Abstract
Recently, the human telomeric d[TAGGG(TTAGGG)3] sequence has been shown to form in K+ solution an intramolecular (3+1) G-quadruplex structure, whose G-tetrad core contains three strands oriented in one direction and the fourth in the opposite direction. Here we present a study on the structure of the Bombyx mori telomeric d[TAGG(TTAGG)3] sequence, which differs from the human counterpart only by one G deletion in each repeat. We found that this sequence adopted multiple G-quadruplex structures in K+ solution. We have favored a major G-quadruplex form by a judicious U-for-T substitution in the sequence and determined the folding topology of this form. We showed by NMR that this was a new chair-type intramolecular G-quadruplex which involved a two-layer antiparallel G-tetrad core and three edgewise loops. Our result highlights the effect of G-tract length on the folding topology of G-quadruplexes, but also poses the question of whether a similar chair-type G-quadruplex fold exists in the human telomeric sequences.
INTRODUCTION
Guanine-rich DNA sequences can form four-stranded G-quadruplex structures based on stacking of G•G•G•G tetrads (1–5). G-rich sequences are found at the ends (telomeres) of eukaryotic chromosomes, e.g. (TTAGGG)n repeats in human, and G-quadruplexes formed by these sequences represent attractive anticancer targets (6,7).
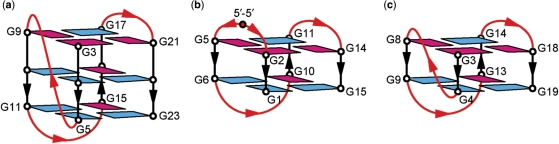
The four-repeat human telomeric d[AGGG(TTAGGG)3] sequence was first shown in 1993 to form an intramolecular G-quadruplex in Na+ solution (8). In this structure, four GGG segments form the G-tetrad core involving three stacked G-tetrads; each strand has both parallel and antiparallel adjacent strands; glycosidic conformations of guanines around each tetrad are syn•syn•anti•anti; three connecting TTA linkers form successively edgewise, diagonal and edgewise loops. In 2002, a K+-containing crystal structure of the same sequence showed a completely different intramolecular G-quadruplex form (9). In this structure, all four strands are parallel; all guanines are anti; the connecting TTA loops are double-chain-reversal. In 2006, four-repeat human telomeric sequences have been shown to form two distinct intramolecular (3+1) G-quadruplexes in K+ solution (10–13). In these structures, the (3+1) core contains three strands oriented in one direction and the fourth in the opposite direction; the glycosidic conformations of G-tetrads are syn•anti•anti•anti and anti•syn•syn•syn; there are one double-chain-reversal and two edgewise TTA loops. These (3+1) G-quadruplex structures differ from each other in the order of loop arrangements (13). In particular, the human telomeric d[TAGGG(TTAGGG)3] sequence forms predominantly a (3+1) G-quadruplex in K+ solution as shown in Figure 1a (12).
Figure 1.
(a) Three-layer intramolecular (3+1) G-quadruplex with one double-chain-reversal and two edgewise loops observed for the human telomeric sequence d[TAGGG(TTAGGG)3] in K+ solution (12). (b) Two-layer intramolecular (3+1) G-quadruplex with three edgewise loops formed by the re-engineered thrombin aptamer d(3′GGT5′-5′TGGTGTGGTTGG3′) sequence (15). (c) A possible model of two-layer intramolecular (3+1) G-quadruplex with one double-chain-reversal and two edgewise loops. This topology was derived from the three-layer G-quadruplex fold shown in (a) by removing the bottom G-tetrad. anti and syn guanines are colored cyan and magenta, respectively.
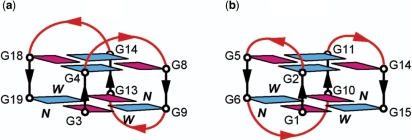
Although many diverse G-quadruplex topologies are known (1–5), so far very few G-quadruplexes with only two G-tetrad layers have been reported. The best studied system was the G-quadruplex of the thrombin aptamer d(GGTTGGTGTGGTTGG) sequence (14), which forms a chair-type G-quadruplex with three edgewise loops (Figure 7b). In this structure, the core consists of two G-tetrad layers, whose glycosidic conformations are syn•anti•syn•anti; each strand is antiparallel to its two neighboring strands (14). Recently, Martino et al. (15) used a 5′–5′ inversion-of-polarity site to re-engineer this sequence to adopt a G-quadruplex topology with the (3+1) core, while maintaining the three edgewise loops (Figure 1b).
