Abstract
Topoisomerases are ubiquitous proteins found in all three domains of life. They change the topology of DNA via transient breaks on either one or two of the DNA strands to allow passage of another single or double DNA strand through the break. Topoisomerases are classified into two types: type I enzymes cleave one DNA strand and pass either one or two DNA strands through the break before resealing it, while type II molecules cleave both DNA strands in concert and pass another double strand through the break followed by religation of the double strand break. Here we review recent work on the structure of type I enzymes. These structural studies are providing atomic details that, together with the existing wealth of biochemical and biophysical data, are bringing our understanding of the mechanism of action of these enzymes to the atomic level.
INTRODUCTION
Topoisomerases are ubiquitous proteins found in all three domains of life (bacteria, archaea and eukarya). They are capable of changing the topology of DNA via transient breaks on one or two of the DNA strands to allow passage of either a single or double DNA strand through the break. Topoisomerases have been involved in several cellular processes, such as transcription, replication and recombination, and the importance of their cellular role is underscored by the fact that they are the target of several cancer chemotherapeutic agents and antibiotics [for a review, see (1)]. Topoisomerases are classified into two types: type I enzymes cleave one DNA strand and pass either one or two strands through the break before resealing it, while type II molecules cleave both DNA strands in concert and pass another double strand through the break followed by religation of the double strand break. Type I enzymes do not require any high-energy cofactor for activity; the reaction is driven by the energy stored in the supercoiled DNA as torsional strain. Type II enzymes also do not require an external source of energy for the cleavage/religation part of the reaction, but they do utilize ATP hydrolysis to drive conformational changes in the protein during the reaction cycle.
Type I enzymes have been further classified into three different families: types IA, IB and IC (2). Family members are similar to each other in sequence, structure and atomic mechanism. However, members of one family show no sequence or structural similarities to members of the other families. The atomic mechanisms employed for cleavage and religation of the DNA by different families have only superficial similarities. Type I enzymes operate by forming a transient phosphotyrosine covalent bond with one end of the broken DNA strand, either the 5′ or the 3′ end, followed by passage of the unbroken strand through the break, and ultimately resealing of the break. In this context, only two possible mechanisms of relaxation are possible: enzyme-bridged strand passage, where the protein mediates the opening of one strand to allow passage of the other, and swiveling, which involves the rotation of the DNA strands. One important difference between these mechanisms is the need to hold one or both ends of the broken DNA strand. During enzyme-bridged strand passage, it is necessary to non-covalently hold one end of the broken strand while the other end is covalently attached to the active site tyrosine. On the other hand, the swiveling mechanism requires the non-covalently bound end of the DNA to move around freely and is not attached to the protein. It is clear that each family of type I enzymes has to employ one of the two relaxation mechanisms and form either a 5′ or 3′ covalent intermediate. It is now established that type IA enzymes form transient 5′ covalent intermediates and relax DNA through an enzyme-bridged strand-passage mechanism, while type IB and IC enzymes both form 3′ covalent intermediates and relax DNA by swiveling, although the details of how this is accomplished are probably very different.
Type I topoisomerases change the topology of DNA without the need of an external source of energy, such as ATP hydrolysis. So far, the only exception is reverse gyrase, which introduces positive supercoils with the aid of ATP hydrolysis, although the way the strand passage and ATP hydrolysis steps are coupled is still not clear. To relax DNA, type I enzymes garner the energy stored as torsional stress in the supercoiled DNA. The way the stored energy in the DNA is transformed into changes in the protein as the reaction proceeds is still not understood and remains one of the major future challenges in understanding not only type I topoisomerases, but all topoisomerases.
TYPE IA TOPOISOMERASES
Type IA enzymes have been found in bacteria, archaea and eukarya and there are clear sequence similarities among all members, suggesting a strong structural and mechanistic conservation (3). They form a phosphotyrosine linkage to the 5′ end of the broken DNA strand while non-covalently holding the free 3′-end hydroxyl group. Relaxation activity occurs in the absence of a high-energy cofactor, but in the presence of divalent cations. The exact role of these cations has not been fully determined, however they are needed for DNA relaxation (4). Although they are an attractive target for chemotherapeutic agents, there are no known inhibitors or drugs that target type IA enzymes. Type IA topoisomerases relax only negatively supercoiled DNA and require single-stranded DNA regions for activity (5). The requirement of single-stranded DNA for activity provides a plausible mechanism for sensing the overall topological state of DNA as single-stranded regions are only found in negatively supercoiled or underwound DNA but not in positively supercoiled or overwound DNA. Thus, the presence of single-stranded regions, a property of DNA that can be monitored locally, could serve as an indicator of the global topological state of DNA.
The core structure of type IA topoisomerases, typically of around 67 kDa in molecular weight, has a characteristic toroidal fold formed by four domains. The toroidal fold was first observed in Escherichia coli topoisomerase I (6) and later in E. coli topoisomerase III (7), Thermotoga maritima topoisomerase I (8), and reverse gyrase from Archaeoglobus fulgidus, a hyperthermophilic archaeal organism (9) (Figure 1). Reverse gyrase is unique as it consists of a topoisomerase domain fused to a helicase domain at the N-terminus of the protein. The core domain is followed by a C-terminal domain of variable size. Sequence comparison of the C-terminal region extending from the core domain of the type IA enzymes shows that this region does not present the same level of conservation as the core. The C-terminal domains can vary greatly in size and play a role in DNA/protein interactions (10–12). In the case of E. coli topoisomerase III, deletion of this C-terminal domain is deleterious, although its removal does not abolish activity completely (13). In the case of E. coli topoisomerase I, the C-terminal domain is essential for activity (14,15). In both cases, the C-terminal domain is responsible for single-stranded DNA binding and confers higher DNA binding affinity (10,12,13). The structures of the C-terminal domain of E. coli topoisomerase I (16) and the intact T. maritima topoisomerase I (8) reveal the presence of zinc-ribbon single-stranded DNA binding motif. The structure of the T. maritima topoisomerase I shows the way the core domain and the C-terminal domain are arranged relative to each other, but the structural bases for the role of this domain in the overall reaction are still unknown.
