Abstract
We present a new methodology for site-specific sensing of peptide–oligonucleotide (ODN) interactions using a solvatochromic fluorescent label based on 3-hydroxychromone (3HC). This label was covalently attached to the N-terminus of a peptide corresponding to the zinc finger domain of the HIV-1 nucleocapsid protein (NC). On interaction with target ODNs, the labeled peptide shows strong changes in the ratio of its two emission bands, indicating an enhanced screening of the 3HC fluorophore from the bulk water by the ODN bases. Remarkably, this two-color response depends on the ODN sequence and correlates with the 3D structure of the corresponding complexes, suggesting that the 3HC label monitors the peptide–ODN interactions site-specifically. By measuring the two-color ratio, we were also able to determine the peptide–ODN-binding parameters and distinguish multiple binding sites in ODNs, which is rather difficult using other fluorescence methods. Moreover, this method was found to be more sensitive than the commonly used steady-state fluorescence anisotropy, especially in the case of small ODNs. The described methodology could become a new universal tool for investigating peptide–ODN interactions.
INTRODUCTION
Fluorescence is a powerful tool for investigating biomolecular interactions. The most widely used technique in this respect is FRET that allows measuring the distance between two fluorophores (1–5). However, FRET shows limitations since it requires double labeling. A single-labeling technique commonly used for the same purpose is steady-state fluorescence anisotropy (6,7), which senses changes in the fluorophore mobility upon interactions. Despite its wide use in bio-sensing technologies, fluorescence anisotropy gives frequently limited response on binding, since it depends on the fluorescence lifetime of the probe and different types of molecular motions such as the local motion of the probe and both segmental and overall motions of the labeled biomolecule.
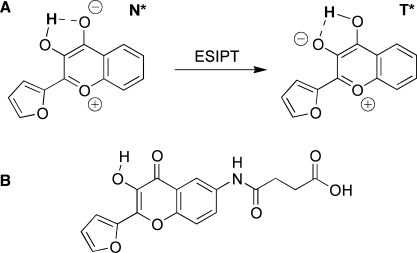
Environment-sensitive (or solvatochromic) fluorescent dyes, which monitor biomolecular interactions by sensing environment changes at the labeled site, become an attractive alternative to fluorescence anisotropy in the recent years. A series of environment-sensitive fluorescent labels, such as Prodan derivatives (8–10), dimethylaminophtalimide (11), dimethylaminonaphtamides (12,13) and others (14) have recently been applied to study protein–protein interactions and protein conformational transitions (8,9,11,12,14). Short peptides labeled by these dyes were successfully used to study phosphorylation-dependent peptide–protein interactions (10), δ-opioid antagonist binding (15) and peptide binding to proteins of a major histocompatibility complex (MHC) at the cell surface (16). Biomolecular interactions commonly decrease the polarity at the labeling site due to screening from water by the binding of the protein partner (16). This change in the polarity can be readily detected by environment-sensitive labels through shifts in their emission maximum or changes in their fluorescence intensity. However, applications of these dyes for sensing interactions of peptides with oligonucleotides (ODNs) have not been described so far, likely because the ODN environment is relatively polar (17,18) so that polarity may not be dramatically affected by the interaction. In addition, most of the mentioned environment-sensitive dyes show relatively low quantum yields in polar protic media (12), and ODN bases frequently play a role of a fluorescence quencher (19,20). Therefore, for studying peptide–ODN interactions we have selected a 3-hydroxychromone (3HC) derivative, 2-(2-furyl)-3HC, which having a satisfactory quantum yield in polar protic solvents, shows a high sensitivity of its dual emission to environment changes in polar media (21,22). 3HC dyes undergo excited-state intramolecular proton transfer (ESIPT) (23) (Figure 1) resulting in the emission of both the normal (N*) excited state and the ESIPT product photo-tautomer (T*). The dual emission of 3HC dyes is highly sensitive to polarity and H-bonding interactions (21,24–29). Increase in the polarity and H-bond donor ability of solvents inhibits the ESIPT reaction and thus decreases the relative intensity of the ESIPT product (T*) (27). Moreover, the position of the maximum of the T* band, being insensitive to solvent polarity, exhibits high sensitivity to H-bond donor ability (21). Thus, the present class of fluorophores provides two independent information channels allowing a thorough characterization of the environment. These unique properties of 3HCs have already been applied for probing proteins, lipid bilayers and cell membranes as well as for monitoring protein–protein and polycation–DNA interactions (30–37).
Figure 1.
Proton transfer reaction of a 3HC dye in the excited state (A) and chemical structure of the used 3HC label (B).
