Abstract
Here we have developed a sensitive DNA amplified detection method based on isothermal strand-displacement polymerization reaction. This method takes advantage of both the hybridization property of DNA and the strand-displacement property of polymerase. Importantly, we demonstrate that our method produces a circular polymerization reaction activated by the target, which essentially allows it to self-detect. Functionally, this DNA system consists of a hairpin fluorescence probe, a short primer and polymerase. Upon recognition and hybridization with the target ssDNA, the stem of the hairpin probe is opened, after which the opened probe anneals with the primer and triggers the polymerization reaction. During this process of the polymerization reaction, a complementary DNA is synthesized and the hybridized target is displaced. Finally, the displaced target recognizes and hybridizes with another probe, triggering the next round of polymerization reaction, reaching a target detection limit of 6.4 × 10−15 M.
INTRODUCTION
Rapid growth of available sequence data has made the detection of nucleic acids critical to the development of modern life sciences (1–3). Among the many methods devised for the detection and analysis of nucleic acids, amplification is one of the most important concepts since it permit the highest analytical sensitivity (4–8). PCR provides a general protocol for the amplified detection of DNA. Although the PCR method is time consuming and not free of limitations, it provides the most versatile method to detect minute amounts of DNA. The design of alternative approaches for the sensitive detection of DNA is in continuous demand. Enzyme conjugates (4,5), DNAzymes (6) and nanoparticles (7,8) have been used as amplifying labels for biorecognition events. Recently, a means of amplified DNA detection methods have been developed based on scission or replication. Since these DNA detection systems can be operated autonomously and repeatedly while they are trigged by the target, large amounts of DNA products are yielded to enhance the signal, the sensitivity of DNA detection is thereby significantly increased (9–14). Several systems have been designed to construct high sensitive DNA detection methods, e.g. autonomous replication of DNA/FokI cutter units (9), autonomous polymerization of a peroxidase-mimicking DNAzyme (10) and autonomous aggregation of Au nanoparticles (11).
Here we report a method for amplified detection of DNA based on the inherent signal-transduction mechanism of the hairpin fluorescence probe and strand-displacement property of polymerase. As such, the hairpin fluorescence probe acts as a template of polymerization reaction and fluorescence signal carrier, while the target acts as a trigger of polymerization reaction. The activation of this DNA detection system is based on the conformational change of the probe induced by hybridization between probe and target DNA. In this method, the target DNA is displaced in the process of the polymerization reaction and then hybridized to another probe. Thus, in essence, this design method allows hybridization, polymerization reaction and displacement to occur cycle-after-cycle, producing, at the same time, an amplified fluorescent signal sufficient to indicate the presence of trace amount of target DNA. Even though, this method is based on a simple design and is easy to use, it has demonstrated a high magnitude of amplified, sensitive detection with a limit of 6.4 × 10–15 M.
MATERIALS AND METHODS
Materials
The hairpin fluorescence probe and oligonucleotide were commercially synthesized by TaKaRa Bio Inc. (Dalian, China). Sequences of the oligos are listed in Table 1. The polymerase Klenow fragment exo− was purchased from New England Biolabs, Inc. The deoxynucleotide solution mixture (dNTPs) was purchased from TaKaRa Bio Inc. (Dalian, China) and the DMSO was obtained from Sigma. All other reagents were of analytical grade. Deionized water was obtained through the Nanopure Infinity™ ultrapure water system (Barnstead/Thermolyne Corp., Dubuque, IA, USA).
Table 1.
The hairpin fluorescence probe and oligonucleotides
| Name | Sequence (5′ to 3′) |
|---|---|
| Hairpin fluorescence probe | FAM-TCTTGGACACA GTAAAGAGAGGTGCGCCCAT TGTGTCCAAGA-DABCYL |
| Target | ATGGGCGCACCTCTCTTTACTGTGTC TTT |
| Random DNA | TGCAAGGTGTCAGTATAATCCGACGT TTT |
| Primer | TCTTGGAC |
The italicized region of the hairpin fluorescence probe identifies the stem sequence and the underlined region identifies the complementary sequence to the primer.
Fluorescence measurement
All fluorescence measurements were carried out on a F2500 fluorometer (Hitachi, Japan) equipped with an aqueous thermostat (Amersham) accurate to 0.1°C. Excitation and emission wavelengths were set at 496 and 517 nm, respectively, with 5-nm bandwidths. The emission spectra were obtained by exciting the samples at 490 nm and scanning the emission from 500 to 600 nm in steps of 1 nm. All samples were incubated at 37°C. When the fluorescence intensity became steady, the target was added into the mixture, and the fluorescence intensity was recorded simultaneously.
