Abstract
Type-I DNA restriction–modification (R/M) systems are important agents in limiting the transmission of mobile genetic elements responsible for spreading bacterial resistance to antibiotics. EcoKI, a Type I R/M enzyme from Escherichia coli, acts by methylation- and sequence-specific recognition, leading to either methylation of DNA or translocation and cutting at a random site, often hundreds of base pairs away. Consisting of one specificity subunit, two modification subunits, and two DNA translocase/endonuclease subunits, EcoKI is inhibited by the T7 phage antirestriction protein ocr, a DNA mimic. We present a 3D density map generated by negative-stain electron microscopy and single particle analysis of the central core of the restriction complex, the M.EcoKI M2S1 methyltransferase, bound to ocr. We also present complete atomic models of M.EcoKI in complex with ocr and its cognate DNA giving a clear picture of the overall clamp-like operation of the enzyme. The model is consistent with a large body of experimental data on EcoKI published over 40 years.
INTRODUCTION
The EcoKI Type I DNA restriction–modification (R/M) enzyme was the first R/M enzyme to be discovered (1) and purified (2). It was determined that EcoKI, like all Type I R/M enzymes (3–8), is a complex oligomer formed from a core methyltransferase (MTase, M.EcoKI) comprised one HsdS DNA Specificity subunit and two HsdM DNA Modification subunits, M2S1, with a total molecular weight of ∼169 kDa. This is complexed with two additional HsdR DNA Restriction subunits, giving a complex R2M2S1 with a total molecular weight of ∼440 kDa that is capable of functioning as both MTase and restriction endonuclease (EcoKI or R.EcoKI) (9,10). Other Type I R/M enzymes display more variable subunit structures with some subunits weakly bound or capable of forming larger aggregates (11–13). Nevertheless, the core functions are performed by the M2S1 MTase and the R2M2S1 bifunctional MTase/endonuclease. Despite much subsequent work, it is only recently that crystal structures of HsdS from Methanococcus jannaschi and Mycoplasma genitalium (14,15) and HsdM from Escherichia coli and Bacteroides thetaiotaomicron (pdb code: 2ar0, New York Structural GenomiX Research Consortium, DOI 10.2210/pdb2ar0/pdb and pdb code: 2okc, Joint Center for Structural Genomics, DOI 10.2210/pdb2okc/pdb, respectively) have become available. The overall structure is not known for either the MTase or the complete R/M complex although several models have been proposed (16–20).
The absence of these structures has hindered study of Type I R/M enzymes but their enduring appeal as research targets (21) is due to their extraordinary complexity of operation. This comprises recognition of a bipartite, asymmetric DNA specificity sequence (e.g. EcoKI recognizes AAC[N6]GTGC), the recognition of methylation of the adenines at the bold underlined positions, and switching between an MTase activity on newly replicated, hemimethylated host DNA and an endonuclease activity on unmethylated foreign DNA (3). The latter reaction requires massive ATP hydrolysis to drive translocation of up to 50 kb of DNA (22,23) at rates of up to 1 kb per second (24–29). The DNA is then cleaved at a random sequence remote from the original specificity sequence, with the generation of variable length single-strand overhangs (28,29). DNA-induced dimerization of EcoKI prior to translocation has been observed using atomic force microscopy (30,31). These varied activities have functional analogies with the operation of other complex DNA-manipulating machines (32,33). Of further note is the presence of two target recognition domains (TRDs) in HsdS that can be easily exchanged for other TRDs to generate novel DNA sequence specificities. The Type I R/M systems can be divided into, at present, five related families, IA to IE, defined by genetic complementation, antibody cross reactivity, DNA hybridization and sequence comparisons (34,35).
Of great topical interest due to the spread of drug resistance by horizontal gene transfer in bacteria (36,37), is the susceptibility of the R/M enzymes to antirestriction measures encoded by a variety of mobile genetic elements such as phage, conjugative plasmids and transposons (38–43). These antirestriction measures include the production of proteins that structurally mimic DNA (44). DNA mimics block the binding sites within R/M enzymes and inactivate them (45–47). The dimeric protein ocr from T7 phage matches the shape and surface potential characteristics of double-stranded DNA bent to the degree necessary for recognition by EcoKI.
We now present a model of M.EcoKI bound to ocr at ∼18 Å resolution as determined by negative-stain electron microscopy (EM). Combining the EM model with new atomic models generated from the known crystallographic structures of the subunits allows a detailed structure of the M.EcoKI enzyme to be constructed. This structure is consistent with and rationalizes many experimental results obtained with M.EcoKI and other Type I MTases.
MATERIALS AND METHODS
Protein samples
M.EcoKI and the ocr protein were prepared as previously described (9,10,46). A complex of M.EcoKI and ocr was prepared by mixing the two proteins together at a molar ratio of 1:1.5 (29 μM M2S1 and 44 μM dimeric ocr) in 20 mM Tris, 100 mM NaCl, pH 7 buffer and incubating at room temperature for 5 min. The excess of ocr was not visible in the EM due to its small size.
