Abstract
In the growing field of genomics, multiple alignment programs are confronted with ever increasing amounts of data. To address this growing issue we have dramatically improved the running time and memory requirement of Kalign, while maintaining its high alignment accuracy. Kalign version 2 also supports nucleotide alignment, and a newly introduced extension allows for external sequence annotation to be included into the alignment procedure. We demonstrate that Kalign2 is exceptionally fast and memory-efficient, permitting accurate alignment of very large numbers of sequences. The accuracy of Kalign2 compares well to the best methods in the case of protein alignments while its accuracy on nucleotide alignments is generally superior. In addition, we demonstrate the potential of using known or predicted sequence annotation to improve the alignment accuracy. Kalign2 is freely available for download from the Kalign web site (http://msa.sbc.su.se/).
INTRODUCTION
Multiple alignments are important to a wide spectrum of applications in comparative sequence analysis (1). The focus in the development of alignment programs has been primarily on increasing their accuracy. Unfortunately, this has led to a benchmark race that is generally won by computationally expensive methods that are impractical for large datasets. It is also not for sure that just because a method is slightly better on a benchmark test, it will always produce the most desirable multiple alignment. Efficient computational properties of alignment algorithms have become more and more important in recent years, perhaps even to a point where it is reasonable to sacrifice some accuracy for large gains in running time.
The typical number of sequences to be aligned has increased in recent years. The Pfam database (2,3) now contains families with 389.2 sequences on average, a number that will steadily increase as more genomes become fully sequenced. Also the number of non-coding RNAs (ncRNA) available is constantly evolving, constituting a new challenge in the area of multiple sequence alignments. By now the Rfam database (4) includes sequences of 607 families. So is it necessary to align all members of a sequence family at once? In a review, Cedric Notredame (5) pointed out that often too many sequences are present, and therefore users have to make a selection prior to the actual alignment. Since then alignment programs and computers have become faster and in 2005 Katoh et al. (6) convincingly demonstrated that alignment accuracy can be increased by including high numbers of homologous sequences. In detail, this strategy includes extra sequences in the alignment procedure itself but removes them from the final alignment. Clearly, alignment programs should be able to take advantage of the wealth of information present in the databases without being hindered by computational concerns. Moreover, programs should be able to align hundreds rather than tens of sequences.
We recently introduced the concept of assessing the quality and overall difficulty of alignments by comparing alternate alignments of the same sequences (7). Another approach that uses multiple, multiple sequence alignments is M-Coffee (8) a meta-alignment algorithm. Both approaches add value to the field but their shared requirement for computing not one but several alignments for each set of sequences somewhat limits their practical usage. Here, in particular, fast alignment methods could be the key to making these approaches more feasible.
Perhaps the most compelling reason for fast aligners is related to the persisting problem of selecting appropriate gap penalties (9–11). That is, since individual sequence families are under unique evolutionary constraints, default parameters giving the overall best results on benchmark sets are often inappropriate for individual cases. It is, therefore, required to assess alignment quality by hand or assisted by automatic methods, to tune alignment parameters and to re-run alignments. Such an iterative strategy clearly benefits from fast and accurate alignment methods.
Another issue especially when aligning sequences with low average pairwise sequence identity (APSI) is the inclusion of external information into the alignment process, e.g. secondary structure annotation or Pfam domains. In previous works that apply hidden Markov models (HMM) to the alignment problem (12,13) or aim to refine existing alignments (14), it has successfully been shown that alignment quality can gain from incorporating additional information into the alignment procedure.
Given the rapidly increasing number of sequences to be aligned, computational challenges are becoming a bottleneck. To find a good compromise between accuracy and computational efficiency, the original Kalign program (15) was optimized in many ways. Most notably, we replaced the Wu–Manber algorithm with the faster Muth–Manber string matching algorithm, and implemented the space-saving Myers and Miller algorithm (16) for dynamic programming. In addition we introduced the alignment of nucleotide sequences and an alignment mode that can incorporate data from extrinsic heterogeneous sources to increase alignment accuracy.
Kalign version 2 employs highly efficient algorithms in a combination that previously has not been applied to the multiple alignment problem. Each component was optimized to gain computational efficiency as well as high accuracy. Together, all the improvements resulted in a multiple sequence aligner that is highly suited to emerging large-scale alignment problems.