Target hybridization with hairpin fluorescence probe
Three samples were prepared to identify the hybridization feature of the hairpin fluorescence probe: sample A containing 5.0 × 10–8 M probe only; sample B containing 5.0 × 10–8 M probe and 5.0 × 10–8 M target; sample C containing 5.0 × 10–8 M probe and 1.0 × 10–7 M target. All samples were performed in 80 μl solution containing 50 mM Tris–HCl (pH 8.0) and 5 mM MgCl2 and incubated at 37°C. The difference of fluorescence intensity between samples with and without target indicated the detection capability of the hairpin probe.
The temperature melting curve of the hairpin fluorescence probe and its target
Two samples were prepared to determine the thermal profiles of the hairpin fluorescence probe and its target: Sample A containing 5.0 × 10–8 M probe only and sample B containing 5.0 × 10–8 M probe and 5.0 × 10–8 M target. All samples were performed in 80 μl solution containing 50 mM Tris–HCl (pH 8.0) and 5 mM MgCl2. The temperature was increased from 40°C to 90°C in steps of 2°C, with each step lasting 2 min.
Amplified detection of target
The experiments were performed in 80 μl solution consisting of 5.0 × 10–8 M probe, 5.0 × 10–8 M primer, 15 U polymerase Klenow fragment exo−, 100 μM dNTPs, 6% DMSO, 1 mM DTT and 5 mM MgCl2 in 50 mM Tris–HCl (pH 8.0) and incubated at 37°C. A series of targets at different concentrations from 1.0 × 10–10 M to 1.28 × 10−15 M were then added to the mixture solution and the fluorescence intensities were recorded.
Gel electrophoresis
A 20% non-denaturing PAGE analysis of the products by the isothermal strand-displacement polymerization reaction was carried out in 1 × TBE (pH = 8.3) at 80 V constant voltage for about 3 h. After Sybr green I staining, gels were scanned using an Image Master VDS-CL (Amersham Biosciences).
RESULTS AND DISCUSSION
Principle of the amplified DNA detection method
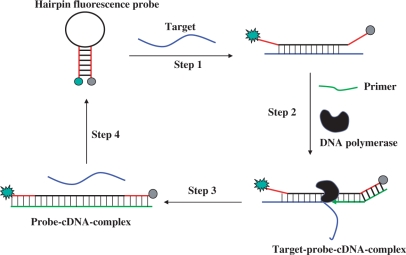
The principle of our isothermal amplified detection DNA method is shown in Figure 1. This DNA detection system consists of a hairpin fluorescence probe, a short primer and polymerase. The hairpin fluorescence probe possesses a stem-loop structure with a fluorophore and a quencher linked to the ends of the stem. The stem is 11-nt sequences long, and the loop is complementary to the target. The primer is an 8-nt sequences long, which is complementary to the stem region of the probe at 3′-end. In the absence of a target, the stem-loop conformational probe is unable to anneal with the primer to induce a polymerization reaction. However, in the presence of target DNA, the probe recognizes and hybridizes with it and undergoes a conformational change, leading to stem separation (Step 1). Following this, the primer anneals with the open stem and triggers a polymerization reaction in the presence of dNTPs/polymerase (Step 2). Next, in the process of primer extension, the target is displaced by the polymerase with strand-displacement activity, after which a complementary DNA is synthesized, forming a probe–cDNA complex (Step 3). Finally, to renew the cycle, the displaced target hybridizes with another probe, which triggers yet another polymerization reaction (Step 4). Throughout this cyclical process, the hairpin fluorescence probe plays a key role as both template of polymerization reaction and fluorescence signal carrier, while the target acts as a trigger of polymerization reaction. Amplified detection results from this activity because the target can be displaced and trigger the polymerization reaction circularly. Under these conditions, even minute amounts of targets can produce obvious fluorescence enhancement based on the circular polymerization reaction triggered by the displaced targets themselves. Therefore, by monitoring the increase of fluorescence intensities, we could detect the target with high sensitivity.
Figure 1.