EM
Grids were made by placing M.EcoKI and ocr complexes diluted 400 times onto UV-treated (48) continuous carbon-coated copper mesh grids, then stained with 1% uranyl acetate solution. Grids were viewed in a Jeol 1200EX electron microscope fitted with a LaB6 electron source operating at 80 kV in low dose mode. Negatives (Kodak SO63) were taken at either 40 000× or 50 000× magnification with defocus ranging from 250 to 850 nm, and digitized at 6.25 µm step size using a Coolscan8000 (Nikon). The defocus and astigmatism were determined with CTFIND3 (49), then 17807 particles were picked from selected micrographs using Boxer (EMAN) (50) in autobox mode. Particles were band-pass filtered using Imagic (ImageScience, Germany), with low frequencies below 1/150 Å and those above the first zero of the contrast transfer function (CTF) being removed. After interpolating the 40 000× and 50 000× magnified particles to a common scale (3.125 Å/pixel), initial class average images without reference bias were generated in Imagic (51) by multi-variate statistical analysis and classification. Six independent starting models consisting of 16 randomly placed Gaussian spheres were made in EMAN and each used in nine rounds of 3D refinement with the dataset. Two of the randomly generated starting models converged on a map similar to others made with some degree of a priori knowledge of the structure using the angular reconstitution method (52). These two 3D maps also showed approximate 2-fold symmetry. One was taken for further refinements in EMAN with C2 point group symmetry imposed. At several steps during refinement, classes that had the best match to the reprojections were selected and new 3D maps made. The final map has a nominal resolution of 18 Å at 0.5 Fourier shell cross-correlation using the odd–even test, and was 3D Fourier filtered at 12 Å. Maps were visualized using UCSF Chimera (53).
Model building
Full details of the model building are described in the Supplementary Data and so are described only briefly here. The model of M.EcoKI bound to DNA or to ocr was built using the following crystal structures: HsdS(MjaXIP) from M. jannaschi (pdb code: 1yf2) (15), HsdM (EcoKI) (pdb code: 2ar0, New York Structural GenomiX Research Consortium, DOI 10.2210/pdb2ar0/pdb) and HsdM from B. thetaiotaomicron VPI-5482 (pdb code: 2okc, Joint Center for Structural Genomics, DOI 10.2210/pdb2okc/pdb), the structure of a Type II MTase bound to its cognate DNA duplex, M.TaqI–DNA (pdb code: 1g38) (54), and the ocr structure (pdb code: 1s7z) (44). The M.TaqI–DNA structure was used for modelling of the HsdS–HsdM orientation. HsdS was modelled on the 1yf2 template by homology modelling and the short C-terminal regions missing from the template were modelled de novo using ROSETTA (55). The crystal structure of the EcoKI HsdM has been solved, but we have remodelled its domain–domain contacts based on the crystal structure of a related HsdM from B. thetaiotaomicron VPI-5482. The missing C-terminal regions of EcoKI HsdM were modelled de novo using ROSETTA and fitted into the EM map for M.EcoKI–ocr described below.
To build the M.EcoKI–DNA model, DNA was added to the HsdS model based on the orientation of DNA in 1g38. HsdM were docked together using HADDOCK (56) to form a dimer via their N-terminal domains. Then, the HsdS–DNA model was combined with the HsdM dimer model so as to satisfy the orientation between the TRD and the catalytic domains of M.TaqI. To find the best fits, normal mode analysis was performed to generate a range of conformations of HsdS and HsdM.
The M.EcoKI–ocr model was constructed by multi-body flexible docking using HADDOCK (56). During the docking procedure, residues of HsdS and HsdM that form contacts with the DNA in the model of M.EcoKI–DNA were restrained to make contact with ocr, based on the assumption that ocr binds to HsdS and HsdM in a similar manner as DNA (44). The alternative models of M.EcoKI–ocr were fitted to the EM map as single rigid bodies using the COLORES module from the SITUS package (57) and the best scoring model was selected.
RESULTS
EM results
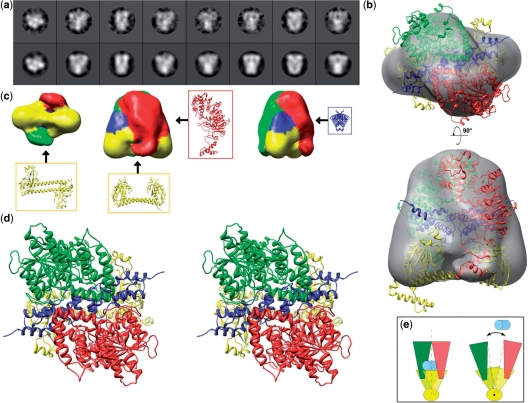
Addition of stoichiometric quantities of the ocr inhibitor was found to stabilize the otherwise labile M.EcoKI complex. Individual molecules of M.EcoKI–ocr complexes embedded in heavy metal stain were imaged by EM and analysed by single particle methods. Particles appeared to be homogeneously sized and were well distributed. Class averages showed a wide variety of views and exhibited considerable detail (Figure 1a) that allowed a unique 3D reconstruction of the complex to a nominal resolution of 18 Å (Figure 1b and c). The atomic model described below has been placed within the EM reconstruction in Figure 1b and has been used to colour the reconstruction surface in Figure 1c. Although at the atomic level, the complex is asymmetric, 2-fold symmetry was imposed since HsdS is known to have strong pseudo-symmetry (14,15,17). The EM map longest dimension was ∼120 Å and 100 Å along the base, and the overall shape was an excellent match to the complex modelled in silico based on biochemical and X-ray data, as described below. As described below and in the Supplementary Data, when one takes the size, shape and symmetry constraints of the HsdS, HsdM and ocr crystal structures and constraints from biochemical data, only one arrangement of these structures within the EM reconstruction was possible. Given this unique solution, density attributable to the two-helix coiled-coil spacer of HsdS could be clearly identified, as well as regions attributable to the two HsdM. In addition to contacting the TRDs of HsdS, the two HsdM wrap around ocr and make contact with each other. An extension projects from the bottom of each HsdM to either side of the HsdS coiled-coil spacer. This density returned in subsequent 3D refinements if masked out of the EM map, supporting the presence of these regions. These extensions could be attributed to the C-terminal regions of HsdM that are not present in the X-ray coordinates, as described below. Density in the centre of the map suggested the presence of the bound inhibitor protein ocr as it is known that ocr is fully enfolded by HsdS and HsdM (47). Sufficient detail was not present to show its exact orientation but its general position could be inferred. Features in the 3D map such as the straight coiled-coil spacer, which was ∼16 Å wide, could also be seen in many class average images (Figure 1a). That features of this size were visible, just below the limit of resolution as measured by FSC, attests to the quality of the map and suggests that the 18 Å estimate of resolution is conservative.
