Abstract
Type II DNA topoisomerases (topos) are essential and ubiquitous enzymes that perform important intracellular roles in chromosome condensation and segregation, and in regulating DNA supercoiling. Eukaryotic topo II, a type II topoisomerase, is a homodimeric enzyme that solves topological entanglement problems by using the energy from ATP hydrolysis to pass one segment of DNA through another by way of a reversible, enzyme-bridged double-stranded break. This DNA break is linked to the protein by a phosphodiester bond between the active site tyrosine of each subunit and backbone phosphate of DNA. The opening and closing of the DNA gate, a critical step for strand passage during the catalytic cycle, is coupled to this enzymatic cleavage/religation of the backbone. This reversible DNA cleavage reaction is the target of a number of anticancer drugs, which can elicit DNA damage by affecting the cleavage/religation equilibrium. Because of its clinical importance, many studies have sought to determine the manner in which topo II interacts with DNA. Here we highlight recent single-molecule fluorescence resonance energy transfer and crystallographic studies that have provided new insight into the dynamics and structure of the topo II DNA gate.
INTRODUCTION
DNA topoisomerases (topos) are molecular machines that regulate the topology of DNA in the cell by allowing DNA strands or double helices to pass through one another. They accomplish this feat via a mechanism involving the breakage and reformation of phosphodiester bonds in the DNA backbone (1–7). Transesterification between an enzyme tyrosyl residue and a DNA phosphate group severs the DNA backbone and results in a covalent enzyme–DNA intermediate; this reaction can be readily reversed to reseal the DNA break. Cells require this activity to resolve topological problems that arise during DNA metabolism because of the double helical nature of DNA and the manner in which DNA is packaged. For such processes as replication or transcription, the helices must separate to allow access to the information stored in the DNA. However, because DNA exists either as circular rings or is associated with proteins, the DNA double-helix supercoils when unwound by cellular machinery. Not only must topos remove these supercoils, they must also decatenate completely replicated DNA prior to cell division, and they play key roles in the maintenance of chromosome structure and genome stability. In the following sections, we describe the architecture of topo II and the proposed mechanism of action that allows it to carry out such DNA transactions.
Architecture of topo II
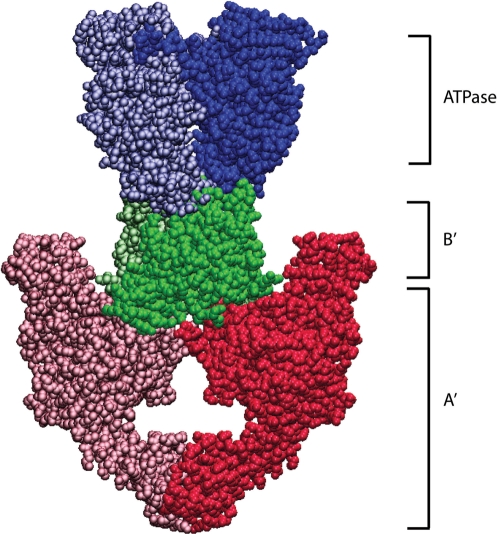
Topoisomerases are classified into two main families, type I and type II, based on whether they break one strand of DNA at a time (type I) or create a double-stranded break in the DNA (type II) (8,9). Here we focus on eukaryotic type II topos, whose dimeric architecture enables them to create such double-stranded breaks. Eukaryotic topo II is composed of three principal domains (ATPase, B′, A′) that are linked by flexible hinge regions (Figure 1) (10–12). Nucleotide binding triggers dimerization of the N-terminal ATPase domains, whose opening and closing is regulated by ATP hydrolysis and product release (10,13,14). The ATPase domain is followed by the central core region, which is composed of the B′ domain and the N-terminal portion of A′. The A′ domain contains the active site tyrosyl residue required for catalysis and is thus referred to as the DNA breakage and rejoining domain. Topo II has a hydrophilic C-terminus (not pictured in Figure 1) that immediately follows the A′ domain, and its sequence is only somewhat similar for closely related species. The C-terminal tail is dispensable for biochemical activity (15–17), but evidence suggests that it serves several important intracellular roles, such as nuclear localization and interaction with other proteins (18–20).
Figure 1.
Structure of yeast topo II. Structures of Saccharomyces cerevisiae topo II (PDB 1PVG and 1BGW) were computationally docked together to illustrate a possible representation of the enzyme. ATPase domains, shown in blue, form the N-gate. The central B′-subfragment forms the DNA gate (green). The C-terminal A′ portion (red) encloses a hole large enough to accommodate T-segment DNA, which is thought to exit through the C-gate.
Over the past two decades, several structures of type II topos have been determined (8,9,21–23). These structures have enabled a deeper understanding of the topo II catalytic mechanism. For eukaryotic type II topos, structures have been determined for N- and C-terminal fragments of Saccharomyces cerevisiae topo II as well as the ATPase domain of human topo IIα. Although a structure has not been solved for a full-length type II topoisomerase, the available structures provide a near-complete picture of the architecture of topo II.