MATERIALS AND METHODS
Distance estimation
The key innovation in Kalign is the use of an approximate string matching algorithm to estimate pairwise sequence distances. The original Kalign algorithm employed the Wu–Manber algorithm (17) for this purpose, primarily because of its flexibility and computational aspects. In our previous study (15) we found that using patterns of length 3 and allowing a single error gave the best results. We here replace the flexible Wu–Manber algorithm with a far quicker, but inflexible algorithm by Muth and Manber (18). This algorithm is limited to find matches containing only single errors but can search a query with thousands of patterns simultaneously. The algorithm is described in detail in Ref. 18.
Just as in the original Kalign algorithm, when a match is reported between two sequences the positions of both are used to determine on which diagonal within a dot-plot representation they fall. Scores are assigned to each diagonal based on the number of matches occurring along the entire length. In the original Kalign algorithm exact and error-containing matches were treated separately. Because of the nature of the Muth–Manber algorithm, exact matches between a pattern and sub-strings of the query are reported multiple times so the two different types of matches do not have to be treated explicitly anymore.
In correspondence with the Fasta program (19,20) we choose to use patterns of length 6 for the alignment of nucleotide sequences.
Dynamic programming
It is widely recognized that the standard dynamic programming algorithm with affine gap penalties (21) is not appropriate for the alignment of long genomic sequences, due to its quadratic space complexity. The memory efficiency of protein alignment programs is often overlooked since it is assumed that the alignment of relatively short protein sequences will require little memory. However, many of the new algorithms published recently, especially the consistency based ones (22–24), require large amounts of memory. Even progressive methods such as ours start to use too much memory when aligning hundreds of sequences because profiles generated close to the root of the guide tree can become very long, especially when low gap penalties are used, and the quadratic space requirement becomes just as limiting as for long genomic sequences. Unfortunately, it is very hard to predict the memory usage in advance since conserved protein families will give alignments where the number of columns corresponds to the length of the longest member sequence while a divergent family may lead to much longer alignments.
To address this growing issue for protein sequences and facilitate the alignment of long nucleotide sequences we followed ClustalW's (25) example and replaced the dynamic programming approach outlined in our first paper by the space saving Myers and Miller algorithm (16). For two sequences of length n and m the Myers and Miller algorithm runs in O(nm) time and requires O(n) space.
Early on in the progressive strategy, particularly when single sequences are aligned, it is very common for alternate alignments to achieve the same score. Previously, Kalign arbitrarily picked one of these alignments. Kalign2 now takes advantage of the divide and conquer approach used in the Myers and Miller algorithm, and picks in the case of equally scoring alignments the one closer to the center of the dynamic programming matrix.
For optimization reasons we adopted a strategy outlined by Robert Edgar (26) whereby separate subroutines are used for the alignment of two sequences, a sequence to a profile and the alignment of two profiles.
Alignment parameters
In addition to the standard gap open and close penalties (which are equal for symmetry reasons) Kalign2 allows users to specify three additional parameters: a terminal gap penalty, a gap_inc parameter, and a bonus score to be added to each field of the substitution matrix. The first one is used to penalize N/C-terminal gaps in proteins or 5′/3′ gaps in nucleotide sequences. The second one can be used to increase gap open and extension penalties depending on the number of existing gaps, i.e. gaps will be penalized harder if there are no or only few gaps. The new gap costs are calculated according to the following formula:
where GAP_default is equivalent to the default penalties for opening gaps, extending gaps, or terminal gaps. The parameter count_res equals the number of residues at a certain position, count_seq denotes the actual number of sequences, and gap_inc can be set by the user to define the degree of gap increase. The partial increase of gap penalties was not observed to significantly increase alignment quality on benchmark sets in terms of alignment scores; however, this parameter might help to make large sequence alignments more compact and readable. The bonus score can be used to force Kalign2 to align distantly related sequences. Essentially, the higher this bonus score the more residues will be aligned. Kalign2 uses a default bonus score of 28.3 for nucleotide alignments and a bonus of 0.02 for protein alignments. These values were derived by cross validation as given below.