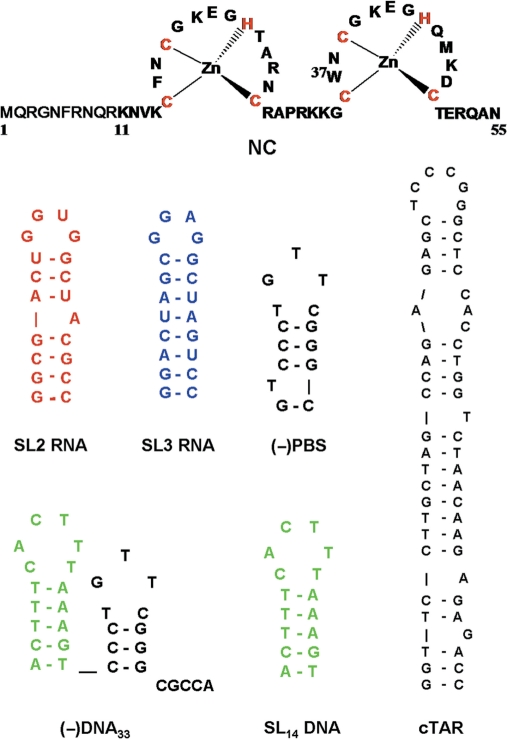
As a target protein for labeling with our dye, we selected the nucleocapsid protein (NC) of the human immunodeficiency virus, type 1 (HIV-1). NC is a small (55 amino acids) basic protein, characterized by two rigid retroviral-type zinc fingers connected by a flexible basic linker and flanked by poorly folded N- and C-terminal basic domains (Figure 2) (38–40). NC is thought to be critically involved in the reverse transcription, integration and encapsidation steps of the viral life cycle, mainly through interaction with nucleic acids (41). NC binds both specifically (42–45) and non-specifically (46,47), to a large range of nucleic-acid sequences. Specific binding to the ψ encapsidation sequence is required for the selective recognition of viral RNA among the large excess of cellular RNAs (48,49). Selectivity is supported by the interaction of the hydrophobic platform at the top of NC folded fingers with the GXG- containing loops of the SL2 and SL3 stem loop sequences of ψ RNA (50–53). In contrast, the coating of the viral RNA by about 2000 copies of NC relies largely on NC ability to bind to nearly any sequence of five to seven nucleotides length (44,47,50,54,55). NC also exhibits nucleic-acid chaperone properties (56,57), which rely on its ability to transiently melt the secondary structure and to activate the annealing of complementary ODNs (58–70). These properties are thought to be essential during reverse transcription, to anneal the primer tRNALys,3 to the RNA primer-binding site (PBS) and to promote the two obligatory strand transfers (71,72) which are required for copying the HIV-1 RNA genome into double-stranded DNA.
Figure 2.
Sequences of the NC protein and the ODNs used in this study. Secondary structures are based on ref. (52,62,66,75,78,79) and m-fold predictions.
In the present work, to explore the potency of the environment sensitive 3HC dyes for sensing peptide–ODN interactions, we coupled a newly designed 3HC label selectively to the N-terminus of the NC (11–55) peptide using solid phase peptide synthesis. This peptide corresponding to the domain of NC fingers was preferred to the native NC since it preserves the nucleic-acid binding and chaperone properties of the protein (65–67,73) but does not aggregate the ODNs (74). The response of the labeled peptide on interaction with SL2 RNA, SL3 RNA, (−)PBS DNA and d(ACGCC) (Figure 2) was investigated and correlated with the known 3D structure of these complexes (52,53,76,77) as well as with the fluorescence anisotropy response of fluorescein-labeled NC(11–55) in the same complexes. Finally, the probe was applied to demonstrate the existence of preferential binding sites on ODNs with multiple NC-binding sites.
MATERIALS AND METHODS
Materials
Reagents were from Merck, Sigma-Aldrich or Applied Bio Systems (Foster City, USA). ODNs were synthesized and HPLC-purified by IBA GmbH (Germany). Their sequences are given in Figure 2. ODN concentrations were determined using the following extinction coefficients at 260 nm (ε260, M−1 × cm−1): 48 360, 58 050, 231 000, 246 000, 168 000, 178 000, 325 000 and 521 900 M−1 × cm−1 for d(ACGCC), d(AACGCC), SL2 RNA, SL3 RNA, SL14 DNA, (−)PBS DNA, (−)DNA33 and cTAR DNA, respectively.
Synthesis of N-(2-furan-2-yl-3-hydroxychromon-6-yl)-succinamic acid
Total 0.2 g (0.82 mmol) of 6-amino-2-furan-2-yl-3HC (37) was dissolved in 10 ml dry DMF and 0.09 g (0.9 mmol) of succinic anhydride was added to this solution. The mixture was left for stirring overnight. Then, it was poured into water and filtrated to give 0.235 g (84%) of the final acid. 1H NMR (300 MHz, DMSO-d6) δ 10.27 (s, 1H, NH), 8.44 (d, J = 3.5 Hz, 1H, ArH), 8.01 (s, 1H, ArH), 7.89 (d, J = 9 Hz, 1H, ArH), 7,67 (d, J = 9 Hz, 1H, ArH), 7.28 (d, J = 2.5 Hz, 1H, ArH), 6.78 (s, 1H, ArH), 2.59–2.50 (m, 4H, CH2CH2).
3HC-NC(11–55)
The NC(11–55) peptide (Figure 2) was synthesized by solid phase peptide synthesis on a 433A synthesizer (ABI, Foster City, CA) as previously described (80). The synthesis was performed at a 0.1 mmol scale using the standard fluorenylmethoxycarbonyl (Fmoc)-amino-acid-coupling protocol starting from 0.54 mmol/g HMP Asn-preloaded resin (ABI). At the end of the synthesis, 100 mg of Fmoc-deprotected peptidylresin was isolated, and washed twice by NMP. Four equivalents of the label [N-(2-furan-2-yl-3-hydroxychromon-6-yl)-succinamic acid] were mixed with 4 eq. of HBTU/HOBt coupling solution (in DMF) and 5 eq. of DIEA. This mixture was immediately added to the peptidylresin and stirred at 40°C for 40 min. Resin was filtrated and washed by NMP and methanol.
Cleavage of the peptidylresin and deprotection was performed for 2 h using a 10 ml trifluoroacetic acid (TFA) solution containing water (5%, v/v), phenol (2%, w/v), thioanisole (5%, v/v) and ethanedithiol (2.5%, v/v). Solution was concentrated in vacuo and the peptide was precipitated using ice-cold diethyl ether and pelleted by centrifugation. The pellet was then washed with diethyl ether and dried before solubilization with aqueous TFA (0.05%, v/v). Purification by HPLC was carried out on a C8 column (uptisphere 300A, 5 µm; 250 × 10, Interchim, France) in water/acetonitrile mixture containing 0.05% TFA with a linear gradient 10–70% of acetonitrile for 30 min and monitored at 360 nm (3HC dye absorption). Molecular mass found by ion spray mass spectrometry (5463) corresponds to the calculated value.
