of the model (dashed line) is shown in Figure 2d, yielding λ ∼ 650, which indicates that RT drop-off in a complex mixture labelling environment plays a considerable role: <10% of labelled transcripts will reach length 1500, and just over 2% will reach length 2500. Fits of gel images and Bioanalyzer data (Supplementary Material) both support the model shown in Figure 2c, which accounts for random primer placement as well as RT enzyme drop-off.
As will be shown later, with a quantitative model of labelling probability, probe–target positional effects can directly be integrated into a principled combined score. On a log-scale, they affect signal strength about 40% as much as probe secondary structure (Figure 3).
Figure 3.
Impact of different probe properties on signal sensitivity Itm. The y-axis plots on a log 10 scale the typical ranges of the probability scores for target labelling (L), probe accessibility (P), binding site accessibility (A), and probe–target binding (B). Ranges are calculated by considering all probe candidates for a specific target. The values 3.8 and 5.8 for probe accessibility (P) and binding site accessibility (A), for example, indicate that, amongst probe candidates for a typical target, probe accessibilities varied by up to 103.8 times, whereas binding site accessibilities varied by up to 105.8 times. In particular, one can see that the impact of binding site accessibility on probe performance is about 100 times stronger than that of probe accessibility. (a) Shows results for E. coli, (b) for H. sapiens.
Thermodynamic modelling of probe hybridization behaviour
Many traditional probe design tools consider GC-content as a proxy for melting behaviour. Still most modern algorithms characterize probes by predictions of probe–target melting temperatures. Neither of these, however, allows a principled prediction of binding behaviour at the actual hybridization temperature. More sophisticated models of probe hybridization improve our ability to predict microarray probe performance (14), justifying the application of thermodynamic modelling despite its relatively high computational cost.
Observed discrepancies (52) between experiments and thermodynamic predictions of so-called two-state models for probe–target binding (34–36) could be explained by interfering secondary structure (37). While modern probe design algorithms (22,23,25,30) do apply thermodynamic models to consider probe secondary structure (34,36) and the effect of target secondary structure has been recognized (14,53), to our knowledge, target structure has so far not been considered by probe design tools for genome-scale microarrays.
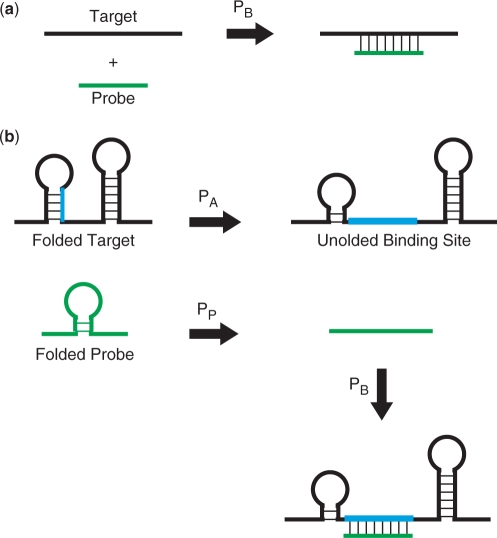
Hybridization behaviour prediction in TherMODO considers binding between a candidate probe and transcripts as well as the probabilities that the probe and its binding sites are accessible rather than folded into stable secondary structures (Figure 4). We have examined the typical contribution of these effects to the predicted binding behaviour of all candidate probes of a target. Figure 3 plots typical log10 ranges for E. coli and Homo sapiens. Note that while actual values vary for different organisms, the relative importance of effects is similar. Probe secondary structure affects log signal intensity 35–40% as much as probe–target binding strength. Recent experimental observations (13) agree with these calculations. This underscores the importance of considering probe self-folding during probe design. Interestingly, binding site accessibility has an even stronger impact than probe secondary structure, affecting log signal intensity about 1/2 to 2/3 as much as probe–target binding strength. This explains that considerable improvements in the quality of probe hybridization prediction is observed when binding site accessibility is also considered (37).
Figure 4.
Models of probe–target hybridization. (a) The two-state model only accounts for the unbound and bound probe–target states. (b) The multi-state model of hybridization behaviour prediction in TherMODO considers binding between a candidate probe and targets as well as the secondary structures of both the probe and its binding site within the target. Redrawn after SantaLucia and Hicks (61).
