Abstract
Small RNAs (sRNAs) are common and effective modulators of gene expression in eukaryotic organisms. To characterize the sRNAs expressed during rice seed development, massively parallel signature sequencing (MPSS) was performed, resulting in the obtainment of 797 399 22-nt sequence signatures, of which 111 161 are distinct ones. Analysis on the distributions of sRNAs on chromosomes showed that most sRNAs originate from interspersed repeats that mainly consist of transposable elements, suggesting the major function of sRNAs in rice seeds is transposon silencing. Through integrative analysis, 26 novel miRNAs and 12 miRNA candidates were identified. Further analysis on the expression profiles of the known and novel miRNAs through hybridizing the generated chips revealed that most miRNAs were expressed preferentially in one or two rice tissues. Detailed comparison of the expression patterns of miRNAs and corresponding target genes revealed the negative correlation between them, while few of them are positively correlated. In addition, differential accumulations of miRNAs and corresponding miRNA*s suggest the functions of miRNA*s other than being passenger strands of mature miRNAs, and in regulating the miRNA functions.
INTRODUCTION
Small RNA (sRNA) molecules are widely recognized as common and effective modulators of gene expression in many eukaryotic organisms (1–3). According to the present knowledge, sRNAs are generally divided into several categories, including microRNAs (miRNAs), short-interfering RNAs (siRNAs), trans-acting siRNAs (ta-siRNAs; 4), natural antisense transcript siRNAs (nat-siRNAs; 5) and Piwi-interacting RNAs (piRNAs) in metazoans (6). In plants, ta-siRNAs and nat-siRNAs guide the cleavage of target mRNAs, whereas heterochromatic siRNAs are involved in DNA and histone modification, leading to the silencing of gene transcription (7). In addition to nat-siRNAs, rice natural antisense miRNAs (nat-miRNAs) were recently identified (8).
miRNAs (∼21 nt) are produced from noncoding imperfectly complementary, stem–loop RNA precursors. The miRNA genes can be transcribed by RNA polymerase II (9) to form the primary miRNAs (pri-miRNAs). In higher plants, pri-miRNAs are processed by dicer-like 1 (DCL1; 10) or DCL4 (11) to form miRNA duplexes through multiple cleavage steps, and then miRNA duplexes are methylated by Hua Enhancer1 (HEN1) at the 3′-end (12) and loaded onto Argonaute 1 (AGO1) where the mature miRNAs guide the cleavage of target mRNAs. In the meanwhile, the passenger strands (referred as miRNA*s) are gradually degraded (13).
Although the first miRNA (lin-4) was discovered by forward genetic screening in Caenorhabditis elegans (14), the majority of currently known plant miRNAs were identified by size-selected cloning and sequencing, especially those in Arabidopsis (15–18) and rice (19–21). Recently developed high-throughput sequencing strategies have expanded the depth of sRNA cloning coverage. In Arabidopsis, analysis through massively parallel signature sequencing (MPSS) identified 67 528 and 27 833 unique 17-nt sequences from inflorescences and seedlings, respectively (22). More than 340 000 unique sRNAs from Arabidopsis seedlings, rosette leaves, flowers and siliques were sequenced using pyrophosphate-based high-throughput sequencing technique (11), and 48 new miRNAs were identified with similar strategy (23). In rice, ∼20 miRNAs were identified by large-scale sequencing of sRNAs in panicles, seedlings and stems (8,24–26).
Several guidelines have been proposed for miRNAs annotation (27). The miRNA precursors should contain stable and conserved stem–loop structures that can be predicted by Mfold (28), and mature miRNAs should be detected by northern blotting or sequencing. In addition, as miRNA genes are transcribed by RNA polymerase II, capped and polyadenylated as normal mRNAs (9), EST analysis is a powerful approach to identify the new miRNAs (29). Identification of a miRNA* sequence, a product of Dicer cleavage corresponding to miRNA (11), also strongly indicates that the corresponding sRNA molecule was indeed processed by Dicer-like RNase III enzyme (11,23,30).
Rice is an important food resource for human daily life and serves as model species of monocotyledon plants. Development and maturation of rice seed, a highly specialized organ of nutrient storage and reproductive development, involve meticulous and fine gene regulations at transcriptional and post-transcriptional levels (31). To further study the complicated regulatory network of rice seed development, and to elucidate the functions of sRNAs during this process, MPSS and integrated bioinformatics analysis were performed, resulting in the identification of novel and candidate miRNAs. Further, expression profiles of miRNAs were analyzed through miRNA microarray hybridization, which have been widely used to study the miRNA expression levels in several species (32–35). Comparison of expression patterns revealed the positive or negative correlations between miRNAs and the corresponding target genes, which greatly expand the understanding of how miRNAs were involved in rice seed development.
MATERIALS AND METHODS
cDNA library construction and MPSS analysis
Rice (Oryza sativa, japonica cultivar Zhonghua 11) immature seeds at 3, 6, 9 and 12 days after anthesis (DAA) were collected and total RNAs were extracted separately using Trizol Reagent (Invitrogene, Nottingham, UK), which were then mixed with equal amounts from individual samples. After separation on denatured polyacrylamide gel electrophoresis (PAGE), the fraction of 18–26-nt small RNAs (sRNAs) was recovered by sRNAs gel extraction Kit (TaKaRa Bio, Otsu, Japan).
The cDNA library construction and MPSS analysis were carried out by TaKaRa Bio Inc. (Otsu, Japan) as described previously (36). In brief, 20 ng of sRNAs fraction was dephosphorylated and ligated with RNA–DNA chimeric 3′-adapter. After purification by denatured PAGE, the RNA fraction with attached adapters was phosphorylated and ligated with RNA–DNA chimeric 5′-adapter. Resultant products were reversely transcribed and amplified by PCR with primers (with a SfaNI site). Then the amplified PCR products were digested by SfaNI and cloned into the Tag vector pMBS I (Solexa). The resultant library, which has more than 1.3 million total complexity, was amplified and loaded onto microbeads. Finally, more than 1 million microbeads were loaded into each flowcell and the signature sequences of 22 bases were determined by a series of enzymatic reactions.
The adaptor sequences of signatures were masked. After removing signatures corresponding to ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs), the total abundances of signatures were around 250 000. sRNAs were normalized to transcripts per quarter million (TPQ) as previously described (22,36). The signatures and abundances of sRNAs in rice seed library can be found in National Center for Biotechnology Information, Gene Expression Omnibus (NCBI, GEO) under series number GSE11974.
Data sets
Genome sequences and annotations of rice (version 5.0) and assembled Zea mays sequence (AZM5) were obtained from TIGR (the Institute for Genomic Research). Sequences of rRNAs, tRNAs, snRNAs and snoRNAs were downloaded from databases including the European ribosomal RNA database (http://www.psb.ugent.be/rRNA/, for rRNA), the Genomic tRNA database (http://lowelab.ucsc.edu/GtRNAdb/, for tRNA) and NONCODE (http://www.bioinfo.org.cn/NONCODE/, for snRNAs and snoRNAs). Mature miRNAs and annoated stem–loop sequences were obtained from miRBase (versions 10.0 and 11.0, http://microrna.sanger.ac.uk/; 37). sRNAs sequences of rice, Arabidopsis and C. elegans were downloaded from rice MPSS database (http://mpss.udel.edu/rice/), Arabidopsis Small RNA Project (ASRP, http://asrp.cgrb.oregonstate.edu/) and GenBank data libraries (GEO accession number GSE5990, sample GSM139137), respectively.
Identification of sRNA clusters and hotspots
The sRNAs were grouped into clusters dependent on their locations on the genome as described previously, i.e. sRNAs within 500 bp of each other were fallen under a cluster (22). To identify the sRNA hotspots, abundance of each signature was firstly normalized by hitting times of signature on the genome, and then the sums of abundances of all signatures in no overlapping 500-bp windows were calculated. The top-ranking windows were used as seeds for extension in both directions until a window hits no signatures (11).
Predictions of miRNAs and corresponding mRNA targets
All the known rice miRNAs, whose precursors contain no repetitive sequences, matched genome for <30 times. Our analysis on the obtained signatures that matched genome for more than 30 times indicated that 72.9% of them (3470 out of 4760) originated from repetitive sequences (TIGR Oryza Repeat Database v3.3). Signatures matched genome for more than 30 times were thus filtered out during miRNA prediction and those corresponding to rRNAs, tRNA, snRNAs and snoRNAs were eliminated. The 400-bp genome sequences on each side of these signatures were extracted and a sliding window of 450 bp with an increment of five bases was scanned along the extracted sequences. All the fragment sequences containing each signature were folded with the help of MFOLD3.2 (28). Structures of the sequences with minimum energy were further analyzed using a Perl script to check whether these structures satisfy the criteria for majority of known plant miRNAs (38). The sequences of candidate precursors were analyzed using RepeatMasker (http://www.repeatmasker.org) to eliminate the repetitive sequences. Finally, signatures whose predicted secondary structures passed the in silico check were grouped by genomic positions. The candidates with miRNA* and/or EST supporting evidences were identified as miRNAs.