One such structure includes the yeast ATPase domain bound to AMPPNP (24). The ATPase region forms a heart-shaped dimer upon nucleotide binding and folds into two discrete domains: the N-terminal region bears the characteristic GHKL ATP-binding fold, while the C-terminal region comprises the transducer domain. The GHKL superfamily is named for its founding members DNA gyrase, Hsp90, histidine kinase and Mut L, and it forms the primary dimer interface of the ATPase region. The C-terminal transducer domain is situated below the ATP-binding fold and is thought to transduce ATP binding and hydrolysis signals to downstream regions of the enzyme, thus coordinating DNA cleavage/religation with DNA transport. Structural support for this proposed transducing role is evidenced by structures of human topo IIα, which were solved with the enzyme in different nucleotide-bound states, either to AMPPNP or ADP (25). Relative to the AMPPNP-bound form, topo II undergoes structural arrangements in the ADP-bound form in which the transducer domain swings out to form a more open and mobile cavity at the base of the structure.
In addition to the structures described for the ATPase domain, structures of a large yeast fragment containing the B′ and A′ domains forming the cleavage core have been solved (22,26,27). The core region of topo II also forms a heart-shaped dimer with a central cavity more than large enough to accommodate duplex DNA, which is thought to exit at the C-terminal dimer interface (C-gate) of the enzyme. The N-terminal portion of the topo II cleavage/core region contains what has been termed the TOPRIM (TOpoisomerase/PRIMase) domain. This region contains the conserved acidic residues believed to promote formation of the topo II/DNA phosphodiester linkage by coordinating Mg2+, which is essential for DNA cleavage. The core of topo II also includes a winged-helix domain (WHD) that contains the active site tyrosyl residue essential for DNA cleavage, and a ‘tower’ domain, which was found to act as a supporting element for DNA (22). The recently determined topo II/DNA structure (described in more detail in succeeding sections) also provides support to previous biochemical evidence that the C-terminal domain (C-gate) of topo II opens to allow expulsion of the transported DNA segment (28). Structures from both N- and C-terminal fragments of topo II have been combined to illustrate a more complete picture of the enzyme, shown in Figure 1. This structure includes the ATPase domain and the B′ and A′ fragments of the yeast enzyme. A detailed view of topo II's structural subdomains can be found in a recent review (23).
Two-gate model for DNA transport
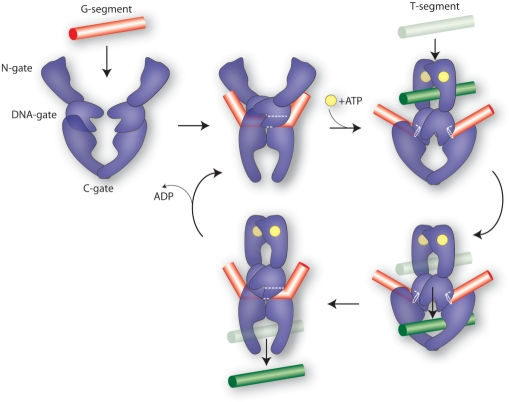
Structural and biochemical evidence have led to the two-gate model for topo II-mediated DNA transport (Figure 2) (22,26,28–30). In this model, topo II utilizes the energy from ATP to transport one intact DNA duplex through an enzyme-mediated 4-bp staggered double-stranded break in a second duplex. In the first step of the reaction, the N-terminal ATPase domains (N-gate) are separated, allowing topo II to bind a DNA duplex within the binding/cleavage core. This DNA has been termed the gate or ‘G-segment’ because it serves as a gate through which another strand must pass. ATP binding to the N-terminal GHKL domains triggers their dimerization and capture of a second DNA duplex. Closure of this N-gate induces a succession of conformational changes that promotes the cleavage and opening of the G-segment, followed by transport of the captured DNA (T-segment DNA) through the break. The T-segment then exits through the opened C-terminal dimerization interface (C-gate), and the DNA gate closes to allow G-segment religation.
Figure 2.
Catalytic cycle of topo II, adapted from Dong and Berger (22). Topo II binds to a DNA duplex, the G-segment (red bar), causing significant DNA bending. ATP (yellow circle) binds to the N-terminal domains, promoting the capture of a second DNA duplex, the T-segment (green bar) and dimerization of the ATPase domains (N-gate). Closure of the N-gate stimulates cleavage of G-segment DNA, coupled with opening of the DNA gate to allow passage of the T-segment through this double-stranded break. The G-segment is religated and the T-segment exits after opening of the C-gate.
Until recently, topo II-mediated G-segment breaks had only been monitored by trapping cleaved DNA with a potent denaturant and analyzing the resultant fragments with techniques such as gel electrophoresis. Because this method is indirect and could possibly perturb the DNA gate equilibrium, new methods were needed to study the topo II DNA gate. Therefore, single-molecule fluorescence resonance energy transfer (FRET) was developed to directly examine the topo II conformational changes accompanying these G-segment breaks (31). Furthermore, new crystallographic work has unlocked important structural details about the topo II/DNA complex (22). This new information is discussed below in terms of the two-gate model.