Considering probe and target structure in terms of accessibility probabilities allows a principled combination of thermodynamic models of individual effects into a combined quantitative score. In contrast, most established tools just exclude all probes with a potential for self-folding. While about half of all targets have <20% of probe candidates excluded by such filtering for probe secondary structure, for 9% of targets, the search space is already reduced by >40%. For the most affected 1% of targets >60% of probes fail this filter, giving a substantially reduced search space. A quantitative incorporation of probe accessibility therefore improves our ability to identify good probes for these difficult targets.
The effect of transcript structure is stronger: only 11% of targets have <20% of probe candidates excluded by filtering for binding site secondary structure. For almost half the targets, the search space is already reduced by >40%. More than a quarter of targets are strongly affected with over 60% of probes failing this filter, giving a substantially reduced search space. For a consideration of binding site accessibility, a quantitative approach instead of threshold-based filtering is therefore even more important.
We note that our combined thermodynamic model is not as elaborate as the most sophisticated multi-state models currently available (37,53,54), which also consider intermolecular probe–probe or target–target complexes or hybrid inter- and intra-molecular structures. By quantitatively integrating a number of important effects, however, we can substantially improve prediction quality over models employed in traditional genome-scale probe design tools and achieve a reasonable compromise between model accuracy and computational complexity (Table 1).
Besides a more sensitive detection of potential cross-hybridization, thermodynamic models allow the selection of probe sets with more uniform binding behaviour. The resulting more accurate microarray readouts support modern analyses methods for the identification of subtle biologically relevant patterns.
Calibrated BLAST heuristic
While thermodynamic models as described above are the best available predictors of hybridization behaviour, their systematic application is expensive in terms of computational resources. Many established probe design tools considerably reduce the computational complexity of assessing all potential binding reactions by applying a sequence similarity-based heuristic as a filter (15). Variants of the BLAST algorithm are the most popular methods for this purpose (15), with smaller ‘word size’ increasing sensitivity as well as run-time (20). When a local sequence match between two potential binding partners identifies a region of interest (16), thermodynamic models can be used to more accurately assess binding strength (23). This can then be used for a quantitative assessment of probe binding.
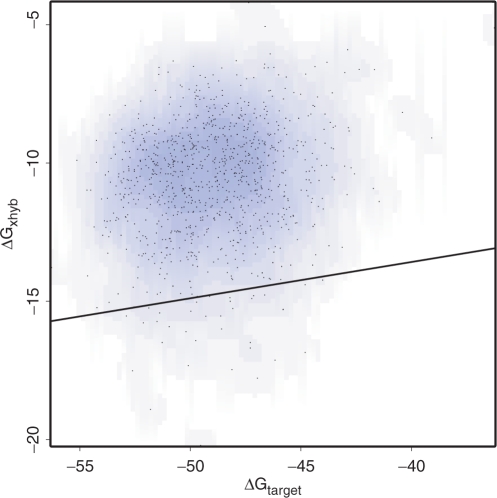
The question arises how the cross-hybridization potential of probe candidates should be assessed for which no sequence similarity to cross-matches could be identified. Examining a random selection of 1000 such probes, for each probe, we applied thermodynamic models to calculate probe–transcript binding strengths for all possible transcripts. For E. coli, Figure 5 plots these probes, with the target binding strength, ΔGtarget, on the x-axis, and the strongest binding strength to cross-matches, ΔGxhyb, on the y-axis. A black line shows the linear regression minus two standard deviations. This forms a conservative estimate of cross-hybridization strength as a function of target-binding strength for probe candidates with no identified sequence similarity to cross-matches. Very similar results were obtained for other organisms, including human (see Supplementary Material). The calibrated heuristic can, therefore, generally be applied to increase the speed of assessing cross-hybridization potential without overestimating the quality of probes for which no cross-matches could be identified by sequence similarity.
Figure 5.
Calibration of the heuristic prediction of cross-hybridization for probes with no crossmatches detected by sequence similarity. The x-axis plots the Gibbs free energy ΔGtarget of probe–target binding and the y-axis shows the Gibbs free binding energy ΔGxhyb of the strongest cross-match predicted by thermodynamic models. The black line represents a linear regression of these values minus two standard deviations, forming a conservatively calibrated heuristic. Data shown are from E. coli; very similar results are obtained for other organisms, including human (see Supplementary Material). The calibrated heuristic can therefore generally be applied.
Using this calibration, the cross-hybridization potential of probe candidates with no sequence similarity to cross-matches can be assessed in a manner consistent with how probes with identified cross-matches are treated. This thus allows a quantitative comparison of all probes.
