Abstract
Mitochondrial DNA (mtDNA) encodes proteins that are essential for cellular ATP production. Reactive oxygen species (ROS) are respiratory byproducts that damage mtDNA and other cellular components. In Saccharomyces cerevisiae, the oxidized base excision-repair enzyme Ntg1 introduces a double-stranded break (DSB) at the mtDNA replication origin ori5; this DSB initiates the rolling-circle mtDNA replication mediated by the homologous DNA pairing protein Mhr1. Thus, ROS may play a role in the regulation of mtDNA copy number. Here, we show that the treatment of isolated mitochondria with low concentrations of hydrogen peroxide increased mtDNA copy number in an Ntg1- and Mhr1-dependent manner. This treatment elevated the DSB levels at ori5 of hypersuppressive [rho–] mtDNA only if Ntg1 was active. In vitro Ntg1-treatment of hypersuppressive [rho–] mtDNA extracted from hydrogen peroxide-treated mitochondria revealed increased oxidative modifications at ori5 loci. We also observed that purified Ntg1 created breaks in single-stranded DNA harboring oxidized bases, and that ori5 loci have single-stranded character. Furthermore, chronic low levels of hydrogen peroxide increased in vivo mtDNA copy number. We therefore propose that ROS act as a regulator of mtDNA copy number, acting through the Mhr1-dependent initiation of rolling-circle replication promoted by Ntg1-induced DSB in the single-stranded regions at ori5.
INTRODUCTION
Mitochondria in eukaryotic cells play a primary role in the production of ATP—the cellular source of energy—through the oxidative phosphorylation system. Mitochondria have their own genome (mtDNA), whose copy number varies in response to the physiological environment surrounding the cell (1). Genes in mtDNA, as well as in nuclear genomes, encode essential protein subunits that participate in oxidative phosphorylation. mtDNA is constantly subjected to oxidative damage due to reactive oxygen species (ROS), which are byproducts of oxidative phosphorylation. This damage results in mutations in the mtDNA, which are associated with aging and various mitochondrial disorders (2). On the other hand, recent evidence suggests that ROS regulate mtDNA replication: e.g. the treatment of human cells with hydrogen peroxide results in a transient increase in mtDNA copy number (3), and the amount of mtDNA correlates with the ROS levels in cell lines carrying different mouse mtDNA haplotypes (4). Because decreased cellular levels of mtDNA clinically correlate with mitochondrial disorders, such as mtDNA depletion syndrome, investigators are seeking to elucidate the mechanisms that regulate mtDNA copy number (5). Studies on yeast and human cells have shown that mtDNA-related nuclear gene expression and the cellular levels of deoxyribonucleoside triphosphates (dNTPs) play critical roles in the regulation of mtDNA copy number (6). Whether mitochondria contain mechanisms that regulate mtDNA copy number in response to ROS, however, is currently unknown (7).
In the yeast Saccharomyces cerevisiae the replication and segregation of mtDNA depend on the homologous DNA pairing protein Mhr1, which is required for mtDNA recombination (8). Generally, DNA recombination is initiated at a double-stranded break (DSB); after the break occurs, a homologous DNA pairing protein directs the formation of a heteroduplex joint (‘the DSB repair model’ of recombination) (9). Our studies revealed that the excision-repair enzyme Ntg1 creates a DSB at ori5, one of the active replication origins in yeast mtDNA; the DSB level increased when yeast cells were exposed to oxidative stress (10). These studies suggested that by recognizing a specific oxidative modification at ori5, Ntg1 induces DSB to initiate rolling-circle mtDNA replication, as well as recombination, through the homologous DNA pairing activity of Mhr1 (10). Ntg1 initiates base-excision repair of oxidative DNA damage by excising damaged bases via its DNA-N-glycosylase activity; subsequently, via its DNA lyase activity, it introduces single-stranded breaks (nicks) in the double-stranded DNA (11). Based on our previous studies showing a role for Ntg1 in the initiation of mtDNA replication, as well as a correlation between the ROS levels in mitochondria and the mtDNA copy number, we sought to uncover the molecular mechanisms underlying ROS-controlled changes in mtDNA copy number.
The Mec1/Rad53 nuclear checkpoint pathway, which is activated by nuclear DNA damage, increases mtDNA copy number by affecting the activity of ribonucleotide reductase and thereby the cellular pool of dNTPs (12). ROS are generated from either the electron transport chain or the metabolic enzyme α-ketoglutarate dehydrogenase in the tricarboxylic acid cycle; both of these ROS sources are found in normally functioning mitochondria (13). In this study, to eliminate the effects of both the Mec1/Rad53 nuclear checkpoint pathway and possible regulatory control by nuclear genes on the amounts of ROS and mtDNA, we employed isolated mitochondria harboring hypersuppressive (HS) [ori5] [rho–] mtDNA, treated with various concentrations of hydrogen peroxide, as a model system to investigate the direct effects of ROS on mtDNA copy number. Hydrogen peroxide has been used as a ROS-generating agent in a variety of studies about the effects of ROS on cellular functions.
In this study, we show that exposing isolated yeast mitochondria to hydrogen peroxide results in simultaneous increases in Ntg1-dependent DSB levels at ori5 and in mtDNA copy number, an effect that is dependent on Ntg1 and Mhr1. It is likely that ROS trigger Ntg1- and Mhr1-dependent mtDNA replication, which leads to an increase in mtDNA copy number.
MATERIALS AND METHODS
Yeast strains
The ntg1-null mutant (YKN1423C-1/▵ntg1), the mhr1-null mutant (YKN1423C-1/▵mhr1), the ogg1-null mutant (YKN1423C-1/▵ogg1) cells and cce1-null mutant (YKN1423C-1/▵cce1) cells were derived from the YKN1423C-1 wild-type strain (nuclear genotype: α leu2, ura3, met3; mitochondrial genotype: HS [ori5] [rho–] mtDNA) as described previously (10). The strains WT/pVT100U and WT/pVT100UGOX were constructed by introducing pVT100U and pVT100UGOX into the OP11c-55R5 wild-type strain (nuclear genotype: a leu2 ura3 trp1; mitochondrial genotype: [rho+] mtDNA). The OP11c-55R5 rho0 strain was constructed as described in (14).
Incubation of isolated mitochondria treated with hydrogen peroxide, the detection of ROS in isolated mitochondria, and the quantification of mtDNA levels
YKN1423C-1, YKN1423C-1/▵ntg1, YKN1423C-1/▵mhr1, YKN1423C-1/▵ogg1 and YKN1423C-1/▵cce1 yeast cells were cultivated to log phase in YPD medium at 30°C for 3 days. Highly enriched mitochondria were prepared and suspended in an import mixture (incubation buffer) [0.6 M mannitol, 20 mM HEPES–KOH (pH 7.4), 1 mM ATP, 1 mM MgCl2, 5 mM phosphoenolpyruvate, 2 units of pyruvate kinase, 40 mM KCl, 5 mM methionine and 3 mg/ml BSA] as described (15), and incubated with various concentrations of hydrogen peroxide at 26°C. The mitochondria were mixed with HincII-linearized pUC119 plasmid DNA (40 μg/ml), which was used as an internal standard. After incubation of the isolated mitochondria with or without hydrogen peroxide, DNA was extracted from the mitochondria as described previously (10). The extracted DNA was digested with BglII, which has a single recognition site in HS [ori5] [rho–] mtDNA and does not recognize pUC119 plasmid DNA. After removing proteins with proteinase K, 6 μg of the digests were analyzed by 1.0% gel electrophoresis as described (10). A 1.0-kbp plus ladder (Invitrogen) was used as a size marker. The signals representing the HS [ori5] [rho–] mtDNA and the linearized pUC119 plasmid DNA were detected by Southern blot analysis, using 32P-labeled HS [ori5] [rho–] mtDNA and linearized pUC119 plasmid DNA fragments as probes (16). Probe B, corresponding to the ori5 region, was amplified with primers Probe B-F (5′-GTTATATATTTATATATTTC-3′) and Probe B-R (5′-CTTTTTATTTTTTATTCTAT-3′). Probe C, corresponding to the segment outside the ori5 region (0.28 kbp) was amplified with the Probe C-F (5′-ATAAGAATTTAATAAGTTAT-3′) and Probe C-R (5′-TCTAGAGGATCTTCTTCATTATA-3′). The 32P signals were qualitatively and quantitatively analyzed with a Fuji BAS2500 image analyzer as described previously (10), and the relative mtDNA copy number was calculated by normalizing the signals for HS [ori5] [rho–] to those for linearized pUC119.