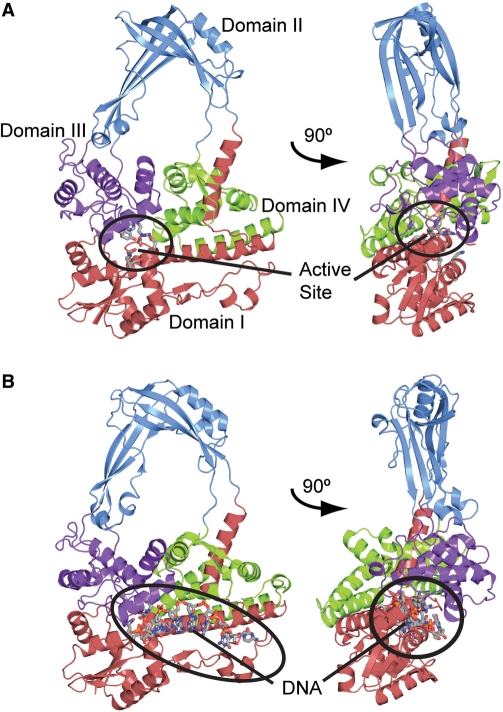
Figure 1.
Structure of a type IA topoisomerase. E. coli topoisomerase III is shown to illustrate the overall structure of a type IA topoisomerase and the typical toroidal fold observed in all members of this type. (A) Diagram showing the structure of the apo-enzyme [PDB 1D6M (7)]. In the absence of DNA, the active site, found at the intersection of domains I and III (encircled), is buried. (B) Diagram showing the structure of a complex with single-stranded DNA [PDB 1I7D (23)]. Note the movement of domains that occurs in order to accommodate DNA. In both diagrams, the four major domains of the protein are colored red, blue, purple and green for domains I, II, III and IV, respectively. The single-stranded DNA binding groove, shown circled in black, extends from domain IV to the active site. The active site residues as well as the single-stranded DNA in the complex are shown in a ball and stick representation.
A combination of biochemical, biophysical and structural studies has helped reveal the atomic mechanism of the type IA enzymes. A detailed model on how type IA enzymes perform complex topological rearrangements was proposed several years ago based on the structure of E. coli topoisomerase I (6). Subsequent biochemical studies using a mutant enzyme capable of forming a disulfide bridge trapped some of the proposed intermediates and confirmed the general features of the mechanism of action (17), in particular that the enzyme opens up through a large conformational change and sequesters the passing DNA strand inside the protein. Recent single molecule experiments (18,19) have further confirmed the general features of the proposed mechanism. These experiments, in conjunction with the wealth of existing biochemical data, confirmed that the enzymes change the linking number in steps of one, as is mandated in an enzyme-bridged strand-passage mechanism.
The proposed mechanism of action involves several steps (Figure 2), and crystal structures for some of these intermediate steps have been obtained (20–22). The structures revealed that the enzyme and the DNA have to change conformation during the catalytic cycle in a coordinated manner. Structures of the apo-state of E. coli topoisomerase I and III (6,7), T. maritima topoisomerase I (8) and A. fulgidus reverse gyrase (9) showed the enzymes in a closed conformation with the active site not fully formed. The structure of E. coli topoisomerase III in complex with a short oligonucleotide shows that type IA enzymes have to undergo conformational changes to form the active site and that these changes are not only confined to the relative movement of the domains, but also involve small but important rearrangements in some of the domains of the molecule (20,23), in particular domain IV, which is directly involved in forming a single-stranded DNA binding groove. Additionally, the structure of a fragment of E. coli topoisomerase I (21) indicated that the enzyme can undergo dramatic rearrangements of one of its domains (domain II), while, the structure of E. coli topoisomerase I in complex with a short oligonucleotide (22) shows the structure of an intermediate where the active site is not yet fully formed, but some movements of secondary structure elements in domain IV have started to occur. There is still no structure of a covalent intermediate that shows the crucial interactions with the 5′ and 3′ ends of the DNA, but recent studies of wild-type E. coli topoisomerase III in complex with DNA showed for the first time a wild-type enzyme in complex with DNA perfectly poised for DNA cleavage (20) (Figure 3). This structure confirms that the formation of a competent active site depends on several rearrangements of the enzyme.
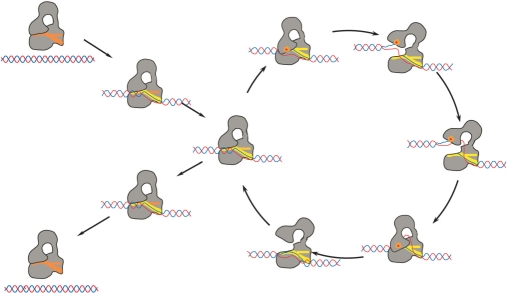
Figure 2.
Diagram showing the proposed mechanism of DNA relaxation by type IA topoisomerases. The mechanism involves several transient conformational intermediates both of the protein and the DNA. The sequence of the steps and the intermediates are hypothetical and more states are likely to be involved in the cycle. Processivity by the enzyme requires that after one relaxation event the protein continues to another relaxation cycle without releasing the DNA. In the diagram, the protein is shown in grey and the DNA in red/blue. The orange dot represents the presence of the covalent protein/DNA complex. The single-stranded DNA binding groove is shown in red or yellow.
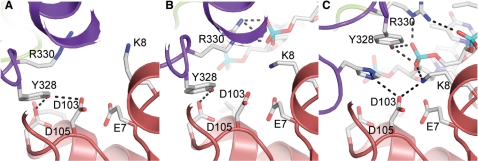
Figure 3.
Active site of E. coli topoisomerase III, a type IA topoisomerase. The diagrams show the active site region in different complexes. (A) Apo-structure, where the protein is in a closed conformation and the active site is buried between domains I and III [PDB 1D6M (7)]. (B) Intermediate structure, where the DNA starts entering the active site but it is not yet positioned properly for cleavage/religation [PDB 2O59 (20)]. (C) Fully formed active site in the wild-type enzyme, with the single-stranded DNA positioned and the active site tyrosine poised for cleavage [PDB 2O19 (20)]. The diagrams illustrate the way the active site is assembled and the interactions with single-stranded DNA. Domain I is shown in red and domain III in purple. Some of the side chains forming the active site are shown, as well as some possible hydrogen bonds. The single-stranded DNA is shown as sticks.