TOPO-II DNA GATE STUDIES
Fluorescence resonance energy transfer (FRET)
FRET has been used extensively for estimating the spatial separation between fluorophores (32). The relationship between FRET efficiency and distance is described by:
| 1 |
where E is the energy transfer efficiency, Ro is the distance for 50% energy transfer (Förster distance), and r is the distance between donor and acceptor fluorophore pairs (33). Because the energy transfer efficiency of two fluorophores depends on the distance between them, this technique is a powerful tool for measuring the internal distance changes of molecules. In the work to be discussed, a nucleic-acid substrate labeled with a FRET pair was used to examine the conformational changes occurring in the topo II DNA gating reaction.
Fluorescent topo II substrate
DNA cleavage, aperture, closure and religation are critical steps in the topo II reaction cycle that are targeted by a host of clinically important anticancer drugs which take advantage of the fact that topo II creates breaks in DNA. Smiley et al. used FRET in an effort to directly study the topo II movements associated with forming these DNA breaks (31). A substrate was created (Figure 3A) that positioned a pair of fluorophores on either side of a Drosophila melanogaster topo II cleavage site (34,35). DNA containing such a sequence has been used extensively in experiments to investigate DNA binding and cleavage by topo II (36–38). While topo II has a modest sequence preference in the cleavage reaction, use of this oligonucleotide substrate with a strongly preferred sequence will result in cleavage at a single specific site. Substrates 28 bp in length were used because earlier footprinting experiments demonstrated that the contact between the enzyme and DNA was limited to ∼25 bp (39,40). Alexa Fluor 488 and Alexa Fluor 555 (or Alexa Fluor 546) were chosen as the FRET pair, and were placed such that they undergo efficient FRET when the substrate is intact, but their FRET efficiency decreases when the DNA gate opens (Figure 3B). Each partner of the FRET pair was conjugated to the thymidine ring at the C5 position and separated from the double helix by a flexible linker arm.
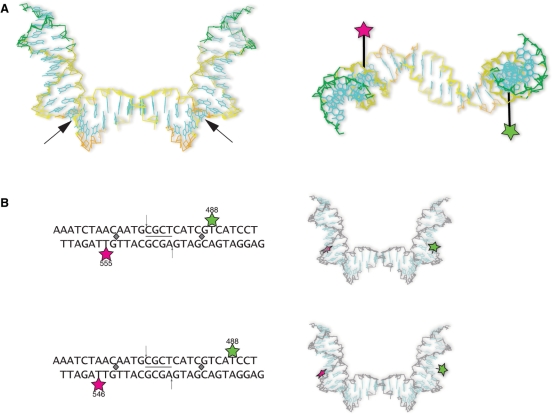
Figure 3.
(A) Design of the fluorescent DNA substrate. A 28-bp oligonucleotide-based DNA substrate was synthesized with a known Drosophila topo II cleavage site (indicated by arrows) flanked by a FRET pair, Alexa Fluor 488 (green) and Alexa Fluor 555 (magenta), separated by ∼1.5 turns on complementary strands. Another substrate was created with Alexa Fluor 488 at the same position, but with Alexa Fluor 546 linked to a different thymidine (red). (B) A schematic diagram of the topo II/DNA complex illustrating the use of FRET in monitoring the opening and closing of the DNA gate. The donor and acceptor fluorophores on the DNA are marked in green and magenta, respectively. ATP is indicated by the yellow circles. (C) Decrease in FRET efficiency is reversible. The FRET efficiency of DNA (blue bar) decreases upon addition of topo II when Mg2+/ATP is present (red bar). When the reaction is reversed with EDTA/NaCl treatment (green bar), the FRET efficiency is restored.
The Förster distance (Ro) for Alexa Fluor 488 and Alexa Fluor 555 is 70 Å, which indicates that the FRET efficiency is 50% when they are 70 Å apart. For the Alexa Fluor 488 and Alexa Fluor 546 pair, Ro is 64 Å. Given that the distance between the fluorophores in the intact substrate is predicted to be ∼54 Å (for Alexa Fluor 488/Alexa Fluor 555) or ∼57 Å (for Alexa Fluor 488/Alexa Fluor 546) based on typical B-helical DNA geometry, FRET between these pairs of fluorophores is efficient when the DNA gate is closed. In the open state, the severed DNA ends are predicted to move apart at least as far as the diameter of B-DNA, or ∼20 Å (27). The fluorophore distances in either the open or closed DNA gate are similar to Ro, and therefore changes in distance associated with the predicted gate opening should result in a relatively large change in FRET.
Ensemble experiments
Ensemble experiments with the fluorescent substrate demonstrate that there is a large decrease in FRET upon addition of Mg2+ (10 mM)/ATP (1 mM). This decrease in FRET was attributed to an increase in distance between the fluorophore pair as the DNA gate opens to physically separate the fluorophores. The FRET efficiency is sensitive to temperature; if the normal reaction temperature (30°C) is significantly lowered, only a small decrease in FRET occurs, suggesting that a majority of the DNA gates are in the closed state at low temperatures. Furthermore, the FRET decrease can be reversed with the addition of high salt and EDTA (Figure 3C), which is known to reverse topoisomerase cleavage reactions.