Detection of newly synthesized mtDNA
Isolated mitochondria were suspended in 100 μl of incubation buffer containing 0.1 mM dATP, 0.1 mM dCTP, 0.1 mM dGTP, 0.065 mM dTTP and 0.035 mM DIG-11-dUTP (Roche) and incubated at 26°C. Aliquots (20 μl) were taken at various time points for mtDNA extraction. Southern dot-blotting analysis was performed using Hybond-N+ membrane as described previously (16). The signals representing newly synthesized mtDNA were detected with anti-DIG-AP conjugates and a DIG detection kit (Roche). Ethidium bromide (EtBr; 10 μg/ml) was used to inhibit mtDNA replication (17).
Assay for oxidative modifications at ori5 by detecting increased DSB levels induced in vitro by purified Ntg1
The standard reaction mixture (12 μl) consisted of 15 μg of BglII-digested DNA, 70 mM 3-(N-morpholino)-propanesulfonic acid (pH 7.5), 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol and 1.6 nM purified Ntg1 protein, as previously described (10). The reaction was carried out at 37°C for 0.5 h as described (11). After proteins were removed with proteinase K, DNA digests electrophoresed on a 2.0% agarose gel at room temperature for 24 h. The signals representing the 0.8-kbp DNA fragment derived from DSBs at ori5 in HS [ori5] [rho–] mtDNA digested with BglII and 1.1-kbp unit length of HS [ori5] [rho–] mtDNA were detected by Southern blot analysis using 32P-labeled 1.1-kbp HS [ori5] [rho–] mtDNA as a probe (10).
Assay for Ntg1-mediated nicking in dsDNA in an ori5-containing region of HS [ori5] [rho–] mtDNA
Isolated mitochondria from wild-type cells were mixed with HincII-linearized pUC119 plasmid DNA (40 μg/ml), which was used as an internal standard. After incubation of the isolated mitochondria with or without hydrogen peroxide at 26°C for 1 h, DNA was extracted from the mitochondria as described previously (10). The BglII–NdeI digested DNA was treated with purified Ntg1 protein (1.6 nM) in the standard reaction mixture at 37°C for 30 min, as described above. After the removal of proteins with proteinase K, the DNA digests were mixed with alkaline loading buffer containing 50 mM NaOH, 1 mM EDTA, 3% Ficoll (w/v), 0.025% bromocresol green (3,3′,5,5′-tetrabromo-m-cresolsulfonphthalein) (w/v) and 0.004% (w/v) xylene cyanol and were then loaded onto a 2.0% alkaline agarose gel. Denaturing agarose gel electrophoresis was performed in 50 mM NaOH and 1 mM EDTA at 30 V/cm for 15 h as described in (18). The signals representing the denatured HS [ori5] [rho–] mtDNA and pUC119 plasmid DNA were detected by Southern blot analysis using 32P-labeled HS [ori5] [rho–] mtDNA and linearized pUC119 plasmid DNA fragments as probes (16).
Assay for the S1-nuclease sensitivity of HS [ori5] [rho–] mtDNA
HS [ori5] [rho–] mtDNA, extracted from isolated mitochondria derived from wild-type cells, was digested with BglII and NdeI. Subsequent treatment with S1 nuclease was performed in a standard reaction mixture (12 μl) consisting of 3 μg of BglII–NdeI digested DNA, 30 mM sodium acetate buffer (pH 4.6), 280 mM NaCl, 1 mM ZnSO4 and various amounts of S1 nuclease at 37°C for 20 min. Digests were separated by electrophoresis on a 2.0% agarose gel. Signals were detected by Southern blot analysis using 32P-labeled 1.1-kbp HS [ori5] [rho–] mtDNA as a probe.
Assay for breakage at an oxidized base in single-stranded DNA by purified Ntg1
The 30-mer oligonucleotides containing either single oxidized base, 5-hydroxyuracil (5-OHU) or thymine at the 14th position (5′-CTCGTCAGCATCTXCATCATACAGTCAGTG-3′) were used as the single-stranded DNA substrates. These oligos were synthesized and purified by Tsukuba Oligo Service and labeled using the DIG 3′-end labeling kit (Roche). The reaction was performed in a buffer suitable for Ntg1 activity as described in (5), which contained various amounts of purified Ntg1 or C▵ntg1 as indicated in Figure 5F, at 37°C for 30 min. The reaction was stopped by the addition of an equal volume of 90% formamide loading dye solution. The samples were heated at 75°C for 2 min and run on a 20% polyacrylamide gel containing 7 M urea. The signals representing each oligo were detected using the DIG detection kit (Roche).
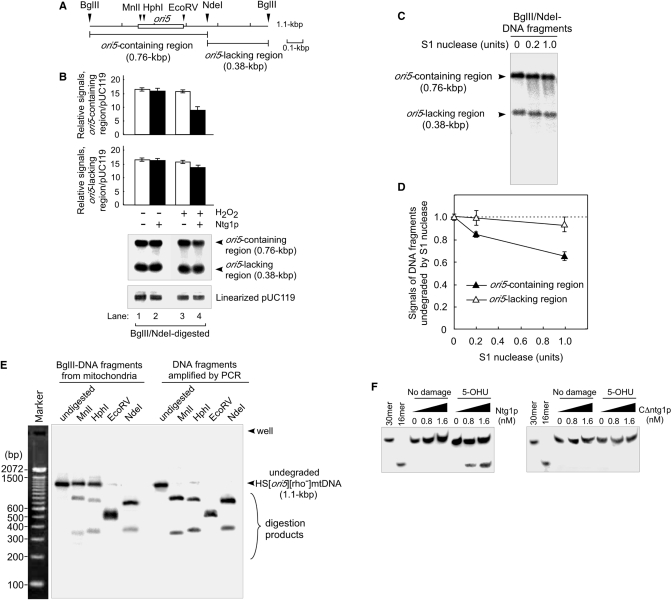
Figure 5.
