Abstract
The LAGLIDADG homing endonuclease (LHE) I-AniI has adopted an extremely efficient secondary RNA splicing activity that is beneficial to its host, balanced against inefficient DNA cleavage. A selection experiment identified point mutations in the enzyme that act synergistically to improve endonuclease activity. The amino-acid substitutions increase target affinity, alter the thermal cleavage profile and significantly increase targeted recombination in transfected cells. The RNA splicing activity is not affected by these mutations. The improvement in DNA cleavage activity is largely focused on one of the enzyme's two active sites, corresponding to a rearrangement of a lysine residue hypothesized to act as a general base. Most of the constructs isolated in the screen contain one or more mutations that revert an amino-acid identity to a residue found in one or more close homologues of I-AniI. This implies that mutations that have previously reduced the endonuclease activity of I-AniI are identified and reversed, sometimes in combination with additional ‘artificial’ mutations, to optimize its in vivo activity.
INTRODUCTION
Homing endonucleases (HEs) are highly specific DNA-binding and cleavage enzymes that are encoded by open reading frames that are embedded within intervening sequences (introns and inteins) in all microbial forms of life, including phage, eubacteria, archaea and single cell eukaryotes (1,2). First discovered in the 1980s as the drivers of mobile introns in yeast (3,4), HEs promote the mobility of the intervening sequences (and their own reading frames) by generating double strand breaks in homologous alleles that lack the intron or intein (5). Break repair leads to transfer of the element via homologous recombination, using the allele that contains the HE gene as a template. Thus, HE genes are selfish DNA sequences that are inherited in a dominant, non-Mendelian manner.
Based on primary sequence homology, five homing enzyme families have been identified, each primarily associated with a unique size and distribution of host genome (2): the LAGLIDADG endonucleases (usually found in archaea and fungal and algal organellar genomes), His-Cys Box family (protist nuclear genomes), the HNH and GIY-YIG endonucleases (usually found in bacteriophage, but with additional rare exceptions in bacteria) and the PD-(D/E)XK family (found in bacterial genomes). The His-Cys Box and HNH family appear to be descended from a common ancestral nuclease (which also gave rise to bacterial colicins and many additional DNA modifying enzymes), while the remaining three families each contain a unique catalytic scaffold and appear to have arisen independently of one another.
The structures of several LHEs bound to their DNA targets have been determined, as summarized in (2). All of these endonucleases display highly modularized structures and DNA-binding scaffolds that are amenable to engineering and selection experiments. Monomeric LHEs (that contain two active sites and two DNA recognition regions on a single peptide chain) are particularly compact proteins (∼200–250 amino acids total) that can recognize very asymmetric DNA target sites (6).
LHEs are the most specific of all known naturally occurring DNA-binding enzymes, recognizing DNA target sites ranging in length from 20 to 24 basepairs with low nanomolar dissociation constants. Their specificity of cognate DNA recognition and cleavage, and the structural basis for this behavior, has been evaluated both for the homodimeric I-CreI (7,8) and for the monomeric I-AniI (9) endonucleases. In both studies, the estimated cleavage specificity of the wild-type (WT) endonuclease under ideal in vitro cleavage conditions approaches 1 in 109. Extensive experimentation by a wide variety of laboratories, mostly with the I-SceI endonuclease, indicate little or no toxicity when the enzyme is transfected and expressed in a variety of cellular experimental contexts (10).
The collaboration between HE genes and their surrounding introns or inteins represent an efficient symbiotic relationship (11,12). The extra-Mendelian inheritance of these elements is driven by the endonuclease, assuring that they rapidly become fixed in a population. In return, the self-splicing intron or intein serves as a refuge for the endonuclease gene, allowing it to invade but not inactivate the host gene. However, after invasion and fixation occurs there is little selective pressure for the maintenance of a functional HE, leading to the gradual accumulation of mutations that reduce its activity. This evolutionary degradation eventually leads to loss of the endonuclease gene and the surrounding intron, followed by subsequent re-invasion. This HE life cycle has been especially well illustrated in a comparative study of analogous HEs in yeast (13).
A small but significant fraction of HE genes has acquired a secondary activity that provides a benefit to the host. As a result of this activity, these proteins are therefore subject to purifying selection that helps maintain their fold and stability. For example, a subset of HEs also facilitates the splicing of their host intron (a behavior termed ‘maturase’ activity) (14–18). While the presence of the intron itself might not benefit the host per se, its splicing is necessary for proper function of the surrounding gene. A HE that is required for intron splicing is therefore under ‘partial’ selection for its DNA cleavage activity. The overall fold of the protein must be maintained in order to maintain splicing activity (which obviously also helps maintain its endonuclease capabilities) but individual mutations that only reduce DNA cleavage are still under minimal purifying selection pressure.
Those naturally-occurring HEs that moonlight as splicing cofactors (or as any other factor that benefits the host) are more persistent in the genome, but often display a chronic disability in their DNA cleavage activity (9,19). Here we study these points using a model HE (I-AniI) that is a very efficient intron splicing cofactor, but a quite inefficient endonuclease. We conducted an in vivo selection experiment for mutated variants of I-AniI that display enhanced cleavage of its DNA target. This experiment yields several reoptimized endonuclease constructs, harboring between one and four point mutations, which display significantly enhanced activity in the screen. Most of these mutations correspond to residues found in homologues of I-AniI, while some are unique. These amino-acid substitutions act synergistically to restore DNA cleavage activity. The construct containing the fewest point mutations of the WT enzyme sequence (and that also displays near 100% recovery in the cleavage activity screen) binds the WT DNA target more tightly, exhibits a catalytic rate enhancement for one of its two active sites, is more active at low physiological temperatures, and is much more efficient at stimulating recombination than its WT parent.
MATERIALS AND METHODS
Proteins
All I-AniI constructs used for in vitro experiments were expressed and purified from Escherichia coli strain BL21-CodonPlus (DE3)-RIL (Stratagene), as previously described (20), with the exception that expression was induced at 15°C for ∼20 h after the culture had achieved early log growth phase (OD600 ∼ 0.6). For all experiments, two point mutations (F80K and L232K) were introduced that improve the solubility of the enzyme and facilitate biochemical studies as previously described (9). Both mutations are far removed from the DNA-binding surface of the enzyme, and are on exposed surfaces of α-helices.
Bacteria-based in vivo cleavage assay and selection
This cleavage assay was performed based on the previous report by Doyon et al. (21). Two copies each of a WT target site or the closely related ‘LIB4’ site (that harbors two basepair substitutions and is bound and cleaved more readily by the endonuclease) were inserted between AflIII and BglII and between NheI and SacII in the reporter (pCcdB) plasmid (four copies total). This plasmid encodes ‘control of cell death B’, a toxic protein to bacteria, and the expression of this gene is induced by isopropyl-β-thio-galactopyranoside (IPTG). The open reading frame (ORF) of the I-AniI construct described above was amplified by PCR, in order to incorporate an NcoI site and NotI site at the 5′ and 3′ termini, and was cloned into the expression (pEndo) plasmid. The HE gene is tightly regulated by pBAD promoter, and addition of L-arabinose promotes the gene transcription. The pEndo plasmid was transformed into DH12S competent cells harboring pCcdB plasmid (containing four copies of WT target site or LIB4 site) by electroporation. The transformants were grown in 2× YT medium at 37°C for 30 min, and were 5-fold diluted with 2× YT medium supplemented with 125 mg/ml carbenicillin and 0.025% L-arabinose. After the culture was grown at 30°C for 4 h, the cells were harvested, resuspended in sterilized water, and spread on both the non-selective plates (1× M9 salt, 1% glycerol, 0.8% tryptone, 1 mM MgSO4, 1 mM CaCl2, 2 μg/ml thiamine and 100 μg/ml carbenicillin) and selective plates (the non-selective plates supplemented with 0.02% L-arabinose and 0.4 mM IPTG). The plates were incubated at 30°C for two days, and the cleavage activity was evaluated by comparing the number of the colonies between the non-selective and the selective plates.