Figure 1.
EM data, 3D reconstructions and model for the M.EcoKI–ocr complex. (a) Eight selected EM class average images (top row) with their corresponding reprojections of the EM map (second row), showing a range of views. (b) Two orthogonal semi-transparent surface representations of the EM 3D reconstruction each with a view of the modelled coordinates fitted as a rigid body (green and red—HsdM; blue—ocr dimer; yellow—HsdS). (c) Three surface views of the M.EcoKI–ocr EM 3D reconstruction coloured according to proximity to the fitted coordinates, with the positions of individual protein chains indicated with arrows. (d) A stereo view of the M.EcoKI–ocr atomic model. (e) Schematic diagram of proposed mechanism of clamping and release of DNA substrate (light blue) facilitated by twisting of the coiled-coil (yellow, viewed end-on).
Model of the M.EcoKI–ocr complex
The general shape of the model of M.EcoKI in complex with ocr dimer is shown in Figure 1d. This atomic model was constructed independently of the EM imaging based upon crystal structures of subunits and biochemical data. The MTase forms a clamp-like structure encircling the ocr dimer. The coiled-coil region of HsdS contains a considerable kink to allow the ocr molecule to fit optimally within this clamp. Each TRD of HsdS contacts one monomer of the ocr molecule and one HsdM. The exact location of the C-terminal helix of the HsdS subunit is speculative but has little effect on the overall outline shape of the molecule. The C-terminal regions of each HsdM, aa ∼470–529, project down to contact the coiled-coil region of HsdS. The location of the C-terminal regions of each HsdM is also speculative but the postulated location does bring conserved regions of HsdM and HsdS into close contact as discussed later and does fit with the EM model. This is important as the other region of contact between HsdM and HsdS uses the TRD regions of HsdS; these are highly variable in sequence in Type I R/M enzymes and not expected to provide a good HsdM–HsdS interface. The interface between the two HsdM is formed by their N-terminal domains, aa 1–153. The clamp-like shape of the MTase model suggests that it must open up along the interface between the two HsdM to allow access for ocr or DNA (Figure 1e). The interfaces between the subunits in the MTase–ocr complex are less extensive than in the MTase–DNA complex described below and the whole MTase appears to be strained to accommodate the ocr molecule that is slightly larger than a DNA duplex. The model structure with ocr bound may represent a ‘partially open clamp’ state, while the model with DNA bound would be a ‘fully closed clamp’ state. Opening of the clamp to allow ocr (or DNA) binding may be accomplished by simply flexing the coiled-coil region of HsdS. Normal mode calculations indicate that such motions should occur with sufficient amplitude to allow binding of ocr or DNA (data not shown). Having constructed the atomic M.EcoKI–ocr model, it is obvious that this can be simply docked into the EM model as shown (Figure 1b). A comparison of the fit of the EcoKI MTase–ocr complex model to the EM map with the fits obtained for models in which subunit structures are randomly docked in to the EM map indicate that our data driven model fits the EM map significantly better than random models (see Supplementary Data).
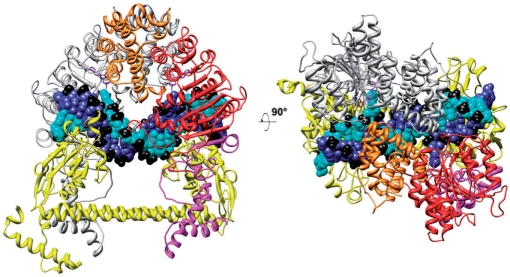
Model of the M.EcoKI–DNA complex
The model of M.EcoKI bound to DNA is shown in Figure 2. The two major differences between this and the partial models of Type I R/M enzymes published previously (16–19) are that the locations of the N-terminal and C-terminal regions of the HsdM are now defined within the overall structure. Most importantly, the N-terminal domains of HsdM can be seen to form a bridge over the DNA duplex, thus forming a clamp-like structure encircling the DNA, which is a key feature that was missing in earlier models. The location of the C-terminal regions of each HsdM with respect to HsdS is slightly different from that in M.EcoKI–ocr model, since the relative orientation of the subunits is altered. The coiled-coil region linking the two TRDs is straighter and the subunit interfaces within the MTase are more extensive in area than in the M.EcoKI–ocr model as the DNA duplex appears to fit better within the MTase allowing the clamp to close fully. The DNA bound to the TRDs contains a sharp bend in the non-specific spacer part of the target sequence and the bend angle of ∼45° matches that observed experimentally (44,58).
Figure 2.
Model of the EcoKI MTase in complex with substrate DNA and AdoMet cofactors. HsdS is shown in yellow, the HsdM at the rear is shown in grey, DNA strands are shown in blue and cyan with phosphates in black, the two AdoMet molecules in purple. The HsdM in the foreground shows the N-terminal domain (aa 1–153) in orange, the catalytic domain (aa 154–469) in red and the C-terminal domain (aa 470–529) in magenta.
DISCUSSION
The proposed in silico models, based upon crystallographic data, fit extremely well with the EM reconstruction and are attractive atomic resolution solutions to the structure of the MTase core of a Type I R/M enzyme. It is important to assess whether the models can accommodate the large body of experimental information concerning M.EcoKI. As most of the published work concerns interaction between EcoKI and DNA, we focus the following discussion primarily on the M.EcoKI–DNA model rather than the M.EcoKI–ocr model. Each section of the discussion describes how experimental data can be accommodated by the EM structure and the derived structural model and indicates new regions of interaction, which could be investigated in the future. Lastly, we compare the structural model of M.EcoKI with those recently proposed for the type IC enzyme, EcoR124I (19,20).