Fl-NC(11–55)
Peptide synthesis was performed as for 3HC-NC(11–55), except that labeling was done with 4 eq. of 5(6)-carboxyfluorescein (Fl) overnight. In the crude mixture, two fluorescein-containing peptides in the ratio 10:1 were found. The main product was isolated by HPLC using the same conditions as for 3HC-NC(11–55). Molecular mass found by ion spray mass spectrometry (5496) corresponds to the calculated value.
Preparation of Zn-bound peptides
Lyophilized peptides were dissolved in water (∼0.5 mg in 500 µl). Then, the peptide concentration was determined using an extinction coefficient of 15 000 M−1 × cm−1 at 350 nm for 3HC-NC(11–55) and 86 000 M−1 × cm−1 at 500 nm for Fl-NC(11–55). Next, 2.2 molar equivalents of ZnSO4 were added to the peptide and pH was raised to its final value, by adding buffer. The increase of pH was done only after zinc addition to avoid oxidization of the zinc-free peptide. Noticeably, addition of a large excess of Zn2+ ions should be avoided since it can affect the 3HC fluorescence properties.
3HC-G5
The pentaglycine peptide was synthesized as described above. N-terminal labeling by N-(2-Furan-2-yl-3-hydroxychromon-6-yl)-succinamic acid was performed with 1.5 eq of the label, 1.5 eq of HBTU/HOBt coupling solution (in DMF) and 3 eq of DIEA overnight. Cleavage was done using 10 ml TFA containing water (10% v/v) and triisopropilsilane (2.5%, v/v). The product was purified by HPLC using the same conditions as for 3HC-NC(11–55) but with a linear gradient 10–50% of acetonitrile for 20 min.
Spectroscopic measurements
Unless otherwise indicated, the experiments were performed in 10 mM phosphate buffer, pH 7.0, 100 mM NaCl, at 20°C.
Absorption spectra were recorded with a Cary 4000 UV-visible spectrophotometer (Varian). Fluorescence spectra were recorded on FluoroMax3 and FluoroLog spectrofluorimeters (Jobin Yvon) equipped with thermostated cell compartments. Steady-state anisotropy was measured on SLM 8000 spectrofluorometer (Aminco, Urbana, IL). Fluorescence spectra were corrected for Raman scattering. Quantum yields were calculated using quinine sulphate in 0.5 M sulphuric acid (quantum yield, φ = 0.577) as a ref. (81). Excitation wavelength was 340 nm for the 3HC-label and 480 nm for the fluorescein label. To determine the affinity of 3HC-NC(11–55) for the ODNs, fixed amounts of the peptide were titrated with ODNs by monitoring the 3HC two-band fluorescence. Affinity constants were determined from direct fitting of the experimental signal to the rewritten Scatchard equation:
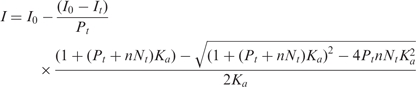 |
1 |
where I and It are the signal (the N*:T* intensity ratio) at a given and a saturating ODN concentration, respectively, I0 is the signal in the absence of ODN, Nt is the total ODN concentration, Pt is the total concentration of peptide, Ka is the apparent affinity constant, n is the number of binding sites. The parameters were recovered from non-linear fits of Equation (1) to experimental datasets by the Microcal OriginTM 6.0 program.
Time-resolved fluorescence measurements were performed with the time-correlated, single-photon counting technique using the excitation pulses at 480 nm provided by a pulse-picked frequency doubled Ti-sapphire laser (Tsunami, Spectra Physics) pumped by a Millenia X laser (Spectra Physics). The emission was collected through a polarizer set at the magic angle and an 8 nm band-pass monochromator (Jobin-Yvon H10) at 520 nm. The instrumental response function was recorded with a polished aluminium reflector, and its full-width at half-maximum was 40 ps.
For time-resolved anisotropy measurements, the fluorescence decay curves were recorded at the vertical and horizontal positions of the polarizer as described (82) and analyzed by the following equation:
| 2 |
where βi are the amplitudes of the rotational correlation times θi; I|| and I⊥ are the intensities collected at emission polarizations parallel and perpendicular, respectively, to the polarization axis of the excitation beam, and G is the geometry factor at the emission wavelength, determined in independent experiments.
Time-resolved intensity and anisotropy data were treated with a non-linear least-square analysis using a home-made software (kindly provided by G. Krishnamoorthy) and the Maximum Entropy Method (MEM) (83). In all cases, the χ2 values were close to 1, and the weighted residuals as well as the autocorrelation of the residuals were distributed randomly around 0, indicating an optimal fit.
RESULTS AND DISCUSSION
Spectroscopic characteristics of the free 3HC label
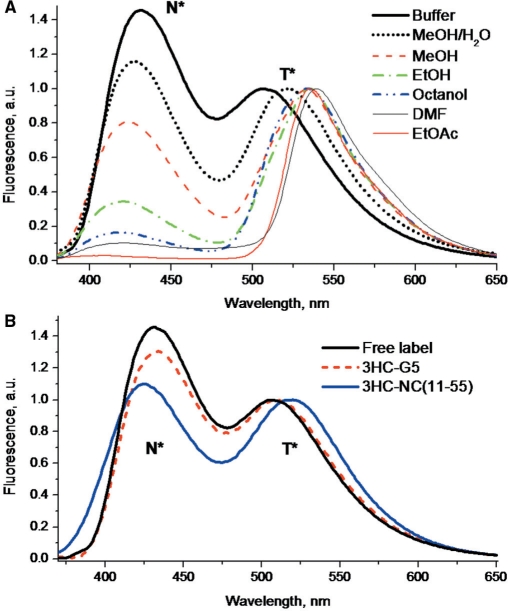
For labeling the N-terminus of NC(11–55), we have synthesized a 3HC derivative bearing a carboxyl functionality. First, we characterized the spectroscopic properties of this label in different organic solvents. The fluorescence spectra of the label are composed of two bands (Figure 3A), where the short- and long-wavelength bands can be unambiguously attributed to the emission of the N* and T* forms of the fluorophore, respectively (Figure 1).
