Abstract
Anti-miRNA antisense inhibitors (AMOs) have demonstrated their utility in miRNA research and potential in miRNA therapy. Here we report a modified AMO approach in which multiple antisense units are engineered into a single unit that is able to simultaneously silence multiple-target miRNAs, the multiple-target AMO or MTg-AMO. We validated the technique with two separate MTg-AMOs: anti-miR-21/anti-miR-155/anti-miR-17-5p and anti-miR-1/anti-miR-133. We first verified the ability of the MTg-AMOs to antagonize the repressive actions of their target miRNAs using luciferase reporter activity assays and to specifically knock down the levels of their target miRNAs using real-time RT-PCR methods. We then used the MTg-AMO approach to identify several tumor suppressors—TGFBI, APC and BCL2L11 as the target genes for oncogenic miR-21, miR-155 and miR-17-5p, respectively, and two cardiac ion channel genes HCN2 (encoding a subunit of cardiac pacemaker channel) and CACNA1C (encoding the α-subunit of cardiac L-type Ca2+ channel) for the muscle-specific miR-1 and miR-133. We further demonstrated that the MTg-AMO targeting miR-21, miR-155 and miR-17-5p produced a greater inhibitory effect on cancer cell growth, compared with the regular single-target AMOs. Moreover, while using the regular single-target AMOs excluded HCN2 as a target gene for either miR-1 or miR-133, the MTg-AMO approach is able to reveal HCN2 as the target for both miR-1 and miR-133. Our findings suggest the MTg-AMO as an improved approach for miRNA target finding and for studying function of miRNAs. This approach may find its broad application for exploring biological processes involving multiple miRNAs and multiple genes.
INTRODUCTION
Discovery of microRNA-1 (miRNA) has revolutionarily advanced our understanding of the molecular mechanisms of gene expression regulation (1,2). MiRNAs are a large, ubiquitous class of endogenous noncoding regulatory mRNAs of ∼22-nt long that post-transcriptionally regulate the expression of numerous eukaryotic genes. These molecules act by binding to their target mRNAs, preferentially to the 3′-untranslated region (3′-UTR) by a partial base-paring mechanism. In order for a miRNA to give rise of functional consequences, its 5′-end 7–8 nt must have exact complementarity to the target mRNA, the so-called ‘seed’ region, and partial complementarity with rest of the sequence. The current model for inhibition of expression by miRNA suggests that a miRNA either inhibits translation or induces degradation of its target mRNA, depending upon the overall degree of complementarity of the binding site, number of binding sites and the accessibility of the bindings sites (3–6). MiRNAs have been increasingly implicated in the control of various biological processes, including cell differentiation, cell proliferation, development and apoptosis, and many pathological processes such as cancer, Alzheimer's disease, cardiovascular disease, etc (7–9). Recent evidence has suggested that miRNAs might be viable therapeutic targets for a wide range of human disease including cancers and cardiovascular disorders (9,10).
One of the indispensable approaches in miRNA research as well as in miRNA therapy is inhibition or loss-of-function of miRNAs. The base-pair interaction between miRNAs and mRNAs is essential for the function of miRNAs; therefore, a logical approach of silencing miRNAs is to use a nucleic acid that is antisense to the miRNA (9,11–15). These anti-miRNA oligonucleotides (AMOs) specifically and stoichiometrically bind, and efficiently and irreversibly silence their target miRNAs. The AMO approach has been used in numerous studies to identify many cellular functions of miRNAs and has been considered a plausible strategy for miRNA therapy of human disease (9,11–15).
However, it has become clear that a particular condition may be associated with multiple miRNAs and a given gene may be regulated by multiple miRNAs. For example, a study directed to the human heart identified 67 significantly upregulated miRNAs and 43 significantly downregulated miRNAs in failing left ventricles versus normal hearts (16). No less than five different miRNAs have been shown to critically involve in cardiac hypertrophy (17–21). Similarly, Volinia et al. (22) conducted a large-scale miRNA analysis on 540 solid tumor samples including lung, breast, stomach, prostate, colon and pancreatic tumors. Their survey revealed 15 miRNAs upregulated and 12 downregulated in human breast cancer tissues. Similar changes of multiple miRNAs were also found in other five solid tumor types. Many of these miRNAs have been reported to regulate cell proliferation or apoptosis and some of them have been considered oncogenic miRNAs or tumor suppressor miRNAs. These properties of miRNA regulation may well create some uncertainties of outcomes by applying the AMO technology to silence miRNAs since knocking down a single miRNA may not be sufficient, and certainly not optimal, to achieve the expected interference of cellular process and gene expression which are regulated by multiple miRNAs. These facts also prompted us to raise several pertinent questions. If targeting a single miRNA is adequate for tackling a pertinent pathological condition? If simultaneously targeting multiple miRNAs relevant to a particular condition offers an improved approach than targeting a single miRNA using the regular AMO techniques? How can we concomitantly silence multiple miRNAs to achieve an interference of a cellular function? To answer these questions, we developed an innovative strategy, the multiple-target AMO technology, which confers a single AMO fragment the capability of targeting multiple miRNAs. We functionally validated the technology with oncogenic miRNAs miR-21, miR-155 and miR-17-5p, and the muscle-specific miRNAs miR-1 and miR-133, by demonstrating the superiority of MTg-AMOs over the regular AMOs in miRNA targeting. Using this new technology, we were also able to identify several new target genes for these miRNAs.
