Abstract
Methylation of CpGs is generally thought to repress transcription without significant influence from the sequence surrounding the methylated dinucleotides. Using the mouse Igf2/H19 imprinting control region (ICR), Igf2r differentially methylated region 2 (DMR2) and bacterial sequences, we addressed how methylation-dependent repression (MDR) from a distance varies with CpG number, density and surrounding sequence. In stably transfected F9 cells, the methylated ICR repressed expression from a CpG-free reporter plasmid more than 1000-fold compared with its unmethylated control. A segment of pBluescript, with a CpG number equal to the ICR's but with a higher density, repressed expression only 70-fold when methylated. A bacteriophage lambda fragment and the Igf2r DMR2 showed minimal MDR activity, despite having CpG numbers and densities similar to or greater than the ICR. By rearranging or deleting CpGs, we identified CpGs associated with three CTCF sites in the ICR that are necessary and sufficient for sequence-specific MDR. In contrast to F9 cells, the methylated ICR and pBS fragments exhibited only 3-fold reporter repression in Hela cells and none in Cos7. Our results show that the strength of MDR from a distance can vary a 1000-fold between different cell types and depends on the sequence surrounding the methylated CpGs, but does not necessarily increase with CpG number or density.
INTRODUCTION
Methylation of CpG dinucleotides plays an important role in the regulation of genomic activity in vertebrates. The majority of DNA methylation is concentrated in transposable elements and centromeric repeats, but low levels are present across the genome (1). Consistent with its concentration on repetitive sequences, DNA methylation inhibits transcription of transposable elements (2) and suppresses chromosomal rearrangements between centromeric and telomeric regions (3,4). DNA methylation is also required for maintaining X-chromosome inactivation and for the allele-specific expression of imprinted genes (5–8).
A large body of work in vivo and in cultured cells has demonstrated a consistent correlation between promoter methylation and gene repression (9,10). The widespread promoter methylation of silenced genes on the inactive X-chromosome is a clear example of this correlation (11). Similarly, aberrant methylation commonly occurs on the promoters of silenced tumor suppressor genes in many types of cancer (12,13). In normal cells, genome-wide and single gene analyses indicate that promoter methylation of autosomal genes is rare and may be restricted to certain pluripotency and testes-specific genes (14,15). As expected, the presence of promoter methylation correlates well with the silencing of these genes. Consistent with a repressive role for methylation, in vitro methylated promoters generally show reduced expression in transfection assays (9). In addition, many methylated genes are induced by treating cultured cells with methyltransferase inhibitors (16). Although transcriptional repression by promoter methylation is well established, methylation is also implicated in gene silencing from a distance by aiding the spread of heterochromatin (9,17). The importance of chromatin in methylation-dependent repression (MDR) is seen in the silencing of methylated plasmids that occurs concurrently with nucleosome deposition (18,19).
DNA methylation appears to mediate repression through multiple mechanisms. At some promoters, methylation directly prevents activation by sterically inhibiting the binding of activator proteins (20,21). Alternatively, the methyl binding domain (MBD) proteins, MeCP2, Mbd1 and Mbd2, are thought to bind to methylated CpGs and recruit proteins capable of forming repressive chromatin structures (22–24). Despite abundant evidence for widespread binding to methylated DNA, the role of the MBD family in the general repression of methylated genes has not been supported by knockout studies. Microarray analyses of tissues from mice with MeCP2 deletions identified relatively few targets of repression (25,26). In addition, the exclusively repressive activity of MeCP2 has been challenged by results showing interaction with a transcriptional activator (26). Beyond the MBD proteins, the DNMT and SRA protein families also have been implicated in recruiting co-repressors to methylated DNA (27,28), but the general relevance of their repressive activity remains to be determined (29).
An important aspect of MDR is whether the repressive activity of methylated CpGs is independent of the sequence surrounding them. It makes conceptual sense that a repression system based on DNA methylation would recognize methylated CpGs independently of their sequence context, and this possibility is supported by the relatively sequence-independent recognition of methylated CpGs by the MBD, SRA and DNMT proteins. Some sequence specificity, however, has been demonstrated for MeCP2 in vitro, whereas Mbd2 showed no specificity in the same assays (30). The zinc finger protein, Kaiso, also shows a limited form of sequence-specific recognition of methylated DNA, as it binds tandem methylated CpGs and ones separated by up to five base pairs (31). The RFX proteins, however, are the best example of transcription factors that recognize specific sequences in a methylation-dependent manner (32). RFX proteins generally bind unmethylated sequences, but one methylated site appears to regulate the collagen α2(I) gene (33). Sequence-specific methylation-dependent (SSMD) factors may also regulate repression by and methylation of certain repetitive elements and imprinting control regions (ICRs) (34,35). In summary, although much evidence supports sequence-independent recognition of methylated CpGs, regulation by SSMD proteins remains a real possibility.
As a model for gene regulation by specific methylated sequences, we focused on the Igf2/H19 ICR. The methylated paternal ICR initiates the silencing of H19 from its position 2 kb upstream of the promoter (34). Notably, no defects in H19 imprinting have been reported in MeCP2 or Mbd2 knockout mice (36). Therefore, we examined whether key methylated CpGs and their surrounding sequence are essential for repression by the methylated ICR. To eliminate effects of both promoter and plasmid methylation, we used a CpG-free reporter plasmid to examine how MDR acts over a distance and found that the methylated ICR exhibits strong repressive activity that depends on the sequence surrounding key methylated CpGs. In contrast, control methylated fragments showed little or moderate repressive activity, despite having CpG numbers and densities similar to or higher than the ICR. Our results suggest that some examples of MDR rely on SSMD repressors for strong MDR activity, whereas general CpG methylation may exhibit only weak or moderate repressive activity.
