Abstract
DNA helicases are motor proteins that play essential roles in DNA replication, repair and recombination. In the replicative hexameric helicase, the fundamental reaction is the unwinding of duplex DNA; however, our understanding of this function remains vague due to insufficient structural information. Here, we report two crystal structures of the DnaB-family replicative helicase from Geobacillus kaustophilus HTA426 (GkDnaC) in the apo-form and bound to single-stranded DNA (ssDNA). The GkDnaC–ssDNA complex structure reveals that three symmetrical basic grooves on the interior surface of the hexamer individually encircle ssDNA. The ssDNA-binding pockets in this structure are directed toward the N-terminal domain collar of the hexameric ring, thus orienting the ssDNA toward the DnaG primase to facilitate the synthesis of short RNA primers. These findings provide insight into the mechanism of ssDNA binding and provide a working model to establish a novel mechanism for DNA translocation at the replication fork.
INTRODUCTION
DNA replication is a complicated process that requires a protein machine known as the replisome, comprising a DNA polymerase, a replicative helicase and a DNA primase (1,2). During DNA replication the leading strand is directly synthesized by DNA polymerase, whereas on the lagging strand the primase interacts with the hexameric helicase to synthesize short RNA primers (3,4) that are used to generate Okazaki fragments that are essential for the progression of the replication fork (5). The most widely studied replication system is that of Escherichia coli, which contains the DnaB replicative helicase and the DnaG primase. DnaG is a specific single-stranded DNA-dependent RNA polymerase (5,6), and DnaB is a multifunctional ATPase that catalyzes the unwinding of double-stranded DNA (dsDNA) into single-stranded DNA (ssDNA) intermediates to provide ssDNA templates for the polymerases at the chromosomal replication forks (7).
DnaB binds transiently with DnaG and thereby limits the size of the nascent primers to ∼9–14 nt (8). During lagging strand synthesis, this cyclic association of DnaG with DnaB is an important driving force to initiate a new cycle of Okazaki fragment synthesis and to terminate preceding fragment synthesis (9). This key event, however, does not always occur in other organisms. For example, the bacteriophage T7 gene 4 protein contains both DNA helicase and primase activies (10). In addition, in Bacillus stearothermophilus the helicase–primase interaction may not be transient, as was shown to be the case in E. coli (11).
In Gram-positive bacteria, such as Bacillus subtilis, the replicative hexameric helicase is named DnaC (12). Notably, DnaC in E. coli is a helicase loader protein rather than a helicase. DnaC and DnaG from B. subtilis were identified by their amino-acid sequence similarity to their E. coli counterparts (12). However, a number of Gram-positive bacterial proteins related to the loading of the replicative helicase differ from those of E. coli (13). Furthermore, unlike in E. coli where the replisome forms a dynamic complex, the B. subtilis replisome can assemble into a stable complex (14), indicating that the DNA-replication mechanisms of the Gram-positive B. subtilis may be similar, but not identical to, those of the Gram-negative E. coli. Therefore, determining the differences in the molecular mechanisms of DNA replication between different organisms will assist in understanding the structure–function relationships of replicative helicases.
From a structural point of view, the replicative helicases can adopt various oligomeric states, including monomers, dimers, hexamers and heptamers (10). All helicases share several common biochemical properties, including nucleic acid binding, nucleoside triphosphate (NTP) binding and hydrolysis and NTP hydrolysis-dependent unwinding of duplex nucleic acids (15). The three-dimensional structure of all helicases reveals at least one conserved domain, called the RecA-like fold, first found in the crystal structure of RecA (16). Recently, the three-dimensional structures of the DnaB-family helicases, T7 gp4, B. stearothermophilus DnaB (BstDnaB), papillomavirus E1 helicase and B. subtilis G40P, have been determined by X-ray crystallography (17–20). All of these enzymes assemble into a ring-shaped hexameric structure. Among these crystal structures, the E1 helicase is the only one to have been determined in complex with ssDNA and ADP. The structure of this complex demonstrates that the DNA-binding hairpins of each subunit form a spiral ‘staircase’ to interact with the ssDNA backbone (20). However, Bailey et al. (21) have pointed out that the ssDNA-binding mechanisms of bacterial DnaB and the E1 helicase may differ because the E1 protein uses different regions of its RecA-like domain for hexamer formation and DNA binding. Furthermore, all characterized superfamily 3 members (including the E1 helicase) display 3′-5′ polarity, but it is believed that the DnaB-like family unwinds the DNA in a 5′-3′ direction (7,22,23).
Despite a common structure/function mechanism for all replicative helicases, it remains intriguing why the helicases of different species might involve different mechanisms of helicase-ssDNA formation. In addition, it is believed that most hexameric helicases exhibit a higher affinity for ssDNA than for dsDNA (24–27). To generate a mechanistic ssDNA-binding model for DnaB-like helicases, it is thus important to elucidate the structure of the DnaB–ssDNA complex. In this study, we present the crystal structures of the DnaB-family protein of the eubacteria Geobacillus kaustophilus HTA426 DnaC (GkDnaC) in both the apo-GkDnaC form and as the GkDnaC–ssDNA complex with a 15-mer oligodeoxythymidylate (dT)15 at 3.6 Å and 4.1 Å, respectively. We also investigated the nature of the GkDnaC–ssDNA interaction in structure-based mutational studies using surface plasmon resonance (SPR) analysis.