MATERIALS AND METHODS
Neonatal rat cardiomyocyte culture
The procedures were essentially the same as previous described (23). One- to two-day-old rats were decapitated, hearts aseptically removed, dissected, minced and exposed overnight to 0.05% trypsin (4°C). The next day, cells were dissociated with collagenase (Worthington), centrifuged (500 g, 4°C), and preplated in culture flasks (60 min, 37°C). Subsequently, nonattached cells (myocytes) were plated on 24-well plates in DMEM/F-12 medium (Invitrogen) containing 10% FBS, 0.1 mM L-ascorbic acid, 0.1 mM bromodeoxyuridine (Sigma), insulin- transferring-sodium selenite (Invitrogen) and 10–9 M triiodothyronine. Over 90% of cells in culture were observed to beat. Use of animals was in accordance with the regulations of the ethic committees of Harbin Medical University and Montreal Heart Institute.
Cell culture
HEK293 cells (human embryonic kidney cell line) and MCF-7 (human breast cancer cell line) were purchased from American Type Culture Collection (ATCC, Manassas, VA). The cells were cultured in Dulbecco's Modified Eagle Medium (DMEM). The culture was supplemented with 10% fetal bovine serum and 100 μg/ml penicillin/streptomycin.
Synthesis of miRNAs and anti-miRNA inhibitor antisense oligonucleotides
Human mature miRNAs (miR-21, miR-155, miR-17-5p, miR-1 and miR-133) were synthesized by Invitrogen. These mature miRNAs were nonchemically modified and carried the 2-nt overhands as defined in miRBase (http://microrna.sanger.ac.uk/registry/; the Wellcome Trust Sanger Institute). The two strands of each miRNA were synthesized separately and underwent annealing steps to a duplex. The sequences of anti-miRNA antisense inhibitor oligodeoxyribonucleotides (AMOs) are the exact antisense copies of their mature miRNA sequences and the multiple-target AMOs (MTg-AMOs) were synthesized by Integrated DNA Technologies, Inc. (IDT). The MTg-AMOs tested in this study were designed to integrate the AMOs against miR-21, miR-155 and miR-17-5p into one AMO (MTg-AMO21/155/17) and those against miR-1 and miR-133 into another antisense fragment (MTg-AMO1/133). An 8-nt linker (underlined letters in Figures 1A and 2A) was inserted to connect the two adjacent antisense units and 5 nt at both ends were locked with methylene bridges (LNA), with the rest of residues at the form of DNA.
Figure 2.
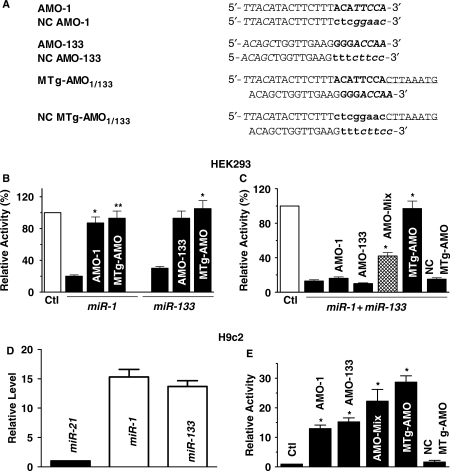
Comparison of anti-miRNA antisense inhibitor oligonucleotides (AMO) and multiple target AMO (MTg-AMO) approaches as tools for miRNA research. (A) Sequences of the AMOs and MTg-AMOs used in the experiment. An 8-nt linker (underlined letters) was inserted to connect the two adjacent antisense units and 5 nt at both ends were locked with methylene bridges. The base substitution mutations (lower bold case) were introduced into the first eight nucleotides from the 5′-end of each antisense units (capital bold letters) to construct a negative control MTg-AMO. (B) Comparison of MTg-AMO1/133 (10 nM) and the regular AMOs (AMO-1 and AMO-133; 10 nM each) in their silencing effects induced by miR-1 and miR-133 (10 nM), respectively, as determined by luciferase reporter activities in HEK293 cells. (C) Comparison of MTg-AMO1/133 and the regular AMOs (AMO-1 and AMO-133) in their silencing effects induced by co-application of miR-1 and miR-133 (miR-1 + miR-133), as determined by luciferase reporter activities in HEK293 cells. AMO-Mix: mixture of AMO-1 and AMO-133, 10 nM each; NC MTg-AMO: negative control MTg-AMO1/133 *P < 0.05 versus Ctl; unpaired Student t-test; n = 5 batches of cells for each group. (D) Verification of presence of miR-1 and miR-133 in H9c2 cells, measured by quantitative real-time RT-PCR. The data were normalized to the value of miR-21. (E) Comparison of effects of MTg-AMO1/133 or the regular AMOs (AMO-1 and AMO-133) alone on luciferase reporter activities in H9c2 cardiac cells. Note that all AMOs significantly enhanced luciferase activity presumably by silencing endogenous miRNAs miR-1 and miR-133. *P < 0.05 versus Ctl; unpaired Student t-test; n = 5 batches of cells for each group.