MATERIALS AND METHODS
Cell culture and transfections
All cells were cultured at 37°C in 5% CO2 in DMEM supplemented with 10% fetal bovine serum. Plasmids were methylated with SssI (NEB) overnight and purified with silica membrane spin columns (Epoch). Mock methylations without enzyme were performed in parallel. Methylation status of plasmids was confirmed by agarose gel electrophoresis and ethidium bromide staining after digestion with BstUI, HhaI and HpaII. Only DNA showing no digestion was used in transfections. Transient transfections of cells in 6-well plates were performed according to the manufacturer's protocol, using 3 µl of Fugene (Roche) or Mirus TransIT-LT1 (Mirus) mixed with 0.9 µg of the reporter plasmids and 0.1 µg of pCMV-Luc. Cells were harvested directly with 1× Reporter Lysis buffer (Promega) after 48 h. β-Galactosidase activity was determined using chlorophenol red β-d-galactopyranoside (CPRG) as substrate. Luciferase reactions were performed according to manufacturers protocol (Promega Bright Glo). Absorbance and luminescence values were measured on a Tecan ULTRA Evolution instrument. Relative repression for each methylated and unmethylated plasmid pair was calculated after normalizing the β-galactosidase levels to luciferase activity. Student's t-test was used to compare results obtained for each methylated and unmethylated plasmid pair.
Stable transfections were performed with Fugene or LT1 at a 3:1 ratio with 1 µg of reporter plasmid in 6 cm dishes. After 2 days, cells were passaged onto 10 cm plates and selected with G418 for 10–14 days. The resulting colonies were passaged onto 6 cm plates and harvested when nearly confluent. Cells were scraped from their plates, pelleted and lysed with 1× Reporter Lysis buffer. β-Galactosidase activities were normalized to total protein in the lysates (Biorad) and used to calculate relative repression of each methylated plamsid compared with its unmethylated version. For azaC treatment, 104 stably transfected cells were passaged onto 12-well plates, cultured for 24 h, and treated with freshly prepared 5-azacytidine at 10 μM for 3 days.
Plasmid constructs
To facilitate cloning, pCpGvitro-neo-lacZ (InvivoGen) was modified by adding an EagI site to the 5′ end of the CMV enhancer and replacing the SpeI site with a BamHI site at the 3′ end of the enhancer to create pCpGfree. To construct pCpGfree2, the CMV enhancer was excised from pCpGfree and replaced with a linker containing a BamHI site. After amplification from NcoI to BamHI (base pairs 1140–3589; Genbank accession number AF049 091), the wild-type ICR and the mutant ICRs, CpG–Shift1 and -Shift2 (described below), were ligated into pGEM-T Easy and sequenced. EagI–BamHI fragments from these three constructs were inserted into EagI and BamHI digested pCpGfree, which excises the CMV enhancer and places the fragments upstream of the EF-1α promoter. To construct pCpGfree-Lam, the 2.3 kb lambda HindIII fragment (base pairs 25 157–27 484; accession number J02459) and pCpGfree digested with EagI and BamHI were made blunt with Klenow and ligated together. To construct pCpGfree-Igf2r, a PvuII fragment from the Igf2r DMR2 (base pairs 5164–8101; accession number AF151 173; a generous gift of Denise Barlow) was excised from pUC19 with EcoRI and BamHI, and a three-part ligation was performed to insert the fragment into EcoRI and BamHI digested pCpGfree. To construct pCpGfree-pBS, a portion of pBluescriptII KS(-) overlapping the bla gene and the f1 origin (base pairs 1982–2961, 1–153; accession number X52 329) was amplified with forward and reverse primers containing EagI and BamHI sites, respectively, and inserted into EagI and BamHI digested pCpGfree. To construct pCpGfree-ΔCpG1 and -ΔCpG2, the AatII and BamHI fragment from pCpGfree-ICR was replaced with the same fragments from the mutant ICRs (generously provided by P. Szabo and M. Bartolomei, respectively). The H19 promoter region (base pairs 4785–5527; accession number AF049 091) was amplified with upstream and downstream primers containing BamHI and HindIII sites, respectively, and inserted into BamHI and HindIII digested pCpGfree2. Multimers of the CTCF sites and their mutants were inserted into the BamHI site of pCpGfree2. Oligonucleotides for each CTCF site are as follow. The lower case letters indicate non-ICR sequence. CTCF sites—Site 1: gatcGGAGTTGCCGCGTGGTGGCAGCAAA; Site 2: gatcCAGGGTTGCCGCACGGCGGCAGTG; Site 3: gatcATGCTACCGCGCGGTGGCAGCC; Site 4: gatcGATGCCGCGTGGTGGCAGTAC Mutant CTCF Site 1—MutA: gatcGGAGTTGCaGCGTGGTGGCAGCAAA; MutB: gatcGGAGTTGCGaGTGGTGGCAGCAAA; MutC: gatcGGAGTgtaCGCGTGGTGGCAGCAAA.
The ICR mutants, CpG-Shift1 and CpG-Shift2, were constructed by a combination of ligating multiple double-strand oligonucleotides containing the appropriate mutations and overlapping PCR with primers containing BbsI sites to create four subfragments of the ICR. After sequencing the four mutant subfragments in pGEM-T Easy, they were excised with BbsI to generate complementary 5′ overhangs of unique ICR sequence and ligated together to generate the complete ICR. ICR-Mut5 was constructed similarly, with the mutations introduced by overlapping PCR that generated three fragments with BbsI ends that when digested and ligated together formed the complete ICR. After subfragment ligation, the three mutants were amplified and inserted into pGEM-T Easy and pCpGfree as described above. All PCR-based constructs were sequenced to ensure fidelity. Further details and primer sequences are available upon request.
Southern blots
Genomic DNA was isolated from cell pellets remaining after extraction of stably transfected cells with 1× Reporter lysis buffer. DNA purification, restriction digests and Southern blotting were performed using standard methods. Hybridization was performed with random primed probes in Rapid-Hyb according to manufacturer's protocol (Amersham).
RESULTS
The Igf2/H19 ICR exhibits sequence-specific MDR
To address the role of CpG number, density and sequence surrounding the CpGs in MDR, we examined the repression activity of different methylated sequences. Most of our effort focused on the ICR from the Igf2/H19 locus because of our interest in genomic imprinting and because ICR deletions suggest that the methylated paternal region represses transcription from a distance (34,37). We also tested fragments from bacteriophage lambda and pBluescript (pBS), and the differentially methylated region 2 (DMR2) located in the second intron of the imprinted Insulin-like growth factorII receptor (Igf2r) gene. DMR2 controls the imprinting of Igf2r and includes a promoter for a non-coding RNA that is silenced by DNA methylation on the maternal allele (38). The lambda and pBS fragments were chosen because they are unlikely to have binding sites for transcription factors or SSMD proteins, except by chance. In addition, the 2.3 kb lambda fragment has almost the same CpG number and density (CpGs/kb) as the 2.4 kb ICR. The 1.1 kb pBluescript fragment also has the same number of CpGs as the ICR, but its CpG density is 2.2-fold higher (Table 1). The Igf2r DMR2 has 45 more CpGs and a 33% higher CpG density than the ICR, which could give it a higher level of MDR activity. The number of tandem and closely spaced CpGs, which are potential binding sites for the repressor Kaiso, also differs between the fragments (Table 1) (31).