An initial identification of the residues comprising the active site was based on the structure of the apo-enzyme (6) and sequence conservation (24). The structure of an E. coli topoisomerase III-single-stranded DNA complex (23) showed that a conformational change occurs upon single-stranded DNA binding and this leads to the creation of a competent active site by bringing together residues that are distant in the apo-enzyme. In the complex structure, the active site tyrosine, an arginine, a lysine and a glutamate form direct contacts to the DNA near the scissile bond. In addition, a second layer of highly conserved amino acids surrounds the active site, including three highly conserved acidic residues that have been shown to be involved in magnesium binding (25). Due to the conformational changes that occur upon binding DNA (Figure 3), some of the residues that in the apo-structure were observed to be distant from the putative active site, they were found to be central to the mechanism in the complex structure. An example is Glu7 in topoisomerase III, which is completely conserved and interacts directly with DNA, but is located several Angstroms away from the active site in the apo-structure and whose exact role could not be surmised from biochemical studies. Several biochemical studies, mostly using site-directed mutagenesis [for example (26–32)] have confirmed the role of many of the residues identified by these structures. Due to their dynamic nature, type IA topoisomerases are a very good example of proteins where understanding the catalytic mechanism in the absence of structures is almost impossible.
There are still many open questions regarding the atomic mechanism of type IA topoisomerases. The chemistry of the mechanism of cleavage and religation, the manner in which the free 3′-end interacts with the protein during the reaction, the role of magnesium, and the atomic mechanism of strand passage are some of the questions that remain to be answered. Our understanding of type IA enzymes will be incomplete until we elucidate the atomic details of the movement of one strand past the other and how the torsional strain in DNA directs conformational changes in the protein. Additional structural information on the intermediate steps in the reaction cycle, especially those with large conformational changes both in the protein and the DNA, is essential to address these difficult questions.
TYPE IB TOPOISOMERASES
Type IB enzymes have been identified in eukaryotes, poxviruses and bacteria, but not in archaea. Eukaryotic type IB molecules are large, typically over 90 kDa in molecular weight, while viral and bacterial type IB molecules are relatively small, around 36 kDa. Despite their differences in size, all type IB molecules share a common fold around the active site region and a common catalytic mechanism (3,33,34) (Figure 4). Interestingly, these similarities extend to the tyrosine recombinase family suggesting a common ancestor for both type IB topoisomerases and tyrosine recombinases (33). Type IB enzymes form a transient 3′ phosphotyrosine covalent linkage with the broken end of the DNA. They can relax both negatively and positively supercoiled DNA without requiring divalent ions or ATP. By breaking transiently one DNA strand, type IB enzymes allow torsionally strained DNA to relax. As long as there is torsional strain to drive the swiveling, the reaction will proceed until the DNA is fully relaxed. Because of this, type IB enzymes do not have to sense directly the global topological state of DNA, they simply provide the means to relieve torsional strain if some is present, even if it is only in a local supercoiled domain.
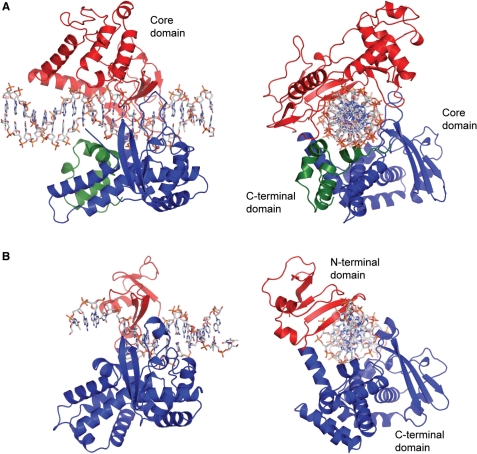
Figure 4.
Structures of type IB topoisomerases. Diagrams of the structures of eukaryotic and viral type IB topoisomerases. (A) Structure of human topoisomerase I in non-covalent complex with DNA [PDB 1A35 (34)]. The protein encircles the DNA by forming a clamp around it. The protein is composed of two domains: a core domain, shown in red and blue, and a C-terminal domain, shown in green. (B) Structure of variola virus topoisomerase I in covalent complex with DNA [PDB 2H7F (51)]. The viral and bacterial type IB enzymes are composed of two domains: an N-terminal domain, shown in red, and a larger, C-terminal domain, shown in blue. Note the marked similarities between the human and poxviral proteins despite their great disparity in size. In the case of human topoisomerase I, the core and C-terminal domains are joined by a linker domain, which is not present in this structure but that has been observed in other structures (44). The DNA in the complexes is shown in a ball and stick representation.
All type IB topoisomerases have many structural and functional characteristics in common. They all contain a highly conserved pentad of residues forming the active site (Tyr, Arg, Arg, Lys and His/Asn) with an identical architecture around the active site region (Figure 5). Eukaryotic and viral enzymes have a histidine in the pentad, while in bacterial type IB enzymes the histidine is replaced by an asparagine. Experiments show that the histidine can be replaced by an asparagine in both viral (35) and eukaryotic (36) topoisomerases, suggesting that a histidine or an asparagine can play the same role and that the overall mechanism of cleavage and religation is identical in all type IB enzymes. The mechanism of cleavage by type IB enzymes has been extensively studied and the role of different residues in catalysis has been characterized in great detail (37). The sequence specificity of the poxviral topoisomerases offers an unique advantage over other topoisomerases for biochemical studies, which has allowed exquisitely detailed biochemical studies that elucidated the role of DNA (38,39) and different amino acids (37,40–42) in the cleavage reaction.
Figure 5.