ori5-containing DNA fragments extracted from isolated mitochondria are sensitive to S1 nuclease, and resistant to single-cut restriction enzymes, within the ori5-region. (A) A physical map of the locations of ori5 and the restriction sites in HS [ori5] [rho–] mtDNA. Digestion of HS [ori5] [rho–] mtDNA with BglII and NdeI gives rise to the ori5-containing region (760 bp) and the remainder of the DNA (380 bp). The ori5 sequence (282 bp) is indicated with an open box. (B) Ntg1-introduced nicks in the ori5-containing region of HS [ori5] [rho–] mtDNA. HS [ori5] [rho–] mtDNA extracted from hydrogen peroxide-untreated (lanes 1 and 2), or treated (lanes 3 and 4) mitochondria isolated from wild-type cells, was digested with BglII and NdeI. The BglII–NdeI digested DNA was treated with purified Ntg1 (1.6 nM) in the standard reaction mixture at 37°C for 30 min (lanes 2 and 4). After the removal of proteins with proteinase K, DNA digests were separated on a 2.0% denaturing (alkaline) agarose gel. The signals resulting from the ori5-containing region, the ori5-lacking region and linearized pUC119 were detected by Southern blot analysis using 32P-labeled HS [ori5] [rho–] mtDNA and linearized pUC119 as probes (bottom panel: Southern blot). The ratios of the signals from the ori5-containing region or the ori5-lacking region to the signal from linearized pUC119 are plotted (upper and middle panels), based on the results from three independent experiments. Error bars shown here are standard deviations. (C) Comparison of the S1 nuclease sensitivities of DNA fragments that contain or lack ori5. HS [ori5] [rho–] mtDNA extracted from wild-type mitochondria was digested with BglII and NdeI, followed by the treatment with S1 nuclease, as indicated. DNA fragments containing ori5 (760 bp), the rest of the mtDNA (380 bp) and the smear caused by S1 nuclease were separated on a 2.0% agarose gel and detected by Southern blot analysis. (D) Quantitative analysis of DNA fragments treated with S1 nuclease. The values of the signals derived from DNA fragments not treated with S1 nuclease were defined as 1. The relative values of signals derived from undegraded DNA fragments, which contain (filled triangles) or lack (open triangles) the ori5 region are plotted. (E) Resistance of HS [ori5] [rho–] mtDNA to single-cut enzymes within or adjacent to the ori5-region. HS [ori5] [rho–] mtDNA from wild-type cells digested with BglII, and ori5-containing DNA fragments amplified by PCR, were treated (or not) with MnlI, HphI, EcoRV or NdeI and then separated on a 2.0% agarose gel. Marker, 100-bp ladder. (F) Removal of oxidized 5-OHU bases located at the middle of the 3′-DIG labeled 30-mer oligos by purified Ntg1. The 3′-DIG-labeled 30-mer oligos containing thymine base instead of 5-OHU were used as a control. The 30-mer oligos harboring 5-OHU or no damage were incubated with indicated amounts of Ntg1 (left panel) or C▵ntg1 (right panel) for 30 min at 37°C. The oligos in the reactions were separated by electrophoresis on a 20% polyacrylamide-urea gel. The signals representing the 3′-DIG-labeled oligos were detected using anti-DIG AP conjugates.
Detection of GOX expression and assays for relative mtDNA copy number and hydrogen peroxide levels
The GOX open reading frame was amplified by PCR from Aspergillus niger as described in (19). The GOX gene was subcloned into the pVT100U vector (20) at a blunted HindIII/XhoI site, where it is under control of the ADH promoter. WT cells harboring pVT100U or pVT100UGOX were cultivated in a 2.0% glucose medium (SD) supplemented with required amino acids at 30°C and harvested before the glucose in the medium is completely consumed. The cell-free extracts were prepared as described in (21). GOX expression was confirmed in the WT cells harboring pVT100UGOX by Western blot analysis using a monoclonal antibody against glucose oxidase (Sigma). As a control, the mitochondrial outer membrane protein porin was detected using a monoclonal anti-porin antibody (molecular probes).
Hydrogen peroxide levels in cells with or without expression of GOX were detected using a Fluoro hydrogen peroxide detection kit (Cell Technology Inc.), after cells suspended in PBS buffer containing 0.5-mm glass beads were disrupted using a Beads Shocker (Yasui Kikai). The amount of fluorescent product resorufin generated from the chemical reaction in which hydrogen peroxide oxidizes 10-acetyl-3,7-dihydroxyphenoxazine was measured at excitation 570 nm and emission 595 nm in fluorescent plate reader (Molecular devices).
Whole cellular DNA was extracted as described in (10). About 20 μg DNA was separated by electrophoresis on a 1.0% agarose gel. [rho+] mtDNA was detected by Southern analysis using highly purified total mtDNA as a probe. NUC1 was detected on the same Southern blot using PCR-amplified DNA fragments of the NUC1 open reading frame as a probe.
RESULTS
Increases in mtDNA copy number in isolated mitochondria harboring HS [ori5] [rho–] mtDNA following the treatment with hydrogen peroxide
Hydrogen peroxide is widely used as an oxidant to investigate the effects of ROS on cellular functions (22). To examine the effects of ROS on mtDNA replication, isolated yeast mitochondria harboring HS [ori5] [rho–] mtDNA were treated with various concentrations of hydrogen peroxide in the incubation buffer. The signals representing mtDNA were detected using Southern analysis with a 282-bp DNA fragment from the ori5 region and a 280-bp DNA fragment outside of the ori5 region, in addition to full-length HS [ori5] [rho–] mtDNA (Figure 1A). As examples, increased and decreased levels of mtDNA detected using full-length HS [ori5] [rho–] mtDNA as a probe are shown in Figure 1B. The relative mtDNA copy number was calculated from the signal from HS [ori5] [rho–] mtDNA relative to that of the extrinsic standard, HincII-linearized pUC119 plasmid. The amount of DNA increased in response to treatment with hydrogen peroxide up to 10 μM (Figure 1C). No significant difference was detected between the relative levels of mtDNA measured using the ori5-specific probe, the probe located outside of ori5, or the full-length probe, when isolated mitochondria were treated with hydrogen peroxide concentrations ranging from 0 to 100 μM (Figure 1C, inserts). This is not a peculiar feature of HS [ori5] [rho–] mtDNA. We have reproduced the major results of the ROS-induced increase in mtDNA copy number using isolated mitochondria containing [rho+] mtDNA (Supplementary Figure 1).
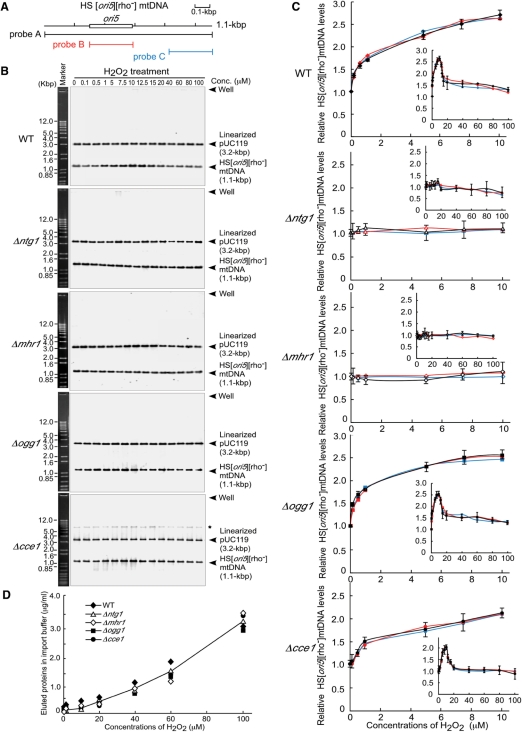
Figure 1.
mtDNA copy number in isolated mitochondria harboring HS [ori5] [rho–] mtDNA, which were treated with various concentrations of hydrogen peroxide. (A) A physical map of the locations of the ori5-region and the probes used for detecting HS [ori5] [rho–] mtDNA. Probe A, full length HS [ori5] [rho–] mtDNA (1.1 kbp); probe B, a 282-bp region of ori5 and probe C, a 280-bp sequence outside of the ori5 region. The ori5 sequence (282 bp) is framed with an open box. (B) HS [ori5] [rho–] mtDNA-containing mitochondria derived from wild-type, ntg1-null, mhr1-null, ogg1-null and cce1-null cells were suspended in incubation buffer and incubated with the indicated concentrations of hydrogen peroxide for 1 h at 26°C. HS [ori5] [rho–] mtDNA and linearized pUC119 plasmid DNA were separated on a 1.0% agarose gel and detected by Southern blot analysis using the sequences described in (A) and linearized pUC119 as probes. Marker, 1.0-kb plus ladder. Asterisk denotes mtDNA molecules containing single-stranded regions resistant to digestion with BglII, likely with a Holliday junction structure, since they mainly appeared in HS [ori5] [rho–] mtDNA extracted form Holliday junction resolvase-lacking cce1-null cells. (C) Quantitative analysis of the mtDNA bands in (B). Upper panel: wild type (filled diamonds); upper middle panel: ntg1-null mutant (open triangles); lower middle panel: mhr1-null mutant (open diamonds); bottom panel: ogg1-null mutant (filled squares) and lower bottom panel: cce1-null mutant (filled circles). In each panel, the colored lines representing the relative mtDNA copy number detected using Southern analysis with various probes. Black lines, probe A; red lines, probe B and blue lines, probe C. Insert in (C), a figure showing the increase of the HS [ori5] [rho–] mtDNA copy number in isolated mitochondria treated with hydrogen peroxide concentrations ranging from 0 to 100 μM. (D) Eluted proteins from inside isolated mitochondria, accompanied by treatment with hydrogen peroxide.