Integrated score and global set optimization
The reduced computational complexity achieved by the tiered structure of the algorithm and exploitation of the calibrated heuristic allows the collection of several relatively expensive model-based probe characteristics. The resulting quantitative prediction of hybridization behaviour yields estimates for probe sensitivity and specificity.
A more sensitive probe responds to its target with higher signal intensity, where the different factors affecting the signal can be integrated by a compound score for the target match,
with probe–target binding probability PB, binding site accessibility PA, probe accessibility PP and PL being the probability of the binding site being part of a labelled transcript. A similar integrated score Ixm can be computed for the strongest cross-match, and Itm/Ixm then reflects the specificity of the probe (cf. Methods section).
To compile a probe set with maximal specificity and uniform sensitivity, a principled trade-off needs to be made. For reasons of symmetry, this is most easily written on a log scale. On one hand, better specificity is obtained for higher log Δ I, where Δ I = Itm/ Ixm. On the other hand, one wishes to minimize the absolute deviation | log Itm - log I0| from the characteristic I0 of the probe set. We propose a joint penalty score
which we need to minimize in a search for probes of good uniformity and high specificity. While differently weighted variants are possible (28,41), this score equally punishes lower specificity as well as deviations from uniformity. From a practical point of view, we further argue that a separation of target and cross-match affinities of Δ I> 1012 does not improve results in a realistic laboratory setting, and we therefore introduce a soft maximum at Δ I ' =1012. At this threshold, cross-talk cannot experimentally be detected even in a worst-case scenario, where the most strongly expressed transcript interferes with the measurement of the most weakly expressed transcript. To motivate the threshold, consider that gene expression has a dynamic range of about 6–8 orders of magnitude (7) and that microarray scanners typically achieve a bandwidth of about 3–4 orders of magnitude. A separation of 12 orders of magnitude, therefore, forms a conservative cut-off point, ensuring probes of practically perfect specificity in any realistic laboratory conditions.
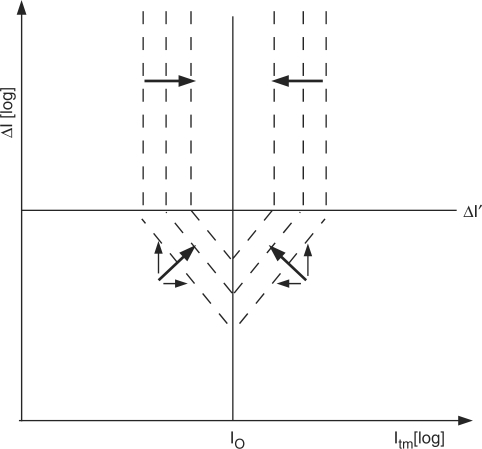
Figure 6 schematically illustrates the optimization of the joint penalty score J. The x-axis plots the integrated compound score Itm for the target match and the y-axis shows Δ I=Itm/Ixm, with Ixm being the integrated score for the strongest cross-match. In general, better probes lie closer to I0, selecting for probe uniformity, and higher in the graph, selecting for better specificity. The dashed lines are isoscores for the joint penalty score J, i.e. probes along these lines are considered equally good. Above Δ I', all probes are considered highly specific, and optimization focuses on probe uniformity.
Figure 6.
Optimization of the joint penalty score J. The x-axis plots the integrated compound score Itm for the target match and the y-axis reflects a measure of specificity, Δ I=Itm/Ixm, with Ixm being the integrated score for the strongest cross-match. Both axes are on log scale. In general, better probes lie closer to I0, selecting for probe uniformity, and higher in the graph, selecting for better specificity. The dashed lines are isoscores for the joint penalty score J, i.e. probes along these lines are considered equally good. This shows that a deviation from the characteristic I0 of the set is allowed if it brings a sufficient increase in specificity. The benefit of an increase of the specificity score above Δ I' is considered to be vanishing, as a further improvement of separation could not be measured experimentally. In the top half of the graph, therefore, all probes are considered highly specific, and optimization focuses on probe uniformity. (Arrows show desired directions of change).
Probe uniformity has traditionally been achieved by constraining individual probe properties to fixed ranges. While most probe design tools will ‘greedily’ accept the first probe candidate meeting the criteria for each target, non-greedy optimization considerably improves probe quality (41). In addition, rather than requiring users to define thresholds to constrain average probe properties, it is preferable to automatically adapt design parameters to the data (40). The compound score introduced here allows us to extend this approach not just to individual probe properties but to an integrated measure of probe sensitivity, I, that is of direct experimental relevance. Moreover, rather than choosing fixed thresholds, the joint penalty score J permits individual probes to deviate from the characteristic I0 if this brings a sufficient increase in probe specificity (Figure 6, diagonal dashed lines).