Figure 3.
Fluorescence spectra of the free 3HC label in different solvents (A) and the labeled peptide 3HC-NC(11–55) in buffer (B). The spectra of the free label and the labeled model peptide (3HC-G5) in buffer are presented for comparison. All the spectra were normalized at the T* band. Excitation wavelength was 340 nm. Buffer was 10 mM phosphate buffer, 100 mM NaCl, pH 7.0.
The nature of the solvent affects strongly the dual emission of the label. In a non-polar aprotic solvent ethyl acetate, the label shows a very low ratio of the N* to T* fluorescence intensities. More polar aprotic solvent DMF induces an increase of the N*:T* band ratio. In protic solvents, the N*:T* intensity ratio is much higher than in aprotic solvents, indicating that the H-bond donor ability of the former inhibits the ESIPT reaction, and thus decreases the emission of the ESIPT product T* (21,84). Moreover, the band ratio increases with polarity in alcohols and reaches its highest value in buffer (Figure 3A, Table 1). The high value of the N*:T* ratio in buffer was independent on NaCl concentration (data not shown), indicating that it is due to water. In addition, due to its strong H-bond donating properties, water induces an outstanding blue shift of the T* band in comparison with alcohols or aprotic solvents. The observed increase of the N*:T* intensity ratio with the polarity and H-bond donor ability of the solvents is fully in line with that reported for the parent non-substituted 2-(2-furyl)-3-HC (21,84). Therefore, being attached to a peptide, the present label is expected to report on the accessibility of the site of labeling to bulk water (which is characterized by high polarity and H-bond donor ability) by its N*:T* ratio and the position of the T* band.
Table 1.
Spectroscopic properties of the free label and the labeled peptidesa
| Solvent | λAbs (nm) | λN* (nm) | λT* (nm) | N*:T* | QY (%) | |
|---|---|---|---|---|---|---|
| Label | Buffer | 357 | 431 | 508 | 1.47 | 2.0 |
| MeOH/H2O | 354 | 427 | 521 | 1.16 | 3.2 | |
| MeOH | 349 | 423 | 533 | 0.81 | 5.6 | |
| EtOH | 351 | 420 | 535 | 0.34 | 6.1 | |
| Octanol | 356 | 419 | 534 | 0.16 | 13.0 | |
| DMF | 344 | 421 | 539 | 0.10 | 6.9 | |
| EtOAc | 345 | 410 | 535 | 0.03 | 13.0 | |
| 3HC-G5 | Buffer | 360 | 432 | 510 | 1.33 | 3.3 |
| 3HC-NC(11–55) | Buffer | 358 | 426 | 517 | 1.12 | 7.8 |
aλAbs, λN* and λT* are the maxima of absorption, N* and T* emission bands respectively. N*:T* is the intensity ratio of the two emission bands measured at the peak maxima; QY is the fluorescence quantum yield. MeOH/H2O is 1/1 methanol–water mixture. EtOAc is ethyl acetate. Excitation wavelength was 340 nm. The peptides were in 10 mM phosphate buffer, 100 mM NaCl, pH 7.0.
Synthesis and characterization of the labeled NC(11–55) peptide
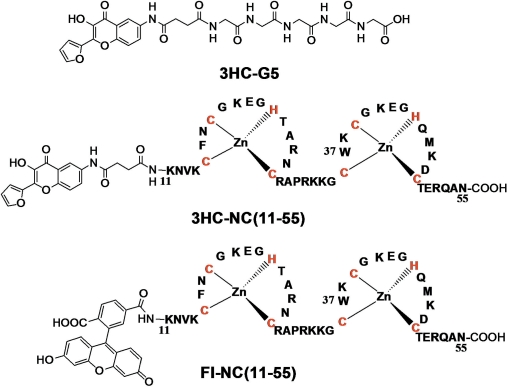
To achieve selective labeling, NC(11–55) peptide was prepared by solid phase synthesis (80) and being attached to the resin was further labeled at its N-terminus with the carboxylic-acid derivative of 3HC. The HPLC-purified labeled peptide was checked by mass spectrometry, confirming that the expected labeled peptide was obtained. The chaperone properties of the labeled peptide were tested using previously described protocols (62,64,67) and were found comparable to those of the native peptide (Figures S1 and S2 in Supplementary Data), indicating that the 3HC label does not interfere with the peptide activity. Moreover, since these chaperone properties are exquisitely sensitive on the proper folding of NC(11–55) (73), it can be further concluded that the 3HC label does not alter the folding of the peptide. Thus, as was expected from its relatively small size and N-terminal location with a four amino-acid separation from the proximal Zn finger, the probe does not perturb the activity and the folding of the peptide.
The emission spectrum of 3HC-NC(11–55) exhibited significant differences with the spectrum of the free label in water, with a decrease of the N*:T* ratio from 1.47 to 1.12 and a 9 nm red-shift of the T* band (Figure 3B, Table 1). Moreover, its fluorescence quantum yield was nearly 4-fold higher than that of the free label in water. According to the data of the probe in model solvents (Table 1) and our previous data (21,84), these changes indicate a decrease in the polarity and/or H-bond donor ability in the probe environment. This decrease was independent of the peptide concentration in the 50–1000 nM range (data not shown) and was thus interpreted as an intramolecular screening of the label from the bulk water in conformations where the probe is in proximity to the KNVK(11–14) sequence and to the zinc finger domain (Figure 4) of the labeled peptide. Indeed, a large collection of conformations is probably explored by the N-terminal domain of NC(11–55) as a consequence of the flexibility of the spacer connecting the label to the peptide and the flexibility of the KNVK(11–14) sequence to which the label is linked (39,40). Noticeably, both the N*:T* band ratio and the T* band position of 3HC-NC were marginally dependent on the NaCl concentration (data not shown), indicating that the intramolecular screening of the dye from water was poorly affected by the ionic strength.