On the other hand, as suggested by previous observations that highly reactive oxygen radicals can damage various cell structures, including mitochondria (23), higher concentrations of hydrogen peroxide (≥20 μM) appeared to rupture the isolated mitochondria, leading to a gradual increase in amount of eluted proteins (Figure 1D); no increase was observed after treatment with ≥20 μM hydrogen peroxide (Figure 1B and C). A higher concentration of hydrogen peroxide may also inhibit enzymes required for mtDNA replication; likewise, an elevated level of oxidative mtDNA damage may slow the progression of mtDNA replication.
No detectable increases in mtDNA copy number were observed in mitochondria isolated from ntg1-null or mhr1-null mutant cells treated with hydrogen peroxide up to 10 μM. In contrast, disruption of the OGG1 gene, which encodes another excision repair enzyme that recognizes 8-oxoguanine, did not suppress the increases in mtDNA copy number (Figure 1B and C). Thus, mtDNA copy number increases in a manner dependent on the functions of Ntg1 and Mhr1.
Previously, we showed that the maintenance of yeast mtDNA depends on the recombination-related genes, MHR1 and CCE1 (14). Nuclear CCE1 gene encodes a mitochondria-localized recombination junction-resolving endonuclease, and likely plays a role in mtDNA recombination (24,25). In response to treatment with hydrogen peroxide up to 10 μM, the amount of DNA in mitochondria isolated from cce1-null mutant cells increased and was only slightly less than that observed in mitochondria isolated from wild-type cells (Figure 1B and C). Thus, the Cce1-dependent recombination system is a minor pathway for the hydrogen peroxide-induced increase in mtDNA copy number.
Treating isolated mitochondria with 10 μM hydrogen peroxide resulted in an increase in mtDNA copy number (Figure 2A), which doubled in the first 0.5 h of incubation (Figure 2B). The rate of this increase slowed after 1 h of incubation (Figure 2B). Since the addition of fresh hydrogen peroxide (10 μM) after 1 h induced a further increase in mtDNA copy number (up to 4.5-fold in another 1 h; data not shown), it is likely that hydrogen peroxide underwent decomposition during incubation.
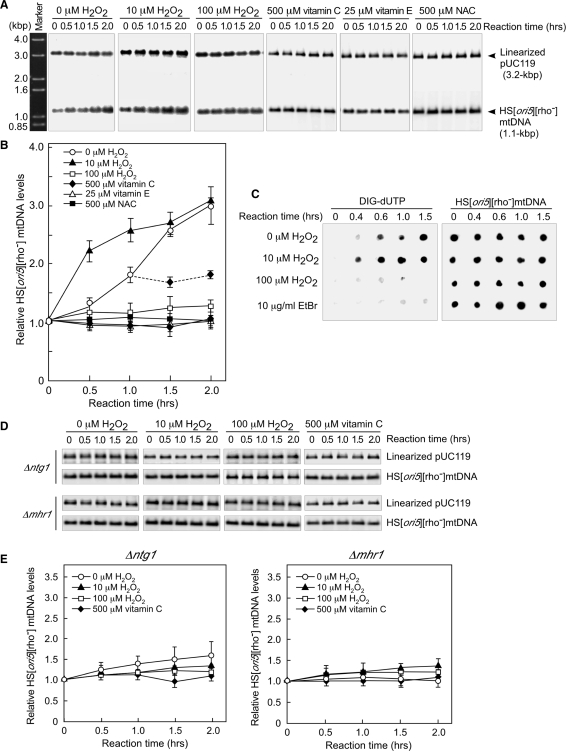
Figure 2.
Changes in mtDNA copy number in isolated mitochondria treated with 10 μM hydrogen peroxide. (A) Treatment with 10 μM hydrogen peroxide increased mtDNA copy number in mitochondria isolated from wild-type cells. Isolated mitochondria were incubated with the indicated concentrations of hydrogen peroxide. mtDNA was digested with BglII and separated by electrophoresis on a 1.0% agarose gel. Marker, 1.0-kbp plus ladder. (B) Quantitative analysis of the relative mtDNA copy number in mitochondria isolated from wild-type cells. Vitamin C (500 μM), 25 μM vitamin E or 500 μM NAC were added to the incubation buffer from the start time point. After 1 h of incubation, mitochondria not treated with hydrogen peroxide were transferred into fresh incubation buffer containing 500 μM vitamin C. The average values from three independent experiments are plotted in (B). (C) Newly synthesized mtDNA (left panel) and total mtDNA (right panel) in isolated mitochondria. Mitochondria isolated from wild-type cells were incubated with DIG-dUTP, which was detected using anti-DIG AP conjugates. Total mtDNA was detected by Southern dot-blotting analysis using 32P-labeled 1.1-kbp HS [ori5] [rho–] mtDNA as the probe. (D) A low concentration (10 μM) of hydrogen peroxide did not induce an increase in mtDNA copy number in mitochondria isolated from either ntg1-null or mhr1-null cells. There was no change in mtDNA copy number when mitochondria isolated from either ntg1-null or mhr1-null cells were incubated with vitamin C. (E) Quantitative analysis of mtDNA copy number in mitochondria isolated from either ntg1-null or mhr1-null mutant cells. The average values from three independent experiments are plotted.
Without hydrogen peroxide treatment, mtDNA copy number in mitochondria isolated from wild-type cells increased only after a lag period of 0.5 h (Figure 2A and B). The difference in the copy number between treated and untreated mitochondria was about 2-fold after 0.5 h of incubation (Figure 2B). Vitamin C, vitamin E and N-acetylcysteine (NAC) are widely used antioxidants that eliminate ROS (26,27). No increase in mtDNA copy number was observed, when mitochondria were incubated with 500 μM vitamin C, 25 μM vitamin E, or 500 μM N-acetylcysteine from the start time point (Figure 2B). In addition, the increase in mtDNA copy number was suppressed by transferring the hydrogen peroxide-untreated mitochondria into fresh incubation buffer containing 500 μM vitamin C after 1 h of incubation (Figure 2B). Thus, it is likely that the increase in mtDNA copy number was induced even in isolated mitochondria not treated with hydrogen peroxide due to ROS generated during the incubation. Isolated mitochondrial α-ketoglutarate dehydrogenase and pyruvate dehydrogenase complexes generate ROS (13). It is likely that ROS generated from these complexes eventually activate mtDNA replication in isolated mitochondria. These observations showed that the increases in mtDNA copy number observed in isolated mitochondria with or without hydrogen peroxide treatment were caused by increased levels of ROS (Figure 2B). In the following experiments, we examined the first 1 h of the incubation, in which the effects of the added hydrogen peroxide is prominent.