To predict the target sites of miRNAs, FASTA Program (39) was used to search the rice cDNA database (TIGR, version 5.0) with +15/–10 scoring matrix. Obtained reverse complement results (maximal 100 000) of each miRNA were used to calculate the mispair scores and Minimum Free Energy (MFE) ratios. In brief, the MFE (ΔGMFE) of a hypothetical duplex containing miRNA target paired with a perfectly complementary sequence and free energy of each actual miRNA-predicted target duplex (ΔGtarget) were determined respectively. MFE ratio was calculated as ΔGtarget/ΔGMFE (4). The mispair score and MFE ratio tolerance limits to ≤4 and ≥0.73 were assigned, and the match up of all miRNAs and predicted targets were checked manually. All the Perl scripts used in miRNA prediction and target prediction were available upon request.
miRNA chip generation and hybridization
The miRNAs in miRBase vesion 8.0, newly identified ones and other candidates in this study, were collected to design the probes. Independent sequences (polyT18 or polyT19) were attached to the full-length miRNA probes at 3′-end to acquire the final 40-nt probes so that they have appropriate extension space at the chip surface. The probes were then attached to the activated slide surface via a C6 5′-amino-modifier (35). Probes for tRNA and U6 RNA were designed and used as internal positive controls. In addition, eight DNA sequences having no homology with any sRNAs were designed and used as external positive controls. All probes were synthesized at MWG Company (Ebersberg, Germany), and dissolved into DNA spotting solution (40 µmol/l), which was spotted onto aldehyde-decorated slides for three repeated dots.
Labeling of sRNAs and chip hybridization were performed as described previously (35). In detail, 50–100 μg of total RNAs from different samples (embryo, endosperm, leaf and root of 7-day-old seedlings, 10-day-old seedling) were size-fractionated using Ambion's miRNA Isolation Kit (Cat#.1560) and labeled with T4 polymerase (32). RNA was dissolved in hybridization buffer containing 15% formamide, 0.2% SDS, 3×SSC and 50× Denhardt's solution. After overnight hybridization at 42°C, slides were washed in 0.2% SDS, 2×SSC for 4 min and 0.2×SSC for another 4 min. Hybridized slides were scanned with CapitalBio® LuxScan™ 10K/A (Beijing, China) and analyzed with the GenePix Pro 4.0 (Axon Instruments, Union City, CA). The maximal signal level of background probes was 200, and 800 was set as detection signal threshold of miRNAs.
Hybridization signals were normalized using medium center method. For each tissue (except embryo) two hybridizations were performed, thus six hybridization signals were finally obtained for each miRNA from one tissue (three signals for embryo). As the immature embryo tissues were difficult to collect and considering that there were three repeat dots of probes on the chip, only one hybridization was performed. At last, mean of hybridization signals was calculated and visualized using TIGR Mev (version 3.1, http://www.tm4.org/mev.html).
Gene expression analysis through microarray hybridization
GeneChip® rice genome array (Affymetrix, Santa Clara, CA) was used for DNA microarray analysis. The RNA samples were same as those for miRNA microarray hybridization (total RNAs were fractioned, the fraction containing large or small RNA molecules were used for DNA and miRNA microarray analysis, respectively). Each sample has two biological repeats, and 10-μg cRNA was used in each hybridization. Washing, staining and scanning of chips were performed according to supplier's protocol. Hybridization signals were normalized using the Affymetrix Microarray Suit program (version 5.0) and visualized using TIGR Mev. The microarray data of Affymetrix arrays are under series number GSE11966 in the database of NCBI, GEO. To check the replicate quality of microarray, least-square linear regression for each tissue was carried out (40) (Table S1).
Expression analysis of mature miRNAs through quantitative real-time RT-PCR (qPCR)
Total RNAs were isolated from various tissues (same as those used in miRNA microarray hybridizations), treated with RNase-free DNaseI (MBI Fermentas, Lithuania), and then polyadenylated (2 μg) by poly(A) polymerase (NEB, USA) at 37°C for 1 h in a 20-μl reaction mixture. After phenol–chloroform extraction and ethanol precipitation, the RNAs were dissolved and reversely transcribed using poly(T) adapter (41). qPCR was performed using SYBR® Green Realtime PCR Master Mix (Toyobo, Japan) and all the primers used were listed in Table S2. The values of threshold cycle (CT, the fractional cycle number at which the fluorescence passes the fixed threshold), were calculated by Rotor-Gene 6 software (Corbett Robotics, Australia). CT values were converted into relative copy numbers using a standard curve (42). As reference gene, 5.8S rRNA was used in qPCR detection of miRNAs in Arabidopsis previously (41). To check whether rice 5.8S rRNA is suitable, we firstly surveyed the expression level of 5.8S rRNA and some other suggested reference genes (43) in GeneChip® rice genome array, and the results showed that the expression levels of these genes were relative stable (Supplementary Figure 1A). Further, expression level of 5.8S rRNA was characterized by qPCR using rice Actin gene (Os03g50890) as reference gene (43), and results confirmed the stable expression of 5.8S rRNA in the samples used in the study (Supplementary Figure 1B). These indicated that it is suitable using rice 5.8S rRNA to normalize the qPCR data.
In addition, reaction efficiencies of qPCR assays for individual miRNAs were determined using Rotor-Gene 6 software. A 10-fold dilution series of purified product of the tested miRNA (or guide genes) were used as the template for qPCR to generate a plot of Log copy numbers of the tested miRNA (or guide genes) versus the corresponding CT as described (41). The slope of the linear plot is defined as –(1/log (E + 1), where E is the reaction efficiency. In Rotor-Gene 6 software, reaction efficiency, whose optimal value is 1, is defined as amplification value –1. The results showed that the reaction efficiency of tested miRNAs approach the optimal value (Supplementary Figure 2).
Sequence alignment and phylogenetic analysis
Precursor sequences of miR169 and miR396 families were aligned with ClustalW (http://www.ebi.ac.uk/Tools/clustalw2; 44), and the conserved fragments were visualized with the help of GeneDoc program (http://www.nrbsc.org/gfx/genedoc). To construct the phylogenetic tree of AGO proteins, the protein sequences were aligned using ClustalW (44), and MEGA4 (45) was employed to generate the phylogenetic tree using neighbor-joining method (46) and Poisson correction method (47). The bootstrap test of 1000 replicates was performed.
RESULTS
sRNAs from rice seeds matched classes of genomic features
RNAs extracted from rice seeds at different developmental stages were used to isolate short RNAs, which were then used to construct the library. Subsequent massively parallel signature sequencing (MPSS) analysis resulted in the obtainment of 797 399 22-nt sequences (or ‘signatures’) after removing low-quality ones, and 111 161 of them were distinct (Figure 1A). Analysis showed that 70.5% of distinct signatures matched the rice genome (the Institute for Genomic Research, TIGR, version 5.0), representing 93.8% of the total signatures (Table S3). There was only a small part of signatures (2.3%) that matched the chloroplast or mitochondria genome (Table S3). Around 9.7% of the distinct signatures matched noncoding RNAs (ncRNAs) including ribosomal rRNAs, tRNAs, snoRNAs and snRNAs, which account for 61.5% of the total signatures (Table 1), similar to the previous report in rice (21). All the genome-matching signatures excluding ncRNAs were then regarded as sRNAs and used for further analysis.
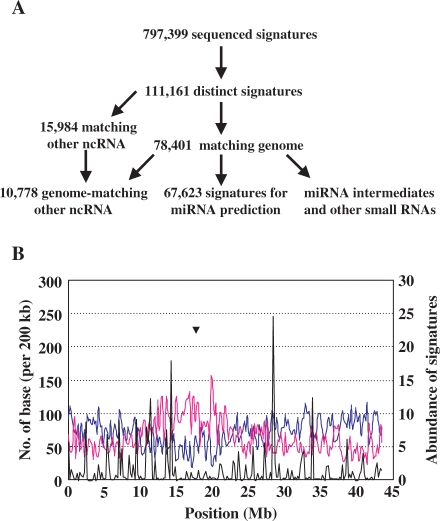
Figure 1.
Analysis approaches and distributions of small RNAs in rice seeds. (A) A flow diagram showing the analysis process of sRNAs in rice seeds. Obtained signatures through MPSS were compared to RNAs from various RNA databases and those do not match the known RNAs were aligned on the genome to extract the flanking sequences for miRNA prediction. (B) Distributions of sRNAs on rice chromosome 1. The x-axis indicates the position on chromosome 1 (Mb). The left y-axis indicates the length of expressed genes (blue line) and repetitive sequence (pink line) per 200 kb of the chromosome. The right y-axis indicates the abundance (per 200 kb) of total sRNAs (black line) on the chromosome. Abundance of each signature was normalized by location numbers on the genome. Triangle indicates the position of centromere.
Table 1.