Table 1.
CpG characteristics of test sequences
| Sequence | Length | CpG number | CpG densitya | Closely spaced CpGsb |
|---|---|---|---|---|
| ICR | 2.4 | 73 | 30.0 | 23 |
| CpG-Shift1 | 2.4 | 73 | 30.0 | 10 |
| CpG-Shift2 | 2.4 | 73 | 30.0 | 1 |
| pBS | 1.1 | 73 | 66.4 | 28 |
| Lambda | 2.3 | 66 | 28.7 | 15 |
| Igf2 DMR2 | 2.9 | 117 | 40.3 | 34 |
| H19 promoter | 0.74 | 28 | 38.0 | 7 |
aNumber of CpGs per 1 kb.
bNumber of CpGs separated by 5 bp or less.
To measure MDR activity, we used derivatives of pCpGvitro-neo-LacZ (pCpGfree), a commercially available reporter plasmid that is completely devoid of CpGs. pCpGfree has a 200 bp human elongation factor-1α (EF-1α) promoter fused to a lacZ reporter gene, the SV40 enhancer/promoter driving a neomycin resistance gene, and three different matrix attachment regions, all of which were modified to be free of CpGs. To simplify inserting test fragments upstream of the promoter, we constructed two versions of pCpGfree with additional restriction sites that include one and two CpGs in their sequence. These CpGs end up distal to the promoter after insertion of the test sequences. Using an essentially CpG-free reporter plasmid allowed us to assess the effects of insert methylation on transcription without interference from plasmid-derived CpGs and without concerns about steric hindrance of promoter factors or the spreading of DNA methylation.
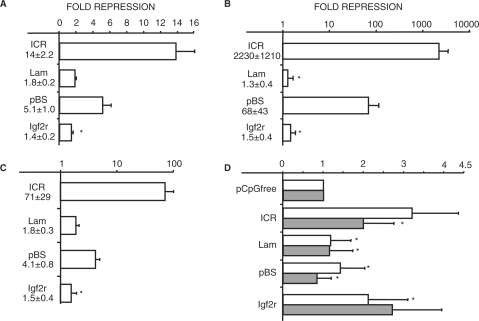
Modified pCpGfree constructs with the four test fragments were fully methylated using the bacterial methylase SssI and transiently transfected into F9 cells, an embryonic carcinoma line. Mock methylated versions of each plasmid were transfected in parallel. Relative to their unmethylated controls, expression from the methylated pCpGfree-ICR and -pBS was repressed 14- and 5-fold, respectively (Figure 1A). In contrast, methylated plasmids containing the lambda fragment and the Igf2r DMR2 showed less than 2-fold repression. When compared with the strongly repressive ICR sequence, the other test sequences with similar or higher CpG numbers and densities showed less inhibition, suggesting that MDR activity depends more on the methylated sequence than CpG number or density.
Figure 1.
Sequence dependence of methylation-mediated repression. (A) Relative repression of methylated reporter plasmids in transient transfections of F9 cells. (B) Logarithmic graph of relative repression of methylated reporter plasmids in stable transfections of F9 cells. (C) Same as in B, except NIH3T3 cells were stably transfected. (D) Expression of unmethylated reporter plasmids normalized to unmethylated pCpGfree in stable transfections of F9 (empty) and 3T3 cells (gray). (A–D) Mean values and standard deviations for each graph were derived from three independent experiments and depict the ratio of expression from the unmethylated and methylated version of each plasmid. All means have P-values <0.05 when compared with unmethylated plasmids, except where indicated by asterisk. Methylated sequences are indicated next to the graphs. Sequences: ICR, Igf2/H19 imprinting control region; Lam, 2.3 kb lambda HindIII fragment; pBS, fragment of pBluescript; Igf2r, DMR2 from the Igf2r gene; pCpGfree, empty reporter plasmid.
The transient transfections indicated that not all methylated sequences are capable of substantial transcriptional repression. Transfected plasmids, however, might not be packaged completely into chromatin, and the apparent lack of repression by the methylated lambda fragment and Igf2r DMR2 could reflect the absence of properly assembled chromatin. To address whether MDR requires normal chromatin, we selected F9 and NIH3T3 transfectants for stable integration of the reporter plasmids and assayed the effects of DNA methylation on transcription. In F9 cells, transcriptional repression by the methylated ICR was dramatic and ranged from ∼900- to over 3000-fold in separate transfection experiments (Figure 1B). Repression by the methylated ICR was nearly complete, and the reporter reactions had to be incubated over 100-fold longer than the unmethylated controls to see significant enzymatic activity. Although considerably less repressive than the ICR, the methylated pBS fragment inhibited reporter expression from 30- to 115-fold compared with the unmethylated plasmid. By contrast, the lambda fragment and the Igf2r DMR2 showed only a modest 1.1–3.1-fold repression when methylated (Figure 1B).
In stable transfections of 3T3 cells, the methylated ICR inhibited transcription 70-fold on average, compared with the 2200-fold seen in F9 cells (Figure 1C). Similarly, relative repression by the methylated pBS fragment was 17-fold lower than in F9 cells. Both the lambda and DMR2 fragments again averaged less than 2-fold relative repression when methylated (Figure 1C). For both F9 and 3T3 cells, we note that the unmethylated ICR and pBS plasmids averaged expression that was roughly 3-fold higher than or equal to the pCpGfree reporter, respectively (Figure 1D). Therefore, most of their decrease in expression upon methylation was due to inhibition of the reporter promoter and not to a loss of activation by either test sequence. The unmethylated lambda and DMR2 fragments also exhibited no or only approximately 2-fold induction, respectively, indicating that much of DMR2's repression may reflect the loss of activation (Figure 1D). To address whether demethylation could explain their different repression activity, methylation sensitive Southern blot analysis was performed on genomic DNA isolated from the stably transfected polyclones. For each methylated sequence and in both cell lines, little or no demethylation was detected (data not shown). Taken together, the dramatically different repressive activity of the four sequences in the context of normal chromatin suggests that the strength of MDR at a distance varies with sequence context of the methylated CpGs and not simply with their number or density.