Figure 7.
Schematic structures of chair-type intramolecular G-quadruplexes observed for (a) the B. mori telomeric sequence Bm-U16 in K+ solution (this work) and (b) thrombin aptamer sequence d(GGTTGGTGTGGTTGG) (Ref. 14). anti and syn guanines are colored cyan and magenta, respectively. W and N represent wide and narrow groove, respectively.
Here we examine the folding topology of the d[TAGG(TTAGG)3] sequence containing telomeric repeats of Bombyx mori (16) and several other insects (16–18), which differs from the human telomeric d[TAGGG(TTAGGG)3] sequence only by one G deletion in each repeat. Previously, it has been shown that some particular intramolecular G-quadruplex topologies can be favored by interactions between terminal and loop residues (13). Because both sequences, d[TAGGG(TTAGGG)3] and d[TAGG(TTAGG)3], contain the same loop and terminal residues and the former forms predominantly a (3+1) G-quadruplex with three G-tetrad layers (Figure 1a), one might expect the latter to form a two-layer (3+1) G-quadruplex (Figure 1c). Instead, we found that a major form of the B. mori telomeric d[TAGG(TTAGG)3] sequence is a new chair-type G-quadruplex with anti-parallel G-tetrad core and three edgewise loops. This result highlights the effect of G-tract length on the folding topology of G-quadruplexes, but also poses the question of whether a similar chair-type G-quadruplex fold exists in the human telomeric sequences.
MATERIALS AND METHODS
DNA sample preparation
Unlabeled and site-specific 2%-15N,13C-labeled DNA oligonucleotides were chemically prepared using products from Glen Research and Cambridge Isotope Laboratories. Samples were dialyzed successively against ∼50 mM KCl solution and against water. DNA concentration was expressed in strand molarity using a nearest-neighbor approximation for the absorption coefficients of the unfolded species (19). Natural and T-to-U-modified DNA oligonugleotides were assumed to have the same extinction coefficient. Unless otherwise stated, all experiments were carried out in a buffer containing 20 mM potassium phosphate (pH 7.0) and 70 mM KCl.
Melting experiments
The thermal stability of different DNA oligonucleotides was characterized by recording the UV absorbance at 295 nm as a function of temperature (20) using a Varian Cary 300 Bio UV-Vis spectrophotometer. The temperature ranged from 20 to 90°C. The heating and cooling rates were 0.25°C per minute. Two baselines corresponding to the completely folded (low temperature) and completely unfolded (high temperature) states were manually drawn in order to determine the fractions of folded and unfolded species during the melting/folding transition. The melting temperature (Tm) was defined as the temperature of the mid-transition point. DNA concentration ranged from 3 to 250 μM. Experiments were performed with quartz cuvettes, 1 cm path length for low DNA concentrations and 0.2 cm path length for high DNA concentrations.
Thermal difference spectra
UV absorbance spectra for the unfolded and folded states of an oligonucleotide were recorded at 90°C and 20°C, temperatures, respectively, above and below its melting temperature (Tm). The difference between these two spectra was defined as the thermal difference spectrum (TDS). The TDS could provide specific signatures of different DNA and RNA structural conformations (21). Spectra (220–320 nm) were recorded on a Varian Cary 300 Bio UV/Vis spectrophotometer using 1 cm path-length quartz cuvettes. DNA concentration was 3 μM.
Circular dichroism (CD)
CD spectra at 20°C were recorded on a Jasco J-810 spectropolarimeter using a 1 cm path-length quartz cuvette in a reaction volume of 800 μl. Concentration of DNA samples was 5 μM. They were annealed at 95°C for 5 min then allowed to cool down to room temperature over several hours. Scans from 220 nm to 320 nm were performed with 200 nm/min scanning speed, 1 nm pitch and 1 nm bandwidth. For each spectrum, an average of three scans was taken, spectral contribution from the buffer was subtracted, and the data were zero-corrected at 320 nm.