Construction of chimeric miRNA binding site–luciferase reporter vectors
The sequence of a fragment containing exact binding sites for all five miRNAs (miR-21/miR-155/miR-17-5p/miR-1/miR-133, Supplementary Figure 1) was synthesized by Invitrogen and the sense and antisense strands of the oligonucleotides were annealed by adding 2 μg of each oligonucleotides to 46 μl of annealing solution (100 mM K-acetate, 30 mM HEPES-KOH, pH 7.4 and 2 mM Mg-acetate) and incubated at 90°C for 5 min and then at 37°C for 1 h. The annealed oligonucleotides were then inserted into HindIII and SpeI sites within the 3′-UTR luciferase gene in the pMIR-REPORT™ luciferase miRNA expression reporter vector (Ambion, Inc.) (23,24).
The 3′-UTR of transforming growth factor BI (TGFBI), a fragment (8590–9590 bp; accession no. NM_000358) containing the putative target site for miR-155 within the 3′-UTR of APC, a fragment (2515–2815 bp; accession no. NM_000038) containing the putative target site for miR17-5p within the 3′-UTR of BCL2L11, a fragment (6685–6885 bp; accession no. NM_138621) containing the predicted miR-133 site within the 3′-UTR of CACNA1C and the 3′-UTR of HCN2 were inserted, respectively, into the 3′-UTR region of the pMIR-REPORT™ luciferase miRNA expression reporter vector (Ambion, Inc.) to create the chimeric luciferase-miRNA vectors (25,26).
Mutagenesis
Nucleotide-substitution mutations (as shown in Figure 1A) were carried out by direct oligomers synthesis by IDT. The substitution nucleotides were so designed to avoid producing new binding sites for other miRNAs. All constructs were sequencing verified.
Figure 1.
Comparison of anti-miRNA antisense inhibitor oligonucleotides (AMO) and multiple target AMO (MTg-AMO) approaches as tools for miRNA research. (A) Sequences of the AMOs and MTg-AMOs used in our experiments. All AMOs and MTg-AMOs were chemically modified to have 5 nt at both ends locked with methylene bridges. An 8-nt linker (underlined letters) was inserted to connect the two adjacent antisense units. The base substitution mutations (lower bold case) were introduced into the first 8 nt from the 5′-end of each antisense units (capital bold letters) to construct a negative control MTg-AMO. (B) Comparison of MTg-AMO21/155/17 (10 nM) and the regular AMOs (AMO-21, AMO-155 and AMO-17; 10 nM each) in their silencing effects induced by miR-21, miR-155, and miR17-5p (10 nM), respectively, as determined by luciferase reporter activities in HEK293 cells. (C) Comparison of MTg-AMO21/155/17 and the regular AMOs (AMO-21 and AMO-155) in their silencing effects induced by coapplication of miR-21 and miR-155 (miR-21 + miR-155), as determined by luciferase reporter activities in HEK293 cells. (D) Comparison of MTg-AMO21/155/17 and the regular AMOs (AMO-21 and AMO-17) in their silencing effects induced by co-application of miR-21 and miR-17-5p (miR-21 + miR-17), as determined by luciferase reporter activities in HEK293 cells. (E) Comparison of MTg-AMO21/155/17 and the regular AMOs (AMO-21, AMO-155 and AMO-17) in their silencing effects induced by co-application of miR-21, miR-155, and miR17-5p (miR-21 + miR-155 + miR-17), as determined by luciferase reporter activities in HEK293 cells. AMO-Mix: mixture of AMO-21, AMO-155 and AMO-17, 10 nM each; NC MTg-AMO: negative control MTg-AMO21/155/17. *P < 0.05 versus Ctl; + P < 0.05 versus AMO-Mix; unpaired Student t-test; n = 5 batches of cells for each group. (F) Verification of presence of miR-21, miR-155, and miR17-5p in HeLa cervical carcinoma cells, measured by quantitative real-time RT-PCR. The data were normalized to the value of miR-1. (G) Comparison of effects of MTg-AMO21/155/17 or the regular AMOs (AMO-21, AMO-155 and AMO-17) alone on luciferase reporter activities in HeLa cells. Note that all AMOs significantly enhanced luciferase activity presumably by silencing endogenous miRNAs miR-21, miR-155, and miR17-5p. *P < 0.05 versus Ctl; unpaired Student t-test; n = 6 batches of cells for each group.
In vitro transfection of miRNAs and luciferase assay
After a 24-h starvation in serum-free medium, cells (1 × 105/well) were transfected with 1 μg the miRNAs, AMOs or MTg-AMOs with lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions (25,26).