Selection of I-AniI variants with efficient cleavage in bacteria by random mutagenesis
Random mutations were induced in I-AniI ORF using GeneMorph II Random Mutagenesis Kit (Stratagene), by following the manufacture's instruction, and the amplified fragments with NcoI/NotI sites were inserted between NcoI and NotI sites in pEndo plasmid. The library with random mutations were transformed into DH12S competent cells harboring pCcdB including WT target site, and the transformants were selected on the selective plates as described above. The survival colonies were streaked on both the selective plates and the negative control plates (the non-selective plates supplemented with 0.02% l-arabinose and 35 μg/ml chloramphenicol), and the mutated endonuclease genes were amplified by PCR from clones that could survive only on the selective plates. The PCR fragments were cloned into pEndo plasmid, and were selected with the same procedures as for the first round selection.
Far-UV circular dichroism
CD spectra were recorded at 25°C between 190 and 260 nm using 0.1 cm path length quartz cuvettes (Hellma) and 6 μM I-AniI variants in 10 mM potassium phosphate buffer (pH 7.0). The spectra were analyzed in the continuous mode with 1 nm bandwidth, 8 s response, and a scan speed of 10 nm/min. Thermal denaturation curves were obtained by measuring the ellipticity of 10 nM protein at 222 nm from 5 to 80°C at 2°C/min. These experiments were independently repeated three times.
Isothermal titration calorimetry
The dissociation constant and thermodynamic values of Y2 I-AniI binding WT target or LIB4 target DNA sequences were analyzed as previously described (9). Briefly, Y2 I-AniI and the target DNA dialyzed in ITC buffer (50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10 mM CaCl2) were quantified by UV spectroscopy, using extinction coefficients to calculate concentrations. Sample concentrations ranged from 5.5 to 6.0 µM I-AniI and 38 to 60 µM dsDNA target. A VP-ITC MicroCalorimeter (µCal) was used for all experiments, with the protein sample in the cell and the DNA target in the auto-pipette. Individual runs consisted of 30 injections of 5-8 µL each, depending on sample concentrations, and were conducted at 30°C with a 351 r.p.m. stirring speed. Data were fit using Origin 7 SR2 software.
In vitro cleavage assays
DNA substrates were amplified by PCR from pBluescript plasmid containing either WT target site or LIB4 target site, with primers designed to generate approximately 700 basepair products. The reaction mixtures (20 μl) contained 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 100 ng (∼11 nM) of the DNA substrates, and the indicated amounts of I-AniI variants. The reactions ran at 37°C for 30 min, and were terminated by adding 5 μl of stop buffer (100 mM EDTA (pH 8.0), 2.5% SDS, 10% glycerol and 0.01% bromophenol blue). The products were separated on a 2% agarose gel with ethidium bromide, and quantified as described above.
For kinetic determination of the relative rates of individual DNA strand cleavage events, the 28 basepair DNA substrate was prepared from two oligonucleotides: 5′-ATTTGAGGAGGTTTCTCTGTAAATAATG and 5′- CATTATTTACAGAGAAACCTCCTCAAAT. Oligonucleotides to be 5′ end-labelled were incubated in buffer with T4 polynucleotide kinase (New England Biolabs Beverly, Massachusetts) and [γ−32P]-ATP (6000 Ci/mmole; Perkin Elmer Boston, Massachusetts). The reactions were incubated at 37°C for 45 min extracted with phenol:chloroform:isoamyl alcohol solution (25:24:1; PCI), and centrifuged through a Sephadex G-50 spun column (Sigma Chemical Co., St. Louis, M0).
The substrate duplex was formed by incubating the labelled oligo (1.25 μM, final) with the corresponding unlabelled partner oligo (5 μM, final) in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2 and 50 mM NaCl (TMN buffer) at 90°C for 2 min and then cooled to 25°C over 20 min. DNA cleavage reactions were carried out at 37°C and contained substrate (6.25 nM) and TMN and the reactions initiated by the addition of WT I-AniI or Y2 I-AniI to a final concentration of 400 nM. Aliquots were removed at specific times (ranging from 0.5 to 40 min) and the reactions terminated by the addition of EDTA to a final concentration of 67 mM, followed by organic extraction. Samples were electrophoresed on 8% acrylamide/7 M urea gels, dried and exposed on a phosphorimager screen and visualized/quantified using a phosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The imager counts in the substrate and product were summed and the fraction product plotted against time. The data fit to a first order rate equation: Fraction of 5′-labelled product = A(1 − e−kt) where A is the amplitude of the product and k represents the pseudo-first order rate constant, kobs. Note that the cleavage rate for the top strand in the small duplex reported here is greater than that reported previously for a larger (∼200 bp) duplex (0.22 min−1) while the rate for the bottom strand is the same (0.11 min−1) (22). As noted in the text, the designation of ‘top’ and ‘bottom’ strand in these methods is opposite that previously described in (22) where the labeling of strands and cleavage products were inadvertantly reversed.
In vivo gene conversion assay
The DR-GFP reporter contains a GFP sequence interrupted by a HE target site and in frame stop codon as a recipient), followed by a 5′ and 3′-truncated GFP as a donor (23). Gene conversion event between these non-functional GFP genes, which is promoted by double strand breaks at the HE target site results in the expression of the intact GFP protein. 3 × 106 of 293T cells were plated 24 h prior to transfection in 12 well plates. The cells were transfected with 1.25 μg each of DR-GFP reporter and HE expression plasmid using calcium phosphate method. Media was replaced at 24 h post transfection, and the GFP positive cells were detected by flow cytometry at 48 h post transfection. Western blots were probed with anti HA monoclonal antibody (Clonetech 6E2) and anti β-actin polyclonal antibody (cell signaling).
RNA splicing assay
The preparations of radiolabelled COBme-pre RNA as well as the kinetic analyses and data fits were carried as described previously (24). The imager counts in the precursor and splicing products were summed and the fraction precursor remaining plotted against time. The protein-dependent splicing data were fit to a first order equation with a double exponential: Fraction RNA bound = A(e−kt) + B(e−kt) where A and B are the amplitude of RNA in each phase and k represents the first order rate constants for each phase. Only the fast phase is reported in Supplementary Table S2. The slow phase, which presumably represents a population of RNA that undergoes a slow folding step required for protein binding or RNA catalysis, (24) was 0.025 (± 0.004) and 0.024 (± 0.009) min−1 for the WT and Y2 proteins.