Nuclear magnetic resonance (NMR)
NMR experiments were performed on 600 MHz and 700 MHz Bruker spectrometers. The strand concentration of the NMR samples was typically 0.5–2.5 mM. Resonance assignments were obtained using site-specific low-enrichment labeling (22) and through-bond correlations at natural abundance (23,24) (JRHMBC, HMQC and TOCSY), and independently verified using NOESY experiments. Inter-proton distances were measured using NOESY experiments.
RESULTS
B. mori telomeric sequences form G-quadruplexes
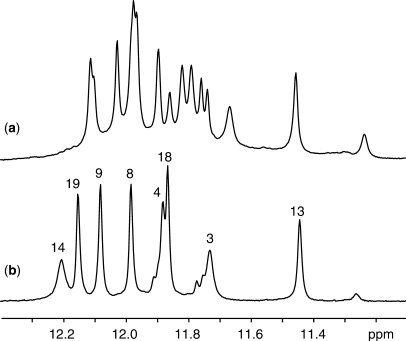
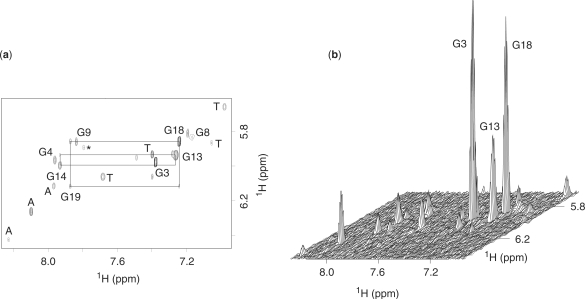
Imino proton NMR spectrum of the B. mori telomeric d(TAGGTTAGGTTAGGTTAGG) sequence (Bm) in K+ solution is plotted in Figure 2a. Sharp imino proton peaks at 10–12 ppm are characteristic of G-quadruplex formation. The number and intensity of peaks in this region indicated that there were multiple G-quadruplexes. We found that the proportions of different forms varied with sequence modifications (U-for-T and BrU-for-T substitutions). This strategy was used previously to favor one conformation among multiple G-quadruplexes (25). Sequence d(TAGGTTAGGTTAGGTUAGG) (Bm-U16) with a U-for-T substitution at position 16 favored a major conformation (Figure 2b) allowing further structural characterization. The imino proton spectrum of Bm-U16 displayed eight major peaks, consistent with formation of a major G-quadruplex conformation involving all guanines in the sequence. Comparison between the spectra of Bm-U16 and Bm (Figure 2) suggested that the structure of Bm-U16 was one of the conformations adopted by Bm.
Figure 2.
Imino proton NMR spectra at 35°C of B. mori telomeric sequences (a) Bm, d(TAGGTTAGGTTAGGTTAGG) and (b) Bm-U16, d(TAGGTTAGGTTAGGTUAGG) in K+ solution. Imino proton assignments of Bm-U16 are labeled with residue numbers. Solution contained 70 mM KCl and 20 mM potassium phosphate, pH 7.
The NMR spectrum of Bm-U16 in Na+ solution (Figure S1a) was significantly different from that in K+ solution (Figure 2b); the number and intensity of imino proton peaks suggested formation of multiple G-quadruplex conformations. In contrast, in a solution mimicking physiological condition containing mixed cations (100 mM K+, 10 mM Na+, 1 mM Ca2+ and 2 mM Mg2+) the spectrum of the same sequence (Figure S1b) was similar to that observed in K+ solution (Figure 2b), indicating the predominance of the K+-containing G-quadruplex form.
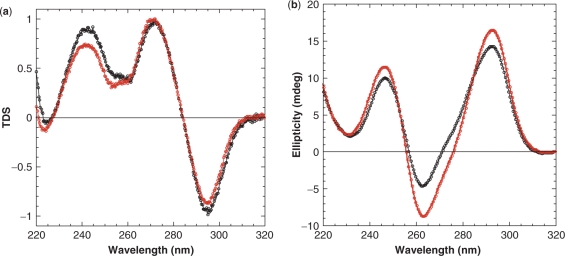
TDS and CD signatures of Bm and Bm-U16
TDS of Bm and Bm-U16 were similar, with two positive maxima at 240 nm and 275 nm and one negative minimum at 295 nm (Figure 3a), consistent with formation of G-quadruplexes (21). Bm and Bm-U16 exhibited very similar CD spectra with a positive peak at 295 nm and a negative peak at 265 nm (Figure 3b), consistent with formation of antiparallel G-quadruplexes (26) in these sequences.