TherMODO implements global set optimization iteratively: at each step, the best probe set for a given characteristic I0 is constructed by minimizing the average joint penalty score J for all targets. Then, I0 is adjusted to improve the set score, in the end yielding the I0 for the globally optimal probe set.
The quality of the final compiled set benefits from a much increased search space as no probe candidates are discarded before the optimization step, yielding more specific and extremely uniform probe sets. In a design run for 4471 annotated transcripts of E. coli K12, we have scored over 11 million probe candidates. The design run covered all transcripts, including protein-coding sequences, pseudo-genes and the 168 structural RNAs. The stable folds of structural RNA transcripts are fully considered during the thermodynamic probe design. The construction of 4381 unique probes achieved the most comprehensive reported coverage of the E. coli transcriptome.
We consider 96.1% of these probes to have both an excellent specificity (Δ I>1012) and extreme binding uniformity (median/mad RT log Itm = -18.000± 0.004, Figure 7). For 2.9% of probes, while being just as specific, sensitivity was reduced by 1% or more because of unusual sequence composition or secondary structure of the target. Less than 1% of probes showed any cross-hybridization potential. Of these, for 30% no binding partners could be identified but cross-hybridization potential was detected by our calibrated heuristic.
Figure 7.
The distribution of the integrated compound score I amongst all probe candidates (a) versus the distribution for the optimized probe set (b). Note the 1:100 difference of scale, which emphasizes the sharpness of the final distribution. Data shown are from E. coli.
Characterization of probe design results
While we appreciate that it is difficult to objectively compare different probe design tools, we have rerun and characterized the results of several popular programs on the full E. coli transcriptome: OligoRankPick (41), OligoArray (20), and Yoda (20). All probe sets were characterized by probe binding strength (PB), probe self-folding (PP), target region accessibility (PA) and positional labelling effects (PL). For this assessment, no compromises were made for the sake of speed and, instead of exploiting sequence-similarity-based heuristics, full model thermodynamic calculations were applied to all probes and their potential binding partners. As a result, these more comprehensive computations give a more accurate joint score J than the approximations used in the actual probe design process which we evaluate.
The three alternate probe design programs examined represent different established approaches to probe design and have complementary features. For instance, YODA incorporates a custom sequence similarity search (SeqMatch) for the identification of potential cross-hybridization that is more sensitive than a BLAST run with typical parameters. OligoArray employs thermodynamic models for the assessment of probe–target duplexes, cross-hybridization, and self-folding. Both YODA and OA use greedy search for selecting probes that match specified design criteria. In contrast, OligoRankPick uses a non-greedy approach, choosing probes from a pool of candidates per target using a weighted rank-sum strategy for a number of probe qualities, such as probe specificity GC-content, self-binding and sequence complexity. With the exception of YODA, the tools employ a BLAST-based filter for identifying cross-hybridization.
Although we make no claims of a systematic comparison, a number of general trends can be observed. For all tools, final probe set properties were more uniform than for the original probe design space. Typically, tools aim for a uniform GC-content or melting temperature. With default parameters, the greedy, threshold-based tools improved uniformity less than 2-fold, while a 4-fold improvement could be achieved with a non-greedy approach (41). Traditionally, GC-content and melting temperature, which are strongly correlated to the free binding energy, have been used as a first proxy of signal strength.
Experiments have since confirmed (8,13,55,56) that the secondary structure of probes and targets as well as labelling effects are important contributions to signal intensity (Figure 3). This is reflected in the broad distributions observed for the predicted hybridization signal sensitivity Itm in probe sets designed by traditional tools, in contrast to the extremely uniform probes constructed by TherMODO. Using a non-greedy approach, uniformity could be improved more than 800-fold over the original design space (Figure 7). It is noteworthy that probe set designs with similarly uniform melting temperatures (20) can have completely different Itm uniformity.