Summary of signatures that match various RNAs
| Locus class | Distinct signatures | Total signatures | Mean frequencya | Mean hitsb |
|---|---|---|---|---|
| Nonprotein-coding RNAs | ||||
| snoRNA | 20 (0.02%) | 56 (0.01%) | 2.8 | 2.1 |
| snRNA | 293 (0.26%) | 5413 (0.68%) | 18.47 | 10.08 |
| tRNA | 1133 (1.02%) | 38 404 (4.82%) | 33.9 | 8.56 |
| rRNA | 9332 (8.40%) | 446 596 (56.01%) | 47.86 | 5.52 |
| Small RNAs matching protein-coding genes | ||||
| Sense | 10 429 (9.38%) | 62 041 (7.78%) | 5.95 | 21.35 |
| Antisense | 5896 (5.30%) | 20 805 (2.61%) | 3.53 | 38.87 |
| Sense and antisense | 1032 (0.93%) | 4885 (0.61%) | 4.73 | 341.69 |
| miRNAs | ||||
| Known | 228 (0.21%) | 4029 (0.50%) | 17.67 | 380.96 |
| Novel | 328 (0.30%) | 7359 (0.92%) | 22.44 | 2.64 |
| Other small RNAs | 82 470 (74.2%) | 207 811 (26.1%) | 2.52 | – |
| Total | 111 161 (100%) | 797 399 (100%) | 7.17 | – |
aThe average frequency of sequenced signatures matching different type RNAs.
bThe average hit numbers on the chromosomes of signatures.
The mean hits of ‘other small RNAs’ and total small RNAs are not calculated as some signatures without hit on the genome, which are indicated as ‘–.’ Numbers in brackets indicate the percentage of the distinct signatures or total signatures.
In rice seeds, signatures matching miRNA precursors account for <1.5% of the total abundance of genome-matching signatures, which is much lower than that in rice inflorescences, seedlings and stems (24), and Arabidopsis flowers and seedlings (22). This may because rice genome contains many interspersed and inverted repeat sequences, which produce abundant siRNAs (24). Additionally, 15.6% of the distinct sRNAs match protein-coding genes in sense and/or antisense orientation (Table 1), and the corresponding encoded proteins involve in various molecular functions, biological processes and cellular components, suggesting the complexity of sRNAs’ origin (Table S4).
In addition, 70.7% of the genome-matching sRNAs locate on genome only once (the location numbers account for 4.5% of the total locations), while ∼3% sRNAs match the genome for more than 100 times (Table S5). Characterization of the distribution of sRNAs, protein-encoding genes and repetitive sequences on chromosome shows that some loci have high propensity to produce sRNAs, and these sRNA hotspots originated preferentially from the regions other than centromeres (Figure 1B, Supplementary Figure 3).
Most sRNAs in rice seeds originated from transposable elements
A total of 81 530 sRNA clusters were identified depending on the distance between every two signatures, and 72 173 of them contain interspersed repeats (annotated by RepeatMasker, http://www.repeatmasker.org; Table S6), 2967 were marked as tandem repeats (by Tandem repeats finder; 48), indicating that repeat-associated siRNAs (rasiRNAs) take up a large portion of sRNAs. The presence of highly abundant rasiRNAs in seeds is consistent with their roles in regulating transposon activity during seed development. Further identification of the top 50 hotspots, according to the normalized abundances of sRNAs, showed that tandem repeat loci inclined to generate sRNAs at high abundance. Nineteen of the top 50 hotspots contain tandem repeats, while only 3.6% of the total sRNA clusters were tandem repeats. This supported the generation mechanism of sRNAs from tandem repeats, proposed by Martienssen (49). In addition, some miRNAs including osa-MIR168a, osa-MIR172b, and osa-MIR159a were also found with high abundance in sRNAs hotspots (Table S7).
Validation of registered miRNAs using sRNA signatures
One of the most critical criteria for miRNA annotation is that the miRNA should originate from a single-strand RNA transcript (27). However, some miRNA precursor sequences of registered miRNAs matched the short signatures in both sense and antisense orientations when the sRNAs sequences were mapped onto the precursor sequences (version 11.0), suggesting that they might not be bona fide miRNAs. In addition, most loci of these miRNAs appear to be repetitive sequences. Among the 269 miRNA loci, 91 precursors were annotated as transposable element repetitive sequences, and most of them were miniature inverted-repeat transposable elements (MITEs, Table S8; 50).
sRNAs that match miRNA precursors can be grouped into several classes (Table S9). For bona fide miRNAs, most of the sRNAs should originate from the positions of mature miRNAs or miRNA*s at sense orientation. Our analysis on the sRNAs sequences from rice seeds (present study), flowers, seedlings and stems (24) showed that the repetitive sequences-containing miRNA loci were different from those without repetitive sequences in at least two aspects. First, the antisense sRNAs generated from the miRNA loci containing repetitive sequences were comparable to sense sRNAs; second, many sRNAs from these loci were generated from the sites other than mature miRNAs or miRNA*s (Table S9). This indicated that the annotated repetitive sequences-containing miRNA loci might be either sRNA clusters producing sRNAs randomly from two strands, or borderline miRNA candidates (8). It is worthy to notice that three miRNAs (osa-MIR531, osa-MIR1426 and osa-MIR1431), which generated sRNAs only from the mature miRNA positions, were also annotated as repetitive sequences (Table S9). Therefore, during the miRNA annotation in the present study, all the sequences containing repetitive sequences or putative precursors matching sRNAs in two orientations at similar abundance were excluded.
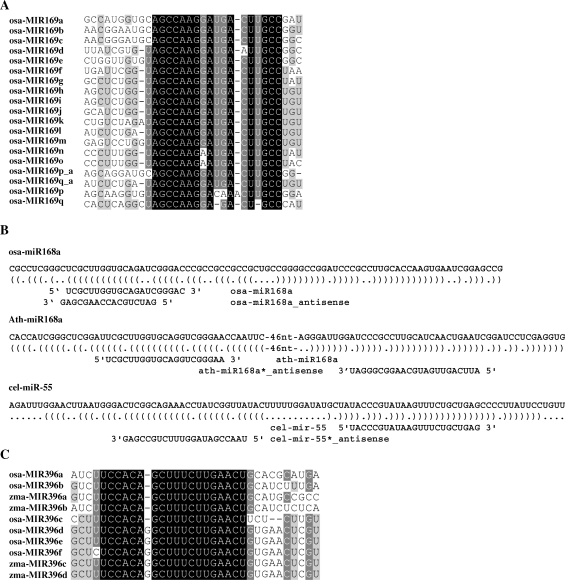
Two annotated miRNA loci, osa-MIR169p and osa-MIR169q, were significantly different from others (Table S9), as they generated many sRNAs in antisense orientation from the sites of mature miRNA and miRNA*. Among the generated sRNAs, some are uniquely mapped on the genome, indicating these two miRNAs may be transcribed from the antisense orientation of the annotated miRNA loci. Alignment on the precursor sequences of all osa-miR169 family members and the reversely complemented sequences of osa-MIR169p and osa-MIR169q showed that the reversely complemented sequences of osa-MIR169p and osa-MIR169q were conserved with osa-MIR169a-o (Figure 2A). This indicated that directions of osa-MIR169p and osa-MIR169q might be mis-annotated in miRBase (version 11.0).
Figure 2.
Small RNAs match miRNA precursors. (A) The directions of osa-MIR169p and osa-MIR169q may be mis-annotated. Annotated precursor sequences of osa-miR169 family, together with reverse-complemented sequences of osa-MIR169p and osa-MIR169q, were aligned. osa-MIR169p_a and osa-MIR169q_a indicated the reverse complemented sequences of osa-MIR169p and osa-MIR169q. ClustalW and GeneDoc were used for sequences alignment and visualizing the conserved fragments. Black, dark gray and light gray shading indicated that the nucleotides were conserved in all, more than 80% (<100%), or more than 60% (<80%), of the sequences, respectively (same for C). (C) sRNAs matched mature miRNA or miRNA* in antisense orientation. The miRNA hairpins were shown in bracket notation. Precursors and mature miRNAs were shown in 5′ to 3′ orientation, whereas antisense sRNAs were shown in 3′ to 5′orientation. A. Subgroups of miR396 family in rice and maize. osa-miR396f and zma-miR396c/d were identified in this study.
Many conserved miRNA families have multiple paralogous loci, and some of them generate same mature miRNAs, making it difficult to identify from which locus the mature miRNAs were generated. In addition, detection of miRNA*s and rare side products of miRNA loci will help to confirm the expression of unique locus (11). Through mapping the signatures obtained from seeds and other tissues (8,24) onto the miRNA precursors, 96 loci from 47 miRNA families were confirmed using signatures uniquely hitting on the genome (Table S10). In addition, our analysis showed that the unique signatures mapping osa-miR156j and osa-miR166j were identical to those mapping osa-miR156h and osa-miR166i, respectively. Considering that osa-miR156j and osa-miR166j were not successfully mapped on the genome (TIGR 4.0), the loci of them might be identical to osa-miR156h and osa-miR166i.