Sensitivity to temperature and EDTA/salt reversal strongly suggests that the observed decrease in FRET is due to topo II DNA gate opening. However, the possibility exists that FRET decreases because of relative orientation changes between the FRET pair. In calculating Ro, a dynamic average of all possible orientations between the donor and acceptor was assumed (41). To determine whether this assumption was valid, fluorescence polarization was measured. Polarization values were consistent with significant rotational freedom in the donor and acceptor pairs, and were unchanged upon addition of Mg2+/ATP. This suggests that changes and/or restrictions in the relative orientations of the donor and acceptor are not responsible for the observed FRET decrease. Taken in combination with several other controls, these lines of evidence are consistent with the measured FRET changes being sensitive to the opening and closing of the topo II DNA gate. To further probe the DNA gates of individual topoisomerase enzymes, the fluorescent substrate was used for single-molecule FRET.
Monitoring the topo II DNA gate with single-molecule FRET
Single-molecule FRET (smFRET) experiments are powerful tools for studying enzyme dynamics and mechanisms (42,43). Examination of the reaction trajectories (time dependence of fluorescence) of single molecules can provide mechanistic information that is not readily available from ensemble experiments. Therefore, smFRET was used to investigate the topo II DNA gate dynamics.
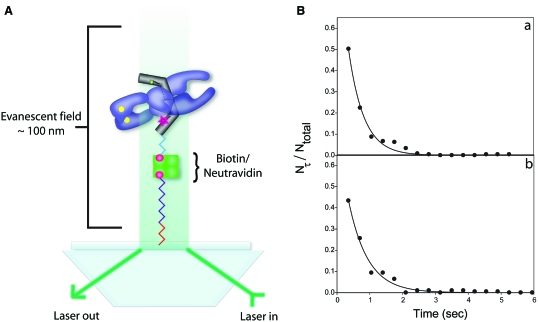
As shown from ensemble experiments, the oligonucleotide substrate with an attached FRET pair was useful for examining the topoisomerase reaction at the DNA gate. For the smFRET experiments, the fluorescent DNA was tagged with biotin at one end and bound to a NeutrAvidin-coated glass slide (Figure 4A). Single molecules of DNA were identified with total internal reflection fluorescence (TIRF) microscopy, and their behavior was observed in the presence of topo II, Mg2+ and ATP. Time trajectories of the individual molecules were monitored, and their fluorescence intensity fluctuated between two distinct states in the presence of 50 nM Drosophila topo II and the cofactors Mg2+/ATP. The fluctuation between the two fluorescent states was dependent on the presence of both ATP and Mg2+, although Mg2+ alone could support occasional gate opening. The critical dependence of the FRET fluctuation on Mg2+/ATP is not due to enzyme binding and dissociation events, because experimental evidence from filter-binding assays indicates that the absence of Mg2+/ATP does not significantly affect either the affinity or specificity of topo II binding to DNA (44). Furthermore, our unpublished results show that topo II remains bound to the fluorescent DNA substrate even upon extensive washing of the slide or addition of external DNA. Therefore, the dependence on Mg2+/ATP is consistent with the FRET fluctuation corresponding to DNA gate opening and closing. We also examined gate movement in the presence of AMPPNP, which will support one round of topo II-mediated DNA strand passage (45). When AMPPNP was substituted for ATP, the single molecules did not fluctuate between two distinct states, which provides further evidence that ATP hydrolysis is required for the gate opening/closing transition.
Figure 4.
(A) Schematic representation of the method of immobilizing and visualizing single DNA molecules. Single DNA molecules are attached to a glass slide using biotin/NeutrAvidin and are separated from the surface via linker arms shown in red, purple and blue. The single DNA molecules were excited using prism-based total internal reflection fluorescence microscopy (TIRF) to create an evanescent field above the surface of the slide. (B) Fraction of single molecules with a lifetime τ, Nτ/Ntotal, as a function of lifetime. These plots were generated from the reaction trajectories (time dependence of FRET) of DNA with Alexa Fluor 488/Alexa Fluor 555 in the presence of 50 nM Drosophila topo II and Mg2+/ATP. The solid lines are weighted non-linear least square fits to Equation (3), from which the ensemble average rate constants can be obtained: kopen from A and kclose from B.
The high FRET state corresponds to the closed DNA gate, while the low FRET state corresponds to the opened DNA gate. The transition between the open and closed states can be represented as:
| 2 |
where kopen and kclose are the rate constants for the topo II-mediated opening and closing reactions at the DNA gate, respectively. The distribution of reaction lifetimes, τ, for a one-step reaction between high- and low-FRET states is given by the relationship (46):
| 3 |
where Nτ is the number of events with a lifetime τ, Ntotal is the total number of events, and k is the ensemble average first-order rate constant [kopen or kclose as defined by Equation (2)]. A plot of the fraction of single molecules with a lifetime τ, Nτ/Ntotal, versus τ follows an exponential decay (Figure 4B) from which the ensemble average rate constants can be determined. A single exponential fits the lifetime decay curves, denoting that the data are consistent with a transition between the closed and open states [Equation (2)].