In addition to probe set uniformity, designs of course aim for high sensitivity and specificity. The binding strength contributes to the sensitivity, and design results were similar for probes of fixed lengths. Allowing variable length probes, the binding strengths were slightly higher for the TherMODO design. TherMODO, however, also optimizes the contributions of other factors to probe sensitivity. Taking probe and target structure as well as labelling effects into account thus improved probe sensitivity for about three out of four genes. Overall, this resulted in an improvement of 11–17% of typical probe sensitivity over traditional designs. Of these, the non-greedy approach (41) outperformed the greedy, threshold-based tools. Longer probe lengths (23) also improved sensitivity.
Last but not least, array quality is determined by probe specificity. For probes of fixed lengths, the non-greedy approaches perform better. It is noteworthy that longer probe lengths contribute considerably to specificity as has also been observed experimentally (49). Specificity in gene expression profiling should not be confused with the need for single-base pair discrimination in single nucleotide polymorphism (SNP) detection that is better served by shorter probes (57). It is noteworthy that all examined tools designed probes of practically perfect specificity for the majority of genes (Table 2). Differences could, however, be observed for more difficult subsets of about 6–12% of design targets. In particular, TherMODO global set optimization gave a (median) 1000-fold improvement in probe specificity for these harder targets. Moreover, only a single TherMODO probe had slightly lower specificity than the corresponding probe from an alternate design: this alternative probe, however, had an unusually low sensitivity. The slight reduction in specificity (RT log Δ I=-1.5), was more than made up by the increased sensitivity (RT log Δ Itm=+5.7) contributing to an improved overall probe set uniformity. All TherMODO probes had a higher joint score J in the assessment. This is a non-trivial observation as the calculated assessment score is more accurate than the approximations used during the probe design process. Complete detailed results of the design runs, comparative statistics and plots for both the calibrated BLAST heuristic and full thermodynamic calculations of probe specificity are provided in the Supplementary Material.
In summary, the extremely high thermodynamic hybridization uniformity observed in TherMODO designed probe sets could be achieved without sacrificing specificity.
SUMMARY AND OUTLOOK
With the ever increasing number of genomes sequenced and custom microarrays now available at prices similar to those ready made, oligonucleotide arrays are often the method of choice for genome-scale quantitative gene expression profiling. While the technology is particularly well suited for the quantification of low copy number transcripts and features high dynamic range, accurate measurements depend on good probe design.
In this article, we have introduced a novel algorithm, TherMODO, that combines a number of advances on the state of the art in probe design for high-performance microarrays. In particular, we include a model of position-dependent labelling effects based on actual experimental data for a quantitative consideration of the trade-off between labelling intensity and probe binding behaviour. The prediction of probe binding behaviour has been improved beyond the traditional two-state models by also considering the probabilities that the probes and the transcript binding sites will be accessible rather than folded into stable secondary structures. These thermodynamic calculations are applied to analyse probe–target binding as well as in the computation of potential cross-hybridization reactions. Cross-hybridization is conservatively assessed by a fast sequence-similarity-based heuristic that has been calibrated by comparison to full thermodynamic models. The different factors affecting the signal were studied for several organisms (data shown for E. coli and human), and could be integrated by a compound score. We optimize designs at the probe set level, jointly maximizing set uniformity and average probe specificity.
The performance of this approach was validated and compared with other popular tools (20,23,41) in a genome-scale probe design for E. coli. A highly specific probe set with extremely uniform hybridization intensities was compiled from a global pool of probe candidates, achieving the best transcript coverage so far reported. To demonstrate the flexibility of the algorithm's; target group structure for handling groups of similar targets, 179 genes with high sequence similarity were collected in 48 target groups for this design. We emphasize that the option of specifying groups of targets that a probe has to bind or may bind is not only useful for the design of comprehensive microarrays probing the genes of an organism with high specificity, but are also valuable for more complex applications. With the increasing number of sequenced strains (58), for example, arrays that allow a direct comparison of multiple strains become of interest (G. Striedner et al., manuscript in preparation).
The open structure of the TherMODO algorithm allows further improvements in a number of areas: in the future, the definition of target groups could conveniently flow from a first pass of the pipeline rather than relying on sequence-similarity-based tools [(50); W. Gish, personal communication]. The general scoring scheme can also easily be extended to consider genome-position-specific effects for the design of tiling arrays. While future support of multiple ‘replicate’ probes per gene is straightforward, the use of multiple probes to discriminate splice forms or other highly similar targets remains an additional challenge (59,60). Lastly, as more sophisticated thermodynamic models become available and compute power increases, the models employed in TherMODO can easily be updated. With the physical delivery of an ordered custom array taking about 3–4 weeks, slightly longer design times on a modern workstation (Table 1) will often be acceptable in return for improved design results.