The mechanism of isothermal amplified detection of DNA based on strand-displacement polymerization reaction. In the presence of target DNA, the hairpin fluorescence probe recognizes and hybridizes with it and undergoes a conformational change, leading to stem separation (Step 1). Following this, the primer anneals with the open stem of the hairpin fluorescence probe and triggers a polymerization reaction in the presence of dNTP/polymerase (Step 2). Next, in the process of primer extension, the target is displaced by the polymerase with strand-displacement activity, after which a complementary DNA is synthesized, forming a probe–cDNA complex (Step 3). Finally, to renew the cycle, the displaced target hybridizes with another hairpin fluorescence probe, which triggers yet another polymerization reaction (Step 4).
Design of the hairpin fluorescence probe
To achieve the degree of amplification desired, our design relies upon the annealing of the primer with the open stem after the hairpin fluorescence probe hybridizes with the target. Traditional hairpin probes, such as molecular beacons (MBs) contain 5–8-nt-long stem sequences and 15–35-nt-long loop sequences, which undergo conformational change upon hybridization with the target (15,16). In contrast, the hairpin fluorescence probe used in this study has a stem long enough to ensure that stem hybridization affinity will be stronger than hybridization affinity with the primer. Therefore, in the absence of target, the primer does not induce polymerization reaction. On the other hand, a stem that is too long would restrain hairpin probe conformational change upon hybridization with target. Hence, in this work, the stem consisted of 11-nt-long sequences, thus forming a structure stable enough to prevent the primer from annealing with duplex stem. In order to open long stem hairpin probe upon hybridization with target, a ‘shared-stem’ (17) structure is designed, i.e. besides the loop region, six bases of the stem at 5′-end is complementary to the target (Table 1). ‘Shared-stem’ structure is a design variant of conventional MB where one arm of the stem participates in either hairpin formation or target hybridization. The hybrid of shared-stem MB with its target has more stable structure than the hybrid of conventional MB with its target (17).
Detection capability of the hairpin fluorescence probe
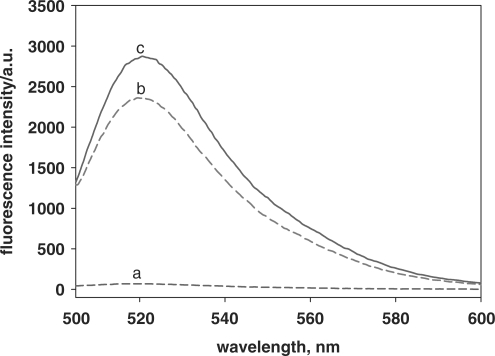
The activation of the DNA system is based on the conformational change of the hairpin fluorescence probe upon hybridization with target DNA. The enhancement of fluorescence intensities, as shown in Figure 2, reveals the conformational change of hairpin fluorescence probe. Curves a–c are fluorescence intensities of 5.0 × 10−8 M probe in the presence of 0, 5.0 × 10–8 and 1.0 × 10–7 M target, respectively. The signal-to-background ratio (SBR) is often used to characterize the detection capability of the hairpin fluorescence probe and is calculated as (Fopen – Fbuffer)/(Fclosed – Fbuffer), where Fbuffer is the fluorescence intensity of buffer solution, Fclosed is the fluorescence intensity of buffer solution by adding probe and Fopen is the fluorescence intensity of buffer solution by adding probe and target (15,16). In this study, SBR of the hairpin fluorescence probe is 39.5 and 45.2, when the concentration of target is 5.0 × 10–8 and 1.0 × 10–7 M, respectively. These results implied that the hairpin fluorescence probe underwent a conformational change upon hybridization with the target and that fluorescence was restored. Therefore, the hairpin fluorescence probe was suitable to act as a template for polymerization reaction in this study.
Figure 2.
Hairpin fluorescence probe hybridization with target. The mixture containing 5.0 × 10−8 M probe with 0 M (curve a), 5.0 × 10−8 M (curve b) and 1.0 × 10–7 M (curve c) target, respectively. All samples were incubated at 37°C.
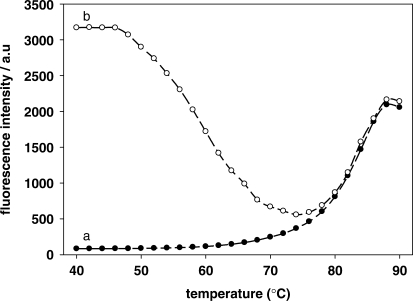
Thermal profiles of the hairpin fluorescence probe (curve a) and the hybrid with its target (curve b) are shown in Figure 3. The Tm value for probe and the probe hybridization to its target was 83°C and 56°C, respectively that are higher than the temperature of the polymerization reaction. This result indicated that the hairpin fluorescence probe, as designed, can be used in this strategy.
Figure 3.