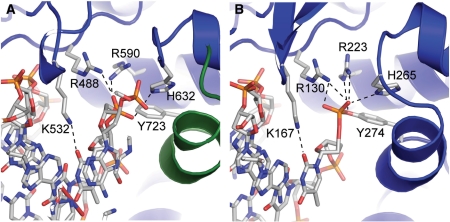
Active site of type IB topoisomerases. The diagrams show the active site region in two different type IB topoisomerases. (A) Active site in the structure of a non-covalent complex of human topoisomerase I with DNA [PDB 1A35 (34)]. Note the presence of the amino acids forming the catalytic pentad, Arg488, Lys532, Arg590, His632 and Tyr723 (in this structure the tyrosine was mutated to a phenylalanine). (B) Active site in the structure of a covalent complex of variola virus topoisomerase I with DNA [PDB 2H7F (51)]. Note the equivalent pentad of amino acids in the active site and the formation of the covalent intermediate between the tyrosine and the 3′ end of the broken strand. The diagrams serve to illustrate the high similarity between the active sites of the smaller and larger type IB enzymes and also the structures of non-covalent and covalent complexes.
Eukaryotic type IB topoisomerases are large proteins that contain multiple structural components that have no counterpart in the viral and bacterial topoisomerases. The N-terminal region of eukaryotic topoisomerases I is dispensable for topoisomerase activity. The central or core domain together with a C-terminal domain can reconstitute a fully functional enzyme (43). These two domains are joined by a short linker domain formed by two long helices (43). The C-terminal domain contains the catalytic tyrosine while all other residues in the catalytic pentad reside in the core domain. Structures of the fully functional enzyme, formed by the core and C-terminal domains and in some instances the linker domain, in complex with DNA shows a C-shaped protein clamp around duplex DNA (34,44) (Figure 4). The two regions that close the C-clamp consist of loops that meet in a non-covalent interaction to envelop the target DNA. In contrast to the eukaryotic enzymes, the poxviral and bacterial type IB topoisomerases contain two distinct domains (45,46). The smaller amino terminal domain is involved in interactions with DNA (45,47,48), while the larger domain contains the active site and retains full enzymatic activity (49). The structures of both domains of vaccinia virus topoisomerase I, the best characterized and studied viral type IB enzyme, are known (33,45) as well as the structure of the intact Deinococcus radiodurans topoisomerase IB (50). The structure of variola virus topoisomerases I in complex with DNA revealed an important point: all type IB enzymes encircle DNA in an identical fashion, despite their differences in size. In addition, poxviral type IB enzymes are unique in that they have a preference for binding to specific DNA sequences. Hence, the structure of variola virus topoisomerase I in complex with DNA (51) helps to understand the atomic basis for the sequence specificity.
Overall, the structure of all these type IB enzymes confirms that eukaryotic, viral and bacterial type IB topoisomerases are similar despite their dissimilarities in size. The structural conservation is particularly striking around the active site region. Even though the N-terminal domain in the bacterial and viral enzymes is less conserved, there are marked commonalities with the equivalent region in the larger enzymes. Clearly, the smaller type IB enzymes represent an abbreviated form of the larger eukaryotic topoisomerases where all the essential regions are present. The mechanisms of cleavage/religation and DNA relaxation are, without any doubt, identical.
Type IB enzymes relax DNA by a mechanism termed ‘controlled rotation’, where one DNA strand swivels around the other. A swiveling mechanism predicts that the enzymes relax DNA in steps of n, and this has been confirmed recently by single-molecule experiments on vaccinia virus, yeast and human topoisomerase IB (52,53). The single-molecule experiments also indicate that the swiveling is slowed down by friction created by the interaction between the enzyme and the DNA, consistent with the embracing of the DNA by the protein as observed in the structures of human and variola virus topoisomerase I in complex with DNA (34,51).
Eukaryotic type IB topoisomerases have received additional attention as important chemotherapeutic targets. They are the target of important anticancer compounds such as camptothecins (54–57), indolocarbazoles (58) and indenoisoquinolines (59–61). Although type IB enzymes are not widespread in bacteria, they are present in some important human pathogens, such as Pseudomonas aeuroginosa and Bordetella parapertussis. Hence, bacterial type IB enzymes could serve as targets of chemotherapeutic agents, especially now that structures are starting to reveal the atomic differences between human, viral and bacterial enzymes. The structures of ternary complexes of the enzyme, DNA and drugs (62–65) reveal the drug interactions at the atomic level and this may be helpful in the development of better type IB poisons. Given the importance of type IB enzymes in cancer therapeutics, it is likely that this will continue to be a fruitful study area and that the information garnered will also shed light on the mechanism of action of the enzymes.
As is the case with the type IA enzymes, there are still many details of the mechanism of type IB enzymes that need to be elucidated. Unlike type IA enzymes, the structures of both covalent and non-covalent intermediates of type IB enzymes are available. Together with the wealth of biochemical information, these structures have helped us understand the cleavage and religation mechanism in great detail. Unfortunately, there is scant atomic information on the way that type IB enzymes drive DNA relaxation. The changes that occur in the protein as the DNA swivels inside the C-clamp are yet to be elucidated. The same is true about the conformational changes in the DNA. Even though the single-molecule experiments suggest the presence of atomic friction due to the interaction of the protein and DNA, the details of this process are largely unknown. Capturing intermediates in the relaxation process is not an easy task, but they are required in order to truly understand the atomic mechanism of action.