Figure 3.
(a) TDS and (b) CD profiles of B. mori telomeric sequences Bm and Bm-U16 in K+ solution. Spectra of Bm and Bm-U16 are shown in red and black, respectively. CD spectra were recorded at 20°C. Solution contained 70 mM KCl and 20 mM potassium phosphate, pH 7.
Stability and stoichiometry of Bm and Bm-U16
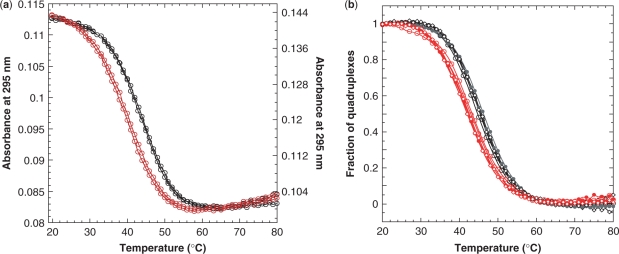
To assess the thermal stability of Bm and Bm-U16, melting experiments were performed by monitoring the UV absorbance at 295 nm (20). Typical denaturation profiles of Bm and Bm-U16 are shown in Figure 4a. The decrease of the 295 nm absorbance as the temperature increases is characteristic of G-quadruplexes. At heating and cooling rates of 0.25°C per minute, the melting and folding profiles were superimposable indicating equilibrium processes. The melting temperature of Bm and Bm-U16 in the presence of 90 mM K+ was 41°C and 45°C, respectively (Figure 4a). These melting temperatures were consistent with NMR measurements (data not shown).
Figure 4.
Melting curves of Bm (red) and Bm-U16 (black). (a) UV absorbance at 295 nm of 3 μM of Bm (right scale) and Bm-U16 (left scale) as a function of temperature. (b) The fractions of G-quadruplexes as a function of temperature for three different DNA concentrations: 3 μM (opened circles), 30 μM (filled circles) and 250 μM (lozenge). Melting curves for different DNA concentrations were superimposable, indicating formation of monomeric intramolecular G-quadruplexes. In each case, both heating and cooling curves are plotted. Solution contained 70 mM KCl and 20 mM potassium phosphate, pH 7.
In order to determine the stoichiometry of the Bm and Bm-U16 G-quadruplexes, we performed melting experiments for different DNA concentrations. The fractions of folded and unfolded species were determined using two baselines corresponding to the completely folded (low temperature) and completely unfolded (high temperature) states. The normalized melting curves of Bm and Bm-U16 for different DNA concentrations, ranging from 3 μM to 250 μM, showed that the melting temperature (Tm) was independent of sample concentrations (Figure 4b). This indicated that the structures were monomeric intramolecular G-quadruplexes, consistent with the observation of narrow linewidths of the some NMR peaks.
NMR spectral assignments of Bm-U16
Guanine imino protons of Bm-U16 were unambiguously assigned by the site-specific low-enrichment approach (22) (Figures S2 and S3). Guanine H8 protons were assigned by natural-abundant through-bond correlations (23,24) to the already assigned imino protons (Figure S4). Aromatic protons (including eight guanine H8 protons) were independently traced in through-bond (HMQC and TOCSY) and through-space (NOESY) correlation experiments (24) (Figures 5 and S5). Peaks from the structured and unfolded forms were identified based on their relative intensities at different temperatures.
Figure 5.
(a) Contour plot and (b) stacked plot of NOESY spectrum of Bm-U16 at 25°C (mixing time, 300 ms). Rectangular H8-H1′ patterns for 5′-syn-anti-3′ steps are highlighted by black lines. Intensity of H8-H1′ cross-peaks indicated the presence of four syn guanines. Strong H8-H1′ cross-peaks are observed for G3, G13 and G18 at 25°C. H8 and H1′ resonances of G8 are broadened at 25°C, reflecting motion(s) in this region. Peaks from the unfolded species were labeled with a star.