Often default gap penalties are adjusted by over training on a particular benchmark set in order to give optimal results. This constitutes a widely recognized problem when benchmarking alignment programs. Direct parameter optimization can, and should be, easily avoided by testing and training on different sets of alignments (6). However, even without this type of direct over-fitting it is difficult to avoid making choices during the development of an alignment program that favor certain benchmark sets—once a new feature is introduced, it is desirable to assess its potential benefits in terms of accuracy. Therefore, choices made during the development phase of an alignment program inadvertently lead to some degree of over-fitting. Unfortunately, it is almost impossible to guard against this type of over-fitting or quantify its overall effect on alignment accuracy.
To estimate alignment parameters, we split benchmark sets into non-overlapping subsets and performed standard cross-validation analysis. Within each set we optimized alignment parameters using an evolutionary algorithm. Final alignment parameters were derived by taking the average of the individual parameter settings for each subset.
Alignment accuracy
The performance of the alignment programs was evaluated using the sum of pairs score (SPS) and the structure conservation index (SCI). The SPS reflects the percentage of correctly aligned residues between a reference alignment and a test alignment, whereas the SCI score accounts for consensus secondary structure information. We used the tools CompalignP, SCIF, and BaliScore that are distributed with the BraliBase2.1 (27) and Balibase3.0 (28) packages to derive SPS and SCI scores.
Feature alignment
We have added support for the inclusion of extrinsic information into the alignment procedure. In particular, we wanted to make this inclusion as general and flexible as possible without incurring an excessive cost to the running time of Kalign.
Our strategy is as follows: each sequence is treated as a profile containing a sequence and an annotation string of arbitrary symbols. During the alignment, the sequence part of the profile is treated as before but the annotation strings are also aligned using a default separate annotation substitution matrix or user-specified scores for matching same or dissimilar features. The score of an alignment then becomes the linear combination of the default sequence-based alignment score (substitution costs minus gap penalties) plus the sum of feature substitution costs of the annotation alignment. The main difference is that the progressive alignment now starts with profiles at the leaves of the guide tree rather than just single sequences.
In our study we used default scores of 75 for matching residues with the same annotation and −5 for matching residues with dissimilar annotation. Other parameter settings were tested as well, but did not affect the results significantly.
A major benefit of this simple approach is that contradictory annotation can be supplied and resolved by the dynamic programming. No extra consistency check, such as the one needed to determine a set of consistent matches in Kalign1, is required. In addition, it is also possible to include a combination of annotations, such as secondary structure, Pfam domains and known active sites. Residues which share all these annotations will be more likely to be aligned than residues sharing only one or two types of annotation.
We tested the influence of secondary structure annotation on alignment accuracy for both RNA sequences and protein sequences. For proteins we used known existing annotations. Whereas for RNA we used minimum free energy structures predicted by the method RNAfold in the ViennaRNA package (29). Structures predicted by RNAfold are provided in the bracket notation. Each structure is represented as a string of length equal to that of the nucleotide sequence consisting of matching brackets (for basepairs) and dots (for unpaired bases).
For aligning sequences in combination with extrinsic information we used the Macsim XML file format (30). In addition to the single sequences each file included available feature annotations. For RNA sequences we simply attached for each sequence position the corresponding annotation, i.e. ‘(‘,’)’, or ‘.’.
Problems might occur when two sequences contain unequal numbers of the same annotation features, possibly due to repeats, the features in the feature-poor sequence can get stretched out to cover all feature in the feature-rich sequence. For example, if sequence A contains two alpha helices and sequence B only one, residues within that helix might be aligned to both helices in sequence A. However, this effect can be easily controlled by manually lowering the scores for matching similar features.
Minor practical improvements
We have added support for Clustal, Pileup, MSF, Stockholm, Uniprot, Swissport and Macsim alignment formats. To allow for easy integration of different data sources, Kalign2 can read sequences from multiple files.
In addition, Kalign2 can align alignments by turning them into profiles.
A relatively minor but much demanded improvement is the sorting of output sequences. Like most other alignment programs, Kalign2 can sort sequences according to the initial guide tree or according to similarity to an individual query sequence.