Crystallographic analysis
The DNA oligonucleotides used for cocrystallization (5′-GCG CGC TGA GGA GGT TTC TCT GTA AAG CGC A-3′ and 5′-GCG CTT TAC AGA GAA ACC TCC TCA GCG CGC T-3′) were purchased from Integrated DNA Technologies (1 mM scale, HPLC purified). The oligonucleotides were dissolved in 10 mM Tris-EDTA (TE) buffer pH 8.0, and the complementary DNA strands were annealed by incubation at 95°C for 10 min and slow cooling to 4°C over a 6 h period. Purified Y2 I-AniI protein (at 10 mg/mL concentration) in 50 mM Hepes-NaOH (pH 7.5), 500 mM NaCl, 20 mM CaCl2, and 5% (v/v) glycerol was mixed with 1.5-fold molar excess of the DNA substrate. The protein-DNA drops were mixed in a 1:1 volume ratio with a reservoir solution containing 80 mM Bis-Tris (pH 6.2), 50 mM CaCl2, and 32% (v/v) polypropylene glycol P400 and equilibrated at 22°C. Crystals grew within two weeks, and were frozen by direct looping and submersion into liquid nitrogen. The crystals diffracted up to approximately 2.7Å resolution at the ALS beamline 5.0.1. Data sets were processed using the DENZO/SCALEPACK software package (25). The structure of Y2 I-AniI/DNA complex was solved by molecular replacement using the PDB coordinates of the WT I-AniI/DNA complex (PDB: 1P8K), and was refined using programs REFMAC5 (26) and CNS (27) (Supplementary Table S3).
RESULTS
Generation of I-AniI variants with optimal in vivo activity towards its WT target sequence
Compared to other well-characterized LHEs, WT I-AniI binds to its physiological, WT target site with reduced affinity (the dissociation constant of I-AniI to its that site is estimated to be ∼100 nM) and displays relatively slow and incomplete cleavage of that site under optimized in vitro reaction conditions (9). The enzyme also displays a temperature optima (∼50°C) that is far removed from the growth optima of its physiological host (this work). As a result, WT I-AniI displays poor cleavage activity in transfected cell lines, including both human cells and in bacteria.
In order to optimize the activity of I-AniI in transfected cells (and to subsequently study the basis for any observed improvement), we conducted a directed evolution experiment to improve cleavage activity of I-AniI against its WT target site. This experiment was conducted using the bacterial screen for HE cleavage activity developed by Doyon et al. (21), that reports on elimination of a reporter plasmid that encodes a cytotoxic protein (see Materials and Methods section). We induced random mutations in the open reading frame of I-AniI using low-fidelity PCR amplification, and screened for the variants that display efficient DNA cleavage using the bacterial selection system and an endonuclease library containing ∼130 000 mutated transformants.
After two rounds of selection, we successfully isolated nine unique endonuclease variants which significantly improved cell growth and survival under selective conditions (Figure 1 and Table 1). These constructs each contained between one and four individual amino-acid substitutions. The variants of I-AniI displayed between 17-fold and 475-fold increases in survival rate and recovery as compared to the WT I-AniI endonuclease (between 3.5% and 95% survival, compared to less than 0.2% for WT I-AniI).
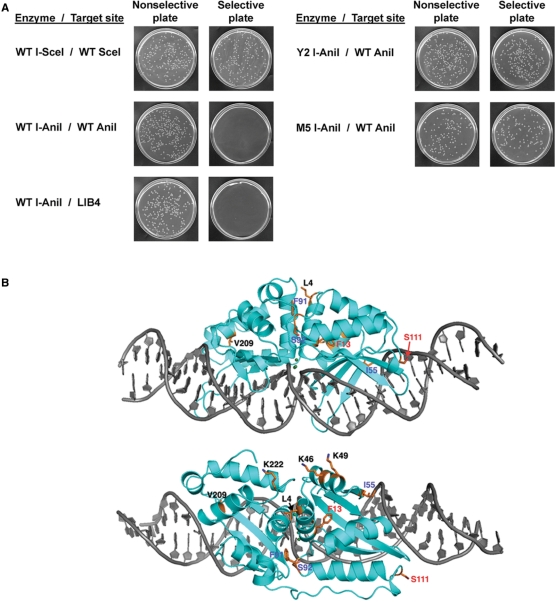
Figure 1.
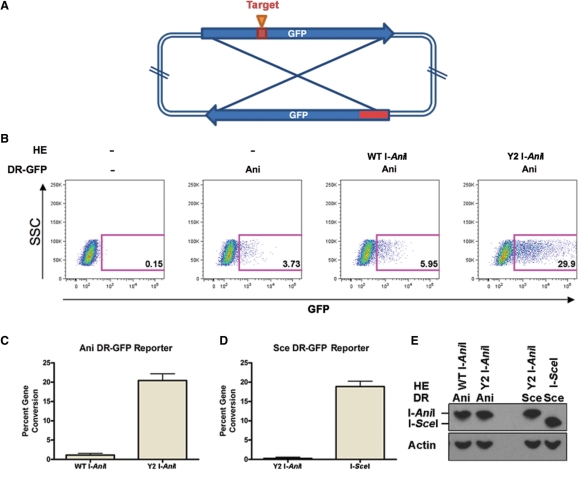
Isolation of optimized I-AniI variants by a bacteria based cleavage assay. (A) The expression of the toxic gene is restricted on the selective plates. WT I-SceI significantly cleaved its wild type target site in bacteria, but WT I-AniI inefficiently did against its physiological (WT I-AniI) and optimal (LIB4) sequences. The Y2 and M5 variants improved the cleavage activity. (B) Ribbon model of WT I-AniI structure. Gray sticks indicates the physiological target DNA. Mutated residues in Y2 construct are labeled in red; remaining mutated residues in M5 construct are labeled in blue.
Table 1.
I-AniI variants identified by a bacteria-based cleavage assay
| I-AniI variants | Survival rate (%)a |
|---|---|
| Wild type | <0.2 |
| F13Y | 36 ± 3.8 |
| F13L, K46Q, S92T | 75 ± 9.6 |
| F13Y, S111Y (Y2) | 95 ± 30 |
| F13Y, I55V | 85 ± 16 |
| F13Y, K46Q, S92T | 93 ± 4.7 |
| F13V, K46Q, S92T | 3.5 ± 1.2 |
| F13Y, S111F, K222R | 94 ± 18 |
| F13L, K46Q, F91I | 71 ± 11 |
| L4I, K49Q, S111F, V209A | 90 ± 17 |
| F13L, S92Tb | 68 ± 11 |
| F13L, K46Qb | 2.7 ± 2.4 |
| S111Yb | 2.6 ± 4.2 |
| S111Fb | 0.26 ± 0.28 |
| F13Y, K222Rb | 37 ± 13 |
| L4I, K49Q, V209Ab | <0.2 |
| F13Y, I55V, F91I, S92T, S111Y (M5)b | 92 ± 10 |
aThe survival rate was determined by dividing the number of colonies on the selective plates by that on the nonselective plates. Errors refer to mean ± S.D. of three replicate plating experiments.
bThese variants were created by site-directed mutagenesis. Boldface correspond to mutations (alone and in combination) found in the Y2 variant of I-AniI.
A total of 10 separate amino-acid positions were mutated (Figure 1), and a total of 13 separate point mutations were distributed across these positions. Two amino-acid positions (F13 and S111) yielded multiple mutations (F13 substituted with Y, L or V and S111 substituted with Y or F). Three of the thirteen mutations (23%) correspond exactly to residues found in one or more close homologues of I-AniI: I55V, S92T and S111Y (Figure 2). Two additional mutations (F13L and K49Q) correspond to similar amino acids also found in those homologues (an isoleucine is found at residue 13 and either an asparagine or an aspartate at residue 49). One construct contained a single mutation (F13Y); this mutation displayed a 180-fold increase in survival and recovery over WT I-AniI. Eight out of nine variants contained a mutation at this position (F13Y, L or V). A variant of I-AniI harboring only two mutations (F13Y/S111Y) yielded nearly 100% survival rates and formation of larger colonies than other variants (Figure 1A). This construct, termed ‘Y2 I-AniI’, represented the simplest mutational route to robust in vivo endonuclease activity and was chosen for detailed biochemical and structural studies.