Next, we investigated whether the hydrogen-peroxide-induced increase in mtDNA copy number was a result of a net increase in newly synthesized mtDNA. We proceeded by examining digoxigenin (DIG)-11-dUTP-labeled mtDNA in isolated mitochondria, whose levels reflect incorporation of the labeled nucleotide throughout the mtDNA (Figure 2C, left panel). As a control, the same amount of DNA extracted from isolated mitochondria at various reaction time points was spotted onto a Hybond-N+ membrane. The signals derived from total mtDNA were detected by Southern dot-blotting analysis using 32P-labeled 1.1-kbp HS [ori5] [rho–] mtDNA as the probe (Figure 2C, right panel). It has been shown that isolated yeast mitochondria incorporate dNTPs into mtDNA (17). The incorporation of DIG-dUTP into mtDNA of isolated mitochondria was observed for 1–1.5 h after the addition of the labeled dNTP, an effect that was enhanced by 10 μM hydrogen peroxide (Figure 2C, left panel). DIG-dUTP-derived signals were not observed in isolated mitochondria in the presence of 10 μg/ml EtBr—an inhibitor of mtDNA polymerase γ—indicating that the increased signals reflected mtDNA synthesis (Figure 2C). Consistent with the preceding experiment, isolated mitochondria treated with a higher concentration of hydrogen peroxide (100 μM) did not incorporate DIG-dUTP into mtDNA (Figure 2C).
In mitochondria isolated from ntg1-null or mhr1-null mutant cells, increases in mtDNA copy number were not observed during a 2-h incubation, regardless of whether the samples were treated with 10 μM hydrogen peroxide, or were incubated in the presence of antioxidants (Figure 2D and E). Furthermore, we did not observe a significant change in mtDNA copy number in isolated mitochondria containing [rho+] mtDNA derived from ntg1-null or mhr1-1 cells (Supplementary Figure 1D and E). These results indicate that the increase in mtDNA copy number requires both Ntg1 and Mhr1.
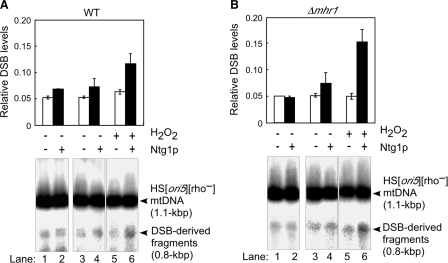
Increased DSB levels at ori5 in hydrogen peroxide-treated mitochondria
We previously showed that Ntg1-induced DSBs initiate mtDNA replication (10). Therefore, we investigated whether the levels of DSBs at ori5 in HS [ori5] [rho–] mtDNA correlated with changes in mtDNA copy number induced by hydrogen peroxide treatment. The 0.8-kbp DNA fragment corresponded to the fragment between the DSB site at ori5 and the BglII site (10). As shown in Figure 3A and B, the increased DSB level correlated with the increase in mtDNA copy number: in mitochondria isolated from wild-type cells, the DSB levels at ori5 increased during 1 h of incubation in 10 μM hydrogen peroxide, but did not change during a 1 h incubation without hydrogen peroxide (Figure 3A and B). The number of DSB at ori5 gradually decreased when hydrogen peroxide-untreated mitochondria were incubated with antioxidants (vitamin C, vitamin E or NAC), suggesting the DSB levels at ori5 are associated with the ROS levels (Figure 3A and B). The addition of antioxidants did not affect mtDNA copy number (Figure 2), suggesting that the content of mtDNA was sustained at a basal level.
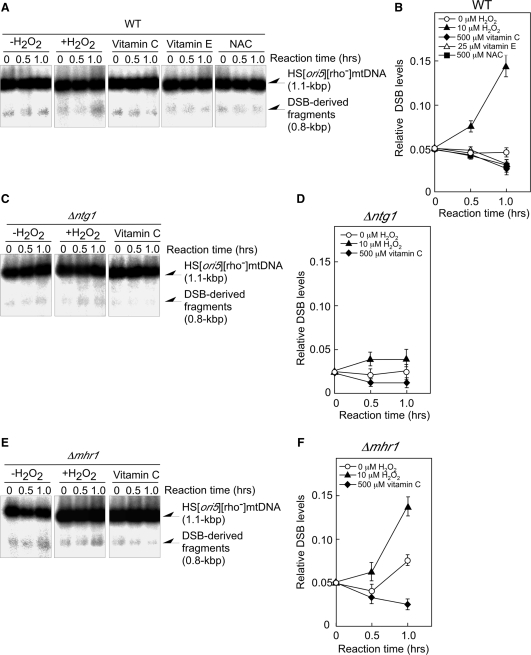
Figure 3.
DSB levels at ori5 in isolated mitochondria treated with or without 10 μM hydrogen peroxide. Isolated mitochondria were treated at 26°C with or without 10 μM hydrogen peroxide for the indicated period of time. In parallel experiments, isolated mitochondria were incubated at 26°C with 500 μM vitamin C, 25 μM vitamin E or 500 μM NAC from the start time point. Mitochondria were isolated from wild-type (A), ntg1-null mutant (C), or mhr1-null (E) mutant cells. After the incubation, mtDNA was extracted, and ∼10 µg of mtDNA was digested with BglII. The digests were separated by electrophoresis on a 2.0% agarose gel. The signals from the 0.8-kbp DNA fragment derived from DSBs at ori5 were normalized to those of the 1.1-kbp unit length of HS [ori5] [rho–] mtDNA as measured by Southern blot analysis using 32P-labeled 1.1-kbp fragments of HS [ori5] [rho–] mtDNA as a probe. The average values from three independent experiments of the signals representing the 0.8-kbp fragments derived from DSBs at ori5, normalized to those from the unit-sized 1.1-kbp fragments of HS [ori5] [rho–] mtDNA, are plotted. DNA was extracted from mitochondria isolated from wild-type (B), ntg1-null (D), or mhr1-null (F) cells.
In mitochondria isolated from ntg1-null mutant cells, the DSB levels at ori5 were about half of those observed in wild-type cells, independent of the hydrogen peroxide treatment or the presence of vitamin C and remained constant during a 1-h incubation (Figure 3C and D); thus, Ntg1 plays an important role in DSB formation at ori5. A slight increase in the DSB levels was observed in 10 μM hydrogen peroxide-treated mitochondria isolated from ntg1-null mutant cells. These data, along with the basal DSB levels at ori5 in mtDNA derived from ntg1-null mutant cells, indicate that there is another mechanism that introduces DSBs at ori5 (12). The Ntg1-independent DSBs may be formed by an unidentified functional homolog of Ntg1, or by the action of an unidentified mitochondrial S1 nuclease-like enzyme that cuts single-stranded structures in double-stranded DNA to create single-strand DNA breaks; during replication, these breaks would then be converted into DSBs in the ori5 region.
The DSB levels at ori5 in HS [ori5] [rho–] mtDNA from hydrogen peroxide-treated mitochondria isolated from mhr1-null mutant cells increased during 1 h of incubation, just as those from MHR1 cells did (Figure 3E and F versus 3A and B). In HS [ori5] [rho–] mtDNA from hydrogen peroxide-untreated MHR1 cells, DSB levels did not change during 1 h of incubation. In contrast, DSB levels increased in HS [ori5] [rho–] mtDNA isolated from hydrogen peroxide-untreated mhr1-null mutant cells; furthermore, the increase in DSB was suppressed by vitamin C (Figure 3E and F versus 3A and B). This difference likely reflects Mhr1-dependent recombination functions such as recombinational DSB repair (9).
Accumulation of oxidative modifications at ori5 that are recognized by Ntg1
To obtain direct evidence that hydrogen peroxide treatment enhances ori5-specific oxidative modifications of mtDNA in isolated mitochondria, we measured the DSB levels at ori5 in HS [ori5] [rho–] mtDNA, as an indication of the unrepaired oxidative damage that accumulated throughout the mtDNA (10). We treated DNA purified from the isolated mitochondria with purified Ntg1, as described previously (10). The resulting increases in the DSB levels served as a measure of the levels of oxidative modifications at ori5. The DSB levels at ori5 in HS [ori5] [rho–] mtDNA purified from untreated mitochondria isolated from wild-type cells did not change during a 0.5-h incubation (Figure 4A, lane 1 versus lane 3), whereas the DSB levels increased slightly (1.3-fold) due to Ntg1 treatment (Figure 4A, lane 1 versus lane 2 and lane 3 versus lane 4). The DSB levels at ori5 in mtDNA isolated from hydrogen peroxide-treated mitochondria doubled following Ntg1 treatment for 0.5 h (Figure 4A, lane 5 versus lane 6), indicating that oxidative modifications accumulated at ori5 as a result of hydrogen peroxide treatment.