Analysis of the M.EcoKI–DNA model
Interface between the two HsdM via their N-terminal domains
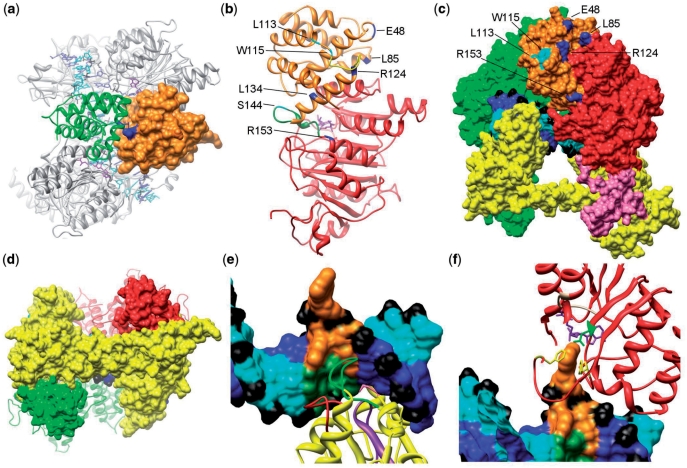
Our model fits each HsdM into the EM structure in such a way as to bring their N-terminal domains into contact with each other (Figure 3a). They effectively form a bridge over the DNA and this bridge must swing open to allow the DNA to enter and be recognized by HsdS. It is known that the ocr protein, which acts as a DNA mimic, is encircled by M.EcoKI (47) supporting the positioning of the N-terminal domains to complete the clamp encircling the DNA. The main interaction between these two domains is via helix G64QEQLQFYRKMLVHL78 contacting, in an antiparallel manner, the equivalent helix within the other HsdM. A second contact region is between a loop H91NVSTTIT98 with the equivalent loop in the other subunit. The helical axes and loops in this interface run approximately parallel to the axis of the coiled-coil in HsdS.
Figure 3.
(a) A top view of the M.EcoKI–DNA model highlighting the interface between the N-terminal domains of HsdM subunits. One N-terminal domain is shown as an orange surface and the second as a green ribbon. The R72Q mutation giving an r+m* phenotype is shown in blue and lies on the HsdM interface. (b) The m* mutations in the HsdM subunit. The N-terminal domain of HsdM is coloured orange and the central methyltransferase catalytic domain in red (the C-terminal domain is omitted). Residues where mutations cause the r−m* and r+m* phenotypes are shown coloured cyan and blue, respectively. Two flexible loops, disordered in the crystal structure, are shown in green and yellow. Both are associated with several m* residues. The cofactor AdoMet, in purple, can be seen in the catalytic domain. (c) The m* mutations in N-terminal domain of HsdM subunit presented in the M.EcoKI–DNA model. The HsdS subunit is yellow, the rear HsdM subunit as a green surface. The domains of the HsdM in the foreground are coloured as in Figure 2. The labelled residues in this HsdM subunit that cause r−m* and r+m* phenotypes are coloured cyan and blue, respectively. DNA is coloured blue and cyan with phosphates in black. (d) The contact between coiled-coil region of HsdS subunit and the C-terminal domain of HsdM subunit in M.EcoKI–DNA model. HsdS subunit surface is shown in yellow with the coiled-coil running horizontally across the image, HsdM subunits are coloured red and green with their C-terminal domains shown as a surface and the remainder as a ribbon. The close contact between the ends of the coiled-coil in HsdS with the C-terminal domains of HsdM can be seen. (e) Recognition of DNA by the N-terminal TRD of HsdS. Non-specific DNA is coloured blue and cyan with phosphates in black. The sequence AAC with the flipped out second A is shown in orange and the complementary sequence in dark green. The HsdS subunit is shown as a yellow ribbon with the previously identified DNA-contacting loops aa 83–91 in red and aa 96–118 in purple. These loops contact the complementary strand. The previously identified contact by Y27 (pink) can just be seen pointing into the minor groove at the rear of the image. The newly identified loop S139AGANINNIK148 is in green and primarily contacts the AAC sequence. (f) Contacts between the catalytic site of HsdM and DNA. DNA is shown as in (e) with the flipped out base sandwiched between HsdM F269 in the ‘NPPF’ motif IV and F345, both shown in yellow. The AdoMet cofactor is shown in purple and the HsdM motif I (beige) containing G177 is contacting the AdoMet. The rest of HsdM is in red. Two phenylalanine residues (F269 and F345, yellow) are available to form stacking interactions with the flipped out DNA base, and N266 (green) is within range for hydrogen bonding.
The N-terminal domain of each HsdM contains the m* mutations (Figure 3a–c) (59), so called because they change the strong preference of M.EcoKI for methylating hemimethylated DNA target sequences (9) to an ability to additionally methylate unmodified targets. It is noteworthy that the disruption caused by the m* mutations does not affect the ability of the enzyme to methylate DNA, only its ability to recognize the methylation state of the target sequence. A single m* mutation, R72Q, lies within the predicted helix G64QEQLQFYRKMLVHL78 and contacts the equivalent helix in the other HsdM (Figure 3a). This mutation has an r+m* phenotype suggesting a small deformation of the interface affecting communication of the methylation status of the two adenines in the target sequence and closure of the proposed clamp around the DNA but without affecting restriction by a complete EcoKI.
The mutations Q48K, L85Q, R124L, R124C, L134V, R153H and R153S also give rise to an r+m* phenotype. These locations in the model lie on the interface between the N-terminal m* domain (aa 1–153) and the middle catalytic domain (aa 154–469) of the same HsdM. It is possible to envisage how these non-conservative changes will disrupt this interface resulting in the observed r+m* phenotype, perhaps by interfering with closure of the clamp.