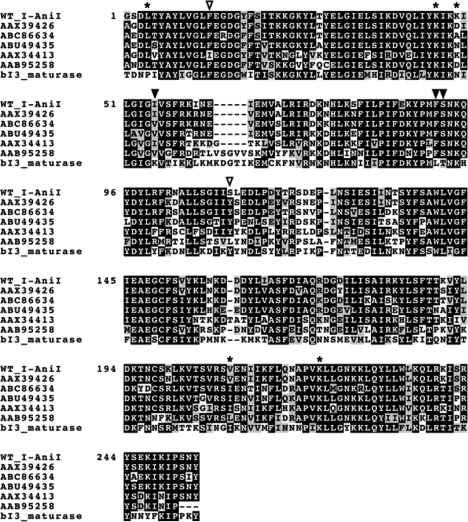
Figure 2.
Sequence alignment of I-AniI homologs. The alignment was performed using CLUSTALW program. Open triangles indicate positions of F13Y and S111Y in the Y2 I-AniI construct. AAX39426, ABC86634, ABU49435, AAX34413, AAB95258 and bI3 maturase are LAGLIDADG proteins from Neosartorya fischeri, Gibberella zeae, Phaeosphaeria nodorum SN15, Agrocybe chaxingu, Venturia inaequalis and Saccharomyces cerevisiae, respectively. Closed triangles represent sites of additional mutations I55V, F91I and S92T in the M5 I-AniI construct. Asterisks indicate remaining positions of point mutations isolated in the cleavage activity screen as listed in Table 1.
Since most of the isolated variants possessed amino-acid substitutions at multiple positions, we deconvoluted several of the mutations via individual site-directed mutations into the WT protein scaffold, to determine their relative contribution of mutations that improve cell growth. The F13Y mutation (located in the first LAGLIDADG helix of the protein scaffold, positioned in the protein's hydrophobic core) was the single amino-acid substitution that contributed most significantly to an increase in the survival rate. The effect of this mutation was enhanced in the presence of several additional mutations, particularly the addition of S111Y (the additional mutation present in the ‘Y2 I-AniI’ constructs). This amino-acid substitution corresponds to the most strongly conserved alternative residue in homologues of I-AniI (Figure 2). On its own, the S111Y substitution increases survival by over 10-fold relative to WT I-AniI (2.7% recovery versus less than 0.2%) (Table 1).
Although the addition of the S111Y mutation to F13Y offered the most direct route to complete optimization of endonuclease activity, a variety of alternate combinations of mutations also increase bacterial survival (including those that echo conserved residues in I-AniI homologues, and others that are unique to the output of the screen). The mutations I55V and S92T, both of which are also recapitulated in one or more naturally occurring homologues of I-AniI, independently increase survival by ∼2-fold and up to 30-fold, respectively when compared to analogous variants that lack those mutations. Similarly, the mutation F91I (which does not correspond to naturally occurring residues in homologues of I-AniI) also enhanced activity and survival.
Three additional mutations found in the survivors from the activity screen (K46Q, K49Q and K222R) alter a series of surface-exposed basic residues that are clustered together on a surface far from the bound DNA (Figure 1B). Various combinations of these three mutations in control experiments indicate that the contribution of those residues to activity and survival is relatively minimal. It is possible that these mutations block a secondary nucleic acid binding activity (perhaps related to its maturase function) in such a way as to reduce a competing source of toxicity caused by over-expression of the protein.
Taken together, it appears that five individual mutations (F13Y, I55V, F91I, S92T and S111Y/F) contribute most significantly, in various combinations, to observed improvements in cleavage of WT I-AniI site in bacteria. Simultaneous incorporation of all of these substitutions (F13Y/I55V/F91I/S92T/S111Y; termed ‘M5 I-AniI’) facilitated significant cell survival (Figure 1 and Table 1) at a level similar to Y2 I-AniI, suggesting that the combination of all five mutations neither impairs nor further improves the activity of the enzyme in transfected cells, relative to the simpler Y2 double mutation (F13Y/S111Y). Like the ‘minimally mutated’ Y2 construct described above, this ‘maximally mutated’ construct was also selected for additional biochemical analyses.
Thermal stability and thermal activity profiles
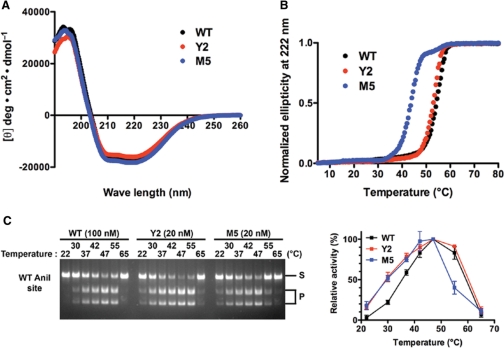
The location of the point mutations in the Y2 and M5 protein variants (three of which are distant from nucleotide bases, and partially or fully buried in the protein core) suggested that the stability and/or dynamic flexibility of the folded proteins might be altered. The circular dichroism spectra of WT, Y2 and M5 variants indicated that all three enzymes possess similar secondary structure content (Figure 3A), and is consistent with the previous observation, the mixed α/β topology of the I-AniI crystal structure. The WT enzyme and the Y2 variant display similar thermal denaturation curves, with observed transition midpoints (TM) of 53.9 and 52.6°C, respectively (Figure 3B). In contrast, M5 variant was partially destabilized, with a reduction in the TM value of approximately 10°C (43.5°C). Because the denaturation and/or aggregation of the protein at high temperature are irreversible, exact changes in the free energy of protein unfolding cannot be determined using this system.
Figure 3.
Secondary structure, thermal stability and thermal activity profile analyses of WT and mutant I-AniI variants. (A) Circular dichroism analysis. Far-UV CD spectra were measured between 190 and 260 nm at 25°C. (B) Temperature-dependent denaturation on the basis of changes in circular dichroism ellipticity at 222 nm. Temperature of 50% transition was 53.9, 52.6 and 43.5°C for WT, Y2 variant and M5 variant, respectively. (C) Thermal profiles of the cleavage activity. The reactions ran at 22, 30, 37, 42, 47, 55 and 65°C. Relative activity was shown by taking cleavage activity at 47°C as 100%. Errors mean ± S.D.
The Y2 and M5 enzymes also display altered thermal reaction profiles, corresponding to enhanced cleavage of the DNA target at the physiological temperature (30°C) that was used in the selection experiment (Figure 3C). Whereas the WT enzyme displays a relatively sharp temperature optima for cleavage at approximately 50°C, the Y2 and M5 enzyme variants respectively display broadened (Y2) or reduced (M5) thermal activity profiles and optima.