MATERIALS AND METHODS
Cloning, expression and purification
The gene GK3476 encoding the GkDnaC helicase was PCR-amplified from G. kaustophilus HTA426 genomic DNA using pfu polymerase (Stratagene). The forward (GGGAATTCCATATGCAACACCATTGGAAAGAGCTG) and downstream (CCCGCTCGAGTGATTCATCACGGTATTTTTTCAGCC) primers were designed to incorporate unique NdeI and XhoI restriction sites, permitting the insertion of the amplified gene into the pET21b vector (Novagen) for protein expression in E. coli. The resultant plasmid, pET21b-GkDnaC, encoded the full-length wild-type GkDnaC helicase fused with a C-terminal His tag (LEHHHHHH). The DnaC mutants were generated according to the QuickChange mutagenesis protocol (Stratagene) using the pET-21b wild-type plasmid as the template. All mutant constructs were verified by DNA sequencing. Escherichia coli BL21(DE3) cells were transformed with these expression vectors and grown to OD600 of 0.7 at 37°C in Luria-Bertani medium containing 50 μg/ml ampicillin. Overexpression of wild-type and mutant GkDnaC constructs was induced with 1 mM isopropyl β-d-thio-galactopyranoside for 3 h at 30°C. The induced cells were harvested by centrifugation (3000 g) for 20 min at 4°C, and resuspended in buffer A (20 mM Tris, 5 mM imidazole, 0.5 M NaCl, pH 7.9) before lysis at 110 MPa using a French press. The wild-type and mutant GkDnaC proteins were then purified from the soluble supernatant by Ni2+-affinity chromatography (HiTrap HP, Amersham Biosciences) followed by size-exclusion chromatography (HiLoad 16/60 SuperdexTM 200 preparation grade column, GE Healthcare). The purified proteins were collected and dialyzed against buffer B (20 mM HEPES, 100 mM NaCl, pH 7.0), and then concentrated to 6 mg/ml for further crystallizations and analyses. Protein purity remains greater than 95% as determined from Coomassie-stained SDS–PAGE (data not shown).
Crystallization, data collection and structure determination
Initial crystallization screening for apo-GkDnaC was performed manually using the hanging-drop vapor diffusion method at 20°C. The protein solution (1 μl, 6 mg/ml) was mixed with KAu(CN)2 (0.2 μl, 1 mM) and 1 μl of reservoir solution for crystallization trials. Apo-GkDnaC crystals were grown reproducibly using a reservoir solution comprised of 1.3 M ammonium sulfate, 20% (v/v) glycerol, 5% (v/v) 1,4-dioxane and 0.1 M MES (pH 6.5). The apo-GkDnaC crystals were cryoprotected in 25% glycerol by quickly transferring the crystal directly from the hanging drop before data collection. GkDnaC was concentrated to 11 mg/ml in buffer B with 1-mM ATPγs and 2-mM MgCl2 for complex preparation. The single-stranded 15-mer dT15 was added to the protein solution at a molar ratio of 1:4 (DnaC monomer:dT15). Prior to crystallization, the complex solution was further purified by Heparin HP column to remove the unbound protein. Using the hanging drop vapor diffusion method at 20°C, the complex crystals grew within 1 week after mixing 1 μl of the protein–ssDNA complex solution with 0.5 μl phenyl mercury acetate (CH3CO2HgC6H5) and 1 μl of 80% reservoir solution containing 0.1 M MES (pH 6.5) and 1.2 M MgSO4. The DnaC–ss(dT15) crystals grew as hexagonal pillars with dimensions of 0.3 mm × 0.3 mm × 0.4 mm. Before the crystals were flash frozen, 30% glycerol was used for dehydration trials. We used electrophoresis to confirm that both the protein and the ssDNA were present in the complex crystal (data not shown). All the crystal data were collected using an ADSC Quantum-315 CCD area detector at the synchrotron radiation X-ray source at Beamline 13B1 of the National Synchrotron Radiation Research Center in Taiwan. All data integration and scaling were performed using the HKL2000 package (28).
SOLVE (29) was used to locate the gold sites and determine the initial multiple-wavelength anomalous diffraction phases at 4.3 Å. The initial phases were extended and further improved to 4.1 Å by RESOLVE (30). The native data set (3.6 Å) was used for further refinement. XtalView (31) was used to examine electron density maps and for model building. Due to the limitations of crystal resolution, the side-chain orientation was built based on the higher-resolution structure of BstDnaB in complex with the HBD of DnaG (17). The sequence identity between GkDnaC and BstDnaB is 96% (Supplementary Figure 2). The model refinement and final analysis were performed with the program CNS (32). The final model had an R-factor of 29.3% for all reflections between 30Å and 3.6 Å resolution and an Rfree of 32.4%. The B-factor of the apo-GkDnaC structure was 167.6 Å2.
The structure of GkDnaC bound to (dT)15 was solved by the molecular replacement software MolRep (33) using the apo-GkDnaC structure as a template. Following minimization refinement with CNS, electron density corresponding to ssDNA was observed in both 2Fo-Fc and Fo-Fc maps. The nucleotides of the ssDNA molecule were gradually built as the map quality improved. Molecular dynamics refinement was performed using the program CNS with a 30- to 4.1-Å-resolution range, and 10% of the data was selected to calculate the Rfree factor to monitor refinement. The final structure was refined to an R-factor of 30.2% and an Rfree of 37.4%. The B-factors were higher in the ssDNA (287 Å2) than in the protein (180 Å2). High B-factors for ssDNA have also been observed in other low-resolution structures (34–36). The Ramachandran plots (37) for both structures did not violate accepted backbone torsion angles. The data collection and structure refinement statistics are listed in Table 1. The PyMol (38) program was used to generate the figures. The atomic coordinates of apo-GKDnaC and ssDNA complex of GKDnaC have been deposited in the Protein Data Bank under accession codes 2VYF and 2VYE, respectively.
Table 1.
Data collection and refinement statistics for apo-form and ssDNA-bound form of GkDnaC
| Crystal | GkDnaC |
GkDnaC (Au-MAD) |
GkDnaC-(dT)15 | ||
|---|---|---|---|---|---|
| Au-derivative | Inflection | High remote | Peak | Native | |
| Data collection | |||||
| Wavelength (Å) | 1.0000 | 1.0401 | 1.0229 | 0.9799 | 1.0000 |
| Resolution (Å) | 30–3.6 | 30-4.1 | 30–4.1 | 30–4.3 | 30–4.1 |
| Space group | P63 | P63 | P63 | ||
| Cell dimension (Å) | a = 176.9 | a = 176.9 | a = 180.8 | ||
| c = 108.8 | c = 109.6 | c = 104.1 | |||
| Completeness (%) | 99.2 (95.5)a | 97.1 (87.1) | 97.8 (89.0) | 90.8 (71.5) | 91.1(91.5) |
| <I/σI> | 36.4 (2.2) | 32.0 (2.8) | 31.8 (3.0) | 21.9 (2.5) | 34.3 (2.4) |
| Rsymb (%) | 6.3 (68.7) | 6.8 (58.0) | 7.1 (54.0) | 6.9 (35.9) | 6.0 (47.5) |
| Redundancy | 5.1 (4.2) | 4.8 (4.6) | 4.8 (4.7) | 2.2 (2.0) | 6.0 (5.8) |
| Refinement | |||||
| Resolution (Å) | 30–3.6 | 30–4.1 | |||
| No. reflections | 18 500 | 11 729 | |||
| Rwork/Rfree | 29.3/32.4 | 30.2/37.4 | |||
| No. atoms | |||||
| Protein | 5913 | 6182 | |||
| DNA | 186 | ||||
| Heteroatoms | 4 | ||||
| R.m.s deviation | |||||
| Bond lengths (Å) | 0.008 | 0.014 | |||
| Bond angles (°) | 1.90 | 2.00 | |||
aValues in parentheses are for the outer shell.