RESULTS
Running time
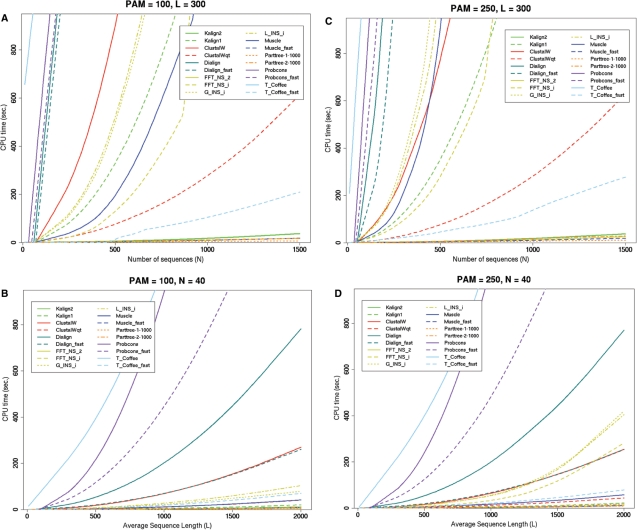
We tested the computational properties of Kalign2 by simulating alignments with varying average sequence length and number of sequences using ROSE (31). The analysis was conducted using the same parameters as used by Katoh et al. (6). Out of the alignment programs tested, Kalign2 was almost as fast as the speed oriented methods like the PartTree option of MAFFT (32) or the fast mode of Muscle (26), but significantly faster than the rest of the methods tested (Figure 1A–C). Just as the previous version of Kalign, the running time of the new version is unaffected by the evolutionary distance of the input sequences.
Figure 1.
Running time of several multiple alignment methods on four scenarios with simulated alignments of varying evolutionary distance (PAM = 100 and PAM = 250), increasing sequence length (L = 10–2000), and number (N = 10–1500). For each case one parameter was varied (x-axis) while two parameters were kept constant (plot heading). Kalign2 scales much better than most of the methods, especially with increasing number of sequences. All tests were carried out on an AMD64 3200+ processor with 2GB of RAM running Linux.
The comparison to the previous version indicates a modest improvement in running time when considering the sequence length and a dramatic reduction in running time when the number of input sequences is considered. The former indicates the efficiency of our implementation of the Myers and Miller algorithm, especially when considering that it should be approximately twice as slow, whereas the latter can be attributed primarily to the Muth and Manber algorithm. Kalign2 aligns 1500 sequences of length 300 in around 30 s (Figure 1A) or 6650 sequences in 1000 s (the upper limit of our test; data not shown).
Memory benchmark
We evaluated the memory requirement of several alignment programs using 11 alignments with increasing number of sequences generated by ROSE (Table 1) using a length of 300 and an evolutionary distance of 100. Among the programs tested, Kalign2 uses the least amount of memory. The improvement in memory requirement over the previous version of Kalign is dramatic. The reason is primarily due to the Myers and Miller algorithm, and also to an oversight in our initial implementation that caused too much memory to be allocated for the storage of the initial string matches.
Table 1.
Memory requirement in megabytes for several alignment programs as a function of the number of sequences (N)
| N | 10 | 50 | 100 | 150 | 200 | 250 | 300 | 350 | 400 | 450 | 500 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kalign2 | 6.6 | 7.1 | 7.4 | 7.7 | 7.8 | 7.9 | 8.3 | 8.5 | 8.5 | 8.8 | 9.3 |
| Kalign1 | 8.6 | 20.6 | 38.0 | 60.6 | 80.5 | 100.3 | 127.1 | 151.6 | 176.0 | 193.5 | 225.6 |
| ClustalW | 7.9 | 8.1 | 8.3 | 8.8 | 9.2 | 9.5 | 10.4 | 10.9 | 11.4 | 12.2 | 13.1 |
| ClustalWqt2 | 7.9 | 8.1 | 8.4 | 8.8 | 9.2 | 9.5 | 10.4 | 10.9 | 11.4 | 12.2 | 13.1 |
| Muscle | 17.6 | 25.1 | 34.3 | 43.0 | 52.7 | 60.2 | 70.4 | 79.8 | 89.2 | 98.3 | 108.2 |
| Muscle_fast | 17.3 | 24.8 | 33.6 | 42.8 | 52.3 | 59.8 | 69.7 | 80.0 | 89.0 | 97.7 | 107.2 |
| T_Coffee_fast | 17.5 | 25.1 | 34.3 | 43.0 | 52.7 | 60.2 | 70.4 | 79.8 | 89.2 | 98.2 | 108.2 |
| G_INS_i | 146.9 | 149.6 | 153.4 | 161.7 | 169.9 | 179.0 | 199.2 | 206.4 | 213.8 | 246.4 | 266.2 |
| L_INS_i | 146.9 | 149.5 | 153.2 | 161.0 | 168.6 | 177.8 | 195.7 | 203.8 | 211.8 | 241.4 | 260.8 |