Among the unique signatures matching miRNA precursors, several ones matched the precursors in antisense orientation. The unique origin of each signature on the genome suggested that it might be produced exclusively from this site (Table S11). Similarly, analysis on the precursor sequences from Arabidopsis (from Arabidopsis Small RNA Project, ASRP, http://asrp.cgrb.oregonstate.edu/) and C. elegans (GenBank data libraries, GEO accession number GSE5990, sample GSM139137) showed that this phenomenon is common in various species, and many sRNAs can match mature miRNAs or miRNA*s in antisense orientation (Figure 2B). Recent report showed that a single Hox locus in Drosophila produces functional miRNAs from opposite DNA strands (51), indicating this phenomenon is conserved throughout the animal and plant kingdoms.
Identification of 26 novel and 12 candidate miRNAs
To identify the novel miRNA loci, the consensus surrounding genomic sequence of each sRNA from this study was extracted, and secondary structure of each sequence was predicted with the help of Mfold program (28). A Perl script was used to evaluate the paired regions of predicted RNA secondary structures according to the criteria by MIRcheck (38). Sequences of putative miRNA loci were then analyzed by RepeatMasker to exclude the repetitive sequences. Those contained no repetitive sequences were regarded as candidate precursors and used for further analysis.
As previously reported, accumulations of miRNA*s were strong supporting evidences for the cleavage by DCLs during the miRNA biogenesis (11), thus the miRNA identification in this study is mainly dependent on the detection of miRNA*s. Eleven novel miRNA loci were finally identified, excluding three miRNAs (osa-miR810b.1, osa-miR1429 and osa-miR1432; references 8 and 26, which were published during the manuscript revision). Interestingly, the abundance of osa-miR1429* (126 TPQ) were higher than osa-miR1429 (15 TPQ) in seed library, suggesting that the accumulation of miRNA*s may differ in different tissues. Further, sRNAs sequences from other six libraries reported by Lu et al. (8) were used to check the presence of miRNA*s, resulting in the identification of eight other miRNA loci (Table 2).
Table 2.
Sequences of novel miRNAs
| miRNA familya | Sequence (5′-3′) | Length | Loci | Abundance | Evidenceb |
|---|---|---|---|---|---|
| osa-miR1866-3p | UGAAAUUCCUGUAAAAUUCUUG | 22 | 1 | 426 | Star |
| osa-miR1846e | CAACGAGGAGGCCGGGACCA | 20 | 1 | 125 | Star, EST |
| osa-miR1428e-5p | UAAGAUAAUGCCAUGAAUUUG | 21 | 2 | 121 | Star, EST |
| osa-miR1846-5p | CAGUGAGGAGGCCGGGGCCGCU | 22 | 3 | 28 | Star, EST |
| osa-miR1874-5p | UAGGGCUACUACACCAUCCAUA | 22 | 1 | 21 | Star |
| osa-miR2090 | AACUCUGAUUCUAGAAUUUUUG | 22 | 1 | 19 | Star |
| osa-miR2091-5p | UCAACCGAGCCGAGGAGGAGG | 21 | 1 | 17 | Star |
| osa-miR2091-3p | CAUACAUUGCCUCCUAGGCUUG | 22 | 1 | 10 | Star |
| osa-miR2092-5p | CAACUGAAGUCGGUGUUUACU | 21 | 1 | 10 | Star |
| osa-miR2092-3p | ACCAGCAUUCCAUUGGCAGAGG | 22 | 1 | 5 | Star |
| osa-miR2093-3p | ACACAUCUUCCAAUUAAUGCAU | 22 | 1 | 9 | Star |
| osa-miR2093-5p | GUGCAUUAAUUGGAAGAACA | 20 | 1 | 7 | Star |
| osa-miR2094-3p | CAGAGCUGUGGCAUCCACGUCG | 22 | 1 | 6 | Star |
| osa-miR2094-5p | UGGCUGCUAGGCUCCUGGGUG | 21 | 1 | 4 | Star |
| osa-miR2095-3p | CUUCCAUUUAUGAUAAGUAU | 20 | 1 | 5 | Star |
| osa-miR2095-5p | CUGAUAAUUUUACGAUGAAUAG | 22 | 1 | 4 | Star |
| osa-miR2096-3p | CCUGAGGGGAAAUCGGCGGGA | 21 | 1 | 5 | Star |
| osa-miR2096-5p | UGCCGAUUUCCCCCUCGGGCG | 21 | 1 | 3 | Star |
| osa-miR1850-5p.2 | UUGUGUGUGAACUAAACGUGG | 21 | 1 | 106 | Star |
| osa-miR1850-3p.2 | CUGUUUAGUUCACAUCAAUCUU | 22 | 1 | 4 | Star |
| osa-miR2097-3p | UUCUCUUCUUCGUGUCGCAUUU | 22 | 1 | 3 | Star |
| osa-miR2097-5p | AGAGAUGGGACGGGCAGGGAAG | 22 | 1 | 1 | Star |
| osa-miR2098-3p | CGGUUUGUCAAGCGGAGUGC | 20 | 1 | 3 | Star |
| osa-miR2098-5p | UCCCGUGGAGGCAGCCGAUG | 20 | 1 | 1 | Star |
| osa-miR2099-5p | UGAAUAUGUUUGUACAAGCUUU | 22 | 1 | 7 | Star |
| osa-miR2099-3p | ACAAAGCUGUAGCGUUAUUC | 20 | 1 | 2 | Star |
| osa-miR2100-5p | UUCUCUCAAGUUGCCAAACAAG | 22 | 1 | 7 | Star |
| osa-miR2100-3p | AACCGCUGUUUAGGCGGAGUGG | 22 | 1 | 2 | Star |
| osa-miR2101-3p | AUUUAACUCAAGUGAGCAUUGU | 22 | 1 | 54 | Star |
| osa-miR2101-5p | ACAUGUUUACAAGUUAAAAUGU | 22 | 1 | 1 | Star |
| osa-miR2102-3p | CAUGGUGCCGGUUCCGGUGGCG | 22 | 1 | 7 | Star |
| osa-miR2102-5p | GGGCAAGCCGCCGCCGCCAC | 20 | 1 | 1 | Star |
| osa-miR396f | UCUCCACAGGCUUUCUUGAACU | 22 | 1 | 138 736 | EST, High |
| osa-miR2103 | UUUCCCUCUCCGUGCGCGCUCG | 22 | 1 | 54 | EST, High |
| osa-miR2104 | GCGGCGAGGGGAUGCGAGCGUG | 22 | 1 | 7 | EST |
| osa-miR2105 | UUGUGAUGUGAAUGAUUCAU | 20 | 1 | 5 | EST |
| osa-miR2106 | CCGAGGUUUUCUGGAUACAUU | 21 | 1 | 5 | EST |
| osa-miR1859 | UUUCCUAUGACGUCCAUUCCAA | 22 | 1 | 164 | High |
| osa-miR827b | UUAGAUGACCAUCAGCAAACA | 21 | 1 | 154 | High |
aSome precursors produce miRNAs from two arms at similar abundance, and both sequences are shown.
bEvidence of identified miRNA. ‘EST’ indicates that the precursor was previously sequenced. ‘Star’ indicates the detection of the corresponding miRNA*. ‘High’ indicates the abundance of the corresponding miRNA is higher than 50 TPQ.
Two candidate precursors producing sRNAs at very high abundance (>180 TPQ) were identified as miRNAs (osa-miR827b and osa-miR1859), of which osa-miR827b was the ortholog of Arabidopsis ath-miR827. Comparison of the candidate precursors with rice ESTs showed that five previously annotated noncoding RNAs were indeed miRNAs. Six precursors, which had matched ESTs and were previously annotated as ‘conserved hypothetical protein’, were identified as miRNA candidates (osa-miRc1-c6). Additionally, six candidate miRNAs with abundance >15 TPQ were identified. As a whole, 26 novel (Table 2) and 12 candidate miRNAs (Table S12) were identified from this study (predicted structures were shown in Supplementary Dataset 1).
Among the novel miRNAs, osa-miR396f was a homolog of osa-miR396 family, which was one of the two novel miRNAs having orthologs in other species. As shown in Figure 2C, miR396 genes can be grouped into two subgroups according to sequences of mature miRNAs. The two annotated novel miRNAs in maize, zma-miR396c from AZM5_13 014 and zma-miR396d from AZM5_11 578, were more conserved with osa-miR396d-e than osa-miR396a-c.
The osa-miR827b was conserved in both rice and Arabidopsis. In miRBase (V11.0), osa-miR827 was recently identified through similarity analysis (52), however, no sRNA from the mature miRNA or miRNA* sites of osa-miR827 precursor was obtained in seed library or other public libraries (8). In contrast, the mature miRNA from osa-miR827b loci was abundant in seed library (150 TPQ), suggesting that osa-miR827b (rather than osa-miR827) has existent physiological functions.