bRsym (I) = ΣhΣi|Ii-I|ΣhΣI I, where I is the mean intensity of the i observations of reflection h.
SPR
The ssDNA-binding experiments were carried out at 293K using a Biacore X apparatus (BIAcore) at 30 μl/min with running buffer that contained 10 mM HEPES (pH 7.4), 5 mM MgCl2, 25 mM NaCl and 0.1 mM ATPγs, with 0.1% Tween-20 to minimize nonspecific electrostatic attraction. A solution (200 nM) of 5′-biotinylated oligonucleotide comprising 30 thymidine residues (5′Bio-(dT)30) in the running buffer was injected over a streptavidin-coated chip surface (Biacore SA chip) for 30 s to yield an increase of 350-resonance units (710 resonance units for the large ligand level) in one of two flow cells. The second flow cell was unmodified and served as a control. When required to remove the tightly bound proteins, regeneration of the flow cells was achieved with 10-s injections of 50 mM NaOH/1 M NaCl. Solutions containing the protein (wild-type and DnaC mutants) at concentrations of 20 nM to 3 μM were injected into the flow cells at 30 μl/min for 2 min. The equilibrium binding constant was estimated by fitting the equilibrium response values at different protein concentrations to the 1:1 Langmuir binding model using BIAevaluation software (BIAcore). Before fitting to the Langmuir model, binding data were corrected by subtraction of the control to account for simple refractive index differences. For concentration-response profiles that did not appear to be saturated within the experimental concentration range, the saturation levels were constrained to the maximal response value expected when the protein molecules bind to all immobilized DNA molecules at 1:1 stoichiometry. Experiments were repeated three times to estimate the uncertainty of the binding constant. The standard deviations of three repeated experiments were used to estimate the error.
RESULTS
Overall structure
We determined the crystal structures of the apo-form and the ssDNA-bound DnaC helicase of G. kaustophilus HTA426 (GkDnaC). The apo-GkDnaC structure was determined by the multiple-wavelength anomalous diffraction (MAD) method, and the phases were solved using an Au-derivative; the structure was refined to 3.6-Å resolution. The structure of the GkDnaC–ssDNA complex was solved using the apo-GkDnaC structure as a probe for molecular replacement and was refined to 4.1-Å resolution. Both crystals belong to the hexagonal space group P63 and share similar unit-cell parameters, as shown in Table 1. Each asymmetric unit contains two GkDnaC molecules to form a dimer in the apo-GkDnaC crystal and one additional (dT)15 oligonucleotide in the GkDnaC–ssDNA complex crystal. In the GkDnaC–ssDNA complex crystal, three dimers are crystallographically related and form an intact ring-shaped hexamer with 3-fold symmetry with three dT15 oligonucleotides bound to the interior surface of the central channel (Figure 1). The shape and dimensions of this hexameric structure in a complex with ssDNA are similar to the structure of BstDnaB in the absence of ssDNA (17). Briefly, the hexameric structure of the GkDnaC helicase forms a double-layered ring structure, which contains an N-terminal domain (NTD) collar with 3-fold symmetry and a C-terminal domain (CTD) collar with a pseudo 6-fold symmetry.
Figure 1.
The GkDnaC-ssDNA complex structure. (A) Side-view of the hexameric GkDnaC–ssDNA complex in ribbon representation. The N-terminal domain (NTD) collar, C-terminal domain (CTD) collar and the bound ssDNA are represented as cartoons and are colored in violet, green and orange, respectively. (B) Top view of the GkDnaC–ssDNA complex. Three binding pockets in the hexameric ring encircle three symmetric ssDNA molecules.
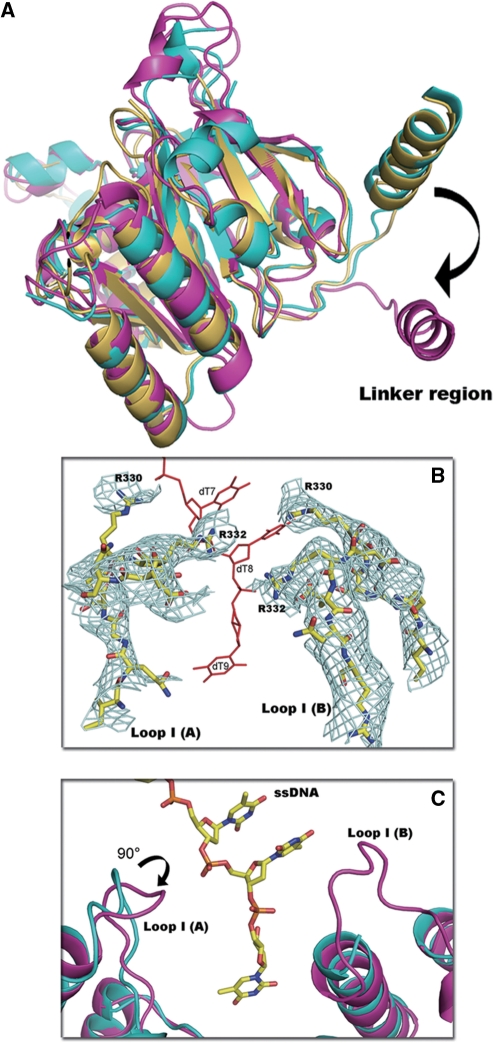
As shown in Figure 1A, the full-length GkDnaC monomer is a two-domain protein that is topologically almost identical to other replicative hexameric helicases (21). The GkDnaC monomer consists of two distinct functional domains: an N-terminal globular domain consisting of residues 1–152, and a C-terminal RecA-like domain. These two domains are connected by a long linker region (residues 153–192) containing one α-helix (α8, residues 163–178) flanked by two loops (residues 152–161 and 179–190). Structural comparison of the two monomers in the asymmetric unit reveals that the most significant structural variations occur in the linker region, and results in two distinct monomer conformations (Figure 2A).