Figure 4.
Labeled peptides.
To further assess the effect of the peptide backbone on the fluorescence properties of the 3HC label, we coupled it to a pentaglycine peptide (3HC-G5) using the same solid phase-based method (Figure 4). As with NC(11–55), labeling of G5 induced a decrease of the N*:T* ratio (from 1.47 to 1.33) and a red-shift (2 nm) of the T* band in respect with the spectrum of the free label in water (Figure 3B, Table 1), as well as a significant fluorescence quantum yield increase. Though less pronounced than with NC(11–55), these changes indicate that interactions of the probe with the flexible backbone of the G5 peptide can reduce the overall accessibility of the probe for water molecules. The more efficient screening from the bulk water seen with NC(11–55) as compared to G5, is likely due to the amino-acid side chains of NC(11–55), which are absent in the G5 peptide.
Interactions of 3HC-NC(11–55) with single binding site ODNs
To explore the applicability of the 3HC label to probe peptide–ODN interactions, we first characterized the interaction of the labeled peptide with d(ACGCC), SL2 RNA, SL3 RNA and (−)PBS DNA (Figure 2). These sequences were selected since they preferentially bind one NC molecule per ODN and the 3D structures of their complexes with NC have been solved (52,53,76,77).
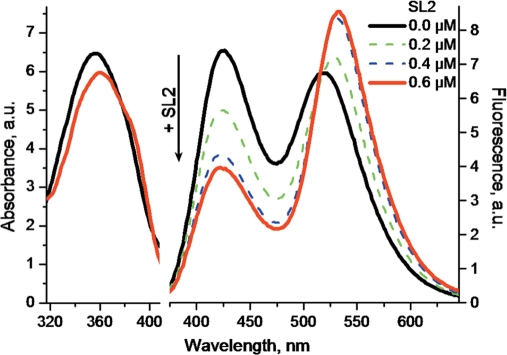
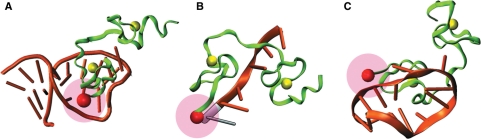
All ODNs induced a substantial decrease of the N*:T* ratio and a red-shift of the T* band in respect with the free 3HC-NC(11–55) peptide (Table 2), indicating an increased screening of the probe from the bulk water. Competition experiments using the SL3 sequence showed that the labeled and the non-labeled peptides bind to the same binding site (Figure S3 in Supplementary Data), suggesting that the 3HC label does not change the binding specificity. Remarkably, large spectroscopic differences were observed among the tested ODNs, underlining differences in the interaction of the N-terminal part of the peptide with these ODNs. The strongest changes were observed with the SL2 stem-loop, which binds strongly NC at the level of its loop (52). With SL2, the N*:T* intensity ratio dropped to 0.46 and the T* band shifted 16 nm to the red (Figure 5, Table 2). The resulting spectrum and quantum yield were close to that of the free probe in ethanol, indicating a strong screening of the label from water in the NC(11–55)/SL2 complex, with marginal quenching of the probe by SL2. Moreover, binding of the peptide to SL2 resulted in a significant absorbance decrease (8% hypochromicity) and a 3 nm red shift of the absorption maximum (Table 1, Figure 5), suggesting a stacking of the label with the SL2 bases. Thus, the 3HC label senses the formation of the 3HC-NC(11–55)/SL2 complex as a decrease in its exposure to water, likely due to its stacking with the ODN bases (37). To strengthen this conclusion, these spectral changes were further related to the known structure of the full-length NC protein bound to SL2. Since NC binds with the SL2 loop mainly through its zinc fingers, and since NC and NC(11–55) exhibit similar binding constants to ODNs (50,73), we reasonably assume that the two proteins bind similarly to SL2. According to the NMR data, the distance of the α-amino group of Lys11 to the SL2 stem in the complex is sufficiently short to allow stacking of the 3HC probe with the ODN bases (Figure 6A).
Table 2.
| Complex with | λAbs (nm) | λN* (nm) | λT* (nm) | N*:T* | QY (%) |
|---|---|---|---|---|---|
| Free | 358 | 426 | 517 | 1.12 | 7.8 |
| SL2 (RNA) | 360 | 425 | 533 | 0.46 | 7.0 |
| SL3 (RNA) | 362 | 424 | 532 | 0.69 | 4.4 |
| (−)PBS | 361 | 423 | 528 | 0.86 | 4.3 |
| d(ACGCC)b | 359 | 424 | 519 | 0.77 | 6.7 |
| d(AACGCC)b | 359 | 426 | 526 | 0.63 | 4.1 |
aλAbs, λN* and λT* are the maxima of absorption, N* and T* emission bands respectively. N*:T* is the intensity ratio of the two emission bands measured at the peak maxima; QY is the fluorescence quantum yield. Excitation wavelength was 340 nm. Measurements were done in 10 mM phosphate buffer, 100 mM NaCl, pH 7.0. bFor measurements with d(ACGCC) and d(AACGCC), NaCl concentration was 30 mM due to their lower binding affinity.
Figure 5.
Changes in the absorption (left) and emission (right) spectra of 3HC-NC(11–55) on binding to SL2 RNA. The spectra of 0.4 µM 3HC-NC(11–55) was recorded in the absence (black) and in the presence of 0.2 (green) 0.4 (blue) and 0.6 µM (red) SL2 RNA in 10 mM phosphate buffer, 100 mM NaCl, pH 7.0. Excitation wavelength was 340 nm.
Figure 6.