Targets of these identified miRNAs were further predicted using the methodology described by Allen et al. (4), and targets of 18 novel miRNAs were captured with assignment of mispair score and MFE ratio tolerance limits to ≤4 and ≥0.73, respectively. Analysis and annotation of the predicted target genes using GO terms showed that they were with diverse functions (Tables 3 and S13), and involved in multiple cellular processes including response to stress, signal transduction and metabolic processes (Table S14).
Table 3.
Predicted target genes of novel miRNAs
| miRNA | Target | MFE | Score | Annotation |
|---|---|---|---|---|
| osa-miR1866-3p | Os09g21110 | 0.775 | 4 | Leucyl-tRNA synthetase, cytoplasmic |
| osa-miR1846e | Os02g04100 | 0.858 | 3 | Proteasome subunit alpha type 1 |
| osa-miR1846-5p | Os10g12320 | 0.977 | 0.5 | Hypothetical protein |
| osa-miR1846-5p | Os08g10350 | 0.841 | 2.5 | Expressed protein |
| osa-miR2091-5p | Os06g48300 | 0.823 | 3.5 | Protein phosphatase 2C isoform epsilon |
| osa-miR2092-5p | Os12g17660 | 0.881 | 2 | Hypothetical protein |
| osa-miR2093-5p | Os02g45054 | 0.786 | 3 | INDETERMINATE-related protein 9 |
| osa-miR2094-5p | Os08g14440 | 0.794 | 3 | Expressed protein |
| osa-miR2096-5p | Os09g37834 | 0.745 | 4 | Serine/threonine-protein kinase receptor precursor |
| osa-miR1850-3p.2 | Os06g05440 | 0.735 | 4 | Expressed protein |
| osa-miR1850-5p.2 | Os07g07530 | 0.812 | 4 | Expressed protein |
| osa-miR2097-5p | Os02g02680 | 0.846 | 3 | NBS-LRR disease resistance protein |
| osa-miR2098-3p | Os05g08950 | 0.873 | 3 | Bifunctional protein tilS/hprT |
| osa-miR2098-3p | Os12g38560 | 0.790 | 3 | Expressed protein |
| osa-miR2098-3p | Os11g05930 | 0.971 | 3.5 | CCT motif family protein |
| osa-miR2098-5p | Os08g37940 | 0.815 | 2.5 | HAD-superfamily hydrolase subfamily IA |
| osa-miR2098-5p | Os02g48880 | 0.841 | 3 | Chloride channel protein CLC-f |
| osa-miR2100-5p | Os01g28810 | 0.913 | 2 | Expressed protein |
| osa-miR2100-5p | Os03g31320 | 0.768 | 3 | RING zinc finger protein-like |
| osa-miR2102-3p | Os08g43160 | 0.884 | 3 | TCP family transcription factor containing protein |
| osa-miR2102-5p | Os04g59470 | 0.932 | 2.5 | NAC domain-containing protein 76 |
| osa-miR2102-5p | Os04g31290 | 0.919 | 2 | Helix-loop-helix DNA-binding domain containing protein |
| osa-miR2102-5p | Os05g25390 | 0.866 | 3 | Protein kinase |
| osa-miR2102-5p | Os01g12280 | 0.852 | 3 | Protein dimerization |
| osa-miR2103 | Os09g20920 | 0.905 | 2 | Hypothetical protein |
| osa-miR2103 | Os01g50190 | 0.905 | 2 | Hypothetical protein |
| osa-miR2103 | Os05g37960 | 0.901 | 1 | Indole-3-acetic acid-amido synthetase GH3.2 |
| osa-miR2103 | Os08g41230 | 0.867 | 3 | BTB/POZ domain containing protein |
| osa-miR2103 | Os03g47920 | 0.783 | 3.5 | Hypothetical protein |
| osa-miR2104 | Os06g11120 | 0.899 | 3 | Conserved hypothetical protein |
| osa-miR2104 | Os10g27380 | 0.826 | 2.5 | Expressed protein |
| osa-miR2104 | Os03g10640 | 0.805 | 3.5 | Calcium-transporting ATPase 2 |
| osa-miR2105 | Os03g39520 | 0.798 | 3.5 | Conserved hypothetical protein |
| osa-miR2105 | Os12g13170 | 0.763 | 3 | Transcription factor HBP-1a |
| osa-miR2105 | Os03g11140 | 0.848 | 3.5 | RHO GTPase activator |
| osa-miR2106 | Os07g41694 | 0.791 | 4 | Acidic leucine-rich nuclear phosphoprotein 32 family |
| osa-miR827b | Os04g48390 | 0.851 | 3 | beta-lactamase, class A |
‘MFE’ indicates the minimum free energy ratio of miRNA:target duplex and target:complementary sequence duplex.
Preferential expression of miRNAs in specific tissues
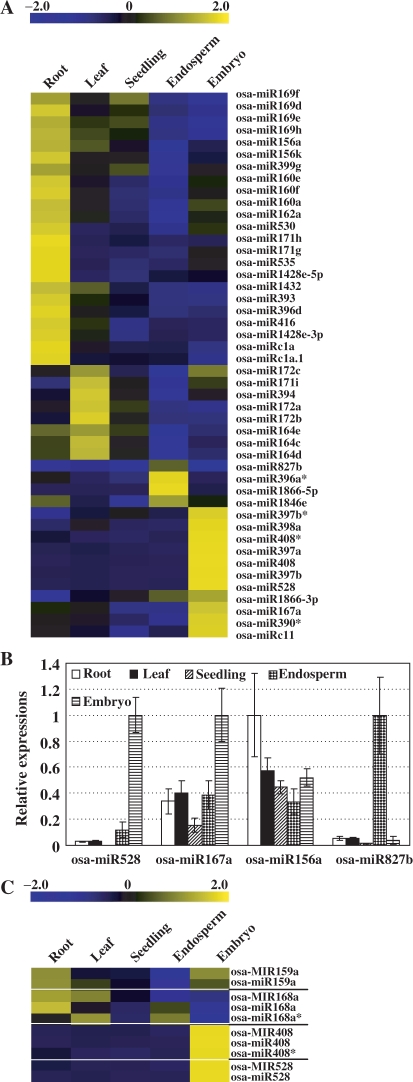
Further, miRNAs chips containing the previously and newly identified miRNAs were generated to study the expression profiles of miRNAs. Several detected miRNA*s by this study were also included on the chips. To analyze the miRNA expression in seeds in detail, RNAs from embryo and endosperm were extracted individually and used for hybridization. In addition, expression profiles of miRNAs in three vegetative tissues (root, leaf and seedlings) were analyzed and compared with those in seed tissues (Table S15). Analysis showed that most miRNAs were expressed preferentially in one or two tissues, and only some of them were highly expressed in most tested tissues (Figure 3A).
Figure 3.
Expression patterns of the known and novel miRNAs. (A) Some miRNAs were preferentially expressed in specific tissues. Several miRNA*s were included. The bar represented the scale of relative expression levels of miRNAs (Log2). (B) Quantitative real-time RT-PCR analysis on the relative expressions of miRNAs in various tissues. The copy numbers of miRNAs were normalized by comparing to that of rice 5.8S rRNA, and expression levels of each miRNA were then normalized by comparing to the highest expression in the tested tissues, which was set as 1.0. The experiments were repeated for three times and error bar represented SD. (C) Close relation of the expression levels of pri-miRNAs (labeled as osa-MIRxxx, by GeneChip® microarray hybridization) and corresponding mature miRNAs (labeled as osa-miRxxx, by miRNA chips). The bar represented the scale of relative expression levels of pri-miRNAs or mature miRNAs (Log2). The white lines compart the pri-miRNAs and the corresponding mature miRNAs into groups.
To corroborate the expression profiles obtained from chip hybridization, quantitative real-time RT-PCR (qPCR) was performed to study the transcripts of several mature miRNAs. In previous report, 5.8S rRNA, which can be polyadenylated by poly(A) polymerase at the 3′-end as mature miRNAs, was used as reference gene in qPCR analysis of miRNAs expression in Arabidopsis (41). Detection on the expression levels of rice 5.8S rRNA from GeneChip® genome array or by qPCR analysis showed that it was relative stable in the tested tissues (Supplementary Figure 1). We thus select rice 5.8S rRNA as reference gene in the analysis. Results showed that the tested miRNAs were indeed preferentially expressed in specific tissues (Figure 3B, Supplementary Figure 4A), being consistent with the data from miRNA chip hybridization. Comparison of the miRNAs expression patterns revealed by chip hybridization with the data of a recent publication (GSE11014; 53), in which sRNAs of four tissues were sequenced using 454 technology, revealed the consistence of the chip data with the differentially expressed miRNAs from 454 dataset, after filtering out the low abundance sequences (Supplementary Figure 4B). These indicated that the expression profiles of miRNAs by microarray hybridization were accurate and reliable.