CpGs associated with CTCF sites are necessary for sequence-specific MDR
The apparent sequence specificity of the MDR could be explained by repressor proteins that bind in a sequence-specific and methylation-dependent manner to the ICR and pBS inserts. To determine whether the strong repression by the ICR is sequence-dependent, we constructed CpG-Shift1 and CpG-Shift2, two mutant ICRs with altered CpG patterns but the same number of CpGs. In each mutant ICR, the positions of CpGs were shifted slightly by mutating either the C or G of a CG dinucleotide and creating a replacement by altering the base next to a nearby C or G. More specifically, the 73 CpGs in the ICR were divided into 13 CpGs associated with the four binding sites for the insulator protein CTCF and the 60 CpGs outside these sites. In CpG-Shift1, the positions of 58 CpGs outside the CTCF sites were changed, whereas CpGs associated the CTCF sites were left unaltered (Figure 2 and Supplementary Figure S1). We specifically separated nearly all CpGs by at least six nucleotides to prevent the binding of Kaiso, which can bind sequences with up to five nucleotides between two methylated CpGs (31). In CpG-Shift2, the 60 CpGs are in the same position as in CpG-Shift1, and 10 of the 13 CTCF site CpGs also were rearranged and separated by more than five bases. It is likely that the CpG rearrangement in CpG-Shift2 destroyed the four CTCF sites, but we mutated each site further to ensure equal elimination (Figure 2) (39). The purpose of the CpG-Shift mutants was to retain the same CpG number and as much of the ICR sequence and local CpG density as possible, while altering potential sites for proteins that bind the ICR in a methylation- and sequence-dependent manner. Thus, these mutants provide comparison sequences that are more than 93% identical to the wild type ICR, but with most or nearly all of the CpGs in different contexts. This is in contrast to simply mutating CpGs, which would make it more difficult to distinguish the effects of reducing CpG number from mutating specific binding sites.
Figure 2.
CTCF sites in the ICR mutants. Wild-type sequence surrounding the four CTCF sites in the Igf2/H19 ICR is shown in black, except for CpGs, which are green. In the mutant ICRs, altered bases are shown in blue, except for new CpGs, which are red. Further description of the ICR mutants is in the text and Supplementary Figure S1. The core of each CTCF site is underlined.
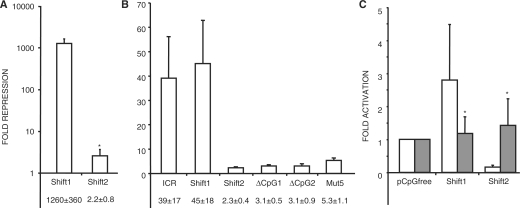
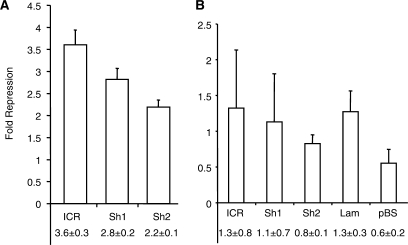
After stable transfection into F9 cells, the methylated CpG-Shift1 plasmid showed dramatically lower reporter expression than its unmethylated version (Figure 3A). In separate experiments, relative repression by the methylated CpG-Shift1 was similar to the wild type ICR and ranged from about 500- to over 1000-fold. Although their ranges of relative repression activity overlapped, the absolute expression from the methylated CpG-Shift1 plasmid was generally higher than that of the methylated ICR plasmid (data not shown). Taken together, these results suggest that altering the CpGs outside of the CTCF sites had only a minimal effect on the ICR's repressive potential. In sharp contrast, reporter expression in F9 cells stably transfected with the methylated CpG-Shift2 plasmid was no more than 3-fold lower than the expression in cells carrying the unmethylated plasmid (Figure 3A), indicating that altering the CpGs associated with the CTCF sites destroyed the ICR's strong repressive potential. Curiously, expression from the unmethylated CpG-Shift2 plasmid in F9 cells averaged about 20-fold lower than that seen with the unmethylated CpG-Shift1 and ICR plasmids (Figure 3C). However, even with its reduced expression potential, activity from the methylated CpG-Shift2 plasmid was on average 25-fold higher than the methylated CpG-Shift1 plasmid. In 3T3 cells, the relative repression activity of methylated CpG-Shift1 and -Shift2 paralleled that seen in F9 cells, but CpG-Shift1's level of repression was much greater in F9 (Figure 3A and B). Also, expression from the unmethylated CpG-Shift2 plasmid in 3T3 cells was comparable to that of the unmethylated ICR and CpG-Shift1 plasmids (Figure 3C). Importantly, expression from the unmethylated CpG-Shift1 plasmid in both cell lines was comparable to that of the ICR and averaged only 2.6-fold more than the empty reporter plasmid (Figure 3C). Thus, the 1000-fold repression by methylated CpG-Shift1 indicates promoter inhibition and not loss of activator protein binding. As with the other test fragments, little or no demethylation was detected by methylation sensitive Southern blot analysis of DNA from the stable transfectants (data not shown). The difference in MDR between CpG-Shift1 and CpG-Shift2 suggests that the sequences surrounding the CpGs associated with the CTCF sites are essential for the strong repression activity of the methylated ICR.
Figure 3.
Sequence-specific repression by CpGs associated with CTCF sites in the ICR. (A) Logarithmic graph of the relative repression of methylated plasmids in stable transfections of F9 cells. (B) Graph of the relative repression of methylated plasmids in stable transfections of NIH3T3 cells. (C) Expression of unmethylated reporter plasmids relative to unmethylated pCpGfree in stable transfections of F9 (empty) and 3T3 cells (gray). (A–C) Mean values and standard deviations below the graphs are from three independent experiments. All means have P-values < 0.05 when compared with unmethylated control plasmids, except where indicated by asterisk. Methylated sequences are indicated below the graphs. Sequences: Shift1, CpG-Shift1; Shift2, CpG-Shift2, ΔCpG1, ICR-ΔCpG1; ΔCpG2, ICR-ΔCpG2; Mut5, ICR-Mut5.