For luciferase assay, cells were co-transfected with 1 μg PGL3–target DNA (firefly luciferase vector) and 0.1 μg PRL-TK (TK-driven Renilla luciferase expression vector) with lipofectamine 2000. Following transfection (48 h), luciferase activities were measured with a dual luciferase reporter assay kit (Promega) on a luminometer (Lumat LB9507).
Quantification of mRNA and miRNA levels
For quantification of transcripts of TGFBI, APC, BCL2L11, HCN2 and CACNA1C, conventional real-time RT-PCR was carried out with total RNA samples extracted from rat ventricular wall of experimental myocardial infarction and treated with DNase I (23,24). TaqMan quantitative assay of transcripts was performed with real-time two-step RT-PCR (GeneAmp 5700, PE Biosystems), involving an initial RT with random primers and subsequent PCR amplification of the targets. Expression level of GAPDH was used as an internal control.
The mirVana™ qRT-PCR miRNA Detection Kit (Ambion) was used in conjunction with real-time PCR with SYBR Green I for quantification of miRNA transcripts in our study, following the manufacturer's instructions. The total RNA samples were isolated with Ambion's mirVana miRNA Isolation Kit, from human left ventricular preparations from patients with myocardial infarction due to coronary artery disease (CAD) and from rat myocardium. Reactions contained mirVana qRT-PCR Primer Sets specific for human or rat miR-1s and human 5S rRNA as positive controls. qRT-PCR was performed on a 96-well StepOnePlus™ system (A&B Applied Biosystems) for 40 cycles. We first determined the appropriate cycle threshold (Ct) using the automatic baseline determination feature. We then performed dissociation analysis (melt-curve) on the reactions to identify the characteristic peak associated with primer-dimers in order to separate from the single prominent peak representing the successful PCR amplification of miRNAs. Fold variations in expression of miRNAs between RNA samples were calculated.
Western blot
The protein samples were extracted from rat left ventricular wall, with the procedures essentially the same as described in detail elsewhere (25,26). The protein content was determined with Bio-Rad Protein Assay Kit (Bio-Rad, Mississauga, ON, Canada) using bovine serum albumin as the standard. Protein sample (∼30 μg) was fractionated by SDS–PAGE (7.5–10% polyacrylamide gels) and transferred to PVDF membrane (Millipore, Bedford, MA). The sample was incubated overnight at 4°C with primary antibodies for TGFBI, APC and BCL2L11 (Santa Cruz), and for HCN2 and Cav1.2 (Alomone Labs). Bound antibodies were detected using the chemiluminescent substrate (Western Blot Chemiluminescence Reagent Plus, NEN Life Science Products, Boston, USA). Western blot bands were quantified using QuantityOne software by measuring the band intensity (Area × OD) for each group and normalizing to GAPDH (anti-GAPDH antibody from Research Diagnostics Inc) as an internal control. The final results are expressed as fold changes by normalizing the data to the control values.
Data analysis
Group data are expressed as mean ± S.E. Statistical comparisons (performed using ANOVA followed by Dunnett's method) were carried out using Microsoft Excel. A two-tailed P < 0.05 was taken to indicate a statistically significant difference. Nonlinear least square curve fitting was performed with GraphPad Prism 3.1.
RESULTS AND DISCUSSION
MTg-AMOs in miRNA target finding
We first designed three regular AMOs (AMO-21, AMO-155 and AMO-17) exactly antisense to miR-21, miR-155, and miR-17-5p, respectively. We choose miR-21, miR-155, and miR-17-5p as the miRNA targets for testing our approach since they are proven oncogenic miRNAs overexpressing in several solid cancers (22,27). We then generated another AMO, which was engineered to contain the three regular AMO units (Figure 1A)that could presumably target all the three miRNAs. For convenience, we named this longer AMO multiple-target AMO21/155/17 or MTg-AMO21/155/17. In addition, a negative control MTg-AMO21/155/17 (Figure 1A) was also constructed as a negative control for the effects of MTg-AMO21/155/17.
We went on to test the ability of these MTg-AMOs to inactivate their respective target miRNAs by luciferase reporter assays. An artificially designed fragment containing the binding sequences for all five target miRNAs (miR-21, miR-155, miR-17-5p, miR-1 and miR-133) was inserted into the 3′-UTR of the luciferase gene in the luciferase reporter vector (Supplementary Figure 1 online). Luciferase activities were measured 48 h after transfection of the vector together with various constructs (as specified) into the HEK293 cells. As shown in Figure 1B, MTg-AMO21/155/17 (10 nM) was able to abolish the inhibitory effect on luciferase activity, induced by any of miR-21, miR-155 and miR-17-5p alone (10 nM) or any combinations of the three miRNAs at 10 nM each (Figure 1C–E), while the negative control MTg-AMO21/155/17 (10 nM) failed to produce any effects. By comparison, the regular AMO targeting a single miRNA affected the luciferase activity only when co-transfected with its target miRNA. Also striking is that MTg-AMO21/155/17 produced significantly greater repressive effects than the mixture of the regular AMOs AMO-21, AMO-155 and AMO-17 at 10 nM each (Figure 1D), which is labeled ‘AMO-Mix’ in the figures. The negative control MTg-AMO21/155/17 did not produce any significant effects (Figure 2C–E). To examine whether the AMOs could also exert silencing effects on endogenous miRNAs in HeLa cervial carcinoma cells, we first confirmed the presence of mature miR-21, miR-155 and miR-17-5p in these cells (Figure 1F), and then we looked at the luciferase activities after co-transfection of the vector and MTg-AMO21/155/17, AMO-21, AMO-155 or AMO-17. As expected, these AMOs all were able to significantly increase luciferase activities (Figure 1G), presumably by knocking down the endogenous miR-21, miR-155 and miR-17-5p in HeLa cells.