G-quadruplex topology of Bm-U16
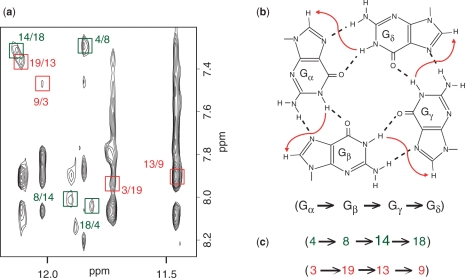
The G-tetrad alignments of Bm-U16 were identified from NOESY spectra (Figure 6a) based on the specific imino-H8 connectivity patterns around individual G-tetrads (Figure 6b). This has established the alignment of two G-tetrads: G4•G8•G14•G18 and G3•G19•G13•G9 (Figure 6a, c). Connecting corners of this G-tetrad core with TTA loop linkers established a chair-type G-quadruplex fold (Figure 7a). The G-tetrad core is of antiparallel-type, in which each G-tract is oriented in the opposite direction with respect to its two neighboring ones. The hydrogen-bond directionalities around the two G-tetrads are of opposite directions, i.e. clockwise and anticlockwise. The G-tetrad core is characterized by two wide and two narrow grooves (Figure 7a). Both wide and narrow grooves in a G-quadruplex are defined between two adjacent antiparallel strands (Figure 7a), however, across the wide groove (denoted W) the G•G base pairing occurs between the Watson–Crick edge of an anti guanine and the Hoogsteen edge of a syn guanine, while across the narrow groove (denoted N) the G•G base pairing occurs between the Watson–Crick edge of a syn guanine and the Hoogsteen edge of an anti guanine. For Bm-U16, the glycosidic conformations of guanines in the G-tetrads are anti•syn•anti•syn. This G-tetrad alignment is consistent with H1′-H8 NOE intensities observed for these residues: four strong H1′-H8 peaks were observed for G3, G8, G13 and G18 (Figure 5 and data not shown). The G-tetrad alignment (Figure 7a) is also consistent with other specific NOE patterns (27) such as those detected for the four 5′-syn-anti-3′ steps in the structure (Figures 5 and S5). All three linkers in the structure, T5-T6-A7, T10-T11-A12 and T15-T16-A17, form edgewise loops. The T5-T6-A7 and T15-T16-A17 loops span two wide grooves, while the central T10-T11-A12 loop spans a narrow groove. Cross-peaks between imino protons of the bottom G-tetrad with loop aromatic protons (Figure 6) indicated the stacking of base(s) below this G-tetrad.
Figure 6.
Determination of G-quadruplex topology for Bm-U16 in K+ solution. (a) NOESY spectrum (mixing time, 200 ms) of Bm-U16 at 35°C. Imino-H8 cross peaks that identify two G-tetrads (colored green and red) are framed and labeled with the number of imino protons in the first position and that of H8 in the second position. (b) Characteristic guanine imino-H8 NOE connectivity patterns around a Gα•Gβ•Gγ•Gδ tetrad as indicated with arrows (connectivity between Gδ and Gα implied). (c) Characteristic guanine imino-H8 NOE connectivities observed for G4•G8•G14•G18 (green) and G3•G19•G13•G9 (red) tetrads.
Spectral broadening and possible motions in loops
At temperatures below 35°C, the spectral broadening was observed for several resonances including H6 protons of thymines in the loops (Figure S5) and imino protons of G3, G4, G13 and G14 in the core (Figure 2). These peaks were sharpened when the temperature increased (Figure S6 and data not shown). These findings were consistent with microsecond-to-millisecond motions in the loops (28).