From the dramatic difference in MDR activity between the wild type and CpG-Shift2 ICRs, it was clear that repression activity centered on the CpGs associated with the CTCF sites. However, the somewhat higher expression from the methylated CpG-Shift1 plasmid suggested that rearranging the ICR's CpGs removed some inhibitory activity recruited by methylation outside of the CTCF sites. To determine if these 60 CpGs have repression activity when methylated and to confirm the importance of the CTCF site CpGs, we examined the MDR activity of two ICRs containing different mutations within the four CTCF binding sites. In ICR-ΔCG1, a total of 10 CpGs and 37 bases within the CTCF sites were mutated, which eliminates CTCF binding (40) (Figure 2). In ICR-ΔCG2, nine CpGs and 14 additional bases within the CTCF sites were mutated, but CTCF binding is retained (41) (Figure 2). After methylation and stable transfection into 3T3 cells, the methylated ICR-ΔCG1 and −ΔCG2 plasmids showed no more than 5-fold lower activity than the unmethylated controls (Figure 3B). Southern analysis confirmed that both mutants maintained their methylation (data not shown). Together with the CpG-Shift1 and -Shift2 results, the substantial loss of MDR activity exhibited by ICR-ΔCG1 and -ΔCG2 further implicates the CTCF site CpGs in the methylated ICR's repressive activity.
One concern not addressed by the CpG-Shift and ΔCG mutants is that the MDR activity of a sequence could depend on particular arrangements or patterns of CpG clusters that may form optimal sites for the binding of multiple methylation-dependent, but sequence non-specific repressors. As all four mutant ICRs have disrupted both CpG patterns and sequence context, we constructed ICR-Mut5 in which only bases surrounding the CTCF site CpGs were altered, but the CpG patterns are intact (Figure 2). Despite these minimal changes, methylated ICR-Mut5 demonstrated low levels of MDR that were similar to ICR-ΔCG1 and -ΔCG2 (Figure 3B). Thus, not only are the CTCF site CpGs necessary for the methylated ICR's repression activity, but their surrounding sequences are necessary as well. These results indicate that together they form part of the recognition site for SSMD repressors. We note, however, that the moderate amount of MDR activity exhibited by ICR-Mut5 and the ΔCG mutants indicates that the CpGs away from the CTCF sites contribute to the full inhibitory activity of the methylated ICR.
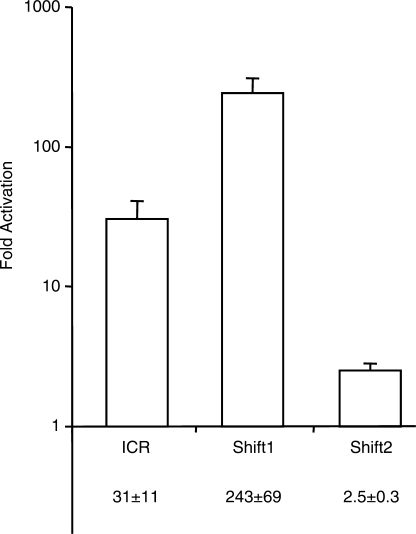
As a control, we confirmed that the strong MDR activity of the methylated ICR and CpG-Shift1 fragments was due to DNA methylation by measuring induction of the reporter plasmid after treating cells with the DNA methyltransferase inhibitor 5-azacytidine (azaC). In polyclones of F9 cells carrying the methylated ICR or CpG-Shift1 plasmids, 3 days of culture with azaC induced β-gal expression 30- and 300-fold, respectively (Figure 4). Only 2.5-fold induction by azaC was seen in cells transfected with the methylated CpG-Shift2 plasmid (Figure 4). Activation of the methylated ICR and CpG-Shift1 plasmids by azaC treatment indicated that at least some of their repression is attributable to DNA methylation. The modest induction of the methylated CpG-Shift2 plasmid by azaC further suggested that methylation has a comparatively small effect on its expression level.
Figure 4.
AzaC treatment induces methylated plasmids. Logarithmic graph of relative activation of the indicated methylated reporter plasmids after treating stable F9 polyclones with 5-azacytidine. Mean values and standard deviation are from three independent experiments. All means have P-values < 0.05 when compared with untreated parallel cultures. Abbreviations are in Figure 1.
CTCF site CpGs and surrounding bases are sufficient for sequence-specific MDR
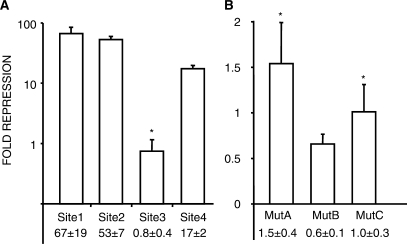
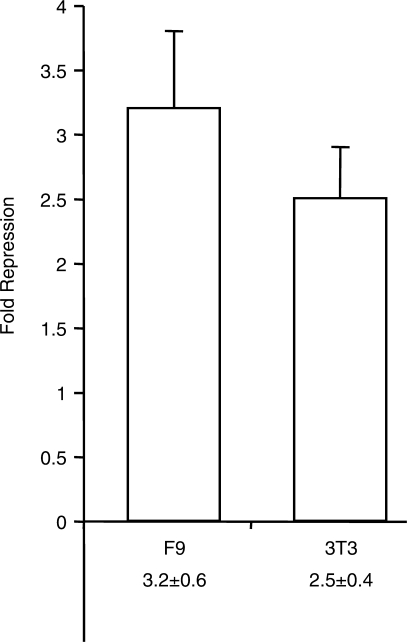
After establishing their necessity, we determined if the CTCF site CpGs are sufficient for MDR from a distance. We inserted five copies of portions of the four CTCF sites into pCpGfree, and stably transfected the methylated and unmethylated plasmids into 3T3 cells. The methylation of sequences from CTCF Site 1, 2 and 4 lead to strong repression of the reporter, while methylated Site 3 showed little repressive activity (Figure 5A). To address sequence specificity of the repression, multimers of three mutant versions of CTCF Site 1 were tested for MDR activity. Mut1A and B have point mutations in one of Site 1's tandem CpGs. In Mut1C, three bases adjacent to the tandem CpGs were altered. In stably transfected 3T3 cells, methylated multimers of the three mutants had essentially no repressive activity (Figure 5B). These results indicate that three of the four elements are sufficient for strong MDR of transcription and that both the methylated CpGs and their surrounding sequence are required for repression.