TYPE IC TOPOISOMERASES
The third family, type IC topoisomerases, was recently defined with the realization that topoisomerase V is distinct from all other topoisomerases. DNA topoisomerase V was originally found in the hyperthermophile Methanopyrus kandleri isolated from a deep-water ‘black smoker’ chimney in the Gulf of California (66). Later on its presence was confirmed in other Methanopyrus isolates from around the world (A. Slesarev, personal communication). In common with type IB enzymes, topoisomerase V cleaves one DNA strand, forms a transient phosphotyrosine bond with the 3′ end of the broken strand, can relax positive and negative supercoils, and does not require the presence of magnesium or ATP (67). Despite these biochemical similarities, it shows no sequence or structural similarity to other topoisomerases. Topoisomerase V is a large enzyme, over 100 kDa in molecular weight, with unusual characteristics. Not only can it relax DNA identically to other topoisomerases (67), but it is also involved in DNA repair (68). The two different catalytic activities are found in the same polypeptide: topoisomerase activity resides in the N-terminus of the protein, while the C-terminus of the protein possesses apurinic/apyrimidinic (AP) site-processing activity, normally associated with base excision DNA repair (69). Sequence analysis indicates that the protein contains 24 helix-hairpin-helix (HhH) DNA binding motifs arranged in 12 tandem (HhH)2 domains (70) that follow the topoisomerase domain (68). N-terminal constructs as small as 30 kDa comprising the topoisomerase domain have full topoisomerase activity. The AP site-processing activity resides in the C-terminal 34 kDa fragment of the protein (69), which also contains several putative HhH motifs (68). The minimal construct with both topoisomerase and DNA repair activities is a 78 kDa N-terminal fragment (69). Topoisomerase V catalyzes DNA relaxation under extreme conditions. It has maximal topoisomerase activity at around 108°C and is still active at 122°C (71). It is also active in a wide range of conditions, up to 0.65 M NaCl or KCl and up to 3.1 M potassium glutamate. These properties have been attributed to the HhH motifs, as fragments containing fewer motifs show optimal activity in a narrower set of conditions and also a marked loss of processivity (69).
The structure of a 61 kDa N-terminal fragment of topoisomerase V comprising the topoisomerase domain and eight HhH motifs reveals a fold that is not related to the folds observed in any other topoisomerase, tyrosine recombinase, or other known protein (72) (Figure 6A). The composition of the active site of topoisomerase V is similar to the one in type IB molecules and includes the active site tyrosine, two arginines, a lysine and a histidine (Figure 6B). The key difference between the active site of type IB and type IC enzymes is that the spatial arrangement of these residues is different. In addition, an acidic residue in topoisomerase V has no equivalent in type IB enzymes, suggesting an alternative mechanism of catalysis. Interestingly, type IA enzymes do contain an acidic residue in the active site, but it is not known whether the acidic amino acids play a similar role in catalysis. Furthermore, the structure suggests that a large conformational change in the protein is needed for activity, which could alter the conformation and position of the residues in the active site, similarly to what was observed in type IA enzymes (see above). The mechanism of DNA cleavage and religation employed by topoisomerase V is not understood. Limited mutagenesis experiments suggest that topoisomerase V employs a different mechanism from type IB enzymes (72) as mutation of equivalent residues in both enzymes does not produce similar reductions in relaxation activity. Further studies are needed to elucidate the exact mechanism of catalysis, the residues involved, and their role in the cleavage/religation reaction. Topoisomerase V also has DNA repair activity, but the structure of the domain containing the AP site processing activity is not known yet and the residues involved in DNA repair have not been identified.
Figure 6.
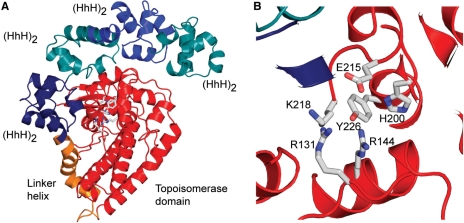
Structure of a type IC topoisomerase. (A) Diagram showing the structure of a 61 kDa fragment of M. kandleri topoisomerase V [PDB 2CSB (72)]. The fragment comprises the topoisomerase domain and four (HhH)2 domains, which are likely to be involved in DNA binding. The topoisomerase domain, shown in red, and the (HhH)2 domains, shown in tones of blue, are joined by a linker helix, shown in orange. The active site, shown as ball and stick representation, is buried at the interface of the topoisomerase domain and one of the (HhH)2 domains. To access the active site, the protein has to change conformation, probably by separating the sub-domains. (B) The putative amino acids forming the topoisomerase V active site are shown. Aside from the tyrosine, two arginines, a lysine, a histidine and a glutamate form the putative active site. Mutagenesis studies show that removal of the arginines has a marked detrimental effect on activity. Removal of the glutamate also changes the activity, although not as markedly. Removal of the histidine and lysine has a modest effect on activity (72).
The relaxation mechanism of this new topoisomerase is not nearly as well understood as that of the other topoisomerases. Recent single-molecule experiments established that the mechanism of action of topoisomerase V is similar to the ‘controlled rotation’ mechanism proposed for type IB enzymes, where relaxation occurs in steps of n (73). Furthermore, the topoisomerase domain is active in the absence of any of the HhH repeats, suggesting that the topoisomerase domain may be able to relax DNA without having to completely encircle it. These observations strongly suggest that although type IB topoisomerases and topoisomerase V have overall mechanistic similarities, the way they perform the reaction may be very different at the atomic level.
The similarities between topoisomerases V and type IB enzymes in the overall relaxation mechanism are striking: both work by swiveling and form 3′ covalent intermediates. Even more remarkable are the differences: no sequence or structural similarity and a different catalytic mechanism for cleavage and religation. This clearly shows that the two types of enzymes represent two different solutions to the same overall problem. This observation is even more interesting when viewed in the context of all other topoisomerases. There appears to be two general mechanisms employed by topoisomerases for DNA relaxation: (a) type IA and type II enzymes employ an enzyme-bridged strand-passage mechanism, with biochemical similarities in the way they cleave/religate DNA, and structural similarities in various domains, such as the toprim domain (3), (b) type IB enzymes all share a common fold and employ the same swiveling mechanism for DNA relaxation. Topoisomerase V illustrates a completely different solution to the same topological problem without any structural, sequence or biochemical similarity to either type IB or type IA/type II enzymes.