Expression patterns of miRNAs were closely related to their functions. osa-miR160, osa-miR171g/h and osa-miR393 were preferentially expressed in root, and all of their Arabidopsis orthologs were involved in root development through targeting auxin-response factors (ARFs) 10 and 16 (ath-miR160; 54), Scarecrow-like gene (ath-miR171; 55) or transport inhibitor response 1 (TIR1, ath-miR393; 1). osa-miR172, whose Arabidopsis ortholog targets APETALA2 (AP2) to regulate leaf development (1), was preferentially expressed in leaf.
Several miRNAs, including osa-miR167, osa-miR397, osa-miR398, osa-miR408, osa-miR528, osa-miR1866-3p and osa-miRc11, were preferentially expressed in rice seeds. Putative targets of these miRNAs involved in diverse developmental and metabolic processes, suggesting the crucial functions of them in rice seed development. In Arabidopsis and rice, miR167 targeted both ARF6 and ARF8 (16,56), suggesting that osa-miR167 might involve in rice seed development through regulating auxin signaling.
Additionally, four miRNA families (osa-miR397, osa-miR398, osa-miR408 and osa-miR528) that were highly expressed in embryo targeted several genes encoding copper-binding proteins (17,20,38; Table S16). miR397, miR398 and miR408 were conserved between Arabidopsis and rice, and those in Arabidopsis were involved in copper homeostasis regulation by guiding the cleavage of mRNAs of plantacyanins, copper/zinc superoxide dismutases and laccases, respectively (57,58). This also suggested the possible roles of copper homeostasis in seed development. Other miRNAs highly expressed in rice seeds targeted proteasome subunit, beta-lactamase (Table 3), suggesting the functions of miRNAs through regulating protein modification, secondary metabolism and other metabolic pathways.
During the study, it was noticed that the precursors of four miRNAs (osa-MIR159a, osa-MIR168a, osa-MIR408 and osa-MIR528) were represented by probes on GeneChip® rice genome array. As miRNAs were transcribed by RNA polymerase II, and capped, polyadenylated as normal mRNAs (9), the pri-miRNAs can hybridize with the probes as normal mRNAs. Comparison of the expression levels of these four mature miRNAs with those of the corresponding pri-miRNAs showed the close correlation of them (Figure 3C), suggesting the consistent transcriptions of pri-miRNA and mature miRNA. In addition, this also confirmed the expression profiles of mature miRNA revealed by chip hybridization.
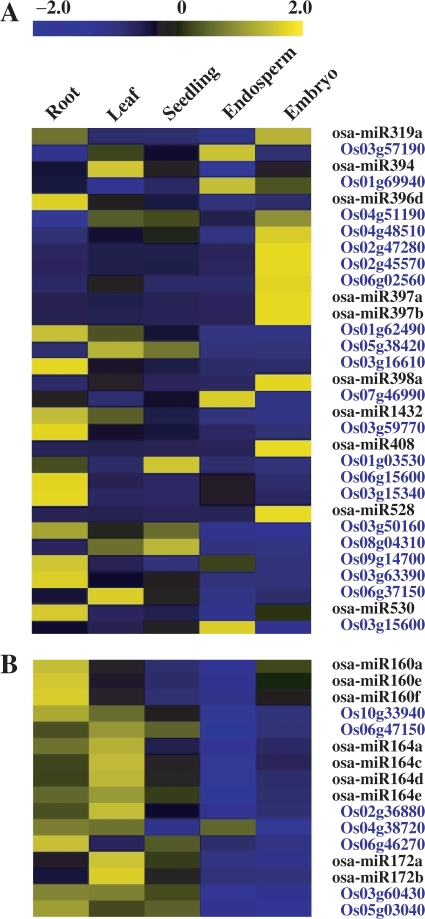
Negative correlation of most miRNAs with the corresponding targets
Expression profiles of miRNAs were compared with those of the corresponding targets (all the targets of known and novel miRNAs used in the analysis were listed in Table S16), which were analyzed through hybridizing the Affymetrix Rice genome DNA chip. The results showed that the expression of most miRNAs was negatively correlated with their targets (Figure 4A), being consistent with their functions in guiding the cleavage of target mRNAs in plants (1,59,60). In addition, some miRNAs (osa-miR160, osa-miR164 and osa-miR172) were positively correlated with their targets (Figure 4B), which may due to the consequence of feedback regulation (61). As reported in Arabidopsis, both MIR164A and CUC2 are transcribed at the margins of young leaf primordia, and the balance between them determines the extent of serration (62). The positive correlations of miRNAs and their targets also suggested that these miRNAs may function by translational repression (60), and indicated that the regulation mechanisms of conserved miRNAs, for example miR164 and miR172, were conserved throughout monocot and dicot plants.
Figure 4.
Expression correlations of miRNAs and the corresponding targets. In each miRNA/target pair (blue characters), the LOC prefixes of TIGR locus identifiers were removed for convenience. The bar represented the scale of relative expression levels of miRNAs or targets (Log2). (A) miRNAs expresssions negatively correlated to those of the corresponding targets. The abundances of targets were low in the tested tissues while miRNAs were highly accumulated. (B) miRNAs expresssions postively correlated to those of the targets. Both the targets and miRNAs were highly accumulated in the same tissue(s).
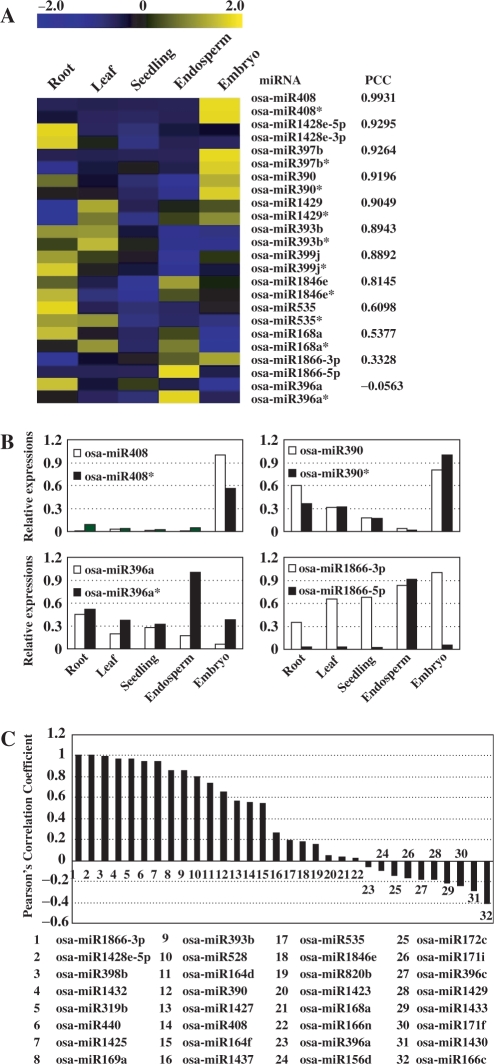
Differential regulations of miRNAs and miRNA*s
Besides being sequenced, some miRNA*s were detected with high-level expressions in several tissues by chip hybridization. As shown in Figure 5A, some mature miRNAs and corresponding miRNA*s accumulated highly in same tissue(s) (osa-miR408, osa-miR1428e-5p and osa-miR397b). This was reasonable as both mature miRNAs and corresponding miRNA*s were produced from same precursors. However, the mature miRNAs and miRNA*s of some miRNAs (osa-miR168a, osa-miR1866-3p and osa-miR396a) accumulated differently in these tissues, suggesting the differential regulation of mature miRNAs and miRNA*s. In addition, for some miRNAs, expression level of miRNA* were even higher than the corresponding miRNAs (Figure 5A and B).
Figure 5.
The correlated expressions of miRNAs and miRNA*s. (A) Correlation of the abundance between miRNAs and miRNA*s was presented by Pearson's correlation coefficient (PCC). The bar represented the scale of the relative expression levels of miRNAs and miRNA*s (Log2). (B) Relative expression levels of miRNAs and miRNA*s. For osa-miR408 and osa-miR390, miRNAs and miRNA*s accumulated highly in same tissues. For osa-miR396a and osa-miR1866-3p, differential accumulation patterns of miRNAs and miRNA*s were detected. Expression levels of miRNAs and miRNA*s were normalized by comparing to the highest expression among them, which was set as 1.0. (C) Pearson's correlation coefficient (PCC) of the abundance between miRNAs and miRNA*s in seven sRNAs libraries. sRNAs originated from multiple precursors were filtered out. The IDs of miRNAs were listed in lower panel.
To confirm our observation, sRNAs from seeds and other six tissues (8) were surveyed to compare the expression profiles of miRNAs and miRNA*s. All the expression abundances of sRNAs were normalized to transcripts per quarter million (TPQ) after filtering out signatures corresponding to rRNAs, tRNAs, snRNAs and snoRNAs, to ensure the normalized values were comparable across the tissues (22,63). Being consistent with the results from chip hybridization, the abundance relations of mature miRNAs and miRNA*s were diverse in these tissues. For osa-miR408 and osa-miR390, mature miRNAs and miRNA*s were closely related, while for osa-miR168a and osa-miR396a, mature miRNAs and miRNA*s accumulated differently in these tissues (Figure 5C).