The remaining mutations, L113R, L113P, L113Q, W115R and S144Y all give an r–m* phenotype. L113, W115 and S144 lie on the surface of the modelled structure and do not contact any other part of the MTase (Figure 3b and c). It is possible that they induce a minor folding problem for the N-terminal domain affecting its interaction with the other HsdM although the mutant enzymes can be purified as normal (our unpublished results). The observation that restriction activity is lost because of these mutations may further suggest that this small region interacts with the HsdR subunit in the complete EcoKI.
Interactions between HsdM and HsdS
The EM density at the bottom of HsdM has a protrusion that passes close to the HsdS coiled-coil and we believe this to be the C-terminal region of HsdM comprising residues ∼470–529. This region is disordered in the two HsdM crystal structures available suggesting that it is flexible in the absence of HsdS. The EM results support the rigidity of the extensions in the complex, since flexible areas are invariably blurred out by the averaging inherent in the single particle methodology. However, the limited resolution of the EM map, and lack of X-ray data, only allows the computational atomic model to be positioned approximately. Sequence analysis indicates that the C-terminal region of HsdM has a propensity to form α-helices and the model suggests that these reach out and contact the coiled-coil in HsdS, as shown in Figure 3d. These contacts would play a central role in the assembly of the MTase providing an interface between known regions of high sequence conservation in both HsdS and HsdM of the Type IA family (5,8). This high degree of conservation in the interface between HsdS and HsdM would be essential to facilitate the exchange of subunits of different Type IA family systems by complementation. Experimental support for this aspect of the EM and computational model comes from proteolysis experiments. It was found that ∼43 residues from the C-terminal region of HsdM could be easily removed, leaving a stable, 55-kDa folded protein (60,61). However, this truncated HsdM was unable to bind to M1S1 complexes indicating that a subunit interface had been removed (60).
In the model, a second HsdS–HsdM interface occurs via a helix-loop structure, N133KISSLSAGANIA144, within the first TRD of HsdS and two loops, aa 430–433 and aa 464–470, on the edge of the catalytic domain within HsdM (not shown in Figure 3). Contacting the same region of the second HsdM, one can identify an equivalent helix and loop in the second TRD of HsdS as A346MMNCVKTTSGQK358. It is known that the TRDs within a family of Type I R/M systems can be exchanged easily to generate new sequence specificities (5,8). However, the TRD amino acid sequences are very variable and would not be expected to offer enough conservation to allow the formation of a particularly stable interface with HsdM. Hence, these regions must be of secondary importance to the regions discussed in the previous paragraph.
HsdS to DNA contacts
The N-terminal part of HsdS forms the first TRD, which recognizes the sequence 5′-AAC-3′. Many mutations in this TRD of HsdS from EcoKI have been catalogued (62,63) but the majority of these were silent. Early modelling studies (64) suggested that these were not on the protein–DNA interface. However, two loops were identified in this analysis, aa 83–91 and aa 96–118, as contacting the DNA and this is corroborated in the current model. Within the second of these loops, mutations of aa 91, 95, 105, 107 and 108 led to a loss of activity although the protein was folded and could bind DNA (62,63). In our model, these appear to contact the complementary strand to the 5′-AAC-3′ sequence (Figure 3e). Chen et al. (65) identified a further amino acid, Y27, by cross-linking experiments in contact with the DNA target. In our map, Y27 is within 4 Å of the minor groove of DNA (not visible in Figure 3e as it is behind the DNA).
In the first TRD of HsdS, our model has an additional loop to those identified previously, S139AGANINNIK148 (Figure 3e). This loop is closest to the major groove DNA bases forming the target sequence 5′-AAC-3′. Mutations S139P, G141V and G141A affected activity but other more conservative substitutions did not alter activity (62,63). N145 is the residue in our model that appears to be able to contact the unpaired thymine base (the partner of the flipped out adenine). In the second TRD, the equivalent loop is T354SGQKGISGK363, and Q357 is able to contact the unpaired thymine in the complementary strand for the second part of the target sequence, 5′-GATC-3′.
HsdM to DNA contacts
Willcock et al. (66) substituted amino acids within the well-characterized, conserved MTase motifs within the catalytic domain of HsdM. The substitution in the AdoMet binding motif I, G177D, affected cofactor binding and the model shows motif I in close contact with the AdoMet (Figure 3f). Changes at locations N266 or F269 in the catalytic motif IV led to a loss of activity. In the model of M.EcoKI with DNA bound, these locations can be seen adjacent to the flipped-out adenine (Figure 3f). In addition, F345 is close to the flipped-out adenine. A similar sandwich of a flipped-out adenine between two aromatic side chains was observed in the crystal structure of the M.TaqI MTase (67).
Analysis of the M.EcoKI–ocr model
This atomic model (Figure 1) is more open than the model with DNA (Figure 2) as there are fewer contacts between the three subunits of M.EcoKI. In particular, the interface between the two HsdM is formed only by helix 64–78, and the C-terminal helices of HsdM, 487–507, appear to have minimal contact with the coiled-coil region of HsdS. It appears that ocr is slightly too large to be as easily accommodated as the DNA molecule. As the subunit interfaces are smaller, one might say that the MTase is almost ‘bursting at its seams’ as it attempts to enwrap the ocr protein (47). This suggests that a clamp-like open and shut mechanism is used by M.EcoKI to bind DNA or ocr.
Mechanism for binding DNA or ocr
The EM structure makes it clear that M.EcoKI can encircle DNA and, to a lesser extent, ocr. Therefore, the structure must open up to allow the DNA (or the ocr protein) access to the TRDs on HsdS and the catalytic site on HsdM. Simple normal mode calculations were performed on HsdS to examine the potential role of the coiled-coil region in allowing such an opening (data not shown). These calculations showed that the lowest mode was a flexing of the coiled-coil moving the TRDs closer together and then further apart. The next lowest mode was a twisting of the coiled-coil around its axis. Noting that the interface between the two HsdM runs approximately parallel to the axis of the coiled-coil in HsdS, then these motions would move the N-terminal domains of the HsdM apart and allow access to circular DNA. A twist of the coiled-coil region of ∼12–14° in our model would produce a gap between HsdM N-terminal domains sufficient to allow passage of DNA or ocr. Obviously, the motions of the complete M.EcoKI will be considerably more complex but this analysis is strongly suggestive of a flexible clamp-like enzyme.