CONCLUSIONS
Over the last few years, the elucidation of the structures of various members of all the topoisomerase types has helped to bring our understanding of the mechanism of action of these important enzymes to the atomic level. In the case of type I enzymes, structures of the apoenzymes, along with enzyme–DNA complexes have provided exquisitely detailed snapshots of the enzymes during their catalytic cycle. The structures of type IA enzymes in different stages of the catalytic cycle illustrate the way these proteins recognize and bind DNA, and also the changes that occur in both substrate and enzyme as they interact with each other. The details of the atomic mechanism of type IB enzymes are also well understood. In particular, the mechanism of cleavage and religation is now very well dissected by a combination of structural and functional studies. Finally, type IC topoisomerases, the newest family of type I enzymes, are not as well understood as only a few structures are available and the biochemical work is not as advanced. Nevertheless, the available information indicates that type IC enzymes evolved a completely different way of performing essentially the same reaction. Overall, the structural work on type I enzymes is now maturing and reaching the stage where many important questions have been answered, but many more remain unanswered and need to be addressed if we truly want to have a complete atomic picture of their mechanism.
FUNDING
National Institutes of Health (GM51350 to A.M.). Funding for open access charge: National Institutes of Health (GM51350 to A.M.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Forterre P. DNA topoisomerase V: a new fold of mysterious origin. Trends Biotechnol. 2006;24:245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 4.Domanico PL, Tse-Dinh YC. Mechanistic studies on E. coli DNA topoisomerase I: divalent ion effects. J. Inorg. Biochem. 1991;42:87–96. doi: 10.1016/0162-0134(91)80035-g. [DOI] [PubMed] [Google Scholar]
- 5.Kirkegaard K, Wang JC. Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single stranded loop. J. Mol. Biol. 1985;185:625–637. doi: 10.1016/0022-2836(85)90075-0. [DOI] [PubMed] [Google Scholar]
- 6.Lima CD, Wang JC, Mondragon A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 7.Mondragon A, DiGate R. Structure of E. coli DNA topoisomerase III. Structure. 1999;7:1373–1383. doi: 10.1016/s0969-2126(00)80027-1. [DOI] [PubMed] [Google Scholar]
- 8.Hansen G, Harrenga A, Wieland B, Schomburg D, Reinemer P. Crystal structure of full length topoisomerase I from Thermotoga maritima. J. Mol. Biol. 2006;358:1328–1340. doi: 10.1016/j.jmb.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez AC, Stock D. Crystal structure of reverse gyrase: insights into the positive supercoiling of DNA. EMBO J. 2002;21:418–426. doi: 10.1093/emboj/21.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beran-Steed RK, Tse-Dinh YC. The carboxy terminal domain of Escherichia coli DNA topoisomerase I confers higher affinity to DNA. Proteins. 1989;6:249–258. doi: 10.1002/prot.340060307. [DOI] [PubMed] [Google Scholar]
- 11.Zhang HL, Malpure S, Li Z, Hiasa H, DiGate RJ. The role of the carboxyl-terminal amino acid residues in Escherichia coli DNA topoisomerase III-mediated catalysis. J. Biol. Chem. 1996;271:9039–9045. doi: 10.1074/jbc.271.15.9039. [DOI] [PubMed] [Google Scholar]
- 12.Ahumada A, Tse-Dinh YC. The Zn(II) binding motifs of E. coli DNA topoisomerase I is part of a high-affinity DNA binding domain. Biochem. Biophys. Res. Commun. 1998;251:509–514. doi: 10.1006/bbrc.1998.9500. [DOI] [PubMed] [Google Scholar]
- 13.Zhang HL, DiGate RJ. The carboxyl-terminal residues of Escherichia coli DNA topoisomerase III are involved in substrate binding. J. Biol. Chem. 1994;269:9052–9059. [PubMed] [Google Scholar]
- 14.Tse-Dinh YC, Beran-Seed RK. Escherichia coli DNA topoisomerase I is a zinc metalloprotein with three repetitive zinc binding domains. J. Biol. Chem. 1988;263:15857–15859. [PubMed] [Google Scholar]
- 15.Lima CD, Wang JC, Mondragon A. Crystallization of a 67 kDa fragment of Escherichia coli DNA topoisomerase I. J. Mol. Biol. 1993;232:1213–1216. doi: 10.1006/jmbi.1993.1474. [DOI] [PubMed] [Google Scholar]
- 16.Yu L, Zhu CX, Tse-Dinh YC, Fesik SW. Solution structure of the C-terminal single-stranded DNA-binding domain of Escherichia coli topoisomerase I. Biochemistry. 1995;34:7622–7628. doi: 10.1021/bi00023a008. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Mondragon A, DiGate RJ. The mechanism of type IA topoisomerase-mediated DNA topological transformations. Mol. Cell. 2001;7:301–307. doi: 10.1016/s1097-2765(01)00178-2. [DOI] [PubMed] [Google Scholar]
- 18.Dekker NH, Rybenkov VV, Duguet M, Crisona NJ, Cozzarelli NR, Bensimon D, Croquette V. The mechanism of type IA topoisomerases. Proc. Natl Acad. Sci. USA. 2002;99:12126–12131. doi: 10.1073/pnas.132378799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekker NH, Viard T, de La Tour CB, Duguet M, Bensimon D, Croquette V. Thermophilic topoisomerase I on a single DNA molecule. J. Mol. Biol. 2003;329:271–282. doi: 10.1016/s0022-2836(03)00320-6. [DOI] [PubMed] [Google Scholar]
- 20.Changela A, DiGate RJ, Mondragon A. Structural studies of E. coli topoisomerase III-DNA complexes reveal a novel type IA topoisomerase-DNA conformational intermediate. J. Mol. Biol. 2007;368:105–118. doi: 10.1016/j.jmb.2007.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg H, Lima CD, Mondragon A. Conformational changes in E. coli DNA topoisomerase I. Nature Struct. Biol. 1999;6:918–922. doi: 10.1038/13283. [DOI] [PubMed] [Google Scholar]