In order to investigate whether the enhanced effects of MTg-AMO could be reproduced on other target miRNAs, we designed another MTg-AMO fragment targeting miR-1 and miR-133 (Figure 2A), the muscle-specific miRNAs crucial to myogenesis and cardiac hypertrophy and ischemic arrhythmogenesis (19–22,24–26,28,29). Here we called this fragment MTg-AMO1/133. As illustrated in Figure 2B and C, MTg-AMO1/133 (10 nM) antagonized the repressive actions produced by either miR-1 or miR-133 (10 nM) in luciferase assays. MTg-AMO1/133 also reversed the inhibitory effects elicited by co-application of miR-1 and miR-133, with a greater potency than the mixture of AMO-1 and AMO-133 (10 nM each, Figure 2C). We then went on to verify the ability of MTg-AMO1/133, AMO-1 and AMO-133 to inhibit the repressing effects induced by endogenous miR-1 and miR-133 in H9c2 rat ventricular cells. We first demonstrated the presence of miR-1 and miR-133 in H9c2 cells (Figure 2D), and then we observed the luciferase activities after co-transfection of the vector and MTg-AMO1/133, AMO-1 and AMO-133. As shown in Figure 2E, these AMOs applied alone each significantly enhanced luciferase activities.
To verify the luciferase results and to search the target genes of the five miRNAs using our MTg-AMO strategy, we first performed computational prediction using the miRBase hosted by the Wellcome Trust Sanger Institute. We identified three tumor suppressor genes—TGFBI, APC and BCL2L11 as potential targets for miR-21, miR-155 and miR-17-5p, respectively, which carry in their 3′-UTRs the binding sites for the three miRNAs (Supplementary Figure 2 online). We then evaluated the effects of MTg-AMO21/155/17 on the protein levels of these three candidate target genes. As depicted in the left and middle panels of Figure 3A, MTg-AMO21/155/17 (10 nM) concordantly increased the levels of tumor suppressor genes TGFBI, APC and BCL2L11 in MCF-7 human breast cancer cells which express abundant endogenous miR-21, miR-155 and miR-17-5p (30) (Supplementary Figure 3 online). As expected, the negative control MTg-AMO21/155/17 failed to elicit any significant effects. The specificities of the regular individual AMOs on their respective target genes were verified with the data shown in the right panel of Figure 3A.
Figure 3.
MTg-AMO as an improved AMO approach for miRNA target finding. (A and B) Western blot analysis of the protein levels of TGFBI, APC BCL2L11, HCN2 and Cav1.2. For MTg-AMO21/155/17, human breast cancer MCF-7 cells were used and for MTg-AMO1/133, cultured neonatal rat ventricular myocytes were used. NC and NC MTg-AMO: negative control MTg-AMO. The relevant miRNA for each target mRNA is specified above the name of the target mRNA in western blot images. *P < 0.05 versus Ctl; +P < 0.05 versus MTg-AMO; unpaired Student t-test; n = 3 for each group. (C and D) Silencing effects of MTg-AMO21/155/17 and MTg-AMO1/133 in HEK293 cells, reported by luciferase vector containing the 3′′-UTRs of the five genes. The cells were co-transfected with the chimeric vectors carrying miR-21, miR-155, and miR17-5p, miR-1 and miR-133 sites, the miRNAs and the MTg-AMOs, and the luciferase activities were measured 48 h after transfection. The concentration of each construct was 10 nM. *P < 0.05 versus Ctl, unpaired Student t-test; n = 5 batches of cells for each group.
Similarly, through theoretical analysis, we identified two ion channel genes HCN2 (encoding a subunit of cardiac pacemaker channel) (26) and CACNA1C (encoding the α-subunit of cardiac L-type Ca2+ channel) (31) as potential targets for miR-1 and miR-133 since the 3′-UTR of HCN2 mRNA contains the putative binding sites for both miR-1 and miR-133, and that of CACNA1C mRNA has a putative binding sequence for miR-133 only (Supplementary Figure 2 online). We observed that transfection of MTg-AMO1/133 remarkably increased the protein levels of the two ion channels in isolated neonatal rat ventricular myocytes which are known to express endogenous miR-1 and miR-133 (23). Notably, neither AMO-1 nor AMO-133 alone produced significant effects on the protein level of HCN2, indicating that use of the regular AMOs can yield false-negative results when a gene is simultaneously regulated by more than one miRNA. Co-transfection of AMO-1 and AMO-133 (AMO-Mix) also increased the HCN2 protein levels, but to a significantly less extent compared to MTg-AMO1/133 (Figure 3B).