Comparison to the Type IC enzyme EcoR124I
Kneale (17) originally recognized the 2-fold symmetry imposed upon Type I enzymes via the structure of the S subunit. This symmetry defines the location of the M subunits and was exploited by Obarska et al. (19) to produce a rather opened-out model of the MTase where the N-terminal domains of each HsdM subunit were speculatively placed some distance from both the DNA and any contact with the other HsdM subunit. Although based upon an erroneous structure of an HsdM subunit (pdb 2ar0 is incomplete and partially incorrectly assigned, see Supplementary Data), the model could, if correct, explain the large compaction of M.EcoR124I observed by small angle neutron scattering when DNA is added to the enzyme (67). This compaction was suggested to be the movement of the HsdM subunits swinging in towards the DNA and closing like a clamp to trap the DNA rather like the closed state we have observed in our structure (Figure 1). Thus, it is probable that the structure of Type I MTases fluctuates between an open form as approximately modelled by Obarska et al. (19) and the closed form observed here and by Taylor et al. (67). Normal mode analyses of our model make it clear that this opening and closing motion, which must open a gap at least 2 nm wide between HsdM subunits to allow double-stranded DNA to pass through, is facilitated by twisting and bending of the coiled-coil region in the HsdS subunit.
More recently, small-angle neutron scattering data and computer modelling of the HsdR subunit of EcoR124I have been presented (20). The HsdR subunit appears to be a disc-shaped protein but a unique atomic model could not be constructed. Presumably, one of these subunits could be placed on either side of the MTase structure so that the ends of the DNA helix would contact the helicase domains in the HsdR subunit. Until a unique structure of the HsdR subunit is presented and more data are acquired on its contacts to the MTase, further speculation on the complete structure of the Type I R/M enzymes seems premature. However, we note that we are currently making progress towards this goal.
CONCLUSION
The new EM structure and computational model of M.EcoKI rationalize, for the first time, a large body of experimental data obtained using many different methods over many years. A mechanistic explanation of the Type I MTase enzymes is suggested by the model, which clearly indicates locations for further analyses such as the HsdM–HsdS and HsdM–HsdM interfaces. When combined with data on M.EcoR124I (67), the model also suggests that a dynamic opening and closing of the protein, driven by a flexing and twisting of the conserved coiled-coil region within HsdS, is required to open up the HsdM–HsdM interface to allow either DNA binding or attack of the ocr antirestriction protein, a protein that ‘disguises itself’ as DNA. The model also provides a basis for completing or constructing models of other Type I MTases from other families such as the Type IC enzyme M.EcoR124I (19) and the Type IB enzyme M.EcoAI.
Data depositions
Atomic coordinates from M.EcoKI–ocr and M.EcoKI–DNA complexes are available from the authors or ftp://ftp.genesilico.pl/iamb/models/MTases/EcoKI/. The EM map is deposited with the EMDB (www.ebi.ac.uk/msd) with code 1543.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
We gratefully acknowledge support from the Biotechnology and Biological Sciences Research Council (BB/D001870/1 to D.T.F.D. and J.T.), the Wellcome Trust (GR080463MA to D.T.F.D.) and the Royal Society of Edinburgh (International Exchange Programme to A.O.-K. and J.M.B.). A.O.-K. and J.M.B. were also supported by the 6FP grant from the European Union (MRTN-CT-2005-019566) and supplementary funds from the Polish Ministry of Science. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
This work was initiated at the Isaac Newton Institute for Mathematical Sciences Workshop on ‘Statistical Mechanics of Molecular and Cellular Biological Systems’, January to July 2004. We thank Professor Noreen Murray and numerous colleagues in her laboratory for countless discussions on the structure of EcoKI.