| FFT_NS_2 | 140.1 | 140.4 | 141.2 | 141.7 | 142.8 | 142.7 | 144.5 | 144.9 | 145.3 | 145.7 | 146.9 |
| FFT_NS_i | 146.0 | 147.5 | 149.8 | 153.2 | 157.0 | 159.1 | 168.1 | 171.2 | 174.0 | 181.0 | 188.7 |
| Parttree-1-1000 | 140.2 | 140.5 | 141.5 | 142.0 | 143.0 | 143.4 | 144.8 | 144.8 | 145.3 | 145.8 | 147.4 |
| Parttree-2-1000 | 140.2 | 140.5 | 141.5 | 142.0 | 143.0 | 143.4 | 144.8 | 145.3 | 145.8 | 146.5 | 147.5 |
| Dialign | 6.7 | 13.1 | 30.5 | 59.6 | 99.7 | 146.0 | – | – | – | – | – |
| Dialign_fast | 8.7 | 13.0 | 30.8 | 60.5 | 101.4 | 147.0 | – | – | – | – | – |
| Probcons_fast | 12.2 | 21.4 | 58.1 | 120.6 | 207.6 | 315.6 | – | – | – | – | – |
| Probcons | 12.2 | 21.3 | 58.1 | 120.6 | 207.6 | – | – | – | – | – | – |
| T_Coffee | 20.9 | 129.1 | 464.5 | – | – | – | – | – | – | – | – |
No measurement could be obtained for some method test set combinations due to excessive time or memory requirements (dashes). Kalign2 requires the least amount of memory followed by ClustalW.
Since desktop computers are commonly equipped with 1 GB of memory or more it is questionable whether this improvement will have any impact when only aligning small numbers of sequences. However, low memory requirement can be advantageous when running Kalign2 on servers shared by many users or when using Kalign2 for a public web server.
Furthermore, most common methods in default mode are only applicable to a couple of hundreds of sequences, and start to allocate huge amounts of memory when applied to extensive datasets. In a more severe test we used the alignment methods MAFFT, Muscle, Probcons, T-Coffee and Kalign2 to align the more than 8000 sequences of the Rfam family Intron-gpII on a standard desktop computer with 2 GB of memory. Most default methods either ran out of memory or aborted after the start due to the large number of sequences. Only fast aligners like Kalign2, Mafft-FFT-NS-2/NS-i or T-Coffee and Muscle in the fast mode were applicable here. We then tried to align the more than 20000 sequences of the Rfam family Intron-gpI. Out of the methods tested only Kalign2 and the Mafft-Parttree variant were able to align the complete dataset. However, Mafft-Parttree was less accurate in our benchmarks. Here Kalign2 clearly benefits from its high speed and its memory efficiency.
Accuracy on protein alignments
The accuracy of Kalign on protein alignments was demonstrated in (15) and is therefore only briefly discussed here. To ensure that the high accuracy of our first version is maintained we evaluated Kalign2 on the Balibase3 (28) benchmark set. The gap penalties for protein alignments were derived via cross validation on Balibase3.0 applying a genetic algorithm approach.
Kalign2 achieved an average SPS score of 82.7% on Balibase which represents a 2.4% improvement over the previous version of Kalign. Compared with other programs Kalign2 is more accurate than other progressive methods, e.g. ClustalW at 75.4% or Muscle in the fast mode at 77.8%. It is about as accurate as some iterative methods, e.g. Muscle at 82.2%, but less accurate than consistency-based methods, e.g. G-INS-i option of Mafft 84.5% (progressive methods < iterative methods, Kalign2 < consistency-based methods; see Table 2 for the individual command line options used for each method). Our measured accuracies of the other methods correspond well to those reported by Katoh et al. (33).
Table 2.