Figure 2.
Interactions between GkDnaC and ssDNA. (A) The structure of the GkDnaC dimer bound to ssDNA. ssDNA (9-mer) shown as sticks within the 2Fo-Fc electron density map is colored pale cyan and contoured at 1σ. The ssDNA occupies a binding pocket fundamentally composed of one α-hairpin and two loops-Loop I(A) and Loop I(B). Molecule A and molecule B are colored in pink and blue, respectively. (B) Side view of surface representation of the GkDnaC dimer. Positive and negative potentials are shown in blue and red, respectively. The bound ssDNA in the binding groove is depicted in stick and colored in yellow. (C) Schematic diagram of the GkDnaC and ssDNA interactions. Hydrogen bonding and electrostatic interactions are shown as black dotted lines, and Van der Waals contacts are represented by gray dotted lines. (D) Basic residues from both subunits interact with ssDNA. Loop I(A) and Loop I(B) are colored in blue, and ssDNA is colored in red. The residues involved in the interaction with ssDNA are labeled.
To analyze the structural deviations of GkDnaC from the G40P helicase and BstDnaB, these structures were superimposed. There is no obvious structural difference in the N-terminal regions of these proteins, indicating that ssDNA binding did not dramatically change the NTD conformation. However, comparison of the C-terminal regions of these molecules shows apparent differences in the linker regions (Figure 3A). In particular, the G40P linker orientation is distinct from that of other helicases. This difference may change the shape of the hexamer assembly and the orientation of the CTD toward the NTD, resulting in a smaller dimension of the central ssDNA-binding channel in G40P.
Figure 3.
Features of the GkDnaC helicase structure. (A) Structural comparison of the C-terminal regions demonstrate apparent differences in the linker regions by superimposing GkDnaC (yellow), BstDnaB (cyan) and G40P (magenta). (B) The final model of DNA-binding loop I (yellow, stick-type) was superimposed on the omitted 2Fo-Fc electron density map contoured at 0.8σ (in cyan mesh). (C) The superimposition of apo-GkDnaC (cyan) and the GkDnaC–ssDNA complex (magenta). Loop I(A) in the GkDnaC–ssDNA complex is facing the 3′-end of ssDNA, and Loop I(B) is disordered in the apo-GkDnaC structure.
To further examine the structural deviation caused by ssDNA binding, we superimposed the apo-GkDnaC and GkDnaC–ssDNA complex structures. Although the overall structure of these two molecules is similar, there are some structural discrepancies located in the loop regions that are involved in DNA binding, particularly in residues 321 to 335. The loops in this region are designated as Loop I(A) and Loop I(B) of subunit A and subunit B, respectively (Figure 3B). The DNA-binding loop I structure in the GkDnaC–ssDNA complex is different compared with the corresponding region in the apo-GkDnaC structure (Figure 3C). Unlike the apo-GkDnaC structure, the position of the Loop I(A) in the GkDnaC–ssDNA complex is close to 3′-end of the ssDNA. In addition, Loop I(B) of the GkDnaC–ssDNA complex also makes contact with the ssDNA, suggesting that both Loops I(A) and I(B) play important roles in stabilizing DNA binding.
The ssDNA-binding site on the hexameric ring
To understand the molecular details of the interaction between the GkDnaC helicase and ssDNA, we studied the structure of GkDnaC in complex with a 15-mer oligodeoxythymidylate (dT)15. The (dT)15 oligonucleotide was observed in the asymmetric unit (Figure 2A). Due to the 3-fold symmetry in the crystal, there were three (dT)15 oligonucleotides bound to the interior surface of the hexameric ring (Figure 1). It should be pointed out that the central channel of hexameric ring helicases only accommodates one unwound ssDNA molecule at a time in vivo (39–41). The NTD and the top region of the CTD comprise the ssDNA-binding site on the hexameric ring, and the ssDNA-binding pocket is formed by subunit A and B. Electrostatic analysis of the dimer surface in the asymmetric unit reveals a groove region with significant positive charge that potentially allows the ssDNA to bind through electrostatic interactions. This finding is consistent with the (dT)15-binding site in the GkDnaC–ssDNA complex structure, as shown schematically in Figure 2B. In addition, this basic groove extends from the NTD to the CTD almost parallel to the central axis.
In the general proposed mechanism, unwound dsDNA threads through the central hole from the CTD to the NTD in the 3′-5′ direction (7). Previous studies have also indicated that the replicative DnaB helicase has two potential ssDNA-binding sites, ‘strong’ and ‘weak’, located at the NTD and CTD, respectively (42,43). However, the major binding sites were observed in the interior surface of the NTD collar in our GkDnaC–(dT)15 complex structure. This may due to a weak affinity of the ssDNA for the CTD or steric constraints in the narrow central cavity of the CTD ring that preclude the approach of ssDNA.