Figure 5.
Methylated CpGs associated with CTCF sites are sufficient for sequence-specific repression. Stable transfections of NIH3T3 cells. (A) Logarithmic graph of relative repression activity of methylated multimers from the four CTCF sites in the ICR. (B) Relative repression activity of methylated multimers of CTCF Site 1 mutants (described in the text). Mean values and standard deviation are from three independent experiments. All means have P-values < 0.05 when compared with unmethylated control plasmids, except where indicated by asterisk.
Repressive activity of the methylated ICR is cell-type specific
To address whether the level of MDR exerted by the different sequences showed any cell-type specificity, we stably transfected Hela and Cos7 cells with methylated and unmethylated reporter plasmids and determined their relative expression levels. In Hela cells, the methylated ICR, CpG-Shift1 and CpG-Shift2 showed moderate repression, but unlike in F9 and 3T3 cells, each fragment repressed within the same 2–4-fold range (Figure 6A). Results from Cos7 cells were even more surprising as reporter expression from the methylated and unmethylated wild-type and mutant ICR plasmids were essentially identical. Similarly, the methylated lambda and pBS fragments showed no repression in Cos7 cells (Figure 6B). Southern analysis demonstrated that the methylated plasmids retained their modification in both cell types, indicating that loss of methylation did not explain their expression (data not shown). These results demonstrate that the strength of repression mediated by upstream DNA methylation is cell-type specific, in addition to sequence-dependent.
Figure 6.
Cell-type dependence of methylation-mediated repression. Graphs of the relative repression of methylated reporter plasmids in stable transfections. (A) Hela stable transfectants. Means have P-values < 0.05 when compared with unmethylated control plasmids. (B) Cos7 transfectants. Means have P-values > 0.05 when compared with unmethylated control plasmids. Mean values and standard deviations are from three independent experiments. Sequences: Sh1, CpG-Shift1; Sh2, CpG-Shift2. Other abbreviations are in Figure 1.
H19 promoter is not silenced by DNA methylation
The ICR's sequence-specific MDR activity exerts its effects over the 200 bp EF-1α promoter, but several methylated control fragments do not. Although these results suggest that only specific methylated squences have strong MDR activity, sequence non-specific MDR could be highly repressive locally, but have little ability to act over even short distances. To test whether the strength of MDR is more independent of sequence when acting locally, we replaced the CpG-less EF-1α promoter in pCpGfree with a fragment of the H19 promoter that has 28 CpGs. Methylation of the paternal H19 promoter appears to be essential for its transcriptional repression in vivo, making it a useful test of local repression by methylation (8). After stable transfection of F9 cells, the methylated H19 promoter plasmid showed roughly 3-fold lower β-gal expression than the unmethylated control (Figure 7). Similar results were seen in 3T3 cells (Figure 7). While this repression represents a roughly 70% reduction in activity, these results show that DNA methylation is not sufficient for silencing of the H19 promoter and could indicate that the strength of local MDR also depends on the sequence being methylated.
Figure 7.
Methylation-mediated repression of the H19 promoter. Graphs of the relative repression of methylated H19 promoter plasmid in stable transfections of F9 and 3T3 cells. The reporter promoter in pGpGfree2 was replaced with the H19 promoter. Mean values and standard deviations are from three independent experiments. Both means have P-values < 0.05 when compared with unmethylated control plasmids.
DISCUSSION
DNA methylation has been proposed to inhibit transcription by sterically blocking the binding of activator proteins and by creating sites for proteins that bind methylated CpGs and recruit transcriptional co-repressors (20–24). Recognition of methylated CpGs is generally thought to show little influence from the sequence surrounding them and to occur in almost all cell types. In support of these ideas, proteins implicated in MDR, such as the MBD, SRA and DNMT families, show limited sequence specificity, and many are broadly expressed (28,36). On the other hand, the extent that these proteins mediate MDR activity in vivo remains controversial for the MBDs and undetermined for the others (26,29). In contrast to predictions of sequence and cell-type independence, we found that the strength of MDR from a distance varied over a 1000-fold between different methylated sequences and between cell lines. These results are consistent with the possibility that sequence-specific MDR may be common amongst genes normally silenced by DNA methylation and that repression levels may vary between both normal and transformed cell types.
Advantages of a CpG-free reporter plasmid
In many studies of MDR, the observed effects of DNA methylation on transcription are likely to be influenced by methylation not only of the test sequence but also of the standard CpG-rich reporter constructs used in most experiments. By employing a reporter that is essentially devoid of CpGs and by placing test sequences upstream of a CpG-less promoter, we avoided any effects of plasmid-derived methylation as well as any steric hindrance of promoter factors. Similar experiments have been performed with standard plasmids by methylating only specific subsets of the plasmid (19,42). In our system, however, the CpG-free plasmid also cannot acquire methylation. Finally, most of our assays involved stable transfections to ensure that repression was measured in the context of complete chromatin, while site of integration effects were minimized by assaying polyclones. These advantages allow a more independent assessment of a methylated sequence's repression potential in normal chromatin. One limitation of the system is that the repressive activity of DNA methylation must overcome any activator proteins binding to the EF-1α promoter. A weaker or partially silenced promoter could detect lower levels of MDR activity.
Sequence-specific repression by the methylated Igf2/H19 ICR
To address the mechanism of MDR from a distance, we focused on the ICR of the Igf2/H19 locus, as the methylated paternal ICR is required to silence H19 expression from its position 2 kb upstream of the promoter (34). Initially, we assumed that the ICR's MDR activity reflected the number or density of methylated CpGs. Compared with the ICR, however, the control lambda and Igf2r DMR2 fragments showed about 1000-fold lower levels of MDR activity in F9 cells, despite having CpG numbers and densities similar to or higher than the ICR. Although these results strongly support a sequence-specific component to the ICR's MDR activity, comparing completely different sequences introduces several caveats to interpreting their relative levels of MDR. One concern is the binding of the repressor Kaiso to methylated tandem or closely spaced CpGs (31), but our results showed no correlation between potential Kaiso sites and MDR. We also found no consistent correlation between MDR activity and the average CpG distance from the TATA box or the number of CpGs within 200 bp of the promoter. The pBluescript fragment, however, scores highest in these two measures of ‘CpG proximity’, which could explain its greater MDR activity than the other controls. Finally, the low MDR of the three control fragments could reflect the binding of methylation-insensitive activators that drove demethylation of the test sequences or activated transcription. Although we cannot exclude localized loss of methylation, Southern analysis of the integrated methylated plasmids did not show significant demethylation. In addition, all of the unmethylated fragments we examined showed no more than 2–3-fold induction of the reporter, suggesting that they do not bind strong activators.