- 22.Perry K, Mondragon A. Structure of a complex between E. coli DNA topoisomerase I and single-stranded DNA. Structure (Camb) 2003;11:1349–1358. doi: 10.1016/j.str.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 24.Caron PR, Wang JC. In: Advances in Pharmacology. Liu LF, editor. 29B. Boca Raton: Academic Press; 1994. pp. 271–297. [DOI] [PubMed] [Google Scholar]
- 25.Zhu CX, Tse-Dinh YC. The acidic triad conserved in type IA DNA topoisomerases is required for binding of Mg(II) and subsequent conformational change. J. Biol. Chem. 2000;275:5318–5322. doi: 10.1074/jbc.275.8.5318. [DOI] [PubMed] [Google Scholar]
- 26.Chen SJ, Wang JC. Identification of active site residues in Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1998;273:6050–6056. doi: 10.1074/jbc.273.11.6050. [DOI] [PubMed] [Google Scholar]
- 27.Zhu CX, Roche CJ, Papanicolaou N, DiPietrantonio A, Tse-Dinh YC. Site-directed mutagenesis of conserved aspartates, glutamates and arginines in the active site region of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1998;273:8783–8789. doi: 10.1074/jbc.273.15.8783. [DOI] [PubMed] [Google Scholar]
- 28.Strahs D, Zhu CX, Cheng B, Chen J, Tse-Dinh YC. Experimental and computational investigations of Ser10 and Lys13 in the binding and cleavage of DNA substrates by Escherichia coli DNA topoisomerase I. Nucleic Acids Res. 2006;34:1785–1797. doi: 10.1093/nar/gkl109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng B, Feng J, Gadgil S, Tse-Dinh YC. Flexibility at Gly-194 is required for DNA cleavage and relaxation activity of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 2004;279:8648–8654. doi: 10.1074/jbc.M312095200. [DOI] [PubMed] [Google Scholar]
- 30.Cheng B, Feng J, Mulay V, Gadgil S, Tse-Dinh YC. Site-directed mutagenesis of residues involved in G Strand DNA binding by Escherichia coli DNA topoisomerase I. J. Biol. Chem. 2004;279:39207–39213. doi: 10.1074/jbc.M405891200. [DOI] [PubMed] [Google Scholar]
- 31.Zhu CX, Roche CJ, Tse-Dinh YC. Effect of Mg(II) binding on the structure and activity of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1997;272:16206–16210. doi: 10.1074/jbc.272.26.16206. [DOI] [PubMed] [Google Scholar]
- 32.Perry K, Mondragon A. Biochemical characterization of an invariant histidine involved in Escherichia coli DNA topoisomerase I catalysis. J. Biol. Chem. 2002;277:13237–13245. doi: 10.1074/jbc.M112019200. [DOI] [PubMed] [Google Scholar]
- 33.Cheng C, Kussie P, Pavletich N, Shuman S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 34.Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 35.Petersen BO, Shuman S. Histidine 265 is important for covalent catalysis by vaccinia topoisomerase and is conserved in all eukaryotic type I enzymes. J. Biol. Chem. 1997;272:3891–3896. doi: 10.1074/jbc.272.7.3891. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, Champoux JJ. The role of histidine 632 in catalysis by human topoisomerase I. J. Biol. Chem. 2001;276:677–685. doi: 10.1074/jbc.M007593200. [DOI] [PubMed] [Google Scholar]
- 37.Krogh BO, Shuman S. Catalytic mechanism of DNA topoisomerase IB. Mol. Cell. 2000;5:1035–1041. doi: 10.1016/s1097-2765(00)80268-3. [DOI] [PubMed] [Google Scholar]
- 38.Tian L, Claeboe CD, Hecht SM, Shuman S. Remote phosphate contacts trigger assembly of the active site of DNA topoisomerase IB. Structure. 2004;12:31–40. doi: 10.1016/j.str.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 39.Tian L, Sayer JM, Jerina DM, Shuman S. Individual nucleotide bases, not base pairs, are critical for triggering site-specific DNA cleavage by vaccinia topoisomerase. J. Biol. Chem. 2004;279:39718–39726. doi: 10.1074/jbc.M407376200. [DOI] [PubMed] [Google Scholar]
- 40.Cheng C, Wang LK, Sekiguchi J, Shuman S. Mutational analysis of 39 residues of vaccinia DNA topoisomerase identifies Lys-220, Arg-223, and Asn-228 as important for covalent catalysis. J. Biol. Chem. 1997;272:8263–8269. doi: 10.1074/jbc.272.13.8263. [DOI] [PubMed] [Google Scholar]
- 41.Wittschieben J, Shuman S. Mutational analysis of vaccinia DNA topoisomerase defines amino acid residues essential for covalent catalysis. J. Biol. Chem. 1994;269:29978–29983. [PubMed] [Google Scholar]
- 42.Wittschieben J, Shuman S. Mechanism of DNA transesterification by vaccinia topoisomerase: catalytic contributions of essential residues Arg-130, Gly-132, Tyr-136 and Lys-167. Nucleic Acids Res. 1997;25:3001–3008. doi: 10.1093/nar/25.15.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart L, Ireton GC, Champoux JJ. The domain organization of human topoisomerase I. J. Biol. Chem. 1996;271:7602–7608. doi: 10.1074/jbc.271.13.7602. [DOI] [PubMed] [Google Scholar]
- 44.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 45.Sharma A, Hanai R, Mondragón A. Crystal structure of the amino terminal fragment of vaccinia virus DNA topoisomerase I at 1.6 A resolution. Structure. 1994;2:767–777. doi: 10.1016/s0969-2126(94)00077-8. [DOI] [PubMed] [Google Scholar]
- 46.Sekiguchi J, Shuman S. Proteolytic footprinting of vaccinia topoisomerase bound to DNA. J. Biol. Chem. 1995;270:11636–11645. doi: 10.1074/jbc.270.19.11636. [DOI] [PubMed] [Google Scholar]
- 47.Sekiguchi J, Shuman S. Identification of contacts between topoisomerase I and its target DNA by site-specific photocrosslinking. EMBO J. 1996;15:3448–3457. [PMC free article] [PubMed] [Google Scholar]