Command-line arguments for each alignment method tested
| Method | Command |
|---|---|
| ClustalW | Clustalw |
| ClustalWqt2 | clustalw –quicktree |
| Dialign | Dialing |
| Dialign_fast | dialign –o |
| FFT_NS_2 | Mafft –retree 2 |
| FFT_NS_i | Mafft –maxiterate 1000 |
| G_INS_i | Mafft –globalair –maxiterate 1000 |
| Kalign2 | kalign2 |
| Kalign1 | kalign1 |
| L_INS_i | Mafft –localpair –maxiterate 1000 |
| Muscle | muscle |
| Muscle_fast | muscle -maxiters 1 -diags -sv -distance1 kbit20_3 |
| Parttree-1-1000 | mafft –retree 1 –parttree –partsize 1000 |
| Parttree-2-1000 | mafft –retree 2 –parttree –partsize 1000 |
| Probcons_fast | probcons -ir 0 |
| Probcons | Probcons |
| ProbconsRNA | probconsRNA |
| T_Coffee | t_coffee |
| T_Coffee_fast | t_coffee -special_mode quickaln |
Accuracy on nucleotide alignments
We used the Bralibase2.1 (27) test set to benchmark Kalign2 against other sequence-based alignment programs for nucleotide sequences. The updated version now contains alignments of 36 RNA families from the Rfam database. Totally, it comprises 18,990 alignments with a varying APSI between 20% and 95%. The alignments are grouped into sets of 2, 3, 5, 7, 10 and 15 sequences. Because we were interested in alignments representing a wide spectrum of test cases we decided to merge all test sets to assess alignment accuracy.
Gap penalties for nucleotide alignments were derived using 5-fold cross validation on a separate data set, i.e. data set 1 of the previous Bralibase benchmark set 2.0. For scoring matches and mismatches Kalign2 uses the HOXD substitution matrix (34).
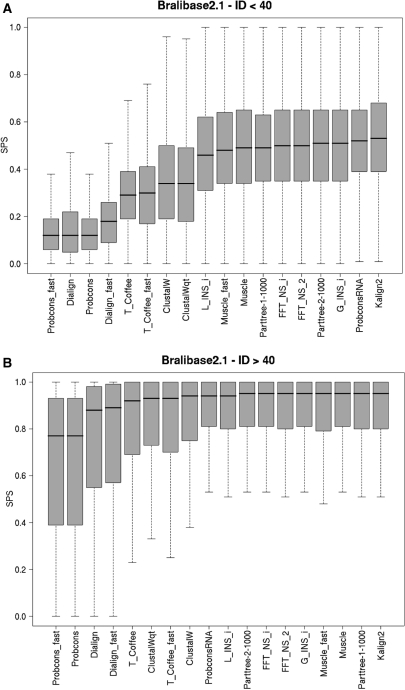
In our tests Kalign2 outperformed many other alignment programs (Figure 2A and 1A, Supplementary Data). In particular, for alignments with low APSI, Kalign2 performed significantly well, both in terms of SPS score as well as SCI score. From this we conclude that Kalign2's fuzzy string matching algorithm is as beneficial in terms of accuracy for nucleotide sequences as it is for protein sequences. With increasing APSI most of the tools tested performed equally well (Figure 2B and 1B, Supplementary Data). Besides Kalign2, especially the tools ProbConsRNA (22), Muscle, and the MAFFT variant G_INS_i performed well.
Figure 2.
Accuracy on RNA alignments using the SPS score. Boxplots for the accuracy measured using the Bralibase2.1 benchmark set. (A) Alignments with an average pairwise sequence identity (APSI) <40%. (B) Alignments with an APSI >40%. Kalign2 was the most accurate method, especially in regions with low APSI.
Accuracy of feature alignments
A new capability in Kalign2 is the use of extrinsic information such as structure or domain annotations to improve alignments. To demonstrate the benefit of Kalign2 feature alignments we discuss two examples of how to increase alignment accuracy with feature alignments.
We used RNAfold to predict secondary structures for all sequences in the Bralibase2.1 benchmark set. Supplying the predicted secondary structure as an additional input to Kalign2 increased the average accuracy from 51.7% to 55.31% for alignments with an APSI below 40%. Above 40% the average accuracy increased slightly from 87.5% to 88.1%.
Further we tested the new feature alignment function using Balibase 3 in combination with the existing secondary structure annotation. With the feature mode enabled Kalign2 obtained an average accuracy of 84.8%, an increase of 2.1 percentage points.