From the lifetime decay curves, rate constants for both DNA gate opening and closing were found to be in the range of 1–2 s−1. This result is consistent with rate constants for the overall topo II strand passage reaction, which are also in the range of 1–3 s−1 when measured with either biochemical or biophysical methods (45,47,48). Surprisingly, the apparent equilibrium constant is close to one since rate constants for DNA gate opening and closing are similar. This suggests that under the conditions tested, the DNA gate spends about half its time in the open state, and half its time in the closed state, thus equally populating both states during steady-state ATP hydrolysis. Additional experiments show that topo II can equally populate both states even with a substrate 38 bp in length, suggesting that at least in the range of 28–38 bp, the rate constants for gate opening and closing do not critically depend on the length of DNA. The ensemble and single-molecule FRET experiments have provided direct measurements of the topo II DNA gate dynamics in the absence of a denaturant. The cleavage/religation equilibrium, a process related to gate opening/closing, has previously been investigated through the use of a denaturant to trap the reaction intermediates. Under these conditions, the equilibrium clearly favors the religated state (34). This has led to the assumption that only a small proportion of topo II/DNA complexes have opened DNA gates at any given time. The topo II DNA gate equilibrium may be changed by specific compounds (many of which comprise clinically important anticancer drugs) that promote the cleaved DNA state and therefore may possibly favor the opened DNA gate (49,50). The smFRET results, however, are not necessarily at odds with experiments using a protein denaturant. Denaturing topo II to trap the cleaved DNA could alter the DNA gate opening/closing equilibrium to selectively favor the closed state and/or specifically capture particular intermediates along the kinetic pathway of gate opening/closing. Furthermore, support that the FRET results provide information regarding DNA gate dynamics comes from estimates of the distance change at the DNA gate. Ensemble and single molecule methods are in reasonable agreement, with both suggesting that the DNA gate can open ∼21–23 Å, which is large enough to accommodate passage of a duplex segment.
Comparison of smFRET substrate with topo II/DNA structure
Dong and Berger determined the crystal structure of the DNA binding and cleavage core fragment of S. cerevisiae topo II bound to a doubly nicked 34-bp oligonucleotide DNA duplex (22). The duplex was found to bind in the previously predicted DNA-binding site (26,51), located in the N-terminal portion of the A′ domain within a deep, positively charged semicircular groove. Approximately 26 bp of the duplex are in direct contact with topo II, although the enzyme makes little specific contact with the nucleotide bases.
The most striking feature of this complex is the dramatic degree to which topo II alters the DNA structure. Each subunit of topo II induces a sharp 75° bend in the DNA backbone, resulting in a global 150° bend. This bend is induced by an isoleucine (Ile 833) residue located on a β-hairpin near the DNA-binding groove. The highly conserved Ile residue intercalates between two DNA basepairs eight nucleotides from the scissile phosphate, thus deforming the DNA minor groove by significantly widening it. This locally induced distortion causes DNA to assume an altered U-like structure, such that the central ∼12 bp (bottom of the ‘U’) remain virtually straight (Figure 5A). The DNA at the base of the ‘U’ adopts an A-form conformation, while the duplex region that bends upward as a result of Ile intercalation (the arms of the ‘U’) remains B-form. The bent 34-bp DNA substrate packs to form a 68-bp DNA minicircle in the crystal lattice. While the bent DNA conformation could be a rare intermediate selected by favorable crystal packing, our previous and new FRET results discussed below suggest that topo II also bends DNA in solution.
Figure 5.
(A) Left panel. Topo II dramatically reshapes DNA. This image depicts the topo II-induced ‘U’-shaped DNA from the topo II/DNA complex (PDB 2RGR) (22). Black arrows indicate the sites of Ile intercalation. Right panel. Top down view of DNA from the topo II/DNA complex. The position of Alexa Fluor 488 (green star) and Alexa Fluor 555 (magenta star) are shown in their respective locations upon topo II bending DNA (linker arm length scaled approximately). The fluorophores are now on opposite sides of the DNA helix. (B) Upper panel. Initial fluorescent DNA substrate. The 28-bp substrate used in initial FRET studies is shown for comparison with the alternate substrate. The cleavage site is flanked by the FRET pair, Alexa Fluor 488 (green star) and Alexa Fluor 555 (magenta star), on complementary strands. The gray diamonds indicate the sites of Ile intercalation, which sharply bends the DNA. The images shown to the right of the substrates depict the relative spatial locations of the fluorophores in the newly determined topo II/DNA structure. Lower panel. Fluorescent DNA substrate with a greater distance separating the FRET pair. The substrate is similar to that in shown in the upper panel, but Alexa Fluor 488 is positioned three nucleotides distal to the cleavage site, and Alexa Fluor 546 is the acceptor.