GkDnaC–ssDNA interaction
Although we used (dT)15 for crystallization, only 9-mer oligonucleotides were observed. The 9-mer ssDNA was seated on a surface-exposed basic pocket formed from two subunits in the asymmetric unit. Interestingly, this finding is generally consistent with previous data demonstrating that the E. coli DnaB hexamer could occlude a 20-mer ssDNA, but only 10-mer ssDNA was strongly protected against nuclease digestion (42–44). Figure 2C presents details of the interaction between GkDnaC and ssDNA. The 5′-end of the ssDNA is directed toward the NTD collar, whereas the 3′-end is directed toward the CTD collar. Thymidine 1 (Thy1) through Thy6 interact mainly with the N-terminal collar, whereas Thy7 to Thy9 interact with the C-terminal collar. The 9-mer ssDNA forms a hook-like shape structure, and the base groups of Thy1 and Thy4 form base interactions. These interactions could further provide a force to stabilize the 5′-end of (dT)15. These insights into the protein–DNA interactions suggest that the basic residues from both the NTD and CTD are critical for ssDNA binding. The ammonium group of Lys50 makes a hydrogen bond with the ribose of Thy4. Arg117 interacts with the ribose of Thy7 and the phosphodiester linkage between Thy6 and Thy7 through hydrogen bonding and electrostatic interactions, respectively. Arg145 also interacts in the same manner with the ribose of Thy8 and the phosphate backbone of Thy9. Lys146 binds to the base of Thy7 via hydrogen bonding. In addition to hydrogen bond and electrostatic interactions, residues Gln41, Gln49, Glu78 and Gln79 form numerous van der Waals contacts with Thy3, Thy4 and Thy6, respectively (Figure 2C). In the CTD collar, Loop I(A) and Loop I(B) from both subunits buttress the 3′-end of ssDNA (Figure 2D). Two basic residues, Arg330 and Arg332, in these flanking loops function like a clamp to mediate the ssDNA binding via hydrogen bonds and a salt-bridge to the phosphate backbone. Notably, these ssDNA-binding loops are not observed in apo-GkDnaC and BstDnaB in the absence of ssDNA. In addition to Arg330 and Arg332, Arg344 is located at the CTD of subunit B and forms a hydrogen bond with the base of Thy 9.
Characteristics of GkDnaC bound to ssDNA
To further characterize the roles of the residues involved in protein–ssDNA interactions, we studied the effect of various mutations on the ability of GkDnaC to bind to ssDNA using SPR. A 5′-biotinylated oligonucleotide comprising 30 thymidine residues [5′Bio-(dT)30] was used as the ligand for GkDnaC in these SPR experiments. Gel filtration chromatography results showed that all the mutants assembled into hexamers (data not shown). Based on the GkDnaC–ssDNA complex structure, we designed single mutants (K50A, R117A, R120A, R145A, R330A, R332A and R344A) and one double mutant (R145K146A) located at the ssDNA-binding site (Figure 2D). The SPR results at various GkDnaC concentrations in the presence of 0.1 mM nonhydrolyzable ATPγs are shown in Supplementary Figure 1. The equilibrium binding isotherm of the wild-type and mutants of GkDnaC fit well to a 1:1 Langmuir binding mode. The dissociation constants (Kd) shown in Table 2 indicate that these positively charged residues located at the ssDNA-binding pocket significantly affect the interaction between the GkDnaC helicase and the ssDNA. The dissociation constants of these mutants are two to three orders of magnitude higher than that of the wild-type GkDnaC.
Table 2.
GkDnaC mutagenesis study in DNA binding site by SPR
| Amino acid substitution | Mutant locations | Dissociation constant Kd (M) |
|---|---|---|
| WT | 4.69 ± 0.4 × 10−8 | |
| K50A | NTD, α3 | 1.41 ± 0.5 × 10−6 |
| R117A | NTD, α-hairpin (α6) | 1.47 ± 0.2 × 10−6 |
| R120A | NTD, α-hairpin (α6) | 1.73 ± 0.5 × 10−6 |
| R145A | NTD, α-hairpin (α7) | 1.39 ± 0.4 × 10−6 |
| R145K146Aa | NTD, α-hairpin (α7) | 2.70 ± 0.6 × 10−5 |
| R330A | CTD, Loop I | 1.78 ± 0.07 × 10−5 |
| R332A | CTD, Loop I | 6.64 ± 0.9 × 10−6 |
| R344A | CTD, α15 | 1.79 ± 0.2 × 10−5 |
aDouble mutant.
The role of flanked loops
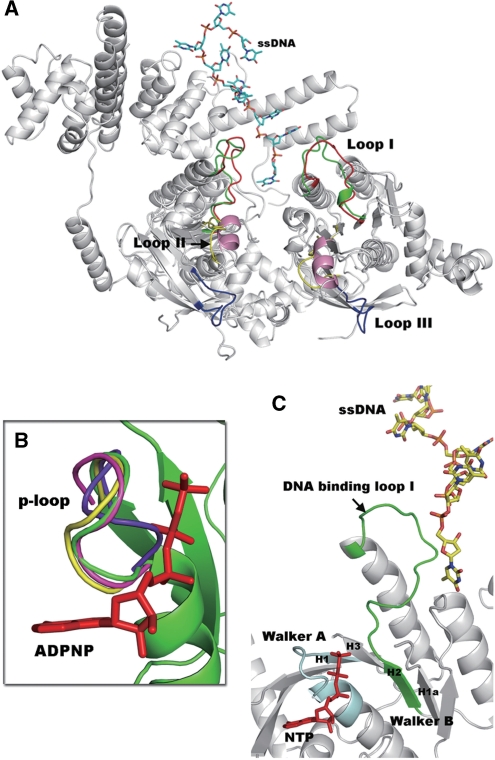
Previous mutational and structural analyses show that there are three loops (Loop I, Loop II and Loop III) of the T7 gp4 helicase that are responsible for DNA binding (19,45,46). Loop I and Loop II are structurally conserved in the RecA-like family (47). We noted that Loops I, II and III are well ordered in both the apo-form and the ADPNP-bound complex structures of gp4 (19). However, these loops are disordered and could not be observed in either the apo-GkDnaC (this work) or in the BstDnaB in complex with the helicase binding domain (HBD) of DnaG (17). This phenomenon may due to a slightly different domain arrangement resulted in close contacts in the RecA-like domain of gp4 that stabilize the loop regions. This also causes the formation of a narrower central channel in gp4 than the channel that is observed in GkDnaC or BstDnaB. However, Loop I and part of Loop II in the GkDnaC–ssDNA complex are well defined in the electron density map. Loop I plays an important role in ssDNA binding in this study. We used structural superimposition to compare the loops of T7 gp4 with those of the GkDnaC–ssDNA complex. As shown in Figure 4A, Loop I of gp4 is superimposed over the corresponding loop I in the GkDnaC–ssDNA complex structure with very little deviation observed, suggesting that the putative DNA-binding loops are flexible and that they may be involved in secondary conformational changes in the partial region upon ssDNA binding.
Figure 4.