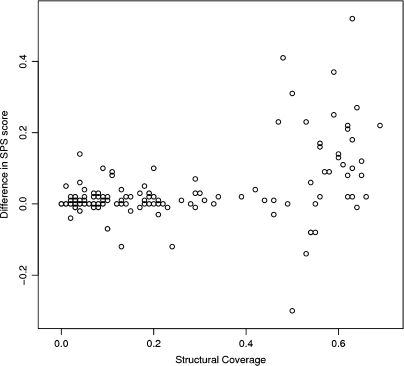
The coverage of structural annotation varies greatly for individual Balibase alignments. To determine whether an increased annotation coverage corresponds to an increase in accuracy we plotted the feature coverage of each alignment case against the difference in accuracy between the default and structure-enhanced Kalign2 mode (Figure 3). There is a noticeable trend supporting the assumption that more annotation results in an increase in accuracy.
Figure 3.
External feature alignment using protein secondary structure generally improves accuracy on the Balibase benchmark. An increase in the SPS score is seen mostly for cases with high structural coverage.
Annotation present in areas that are misaligned with default parameters is clearly more useful than annotation present in areas that are trivial to align. In the most extreme case—BB11025—the sequences are so divergent (average sequence identity 15%) that Kalign2 using sequence alone obtained an SPS score of only 11%, while the structure-enabled Kalign2 scored 62%.
Combining Kalign2 with additional annotations like predicted secondary structure for RNA constitutes an easy but powerful way to enhance alignment accuracy. Since the reference alignments in Balibase3 are derived from the structural annotation themselves, this test is circular and the reported accuracy cannot be used for comparison to other methods. Nevertheless, it underlines the usefulness of the simple method introduced here.
DISCUSSION
This article describes several key improvements to our alignment algorithm Kalign. We have improved the computational properties of Kalign, making the second version a versatile tool for the alignment of multiple biosequences. The combination of high speed and extremely low memory requirement means that Kalign2 can be applied to very big alignment problems, even on small computers. In large scale studies where thousands of alignments are required, Kalign2 can dramatically reduce the computation time while retaining high prediction accuracy.
The accuracy of Kalign2 on nucleotide alignments is notable and the accuracy on protein alignments is better than for comparable algorithms and not much less than the best. We do not believe that the excessive running times of more accurate programs are worth the 1–3% increases in average alignment accuracy reported recently. Another factor to be considered is that high average accuracies on benchmark sets do not necessarily translate into optimal performance for individual cases. Indeed, even the worst method according to a benchmark study can yield the best alignment in a few cases. In our experience the strategy that yields the best result in practice is to re-run an alignment program multiple times, adjusting its parameters along the way. The speed of Kalign2 makes it well suited for this approach.
The feature alignment extension presented here is intriguing. We have shown that combining Kalign2 with structural annotations can obviously increase the prediction accuracy, both for nucleotide sequences as well as for protein sequences. Apart from the obvious benefit of increased accuracy, the feature alignment can also be used to contrast and compare different alignment alternatives. For example, users may wish to compare a structurally motivated alignment to one that is based on Pfam annotation and decide which one is more suitable for their purposes.
Future direction
As far as the authors are concerned, the future of alignment programs lies in two main areas: the combination of heterogeneous data sources to improve accuracy, and taking advantage of alternate alignments of the same sequences, i.e. meta aligners. Kalign2 is ideally suited for both avenues: the newly introduced feature alignment is simple and expandable, and Kalign's speed is crucial for meta aligners.
We can imagine an iterative method which trawls through the search-space by generating several multiple alignments at each step, assessing their accuracy and then alters alignment parameters to generate a more accurate set of alignments in the next set. Kalign2 is particularly suited for such an iterative strategy due to its computational efficiency, which ultimately determines the practical usefulness for large-scale applications in this field.
FUNDING
Funding for open access charges: Swedish Graduate School for Functional Genomics and Bioinformatics.