In summary, TherMODO provides a flexible pipeline for the design of high-performance microarrays. The algorithm benefits from advanced quantitative models and the power of global optimization, and operates without a need for user supplied threshold parameters.
SUPPLEMENTARY DATA
Computed probe designs, comparison statistics and comparative plots, as well as additional experimental data supporting the manuscript are archived online at http://bioinf.boku.ac.at/pub/thermodo2008/.
FUNDING
Vienna Science and Technology Fund, Baxter AG, Austrian Research Centres Seibersdorf, and the Austrian Centre of Biopharmaceutical Technology (‘WWTF Vienna Science Chair of Bioinformatics’ to G.G.L., T.T., P.S., and D.P.K.); Austrian Research Centres Seibersdorf (G.S. and K.B.); Austrian Research Promotion Agency (‘GenAU BIN-II’ to T.T. and I.H.). Funding for open acess charge: WWTF Vienna Science Chair of Bioinformatics.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge helpful discussions with John SantaLucia Jr, Michael Zuker and Ian Korf. We thank Andreas Sommer for assistance with the gel analysis.
REFERENCES
- 1.Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat. Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES. Array of hope. Nat. Genet. 1999;21:3–4. doi: 10.1038/4427. [DOI] [PubMed] [Google Scholar]
- 3.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat. Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 4.Horak CE, Snyder M. ChIP-chip: a genomic approach for identifying transcription factor binding sites. Methods Enzymol. 2002;350:469–483. doi: 10.1016/s0076-6879(02)50979-4. [DOI] [PubMed] [Google Scholar]
- 5.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 6.Derisi J. Overview of nucleic acid arrays. Curr. Protoc. Mol. Biol. 2001 doi: 10.1002/0471142727.mb2201s49. Chapter 22 Unit 22.1. [DOI] [PubMed] [Google Scholar]
- 7.Dudley AM, Aach J, Steffen MA, Church GM. Measuring absolute expression with microarrays with a calibrated reference sample and an extended signal intensity range. Proc. Natl. Acad. Sci. USA. 2002;99:7554–7559. doi: 10.1073/pnas.112683499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kakuhata R, Watanabe M, Yamamoto T, Obana E, Yamazaki N, Kataoka M, Ooie T, Baba Y, Hori T, Shinohara Y. Importance of probe location for quantitative comparison of signal intensities among genes in microarray analysis. J. Biochem. Biophys. Methods. 2008;70:926–931. doi: 10.1016/j.jprot.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Nadon R, Shoemaker J. Statistical issues with microarrays: processing and analysis. Trends Genet. 2002;18:265–271. doi: 10.1016/s0168-9525(02)02665-3. [DOI] [PubMed] [Google Scholar]
- 10.Saidi SA, Holland CM, Kreil DP, MacKay DJ, Charnock-Jones DS, Print CG, Smith SK. Independent component analysis of microarray data in the study of endometrial cancer. Oncogene. 2004;23:6677–6683. doi: 10.1038/sj.onc.1207562. [DOI] [PubMed] [Google Scholar]
- 11.Lee SI, Batzoglou S. Application of independent component analysis to microarrays. Genome Biol. 2003;4:R76. doi: 10.1186/gb-2003-4-11-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koltai H, Weingarten-Baror C. Specificity of DNA microarray hybridization: characterization, effectors and approaches for data correction. Nucleic Acids Res. 2008;36:2395–2405. doi: 10.1093/nar/gkn087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei H, Kuan PF, Tian S, Yang C, Nie J, Sengupta S, Ruotti V, Jonsdottir GA, Keles S, et al. A study of the relationships between oligonucleotide properties and hybridization signal intensities from NimbleGen microarray datasets. Nucleic Acids Res. 2008;36:2926–2938. doi: 10.1093/nar/gkn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luebke KJ, Balog RP, Garner HR. Prioritized selection of oligodeoxyribonucleotide probes for efficient hybridization to RNA transcripts. Nucleic Acids Res. 2003;31:750–758. doi: 10.1093/nar/gkg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreil DP, Russell RR, Russell S. Microarray oligonucleotide probes. Methods Enzymol. 2006;410:73–98. doi: 10.1016/S0076-6879(06)10004-X. [DOI] [PubMed] [Google Scholar]
- 16.Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 2000;28:4552–4557. doi: 10.1093/nar/28.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F, Stormo GD. Selection of optimal DNA oligos for gene expression arrays. Bioinformatics. 2001;17:1067–1076. doi: 10.1093/bioinformatics/17.11.1067. [DOI] [PubMed] [Google Scholar]