Figure 4.
Oxidative modifications of mtDNA, which are represented by in vitro Ntg1-dependent generation of DSBs at ori5. HS [ori5] [rho–] mtDNA was extracted from mitochondria isolated from wild-type (A) or mhr1-null (B) cells, with or without treatment with 10 μM hydrogen peroxide at 26°C for 0.5 h, and digested with BglII. In vitro cleavage by Ntg1 was carried out as described (10). Approximately 15 µg of BglII-digested DNA was incubated without (lanes 1, 3 and 5) or with 1.6 nM purified Ntg1 (lanes 2, 4 and 6) at 37°C for 0.5 h. Lanes 1 and 2, without incubation. After the removal of proteins with proteinase K, the digests were analyzed using electrophoresis on a 2.0% agarose gel. The signals representing 0.8-kbp DNA fragments derived from DSBs at ori5 and 1.1-kbp fragments corresponding to unit-size HS [ori5] [rho–] mtDNA were detected as described in Figure 3B. The average values of the relative DSB levels at ori5 of HS [ori5] [rho–] mtDNA from two independent experiments are shown in the top panels, which correspond to the results from the Southern blot analysis shown in the bottom panel. The ratios of the signals for the 0.8-kbp fragments were normalized against those for the 1.1-kbp fragments.
In agreement with previous results that showed Mhr1 is required for the repair of spontaneously introduced oxidative damage in yeast mtDNA (28), we found that the DSB levels induced by Ntg1 treatment in vitro (i.e. oxidative damage at ori5) were higher (3-fold) in mtDNA purified from hydrogen peroxide-treated mitochondria derived from mhr1-null mutant cells, as compared with the results obtained using MHR1 cells (2-fold) (Figure 4B, lanes 5 and 6, versus Figure 4A, lanes 5 and 6). Thus, treating isolated mitochondria with hydrogen peroxide causes an increase in the number of oxidative modifications at ori5; furthermore, Mhr1 is involved in repair of the DSBs induced at ori5 by Ntg1.
Is there an unusual structure underlying the DSBs introduced by Ntg1 at ori5?
Ntg1 is a DNA N-glycosylase/AP-lyase, which has been shown to remove oxidative bases in double-stranded DNA and to introduce single-stranded breaks in the strand with the abasic site generated (11,29). The known activities of Ntg1 cannot explain the observation that it can also introduce DSBs by itself only in the ori5-region, but not in the rest of HS [ori5] [rho–] mtDNA (10). Ogg1, which specifically recognizes the oxidation-damaged guanine bases (30), did not induce DSBs in the ori5-region (10). DSBs introduced by DNA N-glycosylase/AP-lyase are formed when two or more lesions are very closely positioned in opposite DNA strands to form clustered lesions (31). Thus, we addressed the question of how Ntg1 creates the DSBs and how DSB occurs specifically at ori5, with respect to both the activity of Ntg1 and the structure of the ori5 region.
Since we assumed that ori5-region is more sensitive to oxidative modification by ROS, we first asked whether that Ntg1 introduced a larger number of nicks in the ori5-region than in other regions. HS [ori5] [rho–] mtDNA and the internal standard, HincII-linearized pUC119 plasmid DNA from isolated mitochondria treated or untreated with hydrogen peroxide, were incubated with or without purified Ntg1 in vitro. BglII-NdeI DNA-digests were loaded onto a 2.0% alkaline agarose gel, which was run at a sufficiently high pH to denature double-stranded DNA. After digestion with BglII and NdeI HS [ori5] [rho–], mtDNA gave rise to a 0.74-kbp ori5-containing and a 0.38-kbp ori5-lacking DNA fragments (Figure 5B, bottom panel: Southern blot). The ratio of the ori5-containing DNA fragments to linearized pUC119 decreased by nearly half in samples treated with Ntg1 when compared to samples without Ntg1-treatment (Figure 5B, lane 3 versus lane 4, upper panel). When isolated mitochondria were not treated with hydrogen peroxide, the ratio of ori5-containing DNA fragments to linearized pUC119 decreased only slightly after Ntg1-treatment (Figure 5B, lane 1 versus lane 2, upper panel). On the other hand, the ratio of ori5-lacking DNA fragments to the linearized pUC119 decreased by less than one-tenth after Ntg1-treatment when compared to samples without Ntg1-treatment (Figure 5B, lane 3 versus lane 4, middle panel). When isolated mitochondria were not treated with hydrogen peroxide, we observed no significant difference between the ratio of ori5-lacking DNA fragments to linearized pUC119, regardless of Ntg1-treatment (Figure 5B, lane 1 versus lane 2, middle panel). These results indicate that Ntg1 introduces a larger number of nicks in ori5-containing regions than those in ori5-lacking regions, and thus, that ori5-containing regions have larger numbers of oxidized bases than ori5-lacking regions.
We next asked why more oxidative modifications accumulate at ori5. It is known that single-stranded DNA regions, formed during transcription or replication, are much more sensitive to modifications by ROS than dsDNA, in which bases are more protected inside the double helix (32). Thus, we hypothesized that ori5 accumulates an unusual structure with exposed single-stranded character that confers local sensitivity to ROS, allowing Ntg1 to induce single-strand breaks in both strands, resulting in double-strand breaks specifically at ori5. As we speculated, in comparison with the rest of the mtDNA, the DNA fragments harboring the ori5 region were more sensitive to S1 nuclease, which digests abnormal structures with exposed single-stranded regions in double-stranded DNA (Figure 5C and D). In addition, the ori5 region in HS [ori5] [rho–] mtDNA extracted from isolated mitochondria was more resistant to single-cut restriction enzymes when compared with DNA fragments amplified in PCRs, suggesting that the ori5-region contains single-stranded regions, such as bubble structures formed during transcription or replication (Figure 5E). Obviously, the Ntg1 activity that introduces single-stranded DNA cleavage is required for DSB formation at ori5.
Since the fidelity of DNA repair synthesis would be compromised if template strands were absent, excision repair enzymes are thought to remove oxidize bases only in double-stranded substrates. However, a number of base-excision repair enzymes have been shown to have activity against damaged bases in single-stranded DNA, e.g. in the context of a transcription or replication bubble (33,34). Therefore, we asked whether Ntg1 caused a strand-break by recognizes oxidative bases in single-stranded DNA. As shown in Figure 5F, Ntg1 acted on an oxidized base, 5-OHU, but not on thymine, when these nucleotides were artificially introduced in the middle of 30-mer oligonucleotides labeled with DIG at their 3′ termini. Increased levels of DIG-labeled 16-mer oligos, generated from the sites of strand breakages formed by Ntg1 recognition of oxidized bases in single-stranded DNA, were observed only when the amounts of Ntg1, but not C▵ntg1, were increased (Figure 5F, left panel versus right panel). C▵ntg1 lacks the activity to excise oxidized bases in dsDNA; deletion of the C-terminal 158 amino acid residues removes a part of the active domain (residues 97–318) responsible for the DNA glycosylase activity of Ntg1. Thus, Ntg1 is indeed capable of recognizing oxidized bases and making incisions at these sites in single-stranded DNA.
Taken together, these results suggest that Ntg1 creates DSBs in the ori5 region by two strand breakage events in both complementary strands, by recognizing oxidized base lesions that were preferentially introduced in this site because of the single-strand character of the locus. The ori5-region is rich in A:T base pairs and is therefore likely to be easily melted by both transcription and replication factors.