To verify that TGFBI, APC and BCL2L11 are indeed the cognate targets for miR-21, miR-155 and miR-17-5p, and HCN2 and CACNA1C the cognate targets for miR-1 and miR-133, respectively, we inserted the 3′-UTRs or fragments within the 3′-UTRs containing the predicted miRNA-binding sites of the genes into the 3′-UTR of the luciferase reporter vector. The chimeric vectors containing the 3′-UTRs of TGFBI, APC and BCL2L11, along with other constructs as specified in Figure 3C, were co-transfected into the HEK293 cells. MTg-AMO21/155/17 but not the negative control MTg-AMO21/155/17 abolished the depression of luciferase activities consequent to the interactions between the 3′-UTRs and the miRNAs, whereas none of the AMOs alone was sufficient to reverse completely the inhibition in the presence of miR-21, miR-155 and miR-17-5p together. Similar results were observed with the MTg-AMO1/133 and the 3′′-UTRs of HCN2 and CACNA1C genes (Figure 3D).
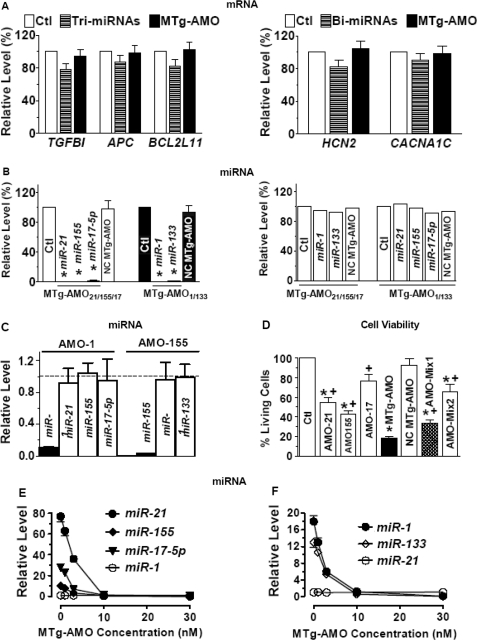
The MTg-AMOs did not significantly affect the mRNA levels of their targets genes (Figure 4A), neither did the regular AMOs (data not shown).
Figure 4.
Verification of the mode of actions of MTg-AMOs. (A) Effects of AMOs and MTg-AMOs on mRNA levels of their targets genes, determined by real-time qRT-PCR methods. Tri-miRNAs: co-transfection of miR-21, miR-155 and miR-17-5p at 10 nM each; Bi-miRNAs: co-transfection of miR-1 and miR-133 at 10 nM each. Note that the mRNA levels of the target genes, as specified, were not affected by the miRNAs. (B) Effects of MTg-AMOs on the activities of their target miRNAs. Note that the MTg-AMOs nearly abolished their target miRNAs but did not affect the non-target miRNAs. *P < 0.05 versus Ctl. n = 5 for each group. (C) Verification of the specificities of individual AMOs against their corresponding miRNAs. Data shown are the effects of AMO-1 and AMO-155 on the activities of various miRNAs. Note that AMO-1 and AMO-155 only knocked down their targeted miRNAs without affecting other miRNAs tested. (D) MTg-AMO as a potential strategy for miRNA therapy. Shown are the effects of each AMO (100 nM), AMO-Mix1 (a mixture of AMO-21, AMO-155 and AMO-17 at 10 nM each), AMO-Mix2 (a mixture of AMO-21, AMO-155 and AMO-17 at 3.3 nM each), MTg-AMO21/155/17 (10 nM) and negative control (NC) MTg-AMO21/155/17 (10 nM) on cell survival, as determined by MTT assay, in MCF-7 cells. *P < 0.05 versus Ctl; +P < 0.05 versus MTg-AMO21/155/17, unpaired Student t-test; n = 5 batches of cells for each group. (E) Measurement of miR-21, miR-155 and miR-17-5p levels by real-time RT-PCR methods. MTg-AMO21/155/17 amounts corresponding to 1-, 3-, 10- and 30-nM transfections were spiked into HeLa cell lysate (one lysate equals one well on a six-well plate) before RNA extraction (33). miR-1 level was used as a control for normalization. (F) Measurement of miR-1 and miR-133 levels by real-time RT-PCR. MTg-AMO1/133 amounts corresponding to 1-, 3-, 10- and 30-nM transfections were spiked into H9c2 cell lysate as described in (E). miR-21 level was used as a control for normalization.
To verify that the effects of MTg-AMO21/155/17 and MTg-AMO1/133 were indeed consequent to interacting with their target miRNAs, we quantified the miRNA levels. Consistent with previous studies showing degradation of target miRNAs by AMOs (11,13), we found that these MTg-AMOs remarkably reduced their target miRNAs (Figure 4B), and so did the regular AMOs (Figure 4C). Noticeably, the MTg-AMOs and the regular AMOs used in our study did not show cross-reactions with nontarget miRNAs. For example, MTg-AMO21/155/17 only reduced miR-21, miR-155 and miR-17-5p levels without altering miR-1 and miR-133 levels, detected by real-time RT-PCR; vice versa, MTg-AMO1/133 reduced only miR-1 and miR-133 levels without altering miR-21, miR-155 and miR-17-5p levels, indicating the specificities of actions of the MTg-AMOs.