3D structure of NC complexes with SL2 (A); d(ACGCC) (B) and (−)PBS (C). Structures are drawn based on NMR data [ref. (53,77,76) corresponding to PDB structures: 1A1T, 1BJ6, 2EXF, respectively]. The red sphere represents the position of the α-amino group of Lys11 (A) and Asn12 (B, C), respectively. The pink sphere corresponds to all possible positions of the 3HC probe, taking into account the length of the linker. Zn atoms are presented as yellow spheres. The nucleotide in gray (B) shows the assumed position of the additional 5′ end A-residue in d(AACGCC).
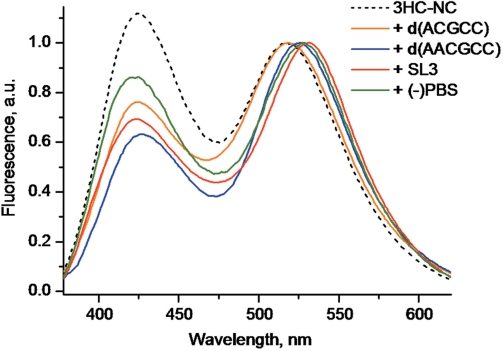
With SL3, the spectral changes of 3HC-NC(11–55) were slightly less pronounced than with SL2, showing a N*:T* ratio value of 0.69 and a 15 nm red-shift of the T* band (Figure 7). These changes were also associated with a significant decrease of the absorbance (8% hypochromicity) and a 4 nm red-shift of the absorption maximum, indicating that the decreased exposure of the probe to water may also be due to its stacking with the bases. As for SL2, the structural model of the NC/SL3 complex (52) suggests that the distance of the N-terminal amino group of NC(11–55) to the SL3 stem allows the stacking of 3HC with one of the bases of the stem (data not shown).
Figure 7.
Normalized fluorescence spectra of 3HC-NC(11–55) complexes with SL3 (red), (−)PBS (green), d(ACGCC) (orange) and d(AACGCC) (blue). The spectrum of the free 3HC-NC(11–55) peptide is given for comparison (dashed black curve). Peptide concentration was 0.2 µM with a 3HC-NC(11–55)/ODN ratio of 1:2 for SL2 and (−)PBS. The buffer was 10 mM phosphate, 100 mM NaCl, pH 7.0. To ensure complete peptide binding with d(ACGCC) and d(AACGCC), the salt concentration was decreased to 30 mM, while the peptide and ODN concentrations were raised to 1 µM and 10 µM, respectively. In these conditions, about 95% of the peptide was saturated by the ODNs (50).
In contrast to SL2 and SL3, only limited spectroscopic changes were observed with d(ACGCC) since its binding to 3HC-NC(11–55) decreased the N*:T* ratio only to a 0.77 value and shifted the T* band by only 2 nm. Moreover, no significant change in the absorption spectrum could be observed, indicating that the stacking of the 3HC probe with the d(ACGCC) bases is negligible. Our data are in line with the NMR-derived structure of the NC(12–53)/d(ACGCC) complex showing that the N-terminus of the peptide does not directly interact with the ODN (Figure 6B) (77). Furthermore, as compared to the NC(12–53) peptide, the additional Lys11 residue of the NC(11–55) peptide is expected to further increase the distance between the peptide N-terminus and the 5′-end of the ODN. Consequently, the limited spectroscopic changes observed with d(ACGCC) are likely due to the poor screening of the 3HC probe from water in conformations where the flexible extremities of the peptide and the ODN contact each other. To check our interpretation, we extended the d(ACGCC) sequence by an additional A residue at its 5′-end. Since the determinant binding region is the central CGC sequence (77), NC(11–55) binds similarly to d(ACGCC) and d(AACGCC) (82). Interaction of 3HC-NC(11–55) with d(AACGCC) resulted in a larger red shift of the T* band (9 nm) than with d(ACGCC) (2 nm), as well as a larger decrease in the N*:T* ratio (Figure 7). According to the NMR-derived NC(12–53)/d(ACGCC) structure (Figure 6B), the additional A nucleotide should come closer to the peptide N-terminus and additionally screen the label from water.
Finally, the smallest decrease in the N*:T* ratio was observed with (−)PBS. However, the shift in the T* band appears rather large, being of 11 nm. As for d(ACGCC), no decrease in the 3HC absorbance and thus, only marginal stacking of the 3HC probe with the bases occurs on binding of (−)PBS. As a consequence, the observed decrease in the N*:T* ratio and the red-shift of the T* band are likely due to transient contacts of the probe with the backbone of the (−)PBS stem (Figure 6C). Since the phosphoribose backbone of the stem is more hydrated than the internal base pair region, only limited shielding of the 3HC label from water can be achieved through this interaction.
Taken together, our data show that the 3HC probe can sensitively differentiate the tested ODNs, both by its N*:T* ratio and the position of its T* band. The response of the probe likely depends on the proximity of the peptide N-terminus with the ODN and the possibility for the probe to stack with the ODN bases or to contact with the phosphoribose backbone.