Increases in mtDNA copy number are sensitive to chronic low levels of hydrogen peroxide in vivo
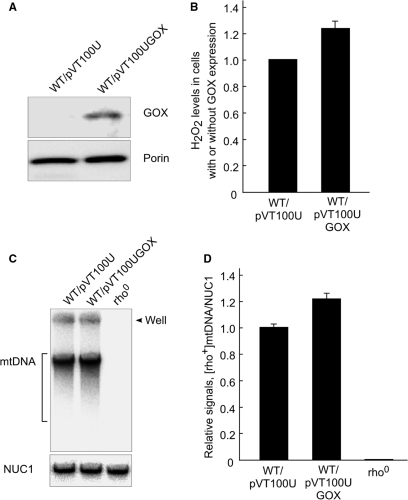
In order to investigate whether in vivo low constant ROS can also increase mtDNA copy number, we expressed the A. niger GOX gene, which encodes glucose oxidase. This enzyme catalyzes the chemical reaction (β-d-glucose + O2 → glucono-δ-lactone + H2O2). Thus, the expression of GOX in yeast cells results in production of a low constant level of ROS, as described in (19). Western blot analysis using monoclonal antibody against glucose oxidase (Sigma) in WT cells harboring pVT100UGOX indicated that GOX gene was detectably expressed (Figure 6A). Southern blot analysis revealed that there was more [rho+] mtDNA in WT cells harboring pVT100UGOX than in WT cells harboring empty vector (Figure 6C). When hydrogen peroxide levels were elevated in the cells with expression of GOX, the relative signals of [rho+] mtDNA versus nuclear NUC1 gene increased 1.3-fold (Figure 6B and D). Since no [rho+] mtDNA was detected from [rho0] cells, which completely lack mtDNA (Figure 6C and D), we conclude that the signals we detected are derived from mtDNA. Although the hydrogen peroxide levels were only slightly elevated, an increase in mtDNA copy number was observed, indicating that the increase in mtDNA copy number is sensitive to even a slight increase of intracellular ROS. The observation was confirmed when we cultivated WT cells at 30°C in YPGlycerol medium supplemented with various concentrations of iron (FeCl2), which also induces ROS generation, likely through the Fenton reaction (Fe2+ + H2O2 → Fe3+ + •OH + OH–) (35). We found that increased concentrations of FeCl2 slightly but significantly enhanced the content of [rho+] mtDNA, e.g. mtDNA copy number increased 1.5-fold at 1.0 mM FeCl2 (Supplementary Figure 2A and B). These results suggest that either chronic low levels of hydrogen peroxide in vivo, or a small, but detectable increase in basal ROS inside cells (whether due to by GOX expression or the presence of iron), cause an increase in mtDNA copy number. These observations support the model wherein ROS act as a regulator of mtDNA copy number to initiate rolling-circle mtDNA replication (Figure 7).
Figure 6.
Effects of GOX expression on mtDNA copy number. (A) Western blot analysis for GOX expression. Cells harboring pVT100U or pVT100UGOX were grown at 30°C in an SD medium supplemented with required amino acid from 1.0 × 105 cells/ml to 2.0 × 107 cells/ml. About 10 μg of total proteins from each strain were analyzed on 10% SDS polyacrylamide gel electrophoresis. GOX and porin were detected by Western blot analysis using monoclonal antibodies. (B) Determination of hydrogen peroxide generation due to GOX expression. Cells harboring pVT100U or pVT100UGOX were grown under the same conditions as described above. The elevated levels of hydrogen peroxide caused by glucose oxidase activity were determined. Averaged values of relative hydrogen peroxide levels, which were obtained from three independent experiments, are plotted. Hydrogen peroxide level in cells harboring an empty vector was set as 1. (C) Southern blot analysis for relative mtDNA copy number. Cells harboring pVT100U or pVT100UGOX were grown under the same conditions as described in A. About 20 μg of total cellular DNA isolated as described (21) was electrophoresed on a 1.0% agarose gel. mtDNA was detected using 32P-labeled [rho+] mtDNA as probes (upper panel). After the 32P-labeled [rho+] mtDNA probe was washed out, the same membrane was probed using a 32P-labeled DNA fragment containing the nuclear NUC1 gene (bottom panel). (D) Quantitative analysis of the relative amounts of mtDNA. The average values of the ratios of the signals from mtDNA versus those from the NUC1 open reading frame, obtained from three independent experiments, are plotted.
Figure 7.
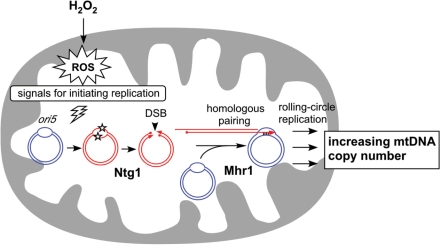
Increases in mtDNA copy number are triggered by ROS. When the levels of ROS increase, mtDNA is oxidatively modified at active replication origins such as ori5. These modifications, which accumulate in DNA structures with more single-stranded character, are recognized by Ntg1, which introduces a DSB at the site. The increased levels of DSBs at replication origins enhance Mhr1-mediated initiation of rolling-circle mtDNA replication, which results in an increase in mtDNA copy number.
DISCUSSION
In this study, we have shown that treating isolated mitochondria with low concentrations (e.g. 10 μM) of hydrogen peroxide increased mtDNA copy number via the functions of Ntg1 and Mhr1. This treatment resulted in accumulation of oxidative modifications preferentially in the ori5-region of mtDNA, and an Ntg1-dependent increase in the DSB level specifically at ori5 in isolated mitochondria and in vitro by use of purified DNA and purified Ntg1. These results led us to propose a model in which elevated levels of ROS (the result of hydrogen peroxide treatment in this study) enhance Ntg1- and Mhr1-dependent rolling-circle-type mtDNA replication to increase mtDNA copy number (Figure 7).
Our previous in vitro and in vivo results demonstrated that the excision-repair enzyme Ntg1, which repairs oxidatively damaged DNA bases, can introduce DSB only at the ori5 locus, and that both Ntg1 and Mhr1 are required for the replication of HS [ori5] [rho–] mtDNA (10). In this study, we further revealed that the ori5 region contains more exposed single-stranded DNA when compared with the rest part of HS [ori5] [rho–] mtDNA, since it is more resistant to the restriction enzyme with a single recognition site in this region, and more sensitive to S1 nuclease (Figure 5C, D and E). This explains the local sensitization of the ori5 region towards oxidative modification by ROS (29). In fact, the ori5-region accumulates a larger number of oxidized bases than other regions of the same DNA, as revealed from its sensitivity toward nicking by Ntg1 in vitro (Figure 5B). In addition, Ntg1 recognizes oxidative bases in single-stranded DNA and cuts single strands (Figure 5F). These observations explain how Ntg1 introduces DSB specifically at ori5-region. By preferentially recognizing oxidative modifications induced by ROS at the ori5-region, Ntg1 induces strand breakage in both complementary strands, resulting in DSB specific to ori5 (Figure 7). Thus, we hypothesized that the observed increase in mtDNA copy number is a result of DSB formation at ori5 mediated by Ntg1.
Supporting this model, the DSB levels at ori5 increased by 3.0-fold during a 1-h incubation with hydrogen peroxide (10 μM) (Figure 3B). MtDNA copy number increased 2.5-fold during this period, and thus was positively correlated with the increase in ori5-specific DSBs (Figure 2B versus Figure 3B). The DSB levels depended on the functions of Ntg1 as well as the hydrogen peroxide treatment (Figure 3C and D). Moreover, oxidative modifications at ori5 increased following a 0.5-h incubation in the presence of hydrogen peroxide, as evidenced by the in vitro generation of DSBs by Ntg1 at ori5 (Figure 4). These results can be explained with our proposed model.