It has recently been noticed that reduction of miRNA level by AMO could be simply due to the interference of AMO with miRNA detection when using northern blot with DNA probes, but not due to miRNA degradation (32,33). To see whether the miRNA reduction detected by real-time RT-PCR in our experiments was also ascribed to the same mechanism, we carried out another study following the procedure described by Fabani and Gait (33). We spiked MTg-AMO21/155/17 into lysate from control untreated HeLa cells at a series of concentrations ranging from 1 to 30 nM before RNA purification involving denaturing conditions, and then measured the levels of miR-21, miR-155 and miR-17-5p with the isolated RNA sample. As shown in Figure 4E, AMO21/155/17 produced concentration-dependent reduction of the levels, indicating that the miRNAs in the AMO21/155/17:miRNA complex were not directed into the RNA fraction during purification. Similar results were obtained with MTg-AMO1/133 on miR-1 and miR-133 in lysate from control untreated H9c2 cells (Figure 4F). Hence, the apparent reduction of miRNA levels revealed by qRT-PCR should not be used as evidence of a miRNA degradation pathway, but merely indicating the binding of the MTg-AMOs to the targeted miRNAs leading to inhibition of miRNA activities (33).
MTg-AMO enhances the efficacy to suppresses cancer cell growth
Since MTg-AMO21/155/17 was designed to target the three most frequently overexpressed oncogenic miRNAs in many cancers (21,22), it was expected to suppress cancer cell growth. To this end, we investigated the effects of MTg-AMO21/155/17 on viability of MCF-7 cells. MTg-AMO21/155/17 (10 nM) diminished the survival rate to ∼18% of the values from control group or negative control MTg-AMO21/155/17 group, whereas in order to reach significant effects with the regular AMOs, the concentration of each AMO had to be elevated to 100 nM (Figure 4D). Most noticeably, co-transfection of AMO-21, AMO-155 and AMO-17 at 10 nM each (a total of 30 nM, AMO-Mix1) or 3.3 nM each (a total of 10 nM, AMO-Mix2) produced significantly smaller decreases in cell survival than 10 nM MTg-AMO21/155/17 (Figure 4D).
In summary, using the MTg-AMO approach, we have been able to identify several tumor suppressors as the target genes for oncogenic miR-21, miR-155 and miR-17-5p and two cardiac ion channel genes for the muscle-specific miR-1 and miR-133. This study also represents the first to reveal the cardiac L-type Ca2+ channel α-subunit gene CACNA1C gene as a target gene for miR-133 action.
The improvement of the MTg-AMO approach over the regular single-target AMO approach is reflected by two findings. First, MTg-AMO21/155/17 demonstrated a strengthened ability to inhibit cancer cell growth compared with the single-target AMOs or combination of these AMOs. Second, the MTg-AMO approach is able to reveal HCN2 gene as a target for miR-1 and miR-133, which is otherwise concealed by the single-target AMO approach (Figure 3). There are two possible explanations for the superiority of the MTg-AMO approach over the regular AMO approach. First, the covalent linkage ensures co-delivery of different AMOs. Given the comparable cell targeting rates for the MTg-AMO and the individual AMOs, only the MTg-AMO approach ensures that the transfected cells received all relevant miRNA antagonists for gene targeting and proliferation inhibition. Another possible explanation is that annealing rate constants describing the annealing reaction between complementary nucleic acids strongly correlate with the length of the two complementary sequences. On the average, longer sequences anneal faster than shorter sequences (34,35). Therefore, the MTg-AMO might anneal faster with the miRNAs compared to the smaller individual AMO. Future studies are needed to verify these notions.
The beauty of this MTg-AMO ‘single-agent, multiple-targets’ strategy is that it can be either homogeneous carrying multiple identical antisense units targeting a specific miRNA to enhance the antagonizing effect or heterogeneous with multiple distinct antisense units targeting different miRNAs to expand targeting capacity. Considering that a given mRNA is regulated by multiple miRNAs and concordantly targeting multiple miRNAs might be required to achieve optimal interference of that gene, the MTg-AMO approach may be superior to the regular AMO technique. Just taking cancer therapy as an example, an MTg-AMO can be designed to target the miRNAs that are involved in tumor cell proliferation, cell death, metastasis and vascular angiogenesis. Targeting any one of these aspects may not be optimal for controlling tumorigenesis and progression. Interfering concomitantly with all these processes should expectedly yield a better anticancer outcome. Our findings therefore suggest the MTg-AMO approach as a useful tool in miRNA target finding and the potential of this novel strategy as a miRNA therapy for cancer and cardiac disorders.