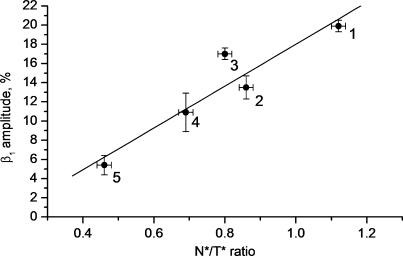
To further characterize the environment changes affecting the N-terminus of NC(11–55) on its interaction with ODNs, we investigated by time-resolved fluorescence anisotropy, the interaction of the same ODNs with NC(11–55) labeled at its N-terminus with fluorescein, Fl-NC(11–55). This technique allows characterizing the changes in the local motion of the probe and the tumbling of the labeled protein, resulting from the ODN binding. Similar to 3HC, the Fl probe did not significantly modify the folding and the chaperone properties of NC(11–55) (data not shown). The time-resolved anisotropy decay of the fluorescein-labeled NC(11–55) was characterized by two correlation times (Table 3). The 2.6 ns correlation time is in excellent agreement with the 2.3 ns correlation time previously reported for the tumbling motion of the NC(12–53) peptide, as measured from the time-resolved anisotropy of the intrinsic Trp37 residue (85). The 0.23 ns component is typical of the probe local motion (85,86). The moderate amplitude (β1=0.20) associated with this short component suggests that Fl rotation is restricted, likely by the proximal finger and the KNVK(11–14) sequence. The restriction of Fl local motion is fully consistent with the partial screening from water observed for 3HC when it is coupled to NC(11–55). Addition of ODNs increased the long correlation time, as expected from the dependence of the tumbling motion on the size and thus, on the molecular weight of the complex. A new intermediate correlation time (1–1.6 ns) appeared in the presence of ODNs, likely due to a segmental motion independent of the overall peptide motion. More interestingly, a sequence-dependent decrease in the amplitude β1 of the short-lived correlation time was observed. This decrease in the β1 value indicates that the bound ODN further restricts the accessible volume in which Fl can rotate. Remarkably, the amplitude associated with the Fl local motion linearly correlates with the N*:T* ratios observed with 3HC-NC(11–55) (Figure 8), further substantiating the established dependence of the 3HC response on the proximity of the probe with the ODN. Moreover, this correlation also suggests that the two dyes behave similarly, so that no specific interaction of 3HC with the ODNs biases its spectroscopic response.
Table 3.
Time-resolved and steady-state fluorescence anisotropy of fluorescein-labeled NC(11–55) in complexes with different ODNsa
| θ1 (ns) | β1 | θ2 (ns) | β2 | θ3, (ns) | β3 | r | N*:T* | |
|---|---|---|---|---|---|---|---|---|
| Free | 0.23 ± 0.01 | 0.20 ± 0.01 | – | – | 2.6 ± 0.2 | 0.80 ± 0.01 | 0.102 ± 0.004 | 1.12 |
| d(ACGCC) | 0.12 ± 0.01 | 0.18 ± 0.01 | 1.0 ± 0.3 | 0.36 ± 0.09 | 3.2 ± 1 | 0.46 ± 0.09 | 0.105 ± 0.004 | 0.80 |
| (−)PBS | 0.12 ± 0.01 | 0.14 ± 0.01 | 1.0 ± 0.3 | 0.39 ± 0.08 | 3.6 ± 1 | 0.47 ± 0.09 | 0.106 ± 0.004 | 0.86 |
| SL3 | 0.12 ± 0.01 | 0.14 ± 0.02 | 1.6 ± 0.2 | 0.46 ± 0.06 | 7.1 ± 1 | 0.40 ± 0.05 | 0.132 ± 0.004 | 0.69 |
| SL2 | 0.13 ± 0.01 | 0.05 ± 0.01 | 1.3 ± 0.2 | 0.43 ± 0.08 | 6.4 ± 0.9 | 0.52 ± 0.08 | 0.141 ± 0.004 | 0.46 |
aResults of at least two independent measurements. θ1 is the correlation time describing the local probe motion and β1 is the contribution of this motion to the fluorescence depolarization. θ2/β2 and θ3/β3 describe segmental and overall motions, respectively. r corresponds to the steady-state fluorescence anisotropy. N*:T* is the band intensity ratio of the 3HC label shown for the comparison.
Figure 8.
Correlation of the N*:T* ratio of 3HC-NC(11–55) with the time-resolved amplitude β1 associated with the fluorescein local motion in Fl-NC(11–55) for their complexes with different ODNs. Bars are corresponding to the experimental errors. Points correspond to free NC(11–55) (‘1’) and complexes with (−)PBS (‘2’), d(ACGCC) (‘3’), SL3 (‘4’) and SL2 (‘5’).
As a consequence, the N*:T* ratio of the 3HC label provides through simple fluorescence intensity measurements, information on the proximity of the labelling site with the ODN, comparable to that obtained from time-resolved measurements with the Fl label. To further show the potency of the proposed 3HC-based methodology, we compared it with steady-state fluorescence anisotropy, which is commonly used as another single-labeling technique for sensing biomolecular interactions. The steady-state fluorescence anisotropy data of the Fl-labeled protein free and bound to the various ODNs are reported in Table 3. No significant change in the Fl-NC(11–55) steady-state anisotropy could be observed with addition of relatively small ODNs, d(ACGCC) or (−)PBS, while with larger ODNs, SL2 and SL3, the Fl anisotropy showed a significant increase. In sharp contrast, significant changes in the N*:T* ratios accompanied the binding of all the studied ODNs to 3HC-NC(11–55), including the small ones, indicating that the 3HC-based approach is not limited by the ODN size. In fact, the environment-sensitive label 3HC reports exclusively on the local properties of the interaction site, so that its signal is not directly affected by variation in the molecular weight of the complex. Thus, labeling peptides with 3HC and monitoring the changes in the N*:T* ratio appears as an interesting and simple methodology for sensing interactions based on the proximity of the labeled site with the ODN, instead of sensing changes in the molecular weight as with anisotropy-based methods. Thus, the methods of anisotropy and the ratiometric measurements with the 3HC label appear complementary.