DISCUSSION
We have shown that the four-repeat B. mori telomeric d[TAGG(TTAGG)3] (Bm) sequence forms a chair-type G-quadruplex which involves a two-layer antiparallel G-tetrad core and three edgewise loops (Figure 7a). Although chair-type G-quadruplexes are among the most intuitive G-quadruplex folding topologies and have been proposed for a long time (1–5), to our knowledge the chair-type topology of Bm (Figure 7a) has not been observed before. A similar topology was initially proposed for the thrombin aptamer sequence d(GGTTGGTGTGGTTGG) (29), but later corrected to another chair-type G-quadruplex topology (Figure 7b) (14). Both these chair-type G-quadruplexes have an antiparallel G-tetrad core and three edgewise loops. However, they differ by loop arrangements: in the structure of Bm (Figure 7a) the central loop (located at the bottom of the G-tetrad core) spans across a narrow groove and the two other loops (first and third, located side by side at the top of the G-tetrad core) span wide grooves, while in the structure of the thrombin aptamer (Figure 7b) the central loop (bottom) spans a wide groove and the two remaining loops (top) span narrow grooves. These loop arrangements could be partially due to the linker lengths: the first and third loops in the thrombin aptamer sequence (TT) might be too short to span wide grooves. Our structural modelling (Supplementary Data) suggested that two-residue (TT) linkers were less favorable to form edgewise loops across wide grooves than narrow grooves. A similar conclusion was obtained from the modelling study of Hazel et al. (30). In a NMR study of G-quadruplexes formed by the two-repeat Tetrahymena telomeric sequence d(TGGGGTTGGGGT) in Na+ solution (31), two-residue (TT) edgewise loops across wide grooves were observed next to edgewise loops across narrow grooves.
However, it should be noted that speculations on loop conformations in G-quadruplexes based solely on loop lengths might be oversimplified. Reported structures of G-quadruplexes (1,2,11–15) including those of the thrombin aptamer sequences (14,15) indicated the contributions of base pairing and stacking in the overall structure stability. Base triads were observed in the G-quadruplex structure formed by the single-repeat Bombyx mori telomeric sequence d(TAGG) in Na+ solution (32). Our NMR data on Bm-U16 indicated that bases from the central loop and terminal residues stacked under the bottom G-tetrad. Spectral broadening suggested motions in this loop. At the top of the structure, the two loops, T5-T6-A7 and T15-T16-A17, can be called ‘adjacent antiparallel loops’ as viewed from the schematic structure (Figure 7a). Spectral broadening in this region can be explained by interconversions (or motions) between different possible base pairing and stacking conformations in the top loops. These could be compared to the motions previously reported for quasi-symmetric loops of an i-motif structure (28).
It is of great interest to be able to predict the G-quadruplex topology based on the DNA sequence. Effect of G-tract length on the topology of G-quadruplexes was recently studied by Rachwal et al. (33) in the context of d(GnT)4 and d(GnT2)4 sequences (n = 3–7) using CD spectroscopy. The B. mori telomeric d[TAGG(TTAGG)3] sequence studied here and the human telomeric d[TAGGG(TTAGGG)3] sequence have a similar loops and termini; they differ from each other only by one G in each repeat. While the human sequence d[TAGGG(TTAGGG)3] forms a (3+1) G-quadruplex with three G-tetrad layers, one may ask whether the B. mori sequence d[TAGG(TTAGG)3] can adopt a similar topology with two G-tetrad layers. We have identified a chair-type G-quadruplex, but did not rule out the co-existence of a (3+1) G-quadruplex with two G-tetrad layers in the B. mori sequenced [TAGG(TTAGG)3]. Our result highlights the effect of G-tract length on the folding topology of G-quadruplexes; conformations the G-tetrad core and the loops are tightly interrelated. Nevertheless, as the chair-type G-quadruplex is identified for the B. mori telomeric sequence, one may also ask the reverse question of whether a similar chair-type conformation also exists in the human telomeric sequences (10,34). Answer to this question needs further investigation.
So far, with a few exceptions (35,36) searches for potential G-quadruplex forming sequences in the genomes have been mainly focused on sequences containing tracts of at least three consecutive guanines (37–44). The observation of stable G-quadruplexes formed by B. mori telomeric sequences advocates for the analysis of genomic sequences containing tracts of two guanines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Ministry of Education grant ARC30/07 and Nanyang Technological University start-up grants (SUG5/06, RG138/06 and M58110032 to A.T.P.). Funding for open access charges: Grant ARC30/07 from Ministry of Education, Singapore.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Jean-Louis Mergny and Dr Patrizia Alberti from the Museum National d’Histoire Naturelle de Paris for sharing their research data and for stimulating discussions. We thank the School of Biological Sciences of NTU, particularly Prof. Yoon Ho Sup and Dr Ye Hong, for granting access to the NMR facility and assisting us with NMR experiments. We thank Prof. Lars Nordenskiöld and Dr Nikolai Korolev for allowing us to use the UV spectrophotometer in their laboratory. R.W.L.A., a student from River Valley High School, and Z.M.T, a student from NUS High School, were participants of Nanyang Research Program. C.L., a student from National Junior College, was a participant of the Science Training and Research program. J.K.C.L. and J.M.W.L., students from Ngee Ann Polytechnic (NP), were on an industrial attachment program supported by NP.