To reduce the caveats of comparing unrelated sequences, we generated the CpG-Shift1 and CpG-Shift2 mutants. By altering only the CpG arrangement, the two mutants retain >93% identity with the ICR, have the same number of total and proximal CpGs, and have nearly the same average CpG distance from the promoter as the wild type ICR. Although we separated CpGs by at least five bases, similar local density and spacing of CpGs was maintained, as both parameters were thought to influence the repressive activity of the ICR. Thus, the ∼600-fold difference in the relative repression between methylated CpG-Shift1 and -Shift2 in F9 cells focused attention on the CpGs associated with the CTCF sites, with fewer of the variables associated with unrelated control sequences. Furthermore, the reduced MDR activity of the ICR-ΔCG1 and ICR-ΔCG2 mutants corroborated these results. We note, however, that the unmethylated CpG-Shift2 plasmid showed reduced expression in F9, but not 3T3 cells. Mutation of the CTCF sites in CpG-Shift2 may have reduced its expression, but this seems unlikely as the wild-type ICR activated the reporter only 3-fold. Alternatively, we may have created sites for repressors expressed in F9 cells. In either case, its reduced expression does not alter our conclusion that SSMD repressors bind to the CTCF site CpGs.
We also considered that specific arrangements of methylated CpGs may be optimal for binding of repressors, and if so, then CpG pattern will dictate repression activity. In the ICR-Mut5, the CpG arrangement of the entire ICR is intact, but the bases surrounding the CTCF site CpGs are altered. This mutant showed greatly reduced MDR activity, indicating that CpG pattern is not critical for repression by the methylated ICR and that bases surrounding the methylated CTCF site CpGs are essential for the binding of SSMD repressors. The sequence-dependent repression by multimers of ∼25 bp sequences derived from the ICR's CTCF sites confirmed these results. Moreover, the methylated multimers have no more than 16 CpGs, suggesting that the additional 57 CpGs in the ICR are not critical for its MDR activity.
The ICR's strong MDR activity is likely due to the binding of SSMD repressors with recognition elements that include unmodified bases and methylated CpGs. The RFX proteins are the best examples of transcription factors that can bind specific sequences containing methylated CpGs (32), although the overall relevance of this activity for RFX function or for methylation-dependent gene regulation is unknown. Similarly, Kaiso and MeCP2 exhibit intermediate sequence specificity, but its relevance to genome-wide MDR and H19 repression remains to be determined (30,31). We found that all three proteins bind at least some of the methylated CTCF site elements in vitro, but they also bind mutant elements that have no repressive activity, suggesting they are not involved in the repression or are not sufficient (data not shown). Identifying the proteins responsible for the ICR's MDR activity will allow us to assess their role in gene repression by DNA methylation.
Repression by the methylated Igf2/H19 ICR in vivo
The strong repression mediated by the three methylated CTCF sites in cultured cells raises the question of whether they also function in the mouse. However, knockin mice with the CTCF site mutant ICRs, ICR-ΔCG1 and -2, show no defects in H19 promoter repression or in paternal ICR methylation (40,43). Although these results indicate that the CTCF site CpGs are not necessary for repression of H19 in normal cells, they do not rule out redundancy with other CpGs in the ICR. Consistent with this possibility, both methylated ICR-ΔCG mutants retain some repressive activity in 3T3 cells, which may reflect binding of SSMD repressors to CpGs outside of the CTCF sites. Similarly, the CTCF site CpGs are not altered in CpG-Shift1, but it has weaker MDR activity than the wild type ICR. Using BLAST analysis, we identified three CpG-containing sequences with similarity to the minimal repressive sequence associated with CTCF Site 1 (data not shown). The three sequences are mutated in CpG-Shift1 and could explain its reduced MDR activity. Conversely, they are not altered in ICR-ΔCG1 and -2, and may provide sufficient activity to repress paternal H19 in mice with these mutant ICRs (40,43). Notably, except for knockdown of Mbd3 (which does not bind methylated DNA), deletion of individual MBD genes does not cause loss of H19 repression (30,44).
Repression of the methylated H19 promoter
The low levels of MDR exhibited by some of the control fragments and mutant ICRs could indicate that DNA methylation generally is not sufficient to nucleate the spreading of transcriptional silencing, but does not exclude that methylation may have strong repressive activity when acting locally. To address this issue, we measured how methylation of the H19 promoter affected its transcriptional activity and found a moderate 2–4-fold repression by complete methylation of the promoter. We have not distinguished whether the CpG methylation has any direct effect on activator binding, but the absence of strong silencing suggests that DNA methylation generally is only moderately inhibitory. Although more promoters will have to be examined in detail, we speculate that many promoters subject to strong MDR are inhibited by the activity of SSMD repressors. In any case, the CpG-free reporter system provides an excellent system to identify SSMD repressor-mediated effects. For example, promoters that are repressed by DNA methylation and also exhibit strong MDR from at distance are likely to be targets for SSMD repressors.
Cell-type-dependent repression by the Igf2/H19 ICR
Given the widespread repression of paternal H19 in vivo, we expected the ICR's potent MDR activity to occur in all cell types. However, repression activity of the methylated ICR was greatly reduced or absent in Hela and Cos7 cells and was equal to that of the control sequences. Thus, cell-type-specific factors appear to be mediating the methylated ICR's repressive activity, which is consistent with the tissue-specific loss of paternal H19 repression in mice with a partial ICR deletion (37). One explanation that we cannot completely exclude is the presence of cell-type-specific promoter binding factors that effectively block the repression by the methylated sequences in Cos7 or Hela cells. We suspect, however, that if the methylated ICR has strong repressive activity in these cells, then it would show measurable repression relative to the control fragments, regardless of bound activator or insulator proteins.