REFERENCES
- 1.Bertani G, Weigle JJ. Host controlled variation in bacterial viruses. J. Bacteriol. 1953;65:113–121. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meselson M, Yuan R. DNA restriction enzyme from E. coli. Nature. 1968;217:1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- 3.Yuan R. Structure and mechanism of multifunctional restriction endonucleases. Ann. Rev. Biochem. 1981;50:285–315. doi: 10.1146/annurev.bi.50.070181.001441. [DOI] [PubMed] [Google Scholar]
- 4.Dryden DTF. Bacterial DNA methyltransferases. In: Cheng X, Blumenthal RM, editors. S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. Singapore: World Scientific Publishing; 1999. pp. 283–340. [Google Scholar]
- 5.Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol. Mol. Biol. Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dryden DTF, Murray NE, Rao DN. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 2001;29:3728–3741. doi: 10.1093/nar/29.18.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourniquel AA, Bickle TA. Complex restriction enzymes: NTP-driven molecular motors. Biochimie. 2002;84:1047–1059. doi: 10.1016/s0300-9084(02)00020-2. [DOI] [PubMed] [Google Scholar]
- 8.Murray NE. Immigration control of DNA in bacteria: self versus non-self. Microbiology. 2002;148:3–20. doi: 10.1099/00221287-148-1-3. [DOI] [PubMed] [Google Scholar]
- 9.Dryden DTF, Cooper LP, Murray NE. Purification and characterization of the methyltransferase from the Type I restriction and modification system of Escherichia coli K12. J. Biol. Chem. 1993;268:13228–13236. [PubMed] [Google Scholar]
- 10.Dryden DTF, Cooper LP, Thorpe PH, Byron O. The in vitro assembly of the EcoKI type I DNA restriction/modification enzyme and its in vivo implications. Biochemistry. 1997;36:1065–1076. doi: 10.1021/bi9619435. [DOI] [PubMed] [Google Scholar]
- 11.Eskin B, Linn S. Deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B.2. Purification, subunit structure, and catalytic properties of restriction endonuclease. J. Biol. Chem. 1972;247:6183–6191. [PubMed] [Google Scholar]
- 12.Suri B, Shepherd JCW, Bickle TA. The EcoA restriction and modification system of Escherichia coli 15T– – enzyme structure and DNA recognition sequence. EMBO J. 1984;3:575–579. doi: 10.1002/j.1460-2075.1984.tb01850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janscak P, Dryden DTF, Firman K. Analysis of the subunit assembly of the type IC restriction–modification enzyme EcoR124I. Nucleic Acids Res. 1998;26:4439–4445. doi: 10.1093/nar/26.19.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calisto RM, Pich OQ, Pinol J, Fita I, Querol E, Carpena X. Crystal structure of a putative type I restriction–modification S subunit from Mycoplasma genitalium. J. Mol. Biol. 2005;351:749–762. doi: 10.1016/j.jmb.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 15.Kim JS, DeGiovanni A, Jancarik J, Adams PD, Yokota H, Kim R, Kim SH. Crystal structure of DNA sequence specificity subunit of a type I restriction–modification enzyme and its functional implications. Proc. Natl Acad. Sci. USA. 2005;102:3248–3253. doi: 10.1073/pnas.0409851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burckhardt J, Weisemann J, Hamilton DL, Yuan R. Complexes formed between the restriction endonuclease EcoK and heteroduplex DNA. J. Mol. Biol. 1981;153:425–440. doi: 10.1016/0022-2836(81)90287-4. [DOI] [PubMed] [Google Scholar]
- 17.Kneale GG. Symmetrical model for the domain structure of Type-I DNA methyltransferases. J. Mol. Biol. 1994;243:1–5. doi: 10.1006/jmbi.1994.1624. [DOI] [PubMed] [Google Scholar]
- 18.Dryden DTF, Sturrock SS, Winter M. Structural modelling of a Type I DNA methyltransferase. Nat. Struct. Biol. 1995;2:632–635. doi: 10.1038/nsb0895-632. [DOI] [PubMed] [Google Scholar]
- 19.Obarska A, Blundell A, Feder M, Vejsadova S, Sisakova E, Weiserova M, Bujnicki JM, Firman K. Structural model for the multisubunit Type IC restriction–modification DNA methyltransferase M.EcoR124I in complex with DNA. Nucleic Acids Res. 2006;34:1992–2005. doi: 10.1093/nar/gkl132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obarska-Kosinska A, Taylor JE, Callow P, Orlowski J, Bujnicki JM, Kneale GG. HsdR subunit of the type I restriction–modification enzyme EcoR1241: biophysical characterisation and structural modelling. J. Mol. Biol. 2008;376:438–452. doi: 10.1016/j.jmb.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loenen WAM. Tracking EcoKI and DNA fifty years on: a golden story full of surprises. Nucleic Acids Res. 2003;31:7059–7069. doi: 10.1093/nar/gkg944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies GP, Kemp P, Molineux IJ, Murray NE. The DNA translocation and ATPase activities of restriction-deficient mutants of EcoKI. J. Mol. Biol. 1999;292:787–796. doi: 10.1006/jmbi.1999.3081. [DOI] [PubMed] [Google Scholar]
- 23.Garcia LR, Molineux IJ. Translocation and specific cleavage of bacteriophage T7 DNA in vivo by EcoKI. Proc. Natl Acad. Sci. USA. 1999;96:12430–12435. doi: 10.1073/pnas.96.22.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Studier FW, Bandyopadhyay PK. Model for how Type I restriction enzymes select cleavage sites in DNA. Proc. Natl Acad. Sci. USA. 1988;85:4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seidel R, van Noort J, van der Scheer C, Bloom JGP, Dekker NH, Dutta CF, Blundell A, Robinson T, Firman K, Dekker C. Real-time observation of DNA translocation by the type I restriction–modification enzyme EcoR124I. Nat. Struct. Mol. Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 26.Seidel R, Bloom JG, Dekker C, Szczelkun MD. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seidel R, Bloom JGP, van Noort J, Dutta CF, Dekker NH, Firman K, Szczelkun MD, Dekker C. Dynamics of initiation, termination and reinitiation of DNA translocation by the motor protein EcoR124I. EMBO J. 2005;24:4188–4197. doi: 10.1038/sj.emboj.7600881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Endlich B, Linn S. The DNA restriction endonuclease of Escherichia-coli-B.2. Further studies of the structure of DNA intermediates and products. J. Biol. Chem. 1985;260:5729–5738. [PubMed] [Google Scholar]
- 29.Jindrova E, Schmid-Nuoffer S, Hamburger F, Janscak P, Bickle TA. On the DNA cleavage mechanism of Type I restriction enzymes. FEBS J. 2005;272:552–552. doi: 10.1093/nar/gki322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis DJ, Dryden DTF, Berge T, Edwardson JM, Henderson RM. Direct observation of DNA translocation and cleavage by the EcoKI endonuclease using atomic force microscopy. Nature Struct. Biol. 1999;6:15–17. doi: 10.1038/4882. [DOI] [PubMed] [Google Scholar]