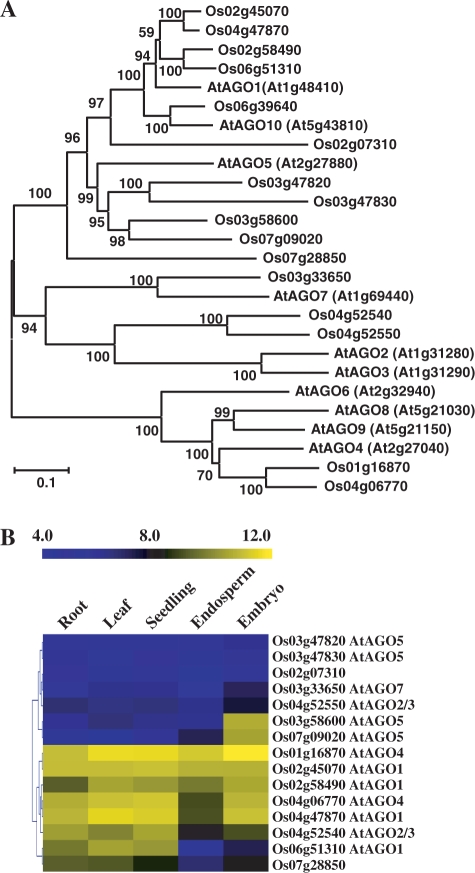
The differential accumulation of miRNAs and miRNA*s in specific tissue may due to the different activities of proteins directly involved in sRNAs biogenesis. Recent report showed that different argonaute (AGO) proteins in Arabidopsis harbor sRNAs with different 5′ terminal nucleotide, and the miRNAs and miRNA*s are directed in different AGO complexes based on their 5′ terminal nucleotide (64). Several homolog AGO proteins were identified in rice (Figure6A; 65) and our analysis showed the different expression levels of them in the tested tissues (Figure 6B). Especially, two AGO-coding genes (Os03g58600, Os07g09020) homologous to AtAGO5 were predominantly expressed in rice embryo, and an AGO-coding gene (Os06g51310) homologous to AtAGO1 was low expressed in rice endosperm and embryo. The variety of AGO gene abundances in these tissues may determine the abundance of miRNA and miRNA*s in RISCs.
Figure 6.
Phylogenetic and expression pattern analysis of rice AGOs. The LOC prefixes of all TIGR locus identifiers were removed for convenience. (A) Phylogenetic analysis of the Arabidopsis and rice AGO proteins. The protein sequences of AtAGO1 to AtAGO10 were obtained from the Arabidopsis Information Resource (http://www.arabidopsis.org). Rice protein sequences were from the Institute for Genomic Research. ClustalW (www.ebi.ac.uk/Tools/clustalw2) was used to align the sequences, and MEGA4 was used to generate the phylogenetic tree using the neighbor-joining and the Poisson correction methods. Bootstrap values were calculated with 1000 permutations. The scale bar represented 0.1-amino-acid substitutions per residue. (B) Expression profiles of rice AGO genes. The most homologous Arabidopsis genes were labeled. The bar represented the scale of relative expression levels of AGO genes and the expression signals were Log2 transformed.
DISCUSSIONS
MPSS analysis using rice seeds at different developmental stages was performed and turned out to be effective in identifying a large number ofsRNAs including novel miRNAs. Several recent studies have reported the large-scale sequencing of sRNAs in rice (8,24–26); however, most of them were obtained from the tissues other than seeds. In the study by Lu et al. (8), only sRNAs with the abundance more than 100 TPQ were used to predict the putative precursors, which may miss some uncharacterized miRNAs. In present study, all the sRNAs (excluding rRNAs, tRNAs, snRNAs and snoRNAs) were analyzed (Figure 1A), and as a result, 26 novel miRNAs were identified mostly dependent on the accumulations of miRNA*s. The targets of 18 novel miRNAs were predicted, and hybridization with the generated miRNA chip provided a detailed expression profile of known and novel miRNAs in vegetative (root, seedling, young leaf) and reproductive (embryo and endosperm) tissues. These will significantly contribute to the discovery and studies of miRNAs in rice, especially rice seeds.
Most rice sRNAs are derived from transposable elements
Of the 81 530 sRNA clusters 72 173 contained interspersed repetitive sequences (Table S6), and of all the identified signatures from rice seeds, miRNAs account for <1.5%, which is much lower than in other rice tissues (8.8%, 36.6% and 12.6% from the inflorescences, seedlings and stems, respectively, 24) or Arabidopsis inflorescences (40%) and seedlings (62%, 22). The low ratios of miRNAs in rice tissues may because that the complex rice genome contains more interspersed and inverted repetitive sequences, and produces many repeat-associated sRNAs (24). The even lower ratio of miRNAs (higher ratio of rasiRNAs) in rice seeds suggested the important roles of rasiRNAs in seed development. As indicated by Girard and Hannon (66), the robust defense of transposition activity coincides with the transposon challenge in the germline, where new insertions of transposons can be transmitted to next generation. High abundance of rasiRNAs in seeds may serve as a suppressor of transposon to prevent its propagation in rice genome. Our results also suggested that miRNAs were low expressed in pre-differentiation tissues including embryo. Indeed, a recent study showed that genes targeted by miRNAs are expressed in a higher level in embryo than in mature tissues in mouse and Drosophila, suggesting that miRNAs are low expressed in embryo (67). Furthermore, analysis on the top 10 highly expressed individual miRNAs revealed that seven of them were with the highest expression in tissues other than embryo or endosperm (Table S15). Although several miRNAs were highly expressed in embryo or endosperm (Figure 3A), the numbers of these miRNAs were less, and the expression levels of them were much lower than those top highly expressed miRNAs in other tissues (Figure 3A, Table S9). As whole, the total abundance of miRNAs was low in rice seeds.
nat-siRNA may be formed when natural antisense transcripts are expressed in same cell (5). After comparing the 687 bidirectional transcript pairs (68), we failed to identify any signatures matching both pairs, which supports the hypothesis that the bidirectional transcripts pairs were mainly to induce the alternative splicing or polyadenylation, other than serving as origins of sRNAs by forming RNA duplex (69,70).
Some registered miRNAs appear to be repetitive sequences
In the guidelines proposed by Ambros et al. (27), miRNAs can be distinguished from siRNAs in several aspects, including genomic origin characters and biogenesis mechanisms. Our study showed that some registered miRNA precursors contain transposon or retrotransposon-like repetitive sequences, even the whole structure of them were annotated as retrotransposon (such as Stowaway) (Table S8). Furthermore, our analysis showed that most of these repeat-associated precursors could generate sRNAs from two strands randomly (Table S9), thus these miRNA precursors might not be bona fide miRNAs (8). Several recently reported miRNAs (26) also contained transposable elements (Table S8), among which three (osa-miR1440, osa-miR1441 and osa-miR1442) can produce sRNAs from two strands randomly (Table S9). During their analysis, repeat-associated borderline precursors were used as positive controls, thus some candidate miRNA precursors similar to repeat elements were also identified as miRNAs (26). This indicates that it is necessary to distinguish the bona fide miRNAs from borderline ones for annotating the novel miRNAs. According to the definition of a miRNA, one of the most critical criteria is that the miRNA should originate from a single-strand RNA transcript (27). Thus, if a locus produces sRNAs from both two strands, this locus should be siRNA locus; if a candidate precursor contains repeat elements, it should be very careful to annotate it as an miRNA, as most of the precursors of this type can produce sRNAs from both strands and many of these sRNAs are not from the site of mature miRNAs or miRNA*s (Table S9).
Diverse regulatory effects of miRNAs in rice seeds
Auxin plays crucial roles during seed development, including pattern formation, cell division and cell expansion (71). The mutation of AUXIN RESPONSE FACTOR 2 (ARF2) leads to dramatic increase of seed size and weight in Arabidopsis (71). osa-miR167, the target genes of which encode transcription factors ARF6 and ARF8 (56), was preferentially expressed in rice seeds (Figure 3B). In addition, osa-miR167 was induced by auxin, resulting in the decrease of ARF8 mRNA abundance. ARF8 positively regulated the expression of OsGH3.2, a rice indole-3-acetic acid (IAA)-conjugating enzyme, which in turn participated in the control of cellular-free IAA levels (56). In rice seeds, the IAA level is around 40-fold higher than in other tissues (72), which is consistent with the high abundance of osa-miR167 in rice seeds and suggests the regulatory roles of auxin-miR167-ARF8-OsGH3.2 pathway in auxin amount and hence the rice seed development.