Binding affinity
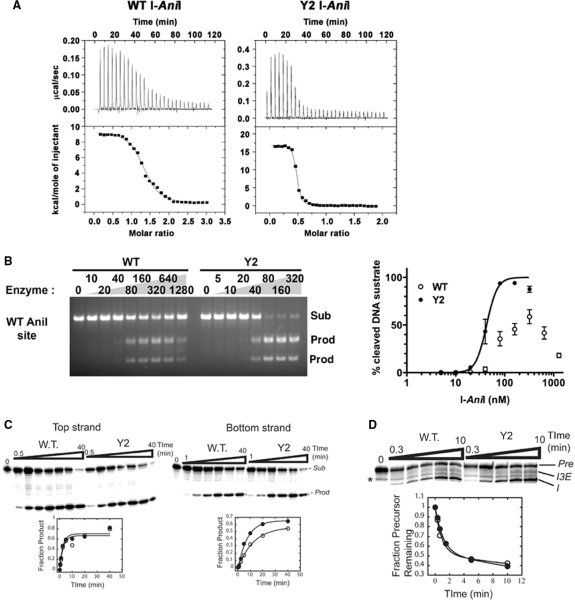
Because the Y2 construct displays the highest survival frequency in the bacterial screen for in vivo cleavage activity, contains fewer mutations and is physically more stable than the M5 mutant, its DNA-binding and endonuclease activity was examined further. The affinity and thermodynamic profile of cognate DNA target binding was measured using isothermal titration calorimetry. The Y2 variant displayed significantly tighter binding to its physiological target site than the WT enzyme, with a measured dissociation constant corresponding to 12 ± 3.0 nM (compared to a KD value for the WT enzyme of approximately 96 nM) (Figure 4A and Supplementary Table S1). The tighter binding for Y2 I-AniI corresponds to a reduction in ΔGbinding (−1.2 kcal/mol) that results from a more favorable entropic change (−TΔS, −7.2 kcal/mol), which was partially offset by a more unfavorable enthalpic change (ΔH, +6.0 kcal/mol). In contrast, the affinity of the Y2 variant to the ‘LIB4’ target site [a sequence differing from the WT site at two basepairs, that is bound and cleaved more efficiently by the WT enzyme (9)] is nearly unchanged (Supplementary Figure S1 and Table S1).
Figure 4.
DNA binding affinity, DNA cleavage activity and intron splicing activity of WT and ‘Y2’ optimized I-AniI variant. (A) Binding assay with WT I-AniI (left) and Y2 variant (right) against WT I-AniI site by isothermal titration calorimetry. The left graph is taken from our previous report (9). Dissociation constants and thermodynamic values are summarized in Supplementary Table S1. (B) In vitro cleavage assay against the WT I-AniI target site. A serial dilution of each I-AniI variant was incubated with PCR fragment containing a single copy of the target site at 37°C. The reaction products were quantified using the NIH ImageJ program. Errors mean ± S.D. Sub: linearized plasmid substrate; prod: cleaved products. (C) Relative cleavage of top and bottom DNA strands by WT and Y2 I-AniI. The top strand includes the coding sequence of the mitochondrial apocytochrome B gene from Aspergillus nidulans (5′-TGAGGAGGATTTCTCTGAAA-3′). Sub = substrate; Prod = product. Open ovals = WT, filled ovals = Y2. The data are fit to an equation that describes a first order reaction (see Materials and Methods section). Note that the Y2 variant cleaves the bottom strand more rapidly that the WT protein. For values, see Supplementary Table S2. The designation of ‘top’ and ‘bottom’ strand in these methods is opposite that previously described in (22) where the labelling of strands and cleavage products were inadvertantly reversed. (D) Relative intron splicing activity of WT and Y2 I-AniI. Pre = precursor RNA (substrate); I3E = splicing intermediate after 5′-splice site cleavage with the 3′-exon still linked to the intron; I = free intron (product). The ligated exon product is not shown. The asterisk demarcates an RNA degradation product. Open ovals = WT; filled ovals = Y2. The data are fit to an equation that describes a first order reaction (see Materials and Methods section). For values, see Supplementary Table S2.
In vitro cleavage activity
A series of in vitro cleavage assays were carried out using a linearized DNA construct containing a single copy of the WT target site as a substrate. WT I-AniI was unable to completely cleave this site under the digest conditions used, and the generation of cleavage products was decreased at higher concentrations of the protein (Figure 4B). In contrast, the Y2 I-AniI variant hydrolyzed the substrate more efficiently, at reduced concentrations of enzyme. Under the same reaction conditions, Y2 and WT enzyme constructs cleave the ‘LIB4’ substrate with similar efficiencies (data not shown). Therefore, the selected enzyme constructs display improved activity against the ‘suboptimal’, WT I-AniI site (which was used in the selection), but not against the closely related LIB4 site (which was not used in the selection). This implies that the specificity of the Y2 I-AniI construct is slightly reduced with respect to discrimination between these two target sequences; additional studies of binding and cleavage specifities across the entire target site are in progress.
I-AniI is an asymmetric LHE that displays a sequential mechanism of DNA cleavage. The C-terminal active site cleaves the top strand of its target site (the coding strand of Aspergillus nidulans mitochondrial apocytochrome B gene) significantly faster than the N-terminal active site cleaves the bottom (non-coding) strand (22). To test whether the Y2 variant changed the relative efficiencies of individual strand cleavage events, the rates of hydrolysis for each strand were measured. In these experiments, a small 28 nt duplex was prepared with either the top or bottom strand end-labeled with 32P. Cleavage reactions were performed under single turnover conditions with saturating concentrations of WT I-AniI or the Y2 variant at 37°C. Under these conditions, the WT enzyme cleaved the top strand ∼5-fold faster than the bottom strand [0.51 (±0.14) min−1 versus 0.10 (±0.01) min−1; Figure 4C, Supplementary Table S2]. While cleavage of the top strand by the Y2 variant was not significantly different than the WT enzyme [0.45 (±0.10) min−1, compared to 0.51 min−1], the bottom strand was cleaved approximately 2-fold faster than the WT enzyme [0.21 (±0.05) min−1, compared to 0.10 min−1]. Thus, the Y2 I-AniI construct appears to specifically enhance the ‘slow’ cleavage by the N-terminal active site of I-AniI.
It should be noted that the designation of ‘top’ and ‘bottom’ strand in the paragraph above is opposite that previously described in (22) where the labeling of strands and cleavage products were inadvertently reversed.
Splicing activity
The effect of the F13Y and S111Y mutations on maturase activity was assessed by in vitro splicing assays. I-AniI stimulates splicing of the COB intron via the rapid formation of an initial RNA/protein complex which is then is slowly resolved into a native, splicing-competent particle (24,28). Importantly, under non-saturating protein concentrations [∼100 nM; (29)] the rate of splicing is dependent on both the bimolecular rate constant for formation of the initial complex and the resolution step (24). Therefore, the maturase activity was measured using trace amounts of COBme-preRNA and 20 nM protein. Under these conditions, the rate of splicing for the Y2 protein was the same within error to the WT protein (1.3 (±0.37) min−1 versus 1.7 (±0.66) min−1; Figure 4D, Supplementary Table S2).
In vivo recombination activity
Homing endonucleases are under intense study as potential reagents for gene application, but to date only the I-SceI and I-CreI LAGLIDADG proteins have been reported to be capable of inducing homologous recombination in mammalian cells by generation of double strand breaks (10,30–32). To measure the relative ability of WT I-AniI and the Y2 variant to induce gene conversion in transfected human cells, we used an assay in which human cells are cotransfected with an expression vector for either protein, in combination with a ‘DR-GFP’ reporter plasmid that encodes two non-functional GFP genes (Figure 5A). One copy of the GFP gene is interrupted with WT I-AniI site and two stop codons in the middle of the open reading frame, while the other is prematurely truncated (23). Gene conversion between these two gene copies, either spontaneously or as a result of a double-strand break, leads to complementation of GFP and production of measurable fluorescent cells.
Figure 5.