Thermal profiles of hairpin fluorescence probe and the hybrid with its target. The mixture containing 5.0 × 10–8 M probe with 0 M (curve a) and 5.0 × 10–8 M (curve b) target, respectively. The temperature was increased from 40°C to 90°C in increment of 2°C, with each increment lasting 2 min.
Verification of the amplified DNA detection method
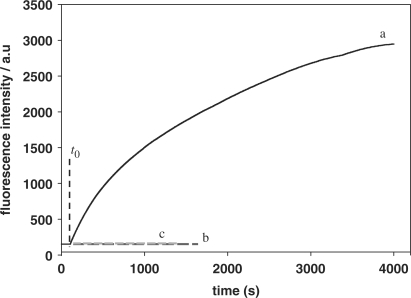
As previously described, the hairpin fluorescence probe undergoes a conformational change upon hybridization with its target and provides an opened stem for primer annealing. Strand-displacement polymerization reaction is then triggered in the presence of polymerase. Having proved the suitability of the hairpin probe, as designed, we now investigated the feasibility of this method for amplification of DNA detection. As shown in Figure 4, the fluorescence intensity increased upon addition of target to the mixture containing dNTPs and polymerase (curve a), indicating that the polymerization reaction was triggered by the target. Furthermore, the fluorescence intensity maintained its increase with time, indicating that the continuous formation of probe–cDNA complex was the result of circular polymerization reaction. The fluorescence intensity of the solution reached maximum within 4000 s, indicating that the target quantitatively converted the probe to the signaling state. In the absence of a target, no fluorescence intensity change was observed, indicating that no polymerization reaction was triggered (curve b). A control experiment performed with addition of random single-stranded DNA revealed that polymerization reaction was not triggered even in the presence of dNTPs/polymerase (curve c), implying that the detection was target-specific.
Figure 4.
Verification of isothermal strand-displacement polymerization reaction. The mixture containing 5.0 × 10−8 M probe and 5.0 × 10–8 M primer with 6.25 × 10–9 M target (curve a), 6.25 × 10–9 M random DNA (curve c) added at t0 in the presence of dNTPs/polymerase respectively; curve b: no target added.
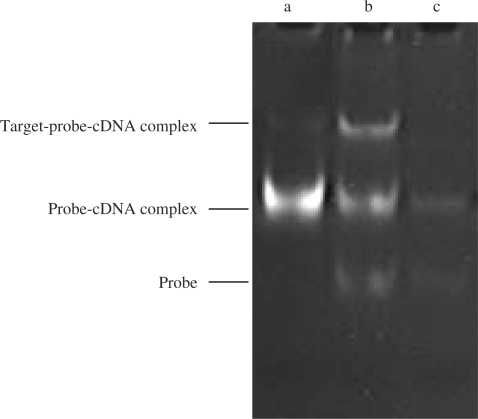
The above results were further confirmed by electrophoresis. Figure 5 shows electrophoresis results of products synthesized by polymerization reaction at various reaction time intervals. Lanes a, b and c are nucleic acid bands generated at 60, 20 and 0 min, respectively. As expected, the content of probe–cDNA complex (the middle band in lane b) increased as the reaction time increased. In addition, a new product (the upper band) with a slower migration speed than that of probe–cDNA complex appeared in lane b. As shown in Figure 1, an intermediate product of probe–target–cDNA complex exists in the process of primer extension before target was displaced (Step 2). Therefore, the new band might be the probe–target–cDNA complex.
Figure 5.
Non-denaturing PAGE analysis of the products by the isothermal strand-displacement polymerization reaction. a–c curves: products generated by the isothermal strand-displacement polymerization reaction containing 2.0 × 10–7 M probe, 2.0 × 10–7 M primer and 2.0 × 10−8 M target at different time intervals in the presence of dNTPs/polymerase, (a) t = 60, (b) t = 20, (c) t = 0 min. All samples were incubated at 37°C.