- 18.Chou HH, Hsia AP, Mooney DL, Schnable PS. Picky: oligo microarray design for large genomes. Bioinformatics. 2004a;20:2893–2902. doi: 10.1093/bioinformatics/bth347. [DOI] [PubMed] [Google Scholar]
- 19.Rahmann S. Fast and sensitive probe selection for DNA chips using jumps in matching statistics. Proc. IEEE Comput. Soc. Bioinform. Conf. 2003;2:57–64. [PubMed] [Google Scholar]
- 20.Nordberg EK. YODA: selecting signature oligonucleotides. Bioinformatics. 2005;21:1365–1370. doi: 10.1093/bioinformatics/bti182. [DOI] [PubMed] [Google Scholar]
- 21.Bozdech Z, Zhu J, Joachimiak MP, Cohen FE, Pulliam B, DeRisi JL. Expression profiling of the schizont and trophozoite stages of plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rimour S, Hill D, Militon C, Peyret P. GoArrays: highly dynamic and efficient microarray probe design. Bioinformatics. 2005;21:1094–1103. doi: 10.1093/bioinformatics/bti112. [DOI] [PubMed] [Google Scholar]
- 23.Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charbonnier Y, Gettler B, Francois P, Bento M, Renzoni A, Vaudaux P, Schlegel W, Schrenzel J. A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus. BMC Genomics. 2005;6:95. doi: 10.1186/1471-2164-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mrowka R, Schuchhardt J, Gille C. Oligodb–interactive design of oligo DNA for transcription profiling of human genes. Bioinformatics. 2002;18:1686–1687. doi: 10.1093/bioinformatics/18.12.1686. [DOI] [PubMed] [Google Scholar]
- 26.Tolstrup N, Nielsen PS, Kolberg JG, Frankel AM, Vissing H, Kauppinen S. OligoDesign: optimal design of lna (locked nucleic acid) oligonucleotide capture probes for gene expression profiling. Nucleic Acids Res. 2003;31:3758–3762. doi: 10.1093/nar/gkg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Seed B. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics. 2003;19:796–802. doi: 10.1093/bioinformatics/btg086. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen HB, Wernersson R, Knudsen S. Design of oligonucleotides for microarrays and perspectives for design of multi-transcriptome arrays. Nucleic Acids Res. 2003;31:3491–3496. doi: 10.1093/nar/gkg622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Sharp BM. Oliz, a suite of perl scripts that assist in the design of microarrays using 50mer oligonucleotides from the 3′ untranslated region. BMC Bioinformatics. 2002;3:27. doi: 10.1186/1471-2105-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon PM, Sensen CW. Osprey: a comprehensive tool employing novel methods for the design of oligonucleotides for dna sequencing and microarrays. Nucleic Acids Res. 2004;32:e133. doi: 10.1093/nar/gnh127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu D, Li G, Wu L, Zhou J, Xu Y. PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics. 2002;18:1432–1437. doi: 10.1093/bioinformatics/18.11.1432. [DOI] [PubMed] [Google Scholar]
- 32.Reymond N, Charles H, Duret L, Calevro F, Beslon G, Fayard JM. ROSO: optimizing oligonucleotide probes for microarrays. Bioinformatics. 2004;20:271–273. doi: 10.1093/bioinformatics/btg401. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka F, Kameda A, Yamamoto M, Ohuchi A. Design of nucleic acid sequences for DNA. Nat. Protoc. 2007;2:2677–2691. [Google Scholar]
- 34.Hofacker I, Fontana W, Stadler P, Bonhoeffer S, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsch. Chem. 1994;125:167–188. [Google Scholar]
- 35.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighborn thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mückstein U, Tafer H, Hackermüller J, Bernhart SH, Stadler PF, Hofacker IL. Thermodynamics of RNA-RNA binding. Bioinformatics. 2006;22:1177–1182. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- 38.Bernhart SH, Tafer H, Mückstein U, Flamm C, Stadler PF, Hofacker IL. Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol. Biol. 2006;1:3. doi: 10.1186/1748-7188-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mückstein U, Tafer H, Bernhard SH, Hernandez-Rosales M, Vogel J, Stadler PF, Hofacker IL. Translational control by RNA-RNA interaction: improved computation of RNA-RNA binding thermodynamics. In: Elloumi M, Küng J, Linial M, Murphy R, Schneider K, Toma C, editors. Bioinformatics Research and Development. Berlin: Springer; 2008. Vol. 13 of Communications in Computer and Information Science. [Google Scholar]