Conflict of interest statement: None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
REFERENCES
- 1.Lecompte O, Thompson JD, Plewniak F, Thierry J, Poch O. Multiple alignment of complete sequences (MACS) in the post-genomic era. Gene. 2001;270:17–30. doi: 10.1016/s0378-1119(01)00461-9. [DOI] [PubMed] [Google Scholar]
- 2.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Notredame C. Recent progress in multiple sequence alignment: a survey. Pharmacogenomics. 2002;3:131–144. doi: 10.1517/14622416.3.1.131. [DOI] [PubMed] [Google Scholar]
- 6.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lassmann T, Sonnhammer ELL. Automatic assessment of alignment quality. Nucleic Acids Res. 2005;33:7120–7128. doi: 10.1093/nar/gki1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace IM, O'Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vingron M, Waterman MS. Sequence alignment and penalty choice. Review of concepts, case studies and implications. J. Mol. Biol. 1994;235:1–12. doi: 10.1016/s0022-2836(05)80006-3. [DOI] [PubMed] [Google Scholar]
- 10.Qian B, Goldstein RA. Distribution of Indel lengths. Proteins. 2001;45:102–104. doi: 10.1002/prot.1129. [DOI] [PubMed] [Google Scholar]
- 11.Qian B, Goldstein RA. Optimization of a new score function for the generation of accurate alignments. Proteins. 2002;48:605–610. doi: 10.1002/prot.10132. [DOI] [PubMed] [Google Scholar]
- 12.Do CB, Gross SS, Batzoglou S. CONTRAlign: discriminative training for protein sequence alignment. In: Apostolico A, et al., editors. RECOMB. Springer-Verlag Berlin, Heidelberg: LNBI 3909; 2006. pp. 160–174. [Google Scholar]
- 13.Karchin R, Cline M, Mandel-Gutfreund Y, Karplus K. Hidden Markov models that use predicted local structure for fold recognition: alphabets of backbone geometry. Proteins: Struct. Funct. Genet. 2003;51:504–514. doi: 10.1002/prot.10369. [DOI] [PubMed] [Google Scholar]
- 14.Chakrabarti S, Lanczycki CJ, Panchenko AR, Przytycka TM, Thiessen PA, Bryant SH. Refining multiple sequence alignments with conserved core regions. Nucleic Acids Res. 2006;34:2598–2606. doi: 10.1093/nar/gkl274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lassmann T, Sonnhammer ELL. Kalign—an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myers EW, Miller W. Optimal alignments in linear space. Comput. Appl. Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 17.Wu S, Manber U. Fast text searching: allowing errors. Commun. ACM. 1992;35:83–91. [Google Scholar]
- 18.Muth R, Manber U. Proceedings of the7th Annual Symposium on Combinatorial Pattern Matching. Vol. 1075. Berlin, Laguna Beach, CA: Springer; 1996. Approximate multiple string search; pp. 75–86. [Google Scholar]
- 19.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson WR. Rapid and sensitive sequence comparison with FASTP and FASTA. Meth. Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 21.Gotoh O. An improved algorithm for matching biological sequences. J. Mol. Biol. 1982;162:705–708. doi: 10.1016/0022-2836(82)90398-9. [DOI] [PubMed] [Google Scholar]
- 22.Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 24.Morgenstern B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 25.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–248. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 26.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilm A, Mainz I, Steger G. An enhanced RNA alignment benchmark for sequence alignment programs. Algorithms Mol. Biol. 2006;1:19. doi: 10.1186/1748-7188-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson JD, Koehl P, Ripp R, Poch O. BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 29.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- 30.Thompson JD, Muller A, Waterhouse A, Procter J, Barton GJ, Plewniak F, Poch O. MACSIMS: multiple alignment of complete sequences information management system. BMC Bioinformatics. 2006;7:318. doi: 10.1186/1471-2105-7-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoye J, Evers D, Meyer F. Rose: generating sequence families. Bioinformatics. 1998;14:157–163. doi: 10.1093/bioinformatics/14.2.157. [DOI] [PubMed] [Google Scholar]
- 32.Katoh K, Toh H. PartTree: an algorithm to build an approximate tree from a large number of unaligned sequences. Bioinformatics. 2007;23:372–374. doi: 10.1093/bioinformatics/btl592. [DOI] [PubMed] [Google Scholar]
- 33.Katoh K, Kuma K, Miyata T, Toh H. Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform Ser Workshop Genome Inform. 2005;16:22–33. [PubMed] [Google Scholar]
- 34.Chiaromonte F, Yap VB, Miller W. Scoring pairwise genomic sequence alignments. Pac. Symp. Biocomput. 2002;7:115–126. doi: 10.1142/9789812799623_0012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.