In light of the new topo II/DNA structure, it is useful to compare the position of the fluorophores in the enzyme-free fluorescent substrate to their analogous positions within the topo II/DNA complex. Because topo II bends DNA, an increased FRET efficiency is predicted to occur upon topo II binding DNA. However, Smiley et al. did not observe an increase in FRET accompanying topo II binding with the fluorescent DNA substrate (31). To help explain this phenomenon, we examined the position of the fluorophores relative to the topo II/DNA structure. Inspection of the complex reveals that Alexa Fluor 488 and Alexa Fluor 555 flank the two DNA bases adjacent to the sites of Ile intercalation. Each fluorophore is positioned at the base of the ‘U’, near the respective Ile residue, and on the second nucleotide that begins to bend upward forming the arms of the ‘U’ (Figure 5B). Being located at the base of the ‘U’ causes their relative spatial positions to be less affected by DNA bending than if they were situated further apart, closer to the ends of the ‘U’ arms. Furthermore, inspection of the complex shows that Alexa Fluor 488 and Alexa Fluor 555 are situated on opposite sides of the DNA helix, instead of on the same face as in the DNA-free substrate (Figure 5A). This positioning helps explain why FRET does not increase upon topo II binding DNA. The distance between donor and acceptor fluorophores was estimated by hand-modeling a linker arm between the nucleotide base and fluorophore. From these coarse estimates, possible distances between donor and acceptor pairs ranged from ∼55 Å to ∼68 Å. The estimated distances are within the range of distances calculated from FRET measurements. FRET values suggest that the fluorophores are ∼51 Å apart in the closed DNA gate and ∼72 Å apart in the open DNA gate. Because of the uncertainty in the locations of the fluorophores when DNA is bound to topo II, it is plausible that the apparent ∼21 Å gate opening is a combination of both topo II-induced DNA distortion and partial DNA gate opening.
Experiments with the fluorophores positioned further apart should result in a FRET increase upon topo II binding DNA, since enzyme binding bends the ends of the DNA toward one another. Consistent with this prediction, our experiments show that the FRET efficiency of such an alternate DNA substrate indeed increases upon topo II binding to DNA. In the new substrate, the FRET pair (Alexa Fluor 488/Alexa Fluor 546) flanks the two sites of Ile intercalation, with Alexa Fluor 488 now located three nucleotides distal to the respective Ile, further up the arms of the ‘U’ (proximal to the 3′-end) (Figure 5B). The distance between the attached FRET pair is expected to become closer by ∼6 Å upon topo II binding the substrate. Initial results demonstrate that topo II increases the FRET efficiency of this substrate by ∼15%, consistent with this distance change (unpublished data). Additional experiments with this and related substrates should further elucidate the molecular motions associated with the topo II DNA gate.
Role of T-segment in gate opening
Previous biochemical studies suggest that a T-segment is required for topo II-mediated G-segment cleavage (37), while more recent work suggests that topo II is competent to cleave DNA in the absence of a T-segment (38). The smFRET experiments were conducted in the absence of a T-segment, and therefore suggest that the topo II DNA gate can open and close without the presence of a second DNA strand. However, other biochemical investigations show that a topo II mutant incapable of cleaving DNA cannot trap a T-segment upon AMPPNP-induced clamping of the N-gate (2,52), supporting the suggestion from the structural studies that the cavity of the N-gate is too small to accommodate duplex DNA (21,53). While the inability of the mutant enzyme to trap a T-segment is consistent with requisite opening of the DNA gate upon T-segment capture, it does not necessarily preclude gate opening in the absence of a T-segment. The possibility exists that the T-segment assists with opening the DNA gate, although ATP alone may be sufficient for complete gate opening in its absence. It is also possible that the FRET decrease is due to the combined effects of partial gate opening in the absence of a T-segment and structural rearrangement of the G-segment. Furthermore, the dynamics of DNA gate opening/closing may be altered in the presence of a T-segment, which has been proposed to affect the DNA gate equilibrium by forcing the DNA gate closed after its passage through the G-segment (52,53). Additional experiments are clearly required to further address the role of the T-segment in the topo II catalytic cycle.
CONCLUSION
The smFRET studies provide a direct observation of the conformational changes at the topo II DNA gate. Furthermore, recent structural studies have offered the first picture of the topo II/DNA complex, allowing a more in-depth understanding of how topo II interacts with DNA. The combination of these studies has further advanced elucidation of the topo II mechanism, while at the same time stimulating additional questions about how the enzyme works. For example, how far can the DNA gate open without a T-segment, and how much of the FRET decrease is due to topo II-induced structural rearrangement of the G-segment? Will the presence of a T-segment affect the DNA gate dynamics and equilibrium? Future smFRET studies with different substrates and better time resolution will help address these questions. Additional structures of topo II/DNA complexes are also needed to advance our knowledge of the complex movements at the topo II DNA gate.
FUNDING
This work was supported by National Institutes of Health (GM29006 to T.-s.H; GM65128 to G.G.H.). T.R.L.C. was supported in part by a grant from the Department of Defense Breast Cancer Research Program. Funding for open access charge: National Institutes of Health (GM29006).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Dr Derike Smiley for conducting initial single molecule experiments. We also thank Dr Jane Richardson for assistance with assembling the topo II structure. Christopher Capp and Stefanie Hartman Chen provided helpful comments on the manuscript.