Comparative in vivo gene conversion activity of WT I-AniI and the Y2 variant. (A) Schematic representation of the DR-GFP reporter plasmid. The reporter plasmid harbors two non-functional copies of the GFP gene, one interrupted by two stop codons followed by a target site for either I-AniI or I-SceI, the other encoding the 5′- and 3′-truncated gene. (B) Representative FACS plot of gene conversion on the DR-GFP reporter with WT AniI site (I-AniI DR-GFP). 293T cells were transiently co-transfected with the reporter and an expression plasmid for either WT I-AniI or Y2 variant, and analyzed by flow cytometry at 48 hours post transfection. In each plot, the fluorescent side scatter ‘SSC’ (as a standard of cell viability) is plotted on the Y axis and the GFP-associated fluorescent signal ‘GFP’ (a measurement of recombiantion of the reporter plasmid in the transfected cells) is plotted on the X-axis. (C,D) graph depicting percent gene conversion of I-AniI DR-GFP (C) and I-SceI DR-GFP (D) induced by either WT I-AniI, Y2 variant or I-SceI. Data represents three independent experiments performed in duplicate normalized to observed background by transfection of the DR-GFP reporter alone, Errors mean + SE. (E) Expression of HEs in the transfected cells. The expression level was analyzed by Western blotting using an antibody against hemagglutinin (HA)-epitope (upper) and β-actin as a control (lower).
Human 293T cells were co-transfected with this reporter plasmid in the presence or absence of the expression construct encoding WT I-AniI or the Y2 variant, and GFP positive cells were detected by flow cytometry (Figure 5B). Transfection of the DR-GFP reporter plasmid alone leads to a measurable but relatively low background of GFP expression, caused by spontaneous conversion between the GFP genes on the reporter, whereas almost 100% of the cells were GFP positive when the GFP expression plasmid (as a positive control) was transfected in the same experimental conditions (data not shown). Co-transfection with WT I-AniI expression plasmid slightly increased the fraction of GFP positive cells over the background. In contrast, transfection with the expression construct for Y2 variant resulted in a dramatic increase in the GFP positive cells (Figure 5B and C).
In a separate control experiment to demonstrate the specificity of action of the Y2 variant, a DR-GFP reporter plasmid containing the target site for the I-SceI site instead of the WT I-AniI site was used, leading to gene conversion only when the gene for I-SceI was introduced in place of Y2 I-AniI (Figure 5D). Finally, the expression level of each HE in transfected human cells was verified by western blotting assay (Figure 5E). These results suggested that Y2 variant induced highly efficient gene conversion by generating double strand breaks at its physiological target site in 293T cells.
Crystal structure
We then solved the crystal structure of the Y2 I-AniI/DNA complex (Y2/DNA) complex to 2.7Å resolution (Figure 6 and Supplementary Figures S2 and S3). The data and refinement statistics are shown in Supplementary Table S3. The crystals were grown in the presence of calcium ions, the DNA substrate was uncleaved, and three independent complexes were present in the asymmetric unit.
Figure 6.
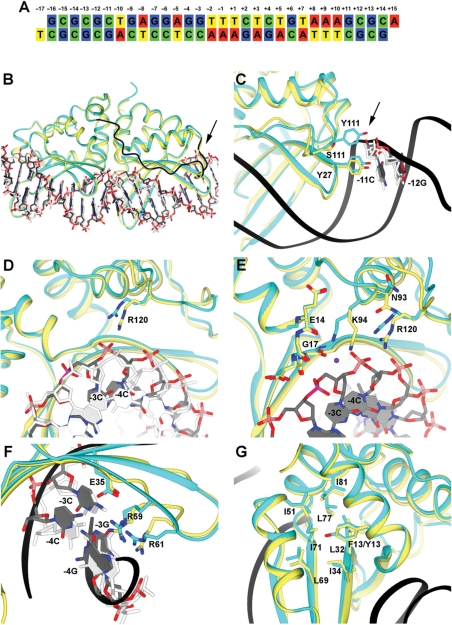
Structural analysis of the Y2 I-AniI construct. Electron density maps for several of the structural features shown here are provided in Supplementary Figure S3. For clarity of viewing, the orientation of the structure shown in panels B–G are rotated 180°C relative to the canonical orientation of the DNA substrate sequence shown in panel A. (A) Sequence of the DNA duplex used for the crystallization. The target site (20 basepairs) is numbered from positions −10 to +10, relative to the center of the four-base overhangs generated by DNA cleavage. The strand breaks are introduced between positions +2 and +3 in the top strand, and between −2 and −3 in the bottom strand. (B) Y2 I-AniI (cyan) in complex with the DNA duplex (grey) superimposed against WT I-AniI (light yellow) bound to the same DNA target sequence (white) (PDB: 2QOJ). The extended loop of Y2 I-AniI linking the N-terminal and the C-terminal domains is shown in dark blue. The arrow in panel B and C points towards S111Y. (C) Magnification of region surrounding residue 111 (S111Y mutation in the Y2 I-AniI construct). (D) Magnification of region surrounding residue 120. (E) Magnification of region surrounding residue 94 and its surrounding active site residues and DNA bases. (F) Magnification of region surrounding basepairs −3 and −4. (G) Magnification of region surrounding residue 13 (F13Y in the Y2 I-AniI construct).
Y2 I-AniI displays a similar overall structure to the structure of WT I-AniI (9,20), except for an extended peptide sequence (residues 108–127) linking the N-terminal and the C-terminal domains. This region contains one of the two amino-acid substitutions in the Y2 construct (S111Y) (Figure 6B and 6C). The overall bend angles of the DNA in the WT and Y2 complexes were also quite similar (Supplementary Figure S4), although with a local shift in the major groove dimensions spanning several basepairs, as described below. The structure indicates that mutation of serine 111 to tyrosine (S111Y) causes a rearrangement of several neighboring residues that leads to a repositioning of the N-terminal active site lysine residue (K94).
Residue Y111 in the Y2 variant makes a new, direct contact with the DNA backbone near base −11C (one position upstream from the protein's specifically recognized target site) (Figure 6C). This contact, and the motion of the surrounding residues, results in a corresponding shift in the neighboring side chain of R120 and a movement of the bottom DNA strand (at positions from −3 to −6) by up to 2.3Å, resulting in a localized narrowing of the DNA major groove (Figure 6D). These conformational changes are augmented by significant rotations of the side chains of N93 (which is a structural neighbor to R120) and its immediate neighbor, K94. The amino nitrogen of this latter side chain, which is hypothesized to act as a general base in one of the two endonuclease active sites, moves by 2.5Å toward the first LAGLIDADG motif, into a conformation that mirrors that of its K227 counterpart in the C-terminal active site (Figure 6E and Supplementary Figure S2).
The base-pairing interactions at positions −3 and −4 in the target site (which display the largest change in position relative to the WT complex, and are the sites of specific, high affinity interactions with several protein residues) are maintained by shift of the guanine bases of the top strand (Figure 6F). Contacts between these bases and the protein (R59 and −4G, R61 and −3G, and E35 and −4C) are also maintained by slight changes in rotameric conformations of those side chains.
In contrast to the direct influence of the S111Y mutation on the structure of the N-terminal active site, residue 13 in I-AniI (the site of a the F13Y substitution from the screen) is buried in the hydrophobic protein core of the protein's N-terminal domain, and does not seem to cause major changes in the hydrophobic residues around Y13, other than the addition of an additional hydroxyl group that is in contact with the neighboring protein backbone (Figure 6G).
A schematic representation of the contacts exhibited by the WT endonuclease and the Y2 variant are shown in Supplementary Figure S5. The Y2 I-AniI/DNA complex is packed in a different space group relative to the WT I-AniI/DNA complex, and the Y2 variant structure was solved at relatively lower resolution (2.7Å). Nevertheless, the Y2 enzyme displays very similar protein-DNA interactions, especially with respect to direct contacts to DNA bases.