- 48.Sekiguchi J, Shuman S. Mutational analysis of vaccinia virus topoisomerase identifies residues involved in DNA binding. Nucleic Acids Res. 1997;25:3649–3656. doi: 10.1093/nar/25.18.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng C, Shuman S. A catalytic domain of eukaryotic DNA topoisomerase I. J. Biol. Chem. 1998;273:11589–11595. doi: 10.1074/jbc.273.19.11589. [DOI] [PubMed] [Google Scholar]
- 50.Patel A, Shuman S, Mondragon A. Crystal structure of a bacterial type IB DNA topoisomerase reveals a preassembled active site in the absence of DNA. J. Biol. Chem. 2006;281:6030–6037. doi: 10.1074/jbc.M512332200. [DOI] [PubMed] [Google Scholar]
- 51.Perry K, Hwang Y, Bushman FD, Van Duyne GD. Structural basis for specificity in the poxvirus topoisomerase. Mol. Cell. 2006;23:343–354. doi: 10.1016/j.molcel.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 52.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 53.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 54.Pommier Y, Kohlhagen G, Kohn KW, Leteurtre F, Wani MC, Wall ME. Interaction of an alkylating camptothecin derivative with a DNA base at topoisomerase I-DNA cleavage sites. Proc. Natl Acad. Sci. USA. 1995;92:8861–8865. doi: 10.1073/pnas.92.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 56.Bjornsti MA, Benedetti P, Viglianti GA, Wang JC. Expression of human DNA topoisomerase I in yeast cells lacking yeast DNA topoisomerase I: restoration of sensitivity of the cells to the antitumor drug camptothecin. Cancer Res. 1989;49:6318–6323. [PubMed] [Google Scholar]
- 57.Wall ME, Wani MC. Camptothecin. Discovery to clinic. Ann. N.Y. Acad. Sci. 1996;803:1–12. doi: 10.1111/j.1749-6632.1996.tb26371.x. [DOI] [PubMed] [Google Scholar]
- 58.Meng LH, Liao ZY, Pommier Y. Non-camptothecin DNA topoisomerase I inhibitors in cancer therapy. Curr. Top. Med. Chem. 2003;3:305–320. doi: 10.2174/1568026033452546. [DOI] [PubMed] [Google Scholar]
- 59.Cushman M, Jayaraman M, Vroman JA, Fukunaga AK, Fox BM, Kohlhagen G, Strumberg D, Pommier Y. Synthesis of new indeno[1,2-c]isoquinolines: cytotoxic non-camptothecin topoisomerase I inhibitors. J. Med. Chem. 2000;43:3688–3698. doi: 10.1021/jm000029d. [DOI] [PubMed] [Google Scholar]
- 60.Jayaraman M, Fox BM, Hollingshead M, Kohlhagen G, Pommier Y, Cushman M. Synthesis of new dihydroindeno[1,2-c]isoquinoline and indenoisoquinolinium chloride topoisomerase I inhibitors having high in vivo anticancer activity in the hollow fiber animal model. J. Med. Chem. 2002;45:242–249. doi: 10.1021/jm000498f. [DOI] [PubMed] [Google Scholar]
- 61.Strumberg D, Pommier Y, Paull K, Jayaraman M, Nagafuji P, Cushman M. Synthesis of cytotoxic indenoisoquinoline topoisomerase I poisons. J. Med. Chem. 1999;42:446–457. doi: 10.1021/jm9803323. [DOI] [PubMed] [Google Scholar]
- 62.Chrencik JE, Burgin AB, Pommier Y, Stewart L, Redinbo MR. Structural impact of the leukemia drug 1-beta-D-arabinofuranosylcytosine (Ara-C) on the covalent human topoisomerase I-DNA complex. J. Biol. Chem. 2003;278:12461–12466. doi: 10.1074/jbc.M212930200. [DOI] [PubMed] [Google Scholar]
- 63.Chrencik JE, Staker BL, Burgin AB, Pourquier P, Pommier Y, Stewart L, Redinbo MR. Mechanisms of camptothecin resistance by human topoisomerase I mutations. J. Mol. Biol. 2004;339:773–784. doi: 10.1016/j.jmb.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 64.Staker BL, Feese MD, Cushman M, Pommier Y, Zembower D, Stewart L, Burgin AB. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005;48:2336–2345. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]
- 65.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl Acad. Sci. USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huber R, Kurr M, Jannasch HW, Stetter KO. A novel group of abyssal methanogenic archaebacteria (Methanopyrus) growing at 110°C. Nature. 1989;342:833–834. [Google Scholar]
- 67.Slesarev AI, Stetter KO, Lake JA, Gellert M, Krah R, Kozyavkin SA. DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature. 1993;364:735–737. doi: 10.1038/364735a0. [DOI] [PubMed] [Google Scholar]
- 68.Belova GI, Prasad R, Kozyavkin SA, Lake JA, Wilson SH, Slesarev AI. A type IB topoisomerase with DNA repair activities. Proc. Natl Acad. Sci. USA. 2001;98:6015–6020. doi: 10.1073/pnas.111040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belova GI, Prasad R, Nazimov IV, Wilson SH, Slesarev AI. The domain organization and properties of individual domains of DNA topoisomerase V, a type 1B topoisomerase with DNA repair activities. J. Biol. Chem. 2002;277:4959–4965. doi: 10.1074/jbc.M110131200. [DOI] [PubMed] [Google Scholar]
- 70.Shao X, Grishin NV. Common fold in helix-hairpin-helix proteins. Nucleic Acids Res. 2000;28:2643–2650. doi: 10.1093/nar/28.14.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kozyavkin SA, Pushkin AV, Eiserling FA, Stetter KO, Lake JA, Slesarev AI. DNA enzymology above 100 degrees C. Topoisomerase V unlinks circular DNA at 80–122 degrees C. J. Biol. Chem. 1995;270:13593–13595. doi: 10.1074/jbc.270.23.13593. [DOI] [PubMed] [Google Scholar]
- 72.Taneja B, Patel A, Slesarev A, Mondragon A. Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J. 2006;25:398–408. doi: 10.1038/sj.emboj.7600922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taneja B, Schnurr B, Slesarev A, Marko JF, Mondragon A. Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc. Natl Acad. Sci. USA. 2007;104:14670–14675. doi: 10.1073/pnas.0701989104. [DOI] [PMC free article] [PubMed] [Google Scholar]