REFERENCES
- 1.Hsieh T-S. DNA topoisomerases. Curr. Opin. Cell Biol. 1992;4:396–400. doi: 10.1016/0955-0674(92)90004-v. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Quart. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 3.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 4.Wang JC. DNA topoisomerases. Ann. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 5.Cozzarelli NR. DNA topoisomerases. Cell. 1980;22:327–328. doi: 10.1016/0092-8674(80)90341-4. [DOI] [PubMed] [Google Scholar]
- 6.Reece RJ, Maxwell A. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 1991;26:335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- 7.Wigley DB. Structure and mechanism of DNA topoisomerases. Ann. Rev. Biophys. Biomol. Struct. 1995;24:185–208. doi: 10.1146/annurev.bb.24.060195.001153. [DOI] [PubMed] [Google Scholar]
- 8.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 9.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 10.Lindsley JE, Wang JC. Proteolysis patterns of epitopically labeled yeast DNA topoisomerase II suggest an allosteric transition in the enzyme induced by ATP binding. Proc. Natl Acad. Sci. USA. 1991;88:10485–10489. doi: 10.1073/pnas.88.23.10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MP, Hsieh TS. Linker insertion mutagenesis of Drosophila topoisomerase II. Probing the structure of eukaryotic topoisomerase II. J. Mol. Biol. 1994;235:436–447. doi: 10.1006/jmbi.1994.1004. [DOI] [PubMed] [Google Scholar]
- 12.Shiozaki K, Yanagida M. A functional 125-kDa core polypeptide of fission yeast DNA topoisomerase II. Mol. Cell Biol. 1991;11:6093–6102. doi: 10.1128/mcb.11.12.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu T, Sage H, Hsieh T.-S. ATPase domain of eukaryotic DNA topoisomerase II. Inhibition of ATPase activity by the anti-cancer drug bisdioxopiperazine and ATP/ADP-induced dimerization. J. Biol. Chem. 2002;277:5944–5951. doi: 10.1074/jbc.M111394200. [DOI] [PubMed] [Google Scholar]
- 14.Lindsley JE, Wang JC. Study of allosteric communication between protomers by immunotagging. Nature. 1993;361:749–750. doi: 10.1038/361749a0. [DOI] [PubMed] [Google Scholar]
- 15.Crenshaw DG, Hsieh T. Function of the hydrophilic carboxyl terminus of type II DNA topoisomerase from Drosophila melanogaster. I. In vitro studies. J. Biol. Chem. 1993;268:21328–21334. [PubMed] [Google Scholar]
- 16.Caron PR, Watt P, Wang JC. The C-terminal domain of Saccharomyces cerevisiae DNA topoisomerase II. Mol. Cell Biol. 1994;14:3197–3207. doi: 10.1128/mcb.14.5.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiozaki K, Yanagida M. Functional dissection of the phosphorylated termini of fission yeast DNA topoisomerase II. J. Cell. Biol. 1992;119:1023–1036. doi: 10.1083/jcb.119.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaiu WL, Hu T, Hsieh TS. The hydrophilic, protease-sensitive terminal domains of eucaryotic DNA topoisomerases have essential intracellular functions. Pacific Symposium on Biocomputing, 1999:578–589. doi: 10.1142/9789814447300_0057. [DOI] [PubMed] [Google Scholar]
- 19.Adachi N, Miyaike M, Kato S, Kanamaru R, Koyama H, Kikuchi A. Cellular distribution of mammalian DNA topoisomerase II is determined by its catalytically dispensable C-terminal domain. Nuclic Acids Res. 1997;25:3135–3142. doi: 10.1093/nar/25.15.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crenshaw DG, Hsieh T. Function of the hydrophilic carboxyl terminus of type II DNA topoisomerase from Drosophila melanogaster. II. In vivo studies. J. Biol. Chem. 1993;268:21335–21343. [PubMed] [Google Scholar]
- 21.Schoeffler AJ, Berger JM. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans. 2005;33:1465–1470. doi: 10.1042/BST0331465. [DOI] [PubMed] [Google Scholar]
- 22.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 23.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Quart. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 24.Classen S, Olland S, Berger JM. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. [Erratum (2003) Proc. Natl Acad. Sci. USA, 100, 14510] Proc. Natl Acad. Sci. USA. 2003;100:10629–10634. doi: 10.1073/pnas.1832879100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei H, Ruthenburg AJ, Bechis SK, Verdine GL. Nucleotide-dependent domain movement in the ATPase domain of a human type IIA DNA topoisomerase. J. Biol. Chem. 2005;280:37041–37047. doi: 10.1074/jbc.M506520200. [DOI] [PubMed] [Google Scholar]
- 26.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 27.Fass D, Bogden CE, Berger JM. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat. Struct. Mol. Biol. 1999;6:322–326. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 28.Roca J, Berger JM, Harrison SC, Wang JC. DNA transport by a type II topoisomerase: Direct evidence for a two-gate mechanism. Proc. Natl Acad. Sci. USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger JM, Wang JC. Recent developments in DNA topoisomerase II structure and mechanism. Curr. Opin. Struct. Biol. 1996;6:84–90. doi: 10.1016/s0959-440x(96)80099-6. [DOI] [PubMed] [Google Scholar]