DISCUSSION
Laboratory reoptimization of homing endonuclease activity partially reverses evolutionary drift
It has been reported that LHEs can accomplish invasion of intron-deficient alleles with much higher frequency than expected from their measured in vitro cleavage efficiencies, suggesting that a significant amount of mutational deterioration can occur before the persistence and propagation of the endonuclease is irreversibly compromised (33). I-AniI clearly displays many of the suboptimal biophysical and enzymatic properties outlined above, that were reoptimized in the laboratory through the identification of amino-acid substitutions that were selected solely on the basis of improved cleavage activity in bacteria. These mutations, when considered separately and in combination, appear to affect the protein scaffold in three ways: by increasing target site affinity, by improving the catalytic efficiency of one active site, and by increasing cleavage activity at 30°C (a temperature well below the thermal optima of the WT enzyme).
The F13Y mutation, which is part of the LAGLIDADG sequence motif and is found in the protein core, clearly represents the most significant mutational ‘hot spot’ identified in the cleavage activity screen (with mutations to both leucine and valine also recovered in addition to tyrosine). This residue is conserved as a phenylalanine in five out of six of the closest homologues of I-AniI (with an isoleucine found in one sequence; Figure 2). When the sequence alignment to I-AniI is extended to more distant homologues (corresponding to BLAST alignment E values of 10−30–10−6) this residue is observed to be heavily diverged, with its position predominantly occupied by small aliphatic and β-branched amino acids (especially valine and threonine).
Given the location of this residue in the completely buried hydrophobic core of the protein, it seems likely that the effect of the mutations recovered at residue 13 (F13Y/L/V) is to increase the global dynamic flexibility of the protein, leading to the reduced thermal temperature profile exhibited by both the Y2 and the M5 enzyme constructs (and the increased survival at the in vivo screening temperature of 30°C). Such effects have been well-documented in both artificially engineered and naturally-occurring enzymes (34–36). If true, this may imply that the ancestral form of I-AniI and its immediate homologues originated in a more thermophilic host organism [prior to transfer to fungal genome(s) such as Aspergillus] and that the variation of this residue position strongly influences overall catalytic efficiency as a function of ambient temperature.
The additional mutation recovered in the Y2 I-AniI variant (S111Y) appears to convert the corresponding amino-acid identity to a residue (tyrosine) that is more strongly conserved in homologous endonucleases (Figure 2). This suggests that ancestors of the I-AniI endonuclease lineage may have possessed a tyrosine residue at this position, which is conserved in most homologues in a multi-sequence alignment. A crystal structure analysis of the Y2 variant, combined with the kinetics of individual strand cleavage events, indicates that this mutation is coupled to conformational rearrangements of both the protein and the DNA substrate that lead to a beneficial rearrangement, and catalytic enhancement, of the N-terminal active site. However, on its own this mutation has less effect on in vivo activity than does F13Y (Table 1).
Therefore, the poor in vivo activity of I-AniI in the bacterial screen appears to primarily result from inadequate activity at low temperature, and secondarily from a slightly malformed active site in one of the two protein domains. It may be that at significantly elevated temperatures, the relative importance of the two mutations in the Y2 I-AniI construct would be reversed, with the S111Y mutation contributing most strongly to the catalytic performance of the enzyme.
The position of two additional mutations that individually synergize with F13Y (I55V and S92T) appears to partially share structural and evolutionary features with the S111Y mutation described above. The side chain of V55 is in proximity to the DNA backbone (positioned to make non-specific contacts to the substrate) and S92T is also found in close proximity to K94 (Figure 2). Thus, the route to recovery of robust in vivo endonuclease activity seems to consistently involve a mutation that alters the thermal profile of the enzyme in combination with at least one additional mutation that alters the structure of the protein-DNA interface and/or the architecture of one of the two active sites.
Although almost all individual mutations described above are readily observed in homologues of I-AniI, their exact combination(s), as recovered in active clones from the screen, are not. This might be explained by two facts. First, the number of potential combinatorial interactions between distant residues in the protein scaffold, that act synergistically to optimize multiple properties and functions (thermal activity, binding affinity and catalytic efficiency) are virtually limitless. Second, the selection pressure applied at each amino-acid position in the screen (to improve DNA cleavage activity) is not a precise counterpart to the selection pressure imposed on those same residues in nature. I-AniI is a bifunctional enzyme that acts both as a highly specific endonuclease and also as an RNA maturase (18). This latter activity (in which the protein facilitates folding and splicing of its cognate intron) is required for proper expression of the mitochondrial apocytochrome b oxidase gene (18,37), and thus provides a benefit to the host that leads to selection pressure for maintenance of the LAGLIDADG protein-fold. While acquisition of this secondary function can therefore indirectly stabilize endonuclease activity during the course of evolution, it would also suppress those mutations that are detrimental to RNA splicing but would otherwise optimize DNA cleavage activity.
If the selection pressures for the two activities of I-AniI and its homologues were strongly balanced, it is possible that certain amino-acid positions in I-AniI and its closest homologues might display elevated frequencies of asynonymous amino-acid substitutions caused by ‘genetic conflict’ – in this case, positions where the requirements for endonuclease and maturase functions produce closely balanced, antagonistic selection pressures in favor of different residues types. This possibility was examined by subjecting the sequences of I-AniI and its homologues (using a BLAST alignment E value cutoff of 10–6) to a ‘Ka/Ks’ analysis (searching for elevated levels of non-synonymous substitutions, relative to silent or synonymous substitutions) (38). Absolutely no residues in the endonuclease were found to display elevated ratios of Ka/Ks; all positions appear to be under either neutral or purifying selection.
Asymmetry in monomeric LHEs active sites
I-AniI, which is a monomeric endonuclease with two separate structural domains, can cleave asymmetric DNA sequences in a highly sequence-specific manner and is an excellent model of an initial protein scaffold for development of a gene-specific reagent. The results described above indicate that during evolution this endonuclease or one of its immediate predecessors suffered a mutation (Y111S) that perturbed one of two active sites.
In addition to I-AniI, several additional homing monomeric LHEs have also been shown to display significant asymmetry in their DNA cleavage properties. The algal endonuclease I-CpaII displays specific DNA nicking behavior that varies as a function of metal concentration, preferentially nicking the bottom strand of its target site at low (<0.5 mM) magnesium concentrations (39). Similarly, the yeast HEs I-SceI recognizes its asymmetrical site with higher affinity binding to the 3′-DNA half-site, leading to accumulation of nicked intermediates during the cleavage reaction (40). Finally, the archaeal endonuclease I-DmoI cleaves the coding strand of its host gene with a slight preference over the non-coding strand (41); this asymmetry is enhanced in engineered variants of this enzyme in which the LAGLIDADG motif residues are exchanged for those of homologous endonucleases (42,43).
We believe the structural changes in the Y2 variant of I-AniI, leading to repositioning of lysine 94 into contact with a putative nucleophilic water molecule, explains the increase in catalytic efficiency in its corresponding active site. This conclusion is in agreement with the hypothesized reaction mechanism for this family of endonucleases (44), which require the presence of one or more positively charged groups to assist in deprotonation of the hydrolytic water and/or to stabilize the phosphoanion transition state. In these enzymes, the usual basic residue in each active site is a lysine residue (K98 and 98′ in I-CreI, K122 and 223 in I-SceI, K43 and 140 in I-DmoI, and K94 and K227 in I-AniI). In the very high resolution structures of the most active LAGLIDADG endonuclease, I-CreI, the positions of K98 and K98′ superimpose quite closely on the position of K94 in the Y2 variant of I-AniI.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R01 GM49857 and RL1 CA133833 to B.L.S., RL1 CA133832 to A.M.S.); the Gates Foundation Grand Challenge Program (to A.M.S. and B.L.S.); National Institutes of Health training grant (T32 GM07270 to M.C.); Japan Society for the Promotion of Science (to R.T.). Funding for open access charge: National Institutes of Health (GM49857 and RL1 CA133833).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Technical assistance and advice was provided by Jill Bolduc, Audrey McConnell-Smith, Lei Zhao and other members of the FHCRC structural biology program.