CONCLUSION
Our results suggest that to understand the how methylated sequences mediate repression, each one may have to be considered individually. Our limited sample indicates that the sequence context of methylated CpGs is a key determinant of MDR strength, while CpG number and density are comparatively poor predictors. Sequence-specific binding of repressors to methylated sequences might explain results showing repression mediated by a single methylated CpG (45,46) or how low-level methylation by site-specific methylases can be repressive (47). Similarly, sequence-specific MDR could explain how a patch of DNA methylation inhibits an unmethylated promoter or blocks elongation (19,48). Sequence specificity is also consistent with the proposal that some promoters have ‘catalyst’ CpGs that, upon methylation, nucleate further promoter methylation (49). Finally, all methylated sequences exhibited at least 1.3–4-fold repression in most cell lines, which may reflect limited sequence-independent repression initiated by DNA methylation. Thus, cells may have two types of methylation-mediated repression, one strong and sequence-specific and one weaker and sequence non-specific.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the National Institutes of Health [CA105017 to C.J.S.]. Funding for open access charge: National Institutes of Health [CA105017].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to D. Barlow for the Igf2r DMR2, and to M. Bartolomei and P. Szabo for the mutant Igf2/H19 ICRs. We thank P. Jones and members of the Schoenherr laboratory for critical reading of the manuscript.
REFERENCES
- 1.Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 3.Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl Acad. Sci. USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 5.Mohandas T, Sparkes RS, Shapiro LJ. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211:393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Taylor SM, Mohandas T, Shapiro LJ. Cell cycle-specific reactivation of an inactive X-chromosome locus by 5-azadeoxycytidine. Proc. Natl Acad. Sci. USA. 1982;79:1215–1219. doi: 10.1073/pnas.79.4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolf SF, Migeon BR. Studies of X chromosome DNA methylation in normal human cells. Nature. 1982;295:667–671. doi: 10.1038/295667a0. [DOI] [PubMed] [Google Scholar]
- 8.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 9.Bird AP, Wolffe AP. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich M. Expression of various genes is controlled by DNA methylation during mammalian development. J. Cell Biochem. 2003;88:899–910. doi: 10.1002/jcb.10464. [DOI] [PubMed] [Google Scholar]
- 11.Migeon BR. X-chromosome inactivation: molecular mechanisms and genetic consequences. Trends Genet. 1994;10:230–235. doi: 10.1016/0168-9525(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 12.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 13.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr. Top. Microbiol. Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 14.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Momparler RL, Bovenzi V. DNA methylation and cancer. J. Cell Physiol. 2000;183:145–154. doi: 10.1002/(SICI)1097-4652(200005)183:2<145::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 17.Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 18.Buschhausen G, Wittig B, Graessmann M, Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc. Natl Acad. Sci. USA. 1987;84:1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr. Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 20.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 21.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 23.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 24.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 25.Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, Schulz R, Lipkowitz B, Ropers HH, Holmes MC, et al. Up-regulation of glucocorticoid-regulated genes in a mouse model of rett syndrome. Hum. Mol. Genet. 2005;14:2247–2256. doi: 10.1093/hmg/ddi229. [DOI] [PubMed] [Google Scholar]
- 26.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 28.Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Damelin M, Bestor TH. Biological functions of DNA methyltransferase 1 require its methyltransferase activity. Mol. Cell Biol. 2007;27:3891–3899. doi: 10.1128/MCB.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Prokhortchouk A, Hendrich B, Jorgensen H, Ruzov A, Wilm M, Georgiev G, Bird A, Prokhortchouk E. The p120 catenin partner kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang XY, Jabrane-Ferrat N, Asiedu CK, Samac S, Peterlin BM, Ehrlich M. The major histocompatibility complex class II promoter-binding protein RFX (NF-X) is a methylated DNA-binding protein. Mol. Cell Biol. 1993;13:6810–6818. doi: 10.1128/mcb.13.11.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sengupta PK, Ehrlich M, Smith BD. A methylation-responsive MDBP/RFX site is in the first exon of the collagen alpha2(I) promoter. J. Biol. Chem. 1999;274:36649–36655. doi: 10.1074/jbc.274.51.36649. [DOI] [PubMed] [Google Scholar]
- 34.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yates PA, Burman RW, Mummaneni P, Krussel S, Turker MS. Tandem B1 elements located in a mouse methylation center provide a target for de novo DNA methylation. J. Biol. Chem. 1999;274:36357–36361. doi: 10.1074/jbc.274.51.36357. [DOI] [PubMed] [Google Scholar]
- 36.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Drewell RA, Brenton JD, Ainscough JF, Barton SC, Hilton KJ, Arney KL, Dandolo L, Surani MA. Deletion of a silencer element disrupts H19 imprinting independently of a DNA methylation epigenetic switch. Development. 2000;127:3419–3428. doi: 10.1242/dev.127.16.3419. [DOI] [PubMed] [Google Scholar]
- 38.Stoger R, Kubicka P, Liu CG, Kafri T, Razin A, Cedar H, Barlow DP. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 39.Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 40.Szabo PE, Tang SH, Silva FJ, Tsark WM, Mann JR. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol. Cell Biol. 2004;24:4791–4800. doi: 10.1128/MCB.24.11.4791-4800.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engel N, West AG, Felsenfeld G, Bartolomei MS. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is uncovered by CpG mutations. Nat. Genet. 2004;36:883–888. doi: 10.1038/ng1399. [DOI] [PubMed] [Google Scholar]
- 42.Hsieh CL. Stability of patch methylation and its impact in regions of transcriptional initiation and elongation. Mol. Cell Biol. 1997;17:5897–5904. doi: 10.1128/mcb.17.10.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 44.Reese KJ, Lin S, Verona RI, Schultz RM, Bartolomei MS. Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3. PLoS Genet. 2007;3:e137. doi: 10.1371/journal.pgen.0030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graessmann A, Sandberg G, Guhl E, Graessmann M. Methylation of single sites within the herpes simplex virus tk coding region and the simian virus 40 T-antigen intron causes gene inactivation. Mol. Cell Biol. 1994;14:2004–2010. doi: 10.1128/mcb.14.3.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santoro R, Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell. 2001;8:719–725. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- 47.Hsieh CL. Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 49.Stirzaker C, Song JZ, Davidson B, Clark SJ. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 2004;64:3871–3877. doi: 10.1158/0008-5472.CAN-03-3690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.