(A) Comparison of the DNA-binding loops between the GkDnaC–ssDNA complex (gray) and the T7 gp4 helicase domain (gray). Loops I, II and III of T7 gp4 helicase are shown in red, yellow and blue, respectively. In addition, the DNA-binding Loop I(A) and Loop I(B) (green) observed in our complex structure fit well to Loop I of T7 gp4, whereas a short helix (pink) in the GkDnaC–ssDNA complex was found in the location of Loop II of T7 gp4. ssDNA is represented as sticks and is colored sky blue. (B) Comparison of the p-loop orientation from the GkDnaC–ssDNA complex (green), the G40P ATPase domain (yellow) and T7 gp4 (magenta) in the presence of the modeled nucleotide ADPNP (red) and apo-GkDnaC (violet). (C) Synergistic effects between the modeled nucleotide (NTP) and ssDNA binding. The Walker A and Walker B motifs are colored in pale cyan and green, respectively. The NTP in the ATP-binding site is colored in red.
DISCUSSION
Nucleotide-binding site neighboring with DNA-interaction site
Universal features of the RecA-like domain include several conserved residues involved in the binding and hydrolysis of the NTP equivalent to the Walker A and B boxes of many ATPases, and an ‘arginine finger’ that participates in energy coupling (48,49). This RecA-like structure in superfamily 4 is defined by five conserved sequence motifs (H1, H1a, H2, H3 and H4) located in the C-terminal region of the protein (50). Motifs H1 and H2 are equivalent to the Walker A and B motifs.
To investigate the synergistic effect between nucleotide and ssDNA binding in the hexameric helicase, we examined the local environment around the NTP-binding pocket. In this study, we found two conformations of the p-loop in the Walker A motif by superimposing DnaB-like helicases from several organisms, including GkDnaC, the G40P ATPase domain and bacteriophage T7 gp4 (Figure 4B). With the exception of apo-GkDnaC, all of these DnaB-like helicases were crystallized in the presence of a nucleotide. However, only bacteriophage T7 gp4 and the G40P ATPase domain show that a nucleotide is present in the ATP binding site (18,19). Surprisingly, in the presence of nucleotide the orientation of the p-loops in the GkDnaC–ssDNA complex is very similar to that of T7 gp4 and the G40P ATPase domain, whereas the p-loops in apo-GkDnaC are oriented in the opposite direction (Figure 4B). This finding suggests that the variable p-loop is a feature associated with nucleotide binding, and thus the p-loop orientation we observed in the complex structure may represent the ATP-bound state. In addition, the Walker B motif, H2, is associated with NTP binding in the RecA-like core (48). Interestingly, our ssDNA–GkDnaC complex crystal structure reveals that loop I, part of the Walker B motif, adjoins the ATP-binding site with the bound ssDNA (Figure 4C). Therefore, we speculate that the orientation of the flanked DNA-binding loop I might be influenced by the Walker B motif upon NTP-binding. This may also explain why NTP binding enhances ssDNA binding (24,51–54).
ssDNA binding mode
Prior to this study, papillomavirus E1 helicase (of superfamily 3) was the only replicative helicase whose structure had been solved in complex with ssDNA; the structure suggested a spiral ‘staircase’ model for interaction with the ssDNA backbone (20). The directionality of translocation in superfamily 3 is 3′-5′. Unlike the E1 helicase, 5′-3′ is the direction of translocation permitted in GkDnaC. Sequence analysis also showed that the identity between GkDnaC and E1 helicase is only 16% (Supplementary Figure 2B). In addition, the hexamer channel in the E1 helicase at both the NTD and CTD collar is much narrower than that of eubacteria hexameric DnaB-like helicases belonging to the superfamily 4 helicases (18,21). As shown in Figure 5, the central channel diameter of the E1 helicase is ∼13 Å, whereas GkDnaC has a wider central channel at the NTD collar (∼50 Å) and a narrower channel at the CTD collar (∼20–30 Å). It should be mentioned that several loop regions expected to be involved in ssDNA binding near the C-terminal channel of GkDnaC were too flexible to be reliably assigned, and therefore the exact dimensions of the CTD collar may be less than that we observed in the current structure. Also, the E1–ssDNA complex structure only showed 7-mer ssDNA from a 13-mer oligo-dT oligonucleotide in the C-terminal collar (Figure 5). In contrast to ssDNA binding in the E1 helicase, the NTD collar and the top region of the CTD collar of GkDnaC were both involved in ssDNA binding. Because of these differences in overall architecture, low sequence identity, size of central hole and biochemical properties, the ssDNA-binding mechanism of eubacteria DnaB-like helicase in superfamily 4 maybe differ from that of the E1 helicase. However, due to the lack of biochemical evidence, we still cannot rule out that the mechanism of ssDNA binding and translocation in the replication fork may be similar to that of E1 helicase.
Figure 5.
Quaternary structure comparison of the GkDnaC–ssDNA and E1–ssDNA complexes. Ribbon representations of GkDnaC and papillomavirus E1 helicases are colored in green and cyan, respectively. The ssDNA is colored in orange. The DNA-binding site observed in each structure is located in a different domain. The diameter of the central hole of the GkDnaC N-terminal collar (∼50 Å) is wider than that of papillomavirus E1 helicase (∼13 Å).
A model for ssDNA binding in the N-terminal region during DNA translocation
In the structure of the E1 helicase–ssDNA complex, the ssDNA forms a right-handed helical arrangement that interacts with the DNA-binding loops of the hexameric channel (20). The structure of the T7 gene 4 helicase ring in complex with a nucleotide suggests that the ssDNA could form a spiral conformation to interact with the ssDNA-binding loops of the channel (19). In addition, previous studies revealed that ssDNA passes through the central hole of the T7 gene 4 helicase ring with a contour length of ∼3 Å per nucleotide (24,40). This indicates that the conformation of the ssDNA in the channel primarily forms a helical shape. Therefore, we speculate that the ssDNA located at the central channel that is surrounded by the helicase domain also forms a right-handed helical arrangement in GkDnaC. The N-terminal region of DnaB is important for helicase activity and DNA-binding affinity (43,55). In addition, our GkDnaC–ssDNA complex structure shows that three ssDNA molecules are bound to the N-terminal DNA-binding sites. Hence, it is possible that the ssDNA that emanates from the helicase domain could interact with the DNA-binding sites of the NTD collar in the process of DNA translocation. Due to the spiral conformation of ssDNA in the E1 helicase, it is also likely that the sequence of ssDNA interacting with three N-terminal DNA-binding sites is dependent on the right-handed helical arrangement of ssDNA in the CTD collar.