We consider the present work still preliminary; to completely validate the MTg-AMO technology as a gene therapy strategy, many important issues are to be unresolved. Future studies are warranted to test more MTg-AMOs for the advantages of this approach and to get insight into more detailed mechanisms for the actions of MTg-AMOs. Dose–response actions were not evaluated in this study due to the difficulties of optimizing the ratio of construct/lipofectamine to ensure efficient transfection. The optimal combination of targets for an MTg-AMO remains unknown. In this study, we tested ‘two-in-one’ and ‘three-in-one’ MTg-AMOs. In theory, ‘N-in-one’ MTg-AMOs (N could be any number of AMO units) can be designed to include more relevant AMOs. More rigorous studies are warranted to define the optimal combination of length of MTg-AMOs to optimize desired effectiveness. This work does not allow us to draw any conclusions as to what the optimal organization is for multiple AMO units (the order of the units) to be placed in a single MTg-AMO molecule.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Nature Science Foundation of China (30430780); the Foundation of National Department of Science and Technology of China (2004CCA06700 to B.Y.); and by the Grant-in-Aid program to Chang Jiang Scholar Professor (to Z.W.). Funding for open access charge: the Foundation of National Department of Science and Technology of China (2004CCA06700 to B.Y.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank XiaoFan Yang for excellent technical support.
REFERENCES
- 1.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 5.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 8.Dalmay T. MicroRNAs and cancer. J. Intern. Med. 2008;263:366–375. doi: 10.1111/j.1365-2796.2008.01926.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Luo X, Lu Y, Yang B. miRNAs at the heart of the matter. J. Mol. Med. 2008 doi: 10.1007/s00109-008-0341-3. doi: 10.1007/s00109-008-0341-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeyaseelan K, Herath WB, Armugam A. MicroRNAs as therapeutic targets in human diseases. Expert. Opin. Ther. Targets. 2007;11:1119–1129. doi: 10.1517/14728222.11.8.1119. [DOI] [PubMed] [Google Scholar]
- 11.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 12.Hammond SM. MicroRNA therapeutics: a new niche for antisense nucleic acids. TiMM. 2006;12:99–101. doi: 10.1016/j.molmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Cheng AM, Byrom MW, Shelton J, Ford LP, Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stenvang J, Kauppinen S. MicroRNAs as targets for antisense-based therapeutics. Expert Opin. Biol. Ther. 2008;8:59–81. doi: 10.1517/14712598.8.1.59. [DOI] [PubMed] [Google Scholar]
- 15.Eckstein F. The versatility of oligonucleotides as potential therapeutics. Expert Opin. Biol. Ther. 2007;7:1021–1034. doi: 10.1517/14712598.7.7.1021. [DOI] [PubMed] [Google Scholar]
- 16.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 17.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl Acad. Sci. USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ. Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 19.Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 20.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart. Do they play a role in cardiac hypertrophy? Am. J. Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J. Mol. Cell. Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang B, Lin H, Xiao J, Lu Y, Luo X, Zhang Y, Xu C, Bai Y, Wang H, Chen G, et al. The muscle-specific microRNA miR-1 causes cardiac arrhythmias by targeting GJA1 and KCNJ2 genes. Nat. Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 24.Xiao J, Luo X, Lin H, Xu C, Gao H, Wang H, Yang B, Wang Z. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J. Biol. Chem. 2007;282:12363–12367. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- 25.Luo X, Lin H, Lu Y, Li B, Xiao J, Yang B, Wang Z. Transcriptional activation by stimulating protein 1 and post-transcriptional repression by muscle-specific microRNAs of IKs-encoding genes and potential implications in regional heterogeneity of their expressions. J. Cell. Physiol. 2007;212:358–367. doi: 10.1002/jcp.21030. [DOI] [PubMed] [Google Scholar]
- 26.Xiao J, Yang B, Lin H, Lu Y, Luo X, Wang Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: Examination on the pacemaker channel genes HCN2 and HCN4. J. Cell. Physiol. 2007;212:285–292. doi: 10.1002/jcp.21062. [DOI] [PubMed] [Google Scholar]
- 27.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 29.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 31.Pang L, Koren G, Wang Z, Nattel S. Tissue-specific expression of two human Cav1.2 isoforms under the control of distinct 5′-flanking regulatory elements. FEBS Lett. 2003;546:349–354. doi: 10.1016/s0014-5793(03)00629-x. [DOI] [PubMed] [Google Scholar]
- 32.Horwich MD, Zamore PD. Design and delivery of antisense oligonucleotides to block microRNA function in cultured Drosophila and human cells. Nat. Protoc. 2008;3:1537–1549. doi: 10.1038/nprot.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabani MM, Gait MJ. miR-122 targeting with LNA/2’-O methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14:336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patzel V, Sczakiel G. Length dependence of RNA-RNA annealing. J. Mol. Biol. 1999;294:1127–1134. doi: 10.1006/jmbi.1999.3330. [DOI] [PubMed] [Google Scholar]
- 35.Lee CH, Wetmur JG. On the kinetics of helix formation between complementary ribohomopolymers and deoxyribohomopolymers. Biopolymers. 1972;11:1485–1497. doi: 10.1002/bip.1972.360110712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.