- 31.Berge T, Ellis DJ, Dryden DTF, Edwardson JM, Henderson RM. Translocation-independent dimerization of the EcoKI endonuclease visualized by atomic force microscopy. Biophys. J. 2000;79:479–484. doi: 10.1016/S0006-3495(00)76309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 2004;14:165–173. doi: 10.1016/j.gde.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Ann. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 34.Titheradge AJB, King J, Ryu J, Murray NE. Families of restriction enzymes: an analysis prompted by molecular and genetic data for type ID restriction and modification systems. Nucleic Acids Res. 2001;29:4195–4205. doi: 10.1093/nar/29.20.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chin V, Valinluck V, Magaki S, Ryu J. KpnBI is the prototype of a new family (IE) of bacterial type I restriction–modification system. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gnh134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature Rev. Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 37.Lindsay JA, Holden MT. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct. Integr. Genomics. 2006;6:186–201. doi: 10.1007/s10142-005-0019-7. [DOI] [PubMed] [Google Scholar]
- 38.Kruger DH, Bickle TA. Bacteriophage survival – multiple mechanisms for avoiding the deoxyribonucleic-acid restriction systems of their hosts. Microbiol. Rev. 1983;47:345–360. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bickle TA, Kruger DH. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkins BM. Plasmid promiscuity: meeting the challenge of DNA immigration control. Environ. Microbiol. 2002;4:495–500. doi: 10.1046/j.1462-2920.2002.00332.x. [DOI] [PubMed] [Google Scholar]
- 41.Zavilgelsky GB. Antirestriction. Mol. Biol. 2000;34:724–732. [Google Scholar]
- 42.Tock MR, Dryden DTF. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 43.Dryden DTF. DNA mimicry by proteins and the control of enzymatic activity on DNA. Trends Biotech. 2006;24:378–382. doi: 10.1016/j.tibtech.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 44.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DTF. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell. 2002;9:187–194. doi: 10.1016/s1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 45.Bandyopadhyay PK, Studier FW, Hamilton DL, Yuan R. Inhibition of the Type I restriction–modification enzymes EcoB And EcoK by the gene 0.3 protein of bacteriophage-T7. J. Mol. Biol. 1985;182:567–578. doi: 10.1016/0022-2836(85)90242-6. [DOI] [PubMed] [Google Scholar]
- 46.Atanasiu C, Byron O, McMiken H, Sturrock SS, Dryden DTF. Characterisation of the structure of ocr, the gene 0.3 protein of bacteriophage T7. Nucleic Acids Res. 2001;29:3059–3068. doi: 10.1093/nar/29.14.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atanasiu C, Su TJ, Sturrock SS, Dryden DTF. Interaction of the ocr gene 0.3 protein of bacteriophage T7 with EcoKI restriction/modification enzyme. Nucleic Acids Res. 2002;30:3936–3944. doi: 10.1093/nar/gkf518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker M, Knight P, Trinick J. Negative staining of myosin molecules. J. Mol. Biol. 1985;184:535–542. doi: 10.1016/0022-2836(85)90300-6. [DOI] [PubMed] [Google Scholar]
- 49.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 50.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 51.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 52.van heel M. Angular reconstitution – a posteriori assignment of projection directions for 3-D reconstruction. Ultramicroscopy. 1987;21:111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 53.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera – a visualization system for exploratory research and analysis. J. Comp. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.Taql in complex with DNA and a cofactor analog. Nat. Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 55.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 56.Dominguez C, Boelens R, Bonvin A. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 57.Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 58.Su TJ, Tock MR, Egelhaaf SU, Poon WCK, Dryden DTF. DNA bending by M.EcoKI methyltransferase is coupled to nucleotide flipping. Nucleic Acids Res. 2005;33:3235–3244. doi: 10.1093/nar/gki618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kelleher JE, Daniel AS, Murray NE. Mutations that confer de novo activity upon a maintenance methyltransferase. J. Mol. Biol. 1991;221:431–440. doi: 10.1016/0022-2836(91)80064-2. [DOI] [PubMed] [Google Scholar]
- 60.Cooper LP, Dryden DTF. The domains of a Type-I DNA methyltransferase – interactions and role in recognition of DNA methylation. J. Mol. Biol. 1994;236:1011–1021. doi: 10.1016/0022-2836(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 61.Powell LM, Lejeune E, Hussain FS, Cronshaw AD, Kelly SM, Price NC, Dryden DTF. Assembly of EcoKI DNA methyltransferase requires the C-terminal region of the HsdM modification subunit. Biophys. Chem. 2003;103:129–137. doi: 10.1016/s0301-4622(02)00251-x. [DOI] [PubMed] [Google Scholar]
- 62.O’Neill M, Dryden DTF, Murray NE. Localization of a protein-DNA interface by random mutagenesis. EMBO J. 1998;17:7118–7127. doi: 10.1093/emboj/17.23.7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Neill M, Powell LM, Murray NE. Target recognition by EcoKI: The recognition domain is robust and restriction-deficiency commonly results from the proteolytic control of enzyme activity. J. Mol. Biol. 2001;307:951–963. doi: 10.1006/jmbi.2001.4543. [DOI] [PubMed] [Google Scholar]
- 64.Sturrock SS, Dryden DTF. A prediction of the amino acids and structures involved in DNA recognition by type I DNA restriction and modification enzymes. Nucleic Acids Res. 1997;25:3408–3414. doi: 10.1093/nar/25.17.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen A, Powell LM, Dryden DTF, Murray NE, Brown T. Tyrosine-27 of the specificity polypeptide of EcoKI can be UV cross-linked to a bromodeoxyuridine-substituted DNA target sequence. Nucleic Acids Res. 1995;23:1177–1183. doi: 10.1093/nar/23.7.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Willcock DF, Dryden DTF, Murray NE. A mutational analysis of the 2 motifs common to adenine methyltransferases. EMBO J. 1994;13:3902–3908. doi: 10.1002/j.1460-2075.1994.tb06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor IA, Davis KG, Watts D, Kneale GG. DNA-binding induces a major structural transition in a type I methyltransferase. EMBO J. 1994;13:5772–5778. doi: 10.1002/j.1460-2075.1994.tb06915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.