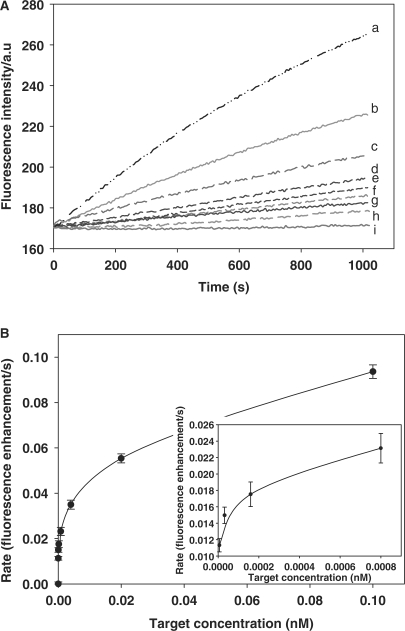
Amplified detection of target with high sensitivity
Figure 6A shows the fluorescence intensities observed upon analyzing different concentrations of targets with this method. The results showed that the number of opened probes increased as the concentration of the targets increased. Figure 6B shows the relationship between the rate of fluorescence enhancement (for 1000 s) and the concentration of target. As the concentration of the target increases, the rate of fluorescence enhancement increases. For this method, when the target is 6.4 × 10–15 and 1.28 × 10–15 M, the (F – F0) is 10.3 and 8.7, the background noise is 3.2, and then the signal-to-noise is 3.2 and 2.7 (F0 is the fluorescence intensity of the solution before the polymerization reaction, and F is the fluorescence intensity of the solution after the polymerization reaction for 1000 s). Therefore, 6.4 × 10–15 M was considered as the detection limit, which is two or three order of magnitude lower than that of other reported DNA amplified detection methods (10,11). The turnover rate of this method is also calculated. As shown in Table S-2, the turnover rate (s–1) of this method was 0.051, 0.211, 0.820 and 2.828, with target concentration of 8.0 × 10–13, 1.6 × 10–13, 3.2 × 10–14 and 6.4 × 10–15 M, respectively. Since the rate of fluorescence enhancement is influenced by multiple factors, including the concentration of input target and reaction time, the turnover rate of this method varies according to different target concentrations.
Figure 6.
Detection of different concentrations of target based on isothermal strand-displacement polymerization reaction. Experiments were performed in the presence of 15U polymerase Klenow fragment exo− and 100 μM dNTPs with 5 × 10–8 M probe, 5 × 10–8 M primer, and different concentrations of target. (A) Monitoring the fluorescence intensity of this amplified DNA detection method over a range of target DNA concentrations. The curves from a to i contain the target with 1.0 × 10–10, 2.0 × 10–11, 4.0 × 10–12, 8.0 × 10–13, 1.6 × 10–13, 3.2 × 10–14, 6.4 × 10–15, 1.28 × 10–15 and 0 M, respectively. All samples were incubated at 37°C. (B) The relationship of the rate of fluorescence enhancement with target DNA concentration.
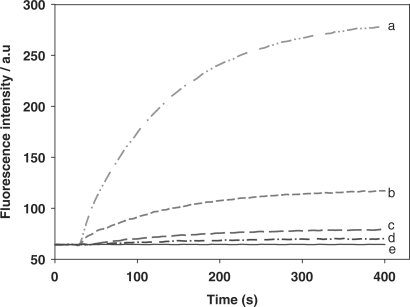
In order to confirm whether the high sensitivity of DNA detection results from the circular strand-displacement polymerization reaction, control experiments with target at various concentrations reacting with the hairpin fluorescence probe in the absence of primer/polymerase were also carried out. As shown in Figure 7, fluorescence intensities also increased with increasing concentration of target, a result obviously produced by the hybridization of target with probe. However, the fluorescence intensities reached a plateau within only a few minutes, implying that no circular strand-displacement polymerization reaction occurred without the inclusion of both primer and polymerase. The detection limit was only 1.0 × 10−10 M.
Figure 7.
The hairpin fluorescence probe response to the target. Experiments were performed in the absence of polymerase and dNTPs with 5 × 10–8 M probe and different concentrations of target. The curves from a to e contain the target with 1.25 × 10–8, 2.5 × 10–9, 5 × 10–10, 1 × 10–10 and 0 M, respectively. All samples were incubated at 37°C.
Moreover, this method is based on an isothermal amplification process, and the product of primer extension is 34-nt sequences long. Consequently only a small interval of time is required for each primer extension. Therefore, although this method is a linear amplification, the circular speed of it is much quicker than that of PCR, and high sensitivity is achieved in a shorter time. Notwithstanding these advances, this method is still not as sensitive as PCR, and, as a result, a pretreatment, such as enrichment, may be necessary for some biological samples. In our case, however, the probe was hybridized with a single-strand DNA, indicating that this method can be mainly used for detection of actual biological samples with single-strand genomic DNA. In nature, many viruses have single-strand genomic DNA, such as Parvoviridae, Geminiviruses, Microviridae or Inoviridae (18). Therefore, this method has potential application to the analysis of pathogens.