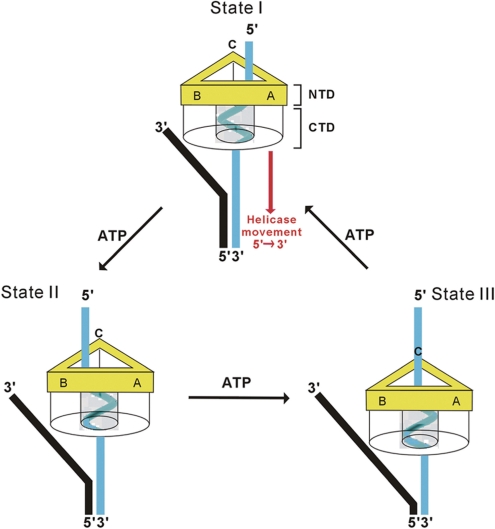
Taken together, we propose a model for the DnaB-like proteins where the ssDNA emanating from the C-terminal helicase channel binds to the N-terminal DNA-binding sites in a clockwise fashion. As shown in Figure 6, GkDnaC binds to the replication fork and moves along the ssDNA in a 5′ to 3′ direction. Three ssDNA-binding sites are located in the N-terminal region of the GkDnaC. In step I, the ssDNA of the replication fork is passed through the hexameric channel. The ssDNA adopts a spiral conformation to interact with the DNA-binding surface of the C-terminal region of the hexameric channel. At this stage, the ssDNA located at the N-terminal region is close to the ssDNA-binding site A. It has been shown that a high-affinity DNA-binding site is close to the N-terminal region of DnaB (43). Thus, this site could allow ssDNA to bind, therefore creating a stable interaction and providing a basic track leading the ssDNA to move toward the position of DnaG primase. When the protein hydrolyzes ATP, this could provide the energy necessary to drive the sequential movement of helicase. Thus, the protein arrives at the state shown in step II, where the ssDNA in the N terminus of the protein approaches ssDNA-binding site B. The cycle of changes would repeat to progress from step II to step III. Consequently, the ssDNA emanating from the helicase domain remains in contact with at least one of the three ssDNA-binding sites in the N-terminal region at all stages in the process, providing increased stability for the ssDNA and accurately directing the ssDNA toward the DnaG primase.
Figure 6.
Model for ssDNA-binding during DNA translocation. A hexameric DnaB-like helicase binds to the replication fork and unwinds it in the 5′ to 3′ direction. The N-terminal collar of the helicase is colored in yellow. For clarity, the C-terminal collar is shown as transparent. The ssDNA that passes through the C-terminal collar is represented by a cyan-colored right-handed spiral. The leading strand and lagging strand of the replication fork are colored in black and cyan, respectively. In State I, the ssDNA emanates from the C-terminal collar and interacts with the DNA-binding pocket A of the N-terminal collar. ATP hydrolysis drives the movement of the helicase toward the 3′ end of the lagging strand and converts the ssDNA into State II. Because the ssDNA (State II) that emanates from the C-terminal collar is close to the DNA-binding pocket B, this pocket is responsible for stabilizing the ssDNA in this state. Following State II, DNA-binding pocket C interacts with the ssDNA to provide stability for the ssDNA (State III).
We have solved the crystal structures of GkDnaC in the apo-form and in a complex with 15-mer-oligodeoxythymidylate ssDNA. In the complex structure, the hexamer consists of three positive electrostatic potential grooves on the interior surface of the helicase ring, and each of which encircles one ssDNA molecule. This ssDNA-binding pocket may guide the ssDNA toward the DnaG primase to synthesize short RNA primers. We also demonstrated by mutagenesis studies and SPR analysis that several key residues surrounding the ssDNA-binding pocket play critical roles in DNA binding and stabilization. In summary, we provide the first ssDNA-bound structure of a eubacterial hexameric ring helicase and propose a possible model for the mechanism of DNA translocation in the replication fork.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academia Sinica and the National Science Council (NSC95-2311-B-001-064–MY3 to C.-D.H.), Taiwan, Republic of China. Funding for open access charge: Academia Sinica.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for access to the synchrotron radiation beamline 13B1 at the National Synchrotron Radiation Research Center in Taiwan.