- 40.Li X, He Z, Zhou J. Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation. Nucleic Acids Res. 2005;33:6114–6123. doi: 10.1093/nar/gki914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu G, Llinas M, Li J, Preiser PR, Bozdech Z. Selection of long oligonucleotides for gene expression microarrays using weighted rank-sum strategy. BMC Bioinformatics. 2007;8:350. doi: 10.1186/1471-2105-8-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratushna VG, Weller JW, Gibas CJ. Secondary structure in the target as a confounding factor in synthetic oligomer microarray design. BMC Genomics. 2005;6:31. doi: 10.1186/1471-2164-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 44.Carninci P, Waki K, Shiraki T, Konno H, Shibata K, Itoh M, Aizawa K, Arakawa T, Ishii Y, Sasaki D, et al. Targeting a complex transcriptome: the construction of the mouse full-length cDNA encyclopedia. Genome Res. 2003;13:1273–1289. doi: 10.1101/gr.1119703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hofacker IL, Priwitzer B, Stadler PF. Prediction of locally stable RNA secondary structures for genome-wide surveys. Bioinformatics. 2004;20:186–190. doi: 10.1093/bioinformatics/btg388. [DOI] [PubMed] [Google Scholar]
- 47.Bompfünewerer AF, Backofen R, Bernhart SH, Hertel J, Hofacker IL, Stadler PF, Will S. Variations on RNA folding and alignment: lessons from benasque. J. Math. Biol. 2008;56:119–144. doi: 10.1007/s00285-007-0107-5. [DOI] [PubMed] [Google Scholar]
- 48.Fotin AV, Drobyshev AL, Proudnikov DY, Perov AN, Mirzabekov AD. Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res. 1998;26:1515–1521. doi: 10.1093/nar/26.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chou CC, Chen CH, Lee TT, Peck K. Optimization of probe length and the number of probes per gene for optimal microarray analysis of gene expression. Nucleic Acids Res. 2004b;32:e99. doi: 10.1093/nar/gnh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Jaroszewski L, Godzik A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics. 2002;18:77–82. doi: 10.1093/bioinformatics/18.1.77. [DOI] [PubMed] [Google Scholar]
- 51.Marko NF, Frank B, Quackenbush J, Lee NH. A robust method for the amplification of RNA in the sense orientation. BMC Genomics. 2005;6:27. doi: 10.1186/1471-2164-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pozhitkov AE, Tautz D, Noble PA. Oligonucleotide microarrays: widely applied–poorly understood. Brief. Funct. Genomic Proteomic. 2007;6:141–148. doi: 10.1093/bfgp/elm014. [DOI] [PubMed] [Google Scholar]
- 53.Mathews DH, Burkard ME, Freier SM, Wyatt JR, Turner DH. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999;5:1458–1469. doi: 10.1017/s1355838299991148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.SantaLucia J. Physical principles and visual-OMP software for optimal PCR design. Methods Mol. Biol. 2007;402:3–34. doi: 10.1007/978-1-59745-528-2_1. [DOI] [PubMed] [Google Scholar]
- 55.Gao Y, Wolf LK, Georgiadis RM. Secondary structure effects on dna hybridization kinetics: a solution versus surface comparison. Nucleic Acids Res. 2006;34:3370–3377. doi: 10.1093/nar/gkl422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu WT, Guo H, Wu JH. Effects of target length on the hybridization efficiency and specificity of rRNA-based oligonucleotide microarrays. Appl. Environ. Microbiol. 2007;73:73–82. doi: 10.1128/AEM.01468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Relogio A, Schwager C, Richter A, Ansorge W, Valcarcel J. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 2002;30:e51. doi: 10.1093/nar/30.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bernal A, Ear U, Kyrpides N. Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res. 2001;29:126–127. doi: 10.1093/nar/29.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Griffith M, Tang MJ, Griffith OL, Morin RD, Chan SY, Asano JK, Zeng T, Flibotte S, Ally A, Baross A, et al. ALEXA: a microarray design platform for alternative expression analysis. Nat. Methods. 2008;5:118. doi: 10.1038/nmeth0208-118. [DOI] [PubMed] [Google Scholar]
- 60.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 61.SantaLucia J, Hicks D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]