CONCLUSION
In summary, a method with a novel platform which amplifies single-strand DNA (ss DNA) detection based on polymerase-induced isothermal strand-displacement polymerization reaction was presented in this paper. The detection limit of this method is 6.4 × 10–15 M, which is five orders of magnitude lower than that of the same hairpin fluorescence probe hybridizing with the target without primer and polymerase. Therefore, this isothermal and rapid analysis of target DNA demonstrated the appealing bioanalytical features of this method, which can be expected to provide a sensitive platform for amplified detection and subsequent analysis of nucleic acids.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Key Basic Research Program of China (2002CB513110); the Major International Joint Research Program of Natural Science Foundation of China (20620120107); the Key Project of Natural Science Foundation of China (90606003); the China National Key Projects (2005EP090026); the Hunan Province Natural Science Foundation of China (08JJ1002). Funding for open access charge: 90606003.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Willner I, Willner B. Biomaterials integrated with electronic elements: enroute to bioelectronics. Trends Biotechnol. 2001;19:222–230. doi: 10.1016/s0167-7799(01)01634-1. [DOI] [PubMed] [Google Scholar]
- 2.Wang J. Survey and summary: from DNA biosensors to gene chips. Nucleic Acids Res. 2000;28:3011–3016. doi: 10.1093/nar/28.16.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tom NG, Lars R, Oliver S. Triplex molecular beacons as modular probes for DNA detection. Angew. Chem. Int. Ed. 2007;46:5223–5225. doi: 10.1002/anie.200700289. [DOI] [PubMed] [Google Scholar]
- 4.Caruana DJ, Heller A. Enzyme-amplified amperometric detection of hybridization and of a single base pair mutation in an 18-base oligonucleotide on a 7-mm-diameter microelectrode. J. Am. Chem. Soc. 1999;121:769–774. [Google Scholar]
- 5.Patolsky F, Lichtenstein A, Willner I. Detection of single-base DNA mutations by enzyme-amplified electronic transduction. Nat. Biotechnol. 2001;19:253–257. doi: 10.1038/85704. [DOI] [PubMed] [Google Scholar]
- 6.Liu J. A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J. Am. Chem. Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Liu G, Merkoc A. Electrochemical coding technology for simultaneous detection of multiple DNA targets. J. Am. Chem. Soc. 2003;125:3214–3215. doi: 10.1021/ja029668z. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Rincon O, Polsky R, Dominguez E. Electrochemical detection of DNA hybridization based on DNA-templated assembly of silver cluster. Electrochem. Commun. 2003;5:83–86. [Google Scholar]
- 9.Weizmann Y, Cheglakov Z, Pavlov V, Willner I. Autonomous fueled mechanical replication of nucleic acid templates for the amplified optical detection of DNA. Angew. Chem. Int. Ed. 2006;45:2238–2242. doi: 10.1002/anie.200503810. [DOI] [PubMed] [Google Scholar]
- 10.Weizmann Y, Beissenhirtz MK, Nowarski ZCR, Kotler M, Willner I. A virus spotlighted by an autonomous DNA machine. Angew. Chem. Int. Ed. 2006;45:7384–7388. doi: 10.1002/anie.200602754. [DOI] [PubMed] [Google Scholar]
- 11.Beissenhirtz MK, Elnathan R, Weizmann Y, Willner I. The aggregation of Au nanoparticles by an autonomous DNA machine detects viruses. Small. 2007;3:375–379. doi: 10.1002/smll.200600450. [DOI] [PubMed] [Google Scholar]
- 12.Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. PNAS. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beissenhirtz MK, Willner I. DNA-based machines. Org. Biomol. Chem. 2006;4:3392–3401. doi: 10.1039/b607033g. [DOI] [PubMed] [Google Scholar]
- 14.Shlyahovsky B, Li D, Weizmann Y, Nowarski R, Kotler M, Willner I. Spotlighting of cocaine by an autonomous aptamer-based machine. J. Am. Chem. Soc. 2007;129:3814–3815. doi: 10.1021/ja069291n. [DOI] [PubMed] [Google Scholar]
- 15.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 16.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 17.Tsourkas A, Behlke1 MA, Bao G. Structure–function relationships of shared-stem and conventional molecular beacons. Nucleic Acids Res. 2002;30:4208–4215. doi: 10.1093/nar/gkf536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jay AL, Heinz FC, Robert AO. Virology. 3rd edn. NJ: Prentice-Hall, Inc. A Paramount Communications Company; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.