- 30.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 31.Smiley RD, Collins TR, Hammes GG, Hsieh TS. Single-molecule measurements of the opening and closing of the DNA gate by eukaryotic topoisomerase II. Proc. Natl Acad. Sci. USA. 2007;104:4840–4845. doi: 10.1073/pnas.0700342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stryer L, Haugland RP. Energy transfer: a spectroscopic ruler. Proc. Natl Acad. Sci. USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Förster T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Annalen. Der. Physik. 1948;437:55–75. [Google Scholar]
- 34.Sander M, Hsieh T.-S. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem. 1983;258:8421–8428. [PubMed] [Google Scholar]
- 35.Sander M, Hsieh T.-S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985;13:1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lund K, Andersen AH, Christiansen K, Svejstrup JQ, Westergaard O. Minimal DNA requirement for topoisomerase II-mediated cleavage in vitro. J. Biol. Chem. 1990;265:13856–13863. [PubMed] [Google Scholar]
- 37.Corbett AH, Zechiedrich EL, Osheroff N. A role for the passage helix in the DNA cleavage reaction of eukaryotic topoisomerase II. A two-site model for enzyme-mediated DNA cleavage. J. Biol. Chem. 1992;267:683–686. [PubMed] [Google Scholar]
- 38.Mueller-Planitz F, Herschlag D. Interdomain communication in DNA topoisomerase II: DNA binding and enzyme activation. J. Biol. Chem. 2006;281:23395–23404. doi: 10.1074/jbc.M604119200. [DOI] [PubMed] [Google Scholar]
- 39.Lee MP, Sander M, Hsieh T.-s. Nuclease protection by Drosophila DNA topoisomerase II. J. Biol. Chem. 1989;264:21779–21787. [PubMed] [Google Scholar]
- 40.Thomsen B, Bendixen C, Lund K, Andersen AH, Sorensen BS, Westergaard O. Characterization of the interaction between topoisomerase II and DNA by transcriptional footprinting. J. Mol. Biol. 1990;215:237–244. doi: 10.1016/S0022-2836(05)80342-0. [DOI] [PubMed] [Google Scholar]
- 41.Dale RE, Eisinger J, Blumberg WE. The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys. J. 1979;26:161–193. doi: 10.1016/S0006-3495(79)85243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Min W, English BP, Luo G, Cherayil BJ, Kou SC, Xie XS. Fluctuating enzymes: lessons from single-molecule studies. Acc. Chem. Res. 2005;38:923–931. doi: 10.1021/ar040133f. [DOI] [PubMed] [Google Scholar]
- 43.Smiley RD, Zhuang Z, Benkovic SJ, Hammes GG. Single-molecule investigation of the T4 bacteriophage DNA polymerase holoenzyme: multiple pathways of holoenzyme formation. Biochemistry. 2006;45:7990–7997. doi: 10.1021/bi0603322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sander M, Hsieh T.-S, Udvardy A, Schedl P. Sequence dependence of Drosophila topoisomerase II in plasmid relaxation and DNA binding. J. Mol. Biol. 1987;194:219–229. doi: 10.1016/0022-2836(87)90370-6. [DOI] [PubMed] [Google Scholar]
- 45.Osheroff N, Shelton ER, Brutlag DL. DNA topoisomerase II from Drosophila melanogaster. J. Biol. Chem. 1983;258:9536–9543. [PubMed] [Google Scholar]
- 46.Antikainen NM, Smiley RD, Benkovic SJ, Hammes GG. Conformation coupled enzyme catalysis: single-molecule and transient kinetics investigation of dihydrofolate reductase. Biochemistry. 2005;44:16835–16843. doi: 10.1021/bi051378i. [DOI] [PubMed] [Google Scholar]
- 47.Lindsley JE, Wang JC. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J. Biol. Chem. 1993;268:8096–8104. [PubMed] [Google Scholar]
- 48.Strick TR, Croquette V, Bensimon D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000;404:901–904. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 49.Liu LF. DNA topoisomerase poisons as antitumor drugs. Ann. Rev. Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 50.Wilsterman AM, Osheroff N. Stabilization of eukaryotic topoisomerase II-DNA cleavage complexes. Curr. Top. Med. Chem. 2003;3:321–338. doi: 10.2174/1568026033452519. [DOI] [PubMed] [Google Scholar]
- 51.Li W, Wang JC. Footprinting of yeast DNA topoisomerase II lysyl side chains involved in substrate binding and interdomainal interactions. J. Biol. Chem. 1997;272:31190–31195. doi: 10.1074/jbc.272.49.31190. [DOI] [PubMed] [Google Scholar]
- 52.Roca J. The path of the DNA along the dimer interface of topoisomerase II. J. Biol. Chem. 2004;279:25783–25788. doi: 10.1074/jbc.M402555200. [DOI] [PubMed] [Google Scholar]
- 53.Wang JC. Unlocking and opening a DNA gate. Proc. Natl Acad. Sci. USA. 2007;104:4773–4774. doi: 10.1073/pnas.0701070104. [DOI] [PMC free article] [PubMed] [Google Scholar]