In order to propose that ROS act as regulators to control mtDNA copy number, we investigated whether mtDNA copy number increases when intracellular ROS levels were elevated, either by expressing A. niger GOX gene, or by adding iron to the medium (Figure 6 and Supplementary Figure 2). As expected, even a slight increase of the ROS level significantly increases mtDNA copy number. Since the optimal amount of ROS required for maximum increase in mtDNA copy number in vivo is still unknown, a greater increase in mtDNA copy number may be observed when we further investigate whether redox/oxidative stress regulates mtDNA replication initiation as another study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported in part by a grant from the RIKEN Bioarchitect Research Program to F.L. and M.Y.; by a grant from the Life Science Foundation of Japan to F.L.; by a Grant-in-Aid for Scientific Research (C) No.18570168 from the Ministry of Education, Culture, Sports, Science and Technology of Japan to F.L.; by a Grant-in-Aid for Scientific Research (C) No. 20570171 from the Ministry of Education, Culture, Sports, Science and Technology of Japan to F.L., and by a grant from the RIKEN Strategic Research Program to F.L. Funding for open access charges: Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Moraes CT. What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 2001;17:199–205. doi: 10.1016/s0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 2.Kang D, Hamasaki N. Alterations of mitochondrial DNA in common diseases and disease states: aging, neurodegeneration, heart failure, diabetes, and cancer. Curr. Med. Chem. 2005;12:429–441. doi: 10.2174/0929867053363081. [DOI] [PubMed] [Google Scholar]
- 3.Wei YH, Lee CF, Lee HC, Ma YS, Wang CW, Lu CY, Pang CY. Increases of mitochondrial mass and mitochondrial genome in association with enhanced oxidative stress in human cells harboring 4,977 BP-deleted mitochondrial DNA. Ann. NY Acad. Sci. 2001;928:97–112. doi: 10.1111/j.1749-6632.2001.tb05640.x. [DOI] [PubMed] [Google Scholar]
- 4.Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, Movilla N, Perez-Martos A, Rodriguez de Cordoba S, Gallardo ME, Enriquez JA. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- 5.Moraes CT, Shanske S, Tritschler HJ, Aprille JR, Andreetta F, Bonilla E, Schon EA, DiMauro S. mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am. J. Hum. Genet. 1991;48:492–501. [PMC free article] [PubMed] [Google Scholar]
- 6.Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 7.Shadel GS. Yeast as a model for human mtDNA replication. Am. J. Hum. Genet. 1999;65:1230–1237. doi: 10.1086/302630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling F, Shibata T. Recombination-dependent mtDNA partitioning: in vivo role of Mhr1p to promote pairing of homologous DNA. EMBO J. 2002;21:4730–4740. doi: 10.1093/emboj/cdf466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 10.Ling F, Hori A, Shibata T. DNA recombination-initiation plays a role in the extremely biased inheritance of yeast [rho–] mitochondrial DNA that contains the replication origin ori5. Mol. Cell. Biol. 2007;27:1133–1145. doi: 10.1128/MCB.00770-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eide L, Bjoras M, Pirovano M, Alseth I, Berdal KG, Seeberg E. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc. Natl Acad. Sci. USA. 1996;93:10735–10740. doi: 10.1073/pnas.93.20.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor SD, Zhang H, Eaton JS, Rodeheffer MS, Lebedeva MA, O’Rourke TW, Siede W, Shadel GS. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:3010–3018. doi: 10.1091/mbc.E05-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starkov AA, Fiskum G, Chinopoulos C, Lorenzo BJ, Browne SE, Patel MS, Beal MF. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. J. Neurosci. 2004;24:7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling F, Makishima F, Morishima N, Shibata T. A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J. 1995;14:4090–4101. doi: 10.1002/j.1460-2075.1995.tb00081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yaffe MP. Analysis of mitochondrial function and assembly. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]
- 16.Southern EM. Base sequence and evolution of guinea-pig alpha-satellite DNA. Nature. 1970;227:794–798. doi: 10.1038/227794a0. [DOI] [PubMed] [Google Scholar]
- 17.Foury F. Repair of mitochondrial DNA in Saccharomyces cerevisiae. Induction of cytoplasmic petite mutants in a nuclear mutant exhibiting thermosensitive mitochondrial deoxyribonuclease activity. J. Biol. Chem. 1982;257:781–787. [PubMed] [Google Scholar]
- 18.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. pp. 5.36–5.39. [Google Scholar]
- 19.Rost D, Welker A, Welker J, Millonig G, Berger I, Autschbach F, Schuppan D, Mueller S. Liver-homing of purified glucose oxidase: a novel in vivo model of physiological hepatic oxidative stress (H2O2) J. Hepatol. 2007;46:482–491. doi: 10.1016/j.jhep.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 20.Vernet T, Dignard D, Thomas DY. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- 21.Ling F, Shibata T. Mhr1p-dependent concatemeric mitochondrial DNA formation for generating yeast mitochondrial homoplasmic cells. Mol. Biol. Cell. 2004;15:310–322. doi: 10.1091/mbc.E03-07-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl Acad. Sci. USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schachter F, Cohen D, Kirkwood T. Prospects for the genetics of human longevity. Hum. Genet. 1993;91:519–526. doi: 10.1007/BF00205074. [DOI] [PubMed] [Google Scholar]
- 24.Kleff S, Kemper B, Sternglanz R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 1992;11:699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ezekiel UR, Zassenhaus HP. Localization of a cruciform cutting endonuclease to yeast mitochondria. Mol. Gen. Genet. 1993;240:414–418. doi: 10.1007/BF00280395. [DOI] [PubMed] [Google Scholar]
- 26.Machida K, Tanaka T, Fujita K, Taniguchi M. Farnesol-induced generation of reactive oxygen species via indirect inhibition of the mitochondrial electron transport chain in the yeast Saccharomyces cerevisiae. J. Bacteriol. 1998;180:4460–4465. doi: 10.1128/jb.180.17.4460-4465.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S, Fu J, Zhou Z. In vitro effect of manganese chloride exposure on reactive oxygen species generation and respiratory chain complexes activities of mitochondria isolated from rat brain. Toxicol. in Vitro. 2004;18:71–77. doi: 10.1016/j.tiv.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Ling F, Morioka H, Ohtsuka E, Shibata T. A role for MHR1, a gene required for mitochondrial genetic recombination, in the repair of damage spontaneously introduced in yeast mtDNA. Nucleic Acids Res. 2000;28:4956–4963. doi: 10.1093/nar/28.24.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You HJ, Swanson RL, Harrington C, Corbett AH, Jinks-Robertson S, Senturker S, Wallace SS, Boiteux S, Dizdaroglu M, Doetsch PW. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry. 1999;38:11298–11306. doi: 10.1021/bi991121i. [DOI] [PubMed] [Google Scholar]
- 30.Karahalil B, Roldan-Arjona T, Dizdaroglu M. Substrate specificity of Schizosaccharomyces pombe Nth protein for products of oxidative DNA damage. Biochemistry. 1998;37:590–595. doi: 10.1021/bi971660s. [DOI] [PubMed] [Google Scholar]
- 31.Harrison L, Brame KL, Geltz LE, Landry AM. Closely opposed apurinic/apyrimidinic sites are converted to double strand breaks in Escherichia coli even in the absence of exonuclease III, endonuclease IV, nucleotide excision repair and AP lyase cleavage. DNA Repair. 2006;5:324–335. doi: 10.1016/j.dnarep.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piedade JA, Oliveira PS, Lopes MC, Oliveira-Brett AM. Voltammetric determination of gamma radiation-induced DNA damage. Anal. Biochem. 2006;355:39–49. doi: 10.1016/j.ab.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 34.Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J. Biol. Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 35.Halliwell B, Gutteridge JM. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.