REFERENCES
- 1.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu. Rev. Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 2.Baker TA, Bell SP. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 3.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 4.Tougu K, Peng H, Marians KJ. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J. Biol. Chem. 1994;269:4675–4682. [PubMed] [Google Scholar]
- 5.Frick DN, Richardson CC. DNA primases. Annu. Rev. Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 6.Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- 7.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 8.Zechner EL, Wu CA, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. III. A polymerase-primase interaction governs primer size. J. Biol. Chem. 1992;267:4054–4063. [PubMed] [Google Scholar]
- 9.Wu CA, Zechner EL, Hughes A.J., Jr, Franden MA, McHenry CS, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. IV. Reconstitution of an asymmetric, dimeric DNA polymerase III holoenzyme. J. Biol. Chem. 1992;267:4064–4073. [PubMed] [Google Scholar]
- 10.Donmez I, Patel SS. Mechanisms of a ring shaped helicase. Nucleic Acids Res. 2006;34:4216–4224. doi: 10.1093/nar/gkl508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bird LE, Pan H, Soultanas P, Wigley DB. Mapping protein-protein interactions within a stable complex of DNA primase and DnaB helicase from Bacillus stearothermophilus. Biochemistry. 2000;39:171–182. doi: 10.1021/bi9918801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaeffer PM, Headlam MJ, Dixon NE. Protein-Protein Interaction in the Eubacterial replisome. IUBMB Life. 2005;57:5–12. doi: 10.1080/15216540500058956. [DOI] [PubMed] [Google Scholar]
- 13.Velten M, McGovern S, Marsin S, Ehrlich SD, Noirot P, Polard P. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol. Cell. 2003;11:1009–1020. doi: 10.1016/s1097-2765(03)00130-8. [DOI] [PubMed] [Google Scholar]
- 14.Soultanas P. A functional interaction between the putative primosomal protein DnaI and the main replicative DNA helicase DnaB in Bacillus. Nucleic Acids Res. 2002;30:966–974. doi: 10.1093/nar/30.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 16.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 17.Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318:459–463. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 18.Wang G, Klein MG, Tokonzaba E, Zhang Y, Holden LG, Chen XS. The structure of a DnaB-family replicative helicase and its interactions with primase. Nat. Struct. Mol. Biol. 2008;15:94–100. doi: 10.1038/nsmb1356. [DOI] [PubMed] [Google Scholar]
- 19.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 20.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 21.Bailey S, Eliason WK, Steitz TA. The crystal structure of the Thermus aquaticus DnaB helicase monomer. Nucleic Acids Res. 2007;35:4728–4736. doi: 10.1093/nar/gkm507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahnert P, Patel SS. Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J. Biol. Chem. 1997;272:32267–32273. doi: 10.1074/jbc.272.51.32267. [DOI] [PubMed] [Google Scholar]
- 23.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 24.Hingorani MM, Patel SS. Interactions of bacteriophage T7 DNA primase/helicase protein with single-stranded and double-stranded DNAs. Biochemistry. 1993;32:12478–12487. doi: 10.1021/bi00097a028. [DOI] [PubMed] [Google Scholar]
- 25.Arai K, Kornberg A. Mechanism of dnaB protein action. III. Allosteric role of ATP in the alteration of DNA structure by dnaB protein in priming replication. J. Biol. Chem. 1981;256:5260–5266. [PubMed] [Google Scholar]
- 26.Arai K, Kornberg A. Mechanism of dnaB protein action. II. ATP hydrolysis by dnaB protein dependent on single- or double-stranded DNA. J. Biol. Chem. 1981;256:5253–5259. [PubMed] [Google Scholar]
- 27.Liu CC, Alberts BM. Characterization of the DNA-dependent GTPase activity of T4 gene 41 protein, an essential component of the T4 bacteriophage DNA replication apparatus. J. Biol. Chem. 1981;256:2813–2820. [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–327. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McRee DE. XtalView/Xfit -a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 34.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao Y, White CL, Luger K. Nucleosome core particles containing a poly(dA.dT) sequence element exhibit a locally distorted DNA structure. J. Mol. Biol. 2006;361:617–624. doi: 10.1016/j.jmb.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 38.DeLano WL. The pyMOL User's Manual. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 39.Konieczny I. Strategies for helicase recruitment and loading in bacteria. EMBO Rep. 2003;4:37–41. doi: 10.1038/sj.embor.embor703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egelman EH, Yu X, Wild R, Hingorani MM, Patel SS. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl Acad. Sci. USA. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaplan DL, O'Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 42.Bujalowski W, Jezewska MJ. Interactions of Escherichia coli primary replicative helicase DnaB protein with single-stranded DNA. The nucleic acid does not wrap around the protein hexamer. Biochemistry. 1995;34:8513–8519. doi: 10.1021/bi00027a001. [DOI] [PubMed] [Google Scholar]
- 43.Jezewska MJ, Rajendran S, Bujalowski W. Functional and structural heterogeneity of the DNA binding site of the Escherichia coli primary replicative helicase DnaB protein. J. Biol. Chem. 1998;273:9058–9069. doi: 10.1074/jbc.273.15.9058. [DOI] [PubMed] [Google Scholar]
- 44.Jezewska MJ, Kim US, Bujalowski W. Binding of Escherichia coli primary replicative helicase DnaB protein to single-stranded DNA. Long-range allosteric conformational changes within the protein hexamer. Biochemistry. 1996;35:2129–2145. doi: 10.1021/bi952345d. [DOI] [PubMed] [Google Scholar]
- 45.Notarnicola SM, Park K, Griffith JD, Richardson CC. A domain of the gene 4 helicase/primase of bacteriophage T7 required for the formation of an active hexamer. J. Biol. Chem. 1995;270:20215–20224. doi: 10.1074/jbc.270.34.20215. [DOI] [PubMed] [Google Scholar]
- 46.Washington MT, Rosenberg AH, Griffin K, Studier FW, Patel SS. Biochemical analysis of mutant T7 primase/helicase proteins defective in DNA binding, nucleotide hydrolysis, and the coupling of hydrolysis with DNA unwinding. J. Biol. Chem. 1996;271:26825–26834. doi: 10.1074/jbc.271.43.26825. [DOI] [PubMed] [Google Scholar]
- 47.Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 48.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 50.Ilyina TV, Gorbalenya AE, Koonin EV. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J. Mol. Evol. 1992;34:351–357. doi: 10.1007/BF00160243. [DOI] [PubMed] [Google Scholar]
- 51.Picha KM, Patel SS. Bacteriophage T7 DNA helicase binds dTTP, forms hexamers, and binds DNA in the absence of Mg2+. The presence of dTTP is sufficient for hexamer formation and DNA binding. J. Biol. Chem. 1998;273:27315–27319. doi: 10.1074/jbc.273.42.27315. [DOI] [PubMed] [Google Scholar]
- 52.Matson SW, Richardson CC. Nucleotide-dependent binding of the gene 4 protein of bacteriophage T7 to single-stranded DNA. J. Biol. Chem. 1985;260:2281–2287. [PubMed] [Google Scholar]
- 53.Yong Y, Romano LJ. Nucleotide and DNA-induced conformational changes in the bacteriophage T7 gene 4 protein. J. Biol. Chem. 1995;270:24509–24517. doi: 10.1074/jbc.270.41.24509. [DOI] [PubMed] [Google Scholar]
- 54.Jezewska MJ, Bujalowski W. Global conformational transitions in Escherichia coli primary replicative helicase DnaB protein induced by ATP, ADP, and single-stranded DNA binding. Multiple conformational states of the helicase hexamer. J. Biol. Chem. 1996;271:4261–4265. doi: 10.1074/jbc.271.8.4261. [DOI] [PubMed] [Google Scholar]
- 55.Biswas EE, Biswas SB. Mechanism of DnaB helicase of Escherichia coli: structural domains involved in ATP hydrolysis, DNA binding, and oligomerization. Biochemistry. 1999;38:10919–10928. doi: 10.1021/bi990048t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.