REFERENCES
- 1.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 5.Simonsson T. G-quadruplex DNA structures – variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 6.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 7.Mergny JL, Hélène C. G-quadruplex DNA: a target for drug design. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 9.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 11.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly JA, Feigon J, Yeates TO. Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 15.Martino L, Virno A, Randazzo A, Virgilio A, Esposito V, Giancola C, Bucci M, Cirino G, Mayol L. A new modified thrombin binding aptamer containing a 5′-5′ inversion of polarity site. Nucleic Acids Res. 2006;34:6653–6662. doi: 10.1093/nar/gkl915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okazaki S, Tsuchida K, Maekawa H, Ishikawa H, Fujiwara H. Identification of a pentanucleotide telomeric sequence, (TTAGG)n, in the silkworm Bombyx mori and in other insects. Mol. Cell Biol. 1993;13:1424–1432. doi: 10.1128/mcb.13.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahara K, Marec F, Traut W. TTAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosome Res. 1999;7:449–460. doi: 10.1023/a:1009297729547. [DOI] [PubMed] [Google Scholar]
- 18.Robertson HM, Gordon KH. Canonical TTAGG-repeat telomeres and telomerase in the honey bee, Apis mellifera. Genome Res. 2006;16:1345–1351. doi: 10.1101/gr.5085606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 20.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 21.Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan AT, Patel DJ. A site-specific low-enrichment 15N,13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- 23.Phan AT. Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA. J. Biomol. NMR. 2000;16:175–178. doi: 10.1023/a:1008355231085. [DOI] [PubMed] [Google Scholar]
- 24.Phan AT, Guéron M, Leroy JL. Investigation of unusual DNA motifs. Methods Enzymol. 2001;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- 25.Phan AT, Patel DJ. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feigon J, Koshlap KM, Smith FW. 1H NMR spectroscopy of DNA triplexes and quadruplexes. Methods Enzymol. 1995;261:225–255. doi: 10.1016/s0076-6879(95)61012-x. [DOI] [PubMed] [Google Scholar]
- 28.Phan AT, Guéron M, Leroy JL. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000;299:123–144. doi: 10.1006/jmbi.2000.3613. [DOI] [PubMed] [Google Scholar]
- 29.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 30.Hazel P, Parkinson GN, Neidle S. Predictive modelling of topology and loop variations in dimeric DNA quadruplex structures. Nucleic Acids Res. 2006;34:2117–2127. doi: 10.1093/nar/gkl182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan AT, Modi YS, Patel DJ. Two-repeat Tetrahymena telomeric d(TGGGGTTGGGGT) sequence interconverts between asymmetric dimeric G-quadruplexes in solution. J. Mol. Biol. 2004;338:93–102. doi: 10.1016/j.jmb.2004.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kettani A, Bouaziz S, Wang W, Jones RA, Patel DJ. Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads. Nat. Struct. Biol. 1997;4:382–389. doi: 10.1038/nsb0597-382. [DOI] [PubMed] [Google Scholar]
- 33.Rachwal PA, Brown T, Fox KR. Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochemistry. 2007;46:3036–3044. doi: 10.1021/bi062118j. [DOI] [PubMed] [Google Scholar]
- 34.He Y, Neumann RD, Panyutin IG. Intramolecular quadruplex conformation of human telomeric DNA assessed with 125I-radioprobing. Nucleic Acids Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scaria V, Hariharan M, Arora A, Maiti S. Quadfinder: server for identification and analysis of quadruplex-forming motifs in nucleotide sequences. Nucleic Acids Res. 2006;34:W683–W685. doi: 10.1093/nar/gkl299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R, Lin Y, Zhang CT. Greglist: a database listing potential G-quadruplex regulated genes. Nucleic Acids Res. 2008;36:D372–D376. doi: 10.1093/nar/gkm787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yadav VK, Abraham JK, Mani P, Kulshrestha R, Chowdhury S. QuadBase: genome-wide database of G4 DNA – occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.