REFERENCES
- 1.Belfort M, Roberts RJ. Homing endonucleases - keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoddard BL. Homing endonuclease structure and function. Quarterly Reviews of Biophysics. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 3.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 4.Kostriken R, Strathern JN, Klar AJ, Hicks JB, Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983;35:167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- 5.Colleaux L, d'Auriol L, Betermier M, Cottarel G, Jacquier A, Galibert F, Dujon B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986;44:521–533. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- 6.Dalgaard JZ, Klar AJ, Moser MJ, Holley WR, Chatterjee A, Mian IS. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res. 1997;25:4626–4638. doi: 10.1093/nar/25.22.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Argast GM, Stephens KM, Emond MJ, Monnat RJ. I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J. Mol. Biol. 1998;280:345–353. doi: 10.1006/jmbi.1998.1886. [DOI] [PubMed] [Google Scholar]
- 8.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 9.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of homing endonuclease specificity and its host target sequence. J. Mol. Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Current Gene Therapy. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 11.Lambowitz AM, Belfort M. Introns as mobile genetic elements. Annu. Rev. Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 12.Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc. Natl Acad. Sci. USA. 1999;96:13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posey KL, Koufopanou V, Burt A, Gimble FS. Evolution of divergent DNA recognition specificities in VDE homing endonucleases from two yeast species. Nucleic Acids Res. 2004;32:3947–3956. doi: 10.1093/nar/gkh734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delahodde A, Goguel V, Becam AM, Creusot F, Perea J, Banroques J, Jacq C. Site-specific DNA endonuclease and RNA maturase activities of two homologous intron-encoded proteins from yeast mitochondria. Cell. 1989;56:431–441. doi: 10.1016/0092-8674(89)90246-8. [DOI] [PubMed] [Google Scholar]
- 15.Wenzlau JM, Saldanha RJ, Butow RA, Perlman PS. A latent intron-encoded maturase is also an endonuclease needed for intron mobility. Cell. 1989;56:421–430. doi: 10.1016/0092-8674(89)90245-6. [DOI] [PubMed] [Google Scholar]
- 16.Mohr G, Perlman PS, Lambowitz AM. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 1993;21:4991–4997. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schafer B, Wilde B, Massardo DR, Manna F, Del Giudice L, Wolf K. A mitochondrial group-I intron in fission yeast encodes a maturase and is mobile in crosses. Curr. Genet. 1994;25:336–341. doi: 10.1007/BF00351487. [DOI] [PubMed] [Google Scholar]
- 18.Ho Y, Kim SJ, Waring RB. A protein encoded by a group I intron in Aspergillus nidulans directly assists RNA splicing and is a DNA endonuclease. Proc. Natl Acad. Sci. USA. 1997;94:8994–8999. doi: 10.1073/pnas.94.17.8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szczepanek T, Jamoussi K, Lazowska J. Critical base substitutions that affect the splicing and/or homing activities of the group I intron bi2 of yeast mitochondria. Mol. Gen. Genet. 2000;264:137–144. doi: 10.1007/s004380000297. [DOI] [PubMed] [Google Scholar]
- 20.Bolduc JM, Spiegel PC, Chatterjee P, Brady KL, Downing ME, Caprara MG, Waring RB, Stoddard BL. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J. Am. Chem. Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 22.Chatterjee P, Brady KL, Solem A, Ho Y, Caprara MG. Functionally distinct nucleic acid binding sites for a group I intron-encoded RNA maturase/DNA homing endonuclease. J. Mol. Biol. 2003;329:239–251. doi: 10.1016/s0022-2836(03)00426-1. [DOI] [PubMed] [Google Scholar]
- 23.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solem A, Chatterjee P, Caprara MG. A novel mechanism for protein-assisted group I intron splicing. RNA. 2002;8:412–425. doi: 10.1017/s1355838202029321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Caprara MG, Chatterjee P, Solem A, Brady-Passerini KL, Kaspar BJ. An allosteric-feedback mechanism for protein-assisted group I intron splicing. RNA. 2007;13:211–222. doi: 10.1261/rna.307907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Downing ME, Brady KL, Caprara MG. A C-terminal fragment of an intron-encoded maturase is sufficient for promoting group I intron splicing. RNA. 2005;11:437–446. doi: 10.1261/rna.7225205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epinat JC, Arnould S, Chames P, rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, Lacroix E. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurokawa S, Yamasaki T, Komatsu T, Watanabe KI, Ohama T. Degenerated recognition property of a mitochondrial homing enzyme in the unicellular green alga Chlamydomonas smithii. Plant Mol. Biol. 2006;62:141–150. doi: 10.1007/s11103-006-9009-y. [DOI] [PubMed] [Google Scholar]
- 34.Prieto J, Epinat JC, Redondo P, Ramos E, Padro D, Cedrone F, Montoya G, Paques F, Blanco FJ. Generation and analysis of mesophilic variants of the thermostable archaeal I-DmoI homing endonuclease. J. Biol. Chem. 2008;283:4364–4374. doi: 10.1074/jbc.M706323200. [DOI] [PubMed] [Google Scholar]
- 35.Siddiqui KS, Cavicchioli R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006;75:403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- 36.Sriprapundh D, Vieille C, Zeikus JG. Directed evolution of Thermotoga neapolitana xylose isomerase: high activity on glucose at low temperature and low pH. Protein Eng. 2003;16:683–690. doi: 10.1093/protein/gzg082. [DOI] [PubMed] [Google Scholar]
- 37.Ho Y, Waring RB. The maturase encoded by a group I intron from Aspergillus nidulans stabilizes RNA tertiary structure and promotes rapid splicing. J. Mol. Biol. 1999;292:987–1001. doi: 10.1006/jmbi.1999.3070. [DOI] [PubMed] [Google Scholar]
- 38.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 39.Turmel M, Mercier JP, Cote V, Otis C, Lemieux C. The site-specific DNA endonuclease encoded by a group I intron in the Chlamydomonas pallidostigmatica chloroplast small subunit rRNA gene introduces a single-strand break at low concentrations of Mg2+ Nucleic Acids Res. 1995;23:2519–2525. doi: 10.1093/nar/23.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perrin A, Buckle M, Dujon B. Asymmetrical recognition and activity of the I-SceI endonuclease on its site and on intron-exon junctions. EMBO Journal. 1993;12:2939–2947. doi: 10.1002/j.1460-2075.1993.tb05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agaard C, Awayez MJ, Garrett RA. Profile of the DNA recognition site of the archaeal homing endonuclease I-DmoI. Nucleic Acids Res. 1997;25:1523–1530. doi: 10.1093/nar/25.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva GH, Belfort M. Analysis of the LAGLIDADG interface of the monomeric homing endonuclease I-DmoI. Nucleic Acids Res. 2004;32:3156–3168. doi: 10.1093/nar/gkh618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva GH, Belfort M, Wende W, Pingoud A. From monomeric to homodimeric endonucleases and back: engineering novel specificity of LAGLIDADG enzymes. J. Mol. Biol. 2006;361:744–754. doi: 10.1016/j.jmb.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 44.Chevalier B, Monnat,J RJ, Stoddard BL. In: Homing Endonucleases and inteins. al M. Be., editor. Vol. 16. Berlin: Springer Verlag; 2005. pp. 34–47. [Google Scholar]