Many target genes other than transcription factor coding genes are also regulated by miRNAs preferentially expressed in seeds, including those encode copper-binding proteins and carbohydrate metabolism enzymes. Such kind of functional relation is similar to miRNAs isolated from developing xylem of stems, which are predicted to target genes involved in biosynthesis of cell wall metabolites in Populus trichocarpa (73). Among the miRNAs that highly expressed in embryo, four miRNA families (osa-miR397, osa-miR398, osa-miR408 and osa-miR528) targeted genes coding copper-binding proteins (17,20,38; Table S16). Interestingly, all these target genes were suppressed in embryo (Figure 4A), suggesting the roles of these miRNAs in maintaining the copper homeostasis (57,58) and/or the ROS levels (74) in rice embryo.
osa-miR397 and osa-miR528 are predicted to target the mRNAs of L-ascorbate oxidases (AO), which catalyze the oxidation of ascorbic acid (AA) to yield dehydroascorbate (DHA) to regulate the apoplastic redox state and modulate plant growth and defense responses by regulating signal transduction cascades (75). It has been reported that the accumulation of DHA oxidized by AO could trigger the arrest of cell division (76). The high amounts of these two miRNAs will result in the low level of AO genes, which may in turn maintain the low level of apoplastic DHA in seeds. This is consistent with the higher cell-division ability of seeds at early development stage.
Differential accumulations of miRNAs and miRNA*s
It is believed that miRNA*s are gradually degraded while mature miRNAs guide the silencing of target mRNAs in miRNA-induced silencing complex (miRISC) (11). Our study indicates that some miRNA*s were also accumulated at certain tissues (Figure 5). Abundance of osa-miR390* was even higher than miR390 in rice embryo (Table S15). The relations between the accumulation abundance of mature miRNAs and miRNA*s are varied: some miRNAs and miRNA*s accumulate at same tissue(s), whereas some accumulate at different tissues (Figure 5). Differential accumulation of miRNAs and miRNA*s may due to the different activities of proteins directly involved in sRNAs biogenesis. Indeed, several rice AGO genes were differently expressed in the tested tissues (Figure 6), and abundances of miRNAs and miRNA*s may be altered due to the variety of AGO gene abundances in these tissues.
The highly abundant miRNA*s may have physiological functions in directing the mRNA cleavage. Similar as previous report, when the miRNA (let-7)/miRNA* duplex is provided, RISC can cleave the antisense RNA of let-7 target genes (77), which may be directed by let-7*. In addition, a recent report in mouse also showed that both strands of miRNAs could target equal number of genes to suppress their expressions (78) and many human miRNAs in miRBase are paired miRNAs (37,78). Therefore, miRNA*s accumulated at tissues different from mature miRNAs may play their roles with the similar mechanism.
Our study also suggests that miRNA*s may regulate mature miRNA functions when both of them are highly expressed in same tissue(s). It was known that during the generation of phased ta-siRNAs, miR390 hits the transcripts of TAS3 loci to trigger the biogenesis of ta-siRNAs (79). However, we did not detect the signatures matching TAS3 transcripts in rice seeds, whereas sRNAs from TAS3 transcripts were sequenced in other tissues (8,24). This, quite possibly, may due to that high level of miR390* in rice seeds suppresses the miR390 function.
The accumulation of miRNA*s may due to the presence of a protein regulator stabling the miRNA/miRNA* duplex. It has been reported that the Beet yellows virus p21 and the Tomato bushy stunt virus p19 can bind the miRNA/miRNA* duplex to increase the miRNA* abundance, resulting in the deduced cleavages of target mRNAs mediated by miRNA (80). Recently, a model showed how mouse miR138-2 is post-transcriptionally regulated (81), of which a certain inhibitor of Dicer was proposed to be present in some tissues like kidney, liver and others, and then mature miR138-2 can only be detected in those tissues where the inhibitor is absent or without function. This inhibitor may function like the regulator of miRNA/miRNA* duplex, as we proposed, to regulate the function of Dicer by binding to the duplex.
FUNDING
State Key Project of Basic Research (2005CB120 803); Chinese Academy of Sciences (KSCX2-YW-N-058); Nature Science Foundation (No. 30 721 061); and High-Tech program (2006AA10A102). Funding for open access charge: Nature Science Foundation.
Conflict of interest statement. None declared
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
REFERENCES
- 1.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006;38:S31–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 2.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 3.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 4.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 7.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 8.Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, German R, Thatcher SR, Maher C, Zhang L, Ware D, et al. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs) Proc. Natl Acad. Sci. USA. 2008;105:4951–4956. doi: 10.1073/pnas.0708743105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl Acad. Sci. USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl Acad. Sci. USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 15.Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JF, Zhou H, Chen YQ, Luo QJ, Qu LH. Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Res. 2004;32:1688–1695. doi: 10.1093/nar/gkh332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Li P, Li X, Liu C, Cao S, Chu C, Cao X. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 2005;139:296–305. doi: 10.1104/pp.105.063420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunkar R, Girke T, Jain PK, Zhu JK. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- 23.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nobuta K, Venu RC, Lu C, Beló A, Vemaraju K, Kulkarni K, Wang W, Pillay M, Green PJ, Wang GL, et al. An expression atlas of rice mRNAs and small RNAs. Nat. Biotechnol. 2007;25:473–477. doi: 10.1038/nbt1291. [DOI] [PubMed] [Google Scholar]
- 25.Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V. CSRDB: a small RNA integrated database and browser resource for cereals. Nucleic Acids Res. 2007;35:D829–D833. doi: 10.1093/nar/gkl991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008;8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 29.Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15:336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- 30.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 31.Duan K, Luo YH, Luo D, Xu ZH, Xue HW. New insights into the complex and coordinated transcriptional regulation networks underlying rice seed development through cDNA chip-based analysis. Plant Mol. Biol. 2005;57:785–804. doi: 10.1007/s11103-005-1803-4. [DOI] [PubMed] [Google Scholar]
- 32.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 33.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 34.Liang RQ, Li W, Li Y, Tan CY, Li JX, Jin YX, Ruan KC. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 2005;33:e17. doi: 10.1093/nar/gni019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 36.Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 39.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 41.Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 42.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S. A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Method. 2007;3:7. doi: 10.1186/1746-4811-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 46.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 47.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. New York: Academic Press; 1965. pp. 97–166. [Google Scholar]
- 48.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martienssen RA. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat. Genet. 2003;35:213–214. doi: 10.1038/ng1252. [DOI] [PubMed] [Google Scholar]
- 50.Wessler SR, Bureau TE, White SE. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 51.Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, Bartel DP, Cohen SM, Kellis M. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu S, Sun YH, Chiang VL. Stress-responsive microRNAs in Populus. Plant J. 2008;55:131–151. doi: 10.1111/j.1365-313X.2008.03497.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 56.Yang JH, Han SJ, Yoon EK, Lee WS. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 2006;34:1892–1899. doi: 10.1093/nar/gkl118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M. Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 2007;282:16369–16378. doi: 10.1074/jbc.M700138200. [DOI] [PubMed] [Google Scholar]
- 58.Abdel-Ghany SE, Pilon M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 2008;283:15932–15945. doi: 10.1074/jbc.M801406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 60.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 61.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 62.Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 2004;14:1641–1653. doi: 10.1101/gr.2275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell. 2007;19:2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu Z, Jian Z, Shen SH, Purisima E, Wang E. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res. 2007;35:152–164. doi: 10.1093/nar/gkl1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osato N, Yamada H, Satoh K, Ooka H, Yamamoto M, Suzuki K, Kawai J, Carninci P, Ohtomo Y, Murakami K, et al. Antisense transcripts with rice full-length cDNAs. Genome. Biol. 2003;5:R5. doi: 10.1186/gb-2003-5-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jen CH, Michalopoulos I, Westhead DR, Meyer P. Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome. Biol. 2005;6:R51. doi: 10.1186/gb-2005-6-6-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faghihi MA, Wahlestedt C. RNA interference is not involved in natural antisense mediated regulation of gene expression in mammals. Genome. Biol. 2006;7:R38. doi: 10.1186/gb-2006-7-5-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- 72.Matsuda F, Miyazawa H, Wakasa K, Miyagawa H. Quantification of indole-3-acetic acid and amino acid conjugates in rice by liquid chromatography-electrospray ionization-tandem mass spectrometry. Biosci. Biotechnol. Biochem. 2005;69:778–783. doi: 10.1271/bbb.69.778. [DOI] [PubMed] [Google Scholar]
- 73.Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL. Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pignocchi C, Kiddle G, Hernández I, Foster SJ, Asensi A, Taybi T, Barnes J, Foyer CH. Ascorbate oxidase-dependent changes in the redox state of the apoplast modulate gene transcript accumulation leading to modified hormone signaling and orchestration of defense processes in tobacco. Plant Physiol. 2006;141:423–435. doi: 10.1104/pp.106.078469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Potters G, Horemans N, Caubergs RJ, Asard H. Ascorbate and dehydroascorbate influence cell cycle progression in a tobacco cell suspension. Plant Physiol. 2000;124:17–20. doi: 10.1104/pp.124.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 78.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 80.Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.