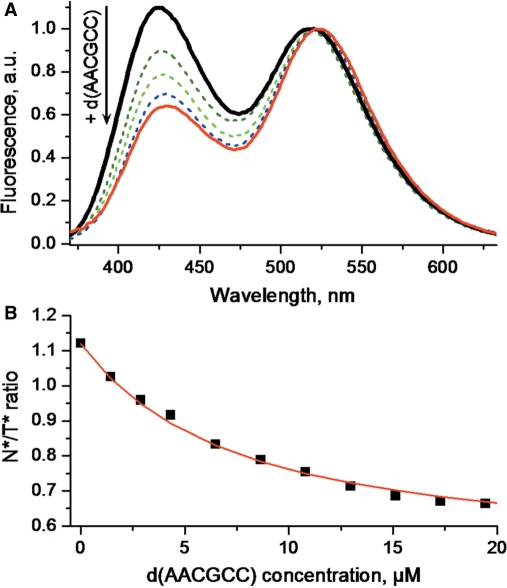
Since large changes in the N*:T* ratio accompanied the binding of the various ODNs to 3HC-NC(11–55), we next explored the possibility to use these changes to determine the binding constants of the NC(11–55)/ODN complexes. The d(AACGCC) and the SL3 RNA were taken as representative examples. Addition of increasing concentrations of d(AACGCC) gradually decreased the N*:T* ratio (Figure 9). Plotting the N*:T* ratio as a function of the 3HC-NC(11–55) concentration and assuming a 1/1 stoichiometry, we could adequately fit the binding curve (Figure 9B) with a binding constant of 1.5(±0.3) × 105 M−1, close to the 1.1(±0.2) × 105 M−1 value reported in the literature for non-labeled NC(12–53) (50). Similarly, titrations of 3HC-NC(11–55) with SL3 (data not shown) provided a binding constant of 2.7(±0.6) × 106 M−1, again in reasonable agreement with the 1.0(±0.2) × 106 M−1 value obtained with the slightly shorter NC(12–53) peptide (50). Both examples showed that the 3HC label does not strongly affect the peptide–ODN interaction and could thus, be used to determine the corresponding binding constants.
Figure 9.
Titration of 3HC-NC(11–55) with d(AACGCC) monitored by 3HC fluorescence. (A) Normalized fluorescence spectra. (B) Titration curve. The solid red line corresponds to the fit of the data points (squares) by a one binding site model. The calculated binding constant was 1.5(±0.3) × 105 M−1. Peptide concentration was 1 µM. Experimental conditions were as for Figure 5.
Interactions of 3HC-NC(11–55) with multiple binding site ODNs
To further characterize the potential use of the 3HC label in peptide–ODN interactions, we next investigated the interaction of 3HC-NC(11–55) with the (−)DNA33 sequence (Figure 2) corresponding to the 3′-terminal 33 nucleotides of the (−)DNA copy of the HIV-1 genome, generated during reverse transcription (65,66).
This sequence is involved in the second strand transfer and contains the (−)PBS stem-loop as well as a second stem-loop of 14 bases, that we call SL14 (Figure 2). Due to its length, (−)DNA33 likely contains multiple binding sites. In this respect, knowing the spectroscopic response associated with the binding of 3HC-NC(11–55) to the (−)PBS loop, we determined whether the peptide preferentially binds to (−)PBS in the (−)DNA33 sequence. Addition of 3HC-NC(11–55) at a 1:1 ratio to (−)DNA33 gives a much lower N*:T* ratio (0.43) than with (−)PBS (0.86), suggesting that the (−)PBS loop does not constitute a preferential binding site. This conclusion was further strengthened by the much higher red-shift of the T* band observed with (−)DNA33 (16 nm) as compared with (−)PBS (11 nm). It thus follows that the peptide may preferentially bind to a different binding site, where the peptide N-terminus is closer to the ODN bases. To determine whether this site is localized in the SL14 stem loop, we characterized the spectroscopic changes of 3HC-NC(11–55) resulting from its binding to the isolated SL14 sequence. Adding the protein at a 1:1 molar ratio, we observed a N*:T* ratio (0.36) and a 13-nm red-shift of the T* band, close to the values obtained with (−)DNA33 (Figure S4 in Supplementary Data), suggesting that the preferential binding site for 3HC-NC(11–55) in (−)DNA33 is located on the SL14 sequence. Nevertheless, a limited binding to the (−)PBS loop is also likely, since the N*:T* ratio for (−)DNA33 is somewhat higher than for SL14. These conclusions on the preferential binding to SL14 and the limited binding to (−)PBS were further supported by titration experiments, revealing that 3HC-NC(11–55) binds with 4-fold higher affinity to SL14 [1.0(±0.3) × 106 M−1] as compared to (−)PBS [2.7(±0.6) × 105 M−1]. The preferential binding of 3HC-NC(11–55) to SL14 was further assessed by the spectrum obtained in the presence of an equimolar mixture of (−)PBS and SL14, which shows a N*:T* ratio of 0.38 very close to that of SL14 alone (0.36) (Figure S4 Supplementary Data). Thus, the proposed environment-sensitive label attached to a peptide can help to localize its preferential binding sites on ODNs, provided the probe responses associated with peptide binding to the individual sites are known from independent measurements.
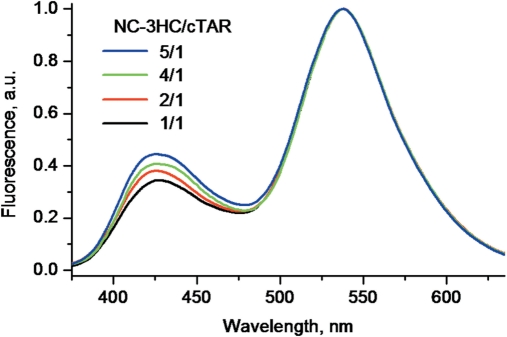
Finally, we characterized the interaction of 3HC-NC(11–55) with cTAR DNA, a stem-loop of 55 nucleotides involved in the NC-promoted first strand transfer, during reverse transcription (61,62,71). This sequence was previously shown to bind eight NC(12–55) molecules with an affinity of 1.7 × 107 M−1 at 30 mM NaCl (73), assuming identical and non-interacting binding sites. Though this assumption was adequate for obtaining good fits of the binding curves and comparing various NC mutants (65,73), it was recognized by the authors to be rather unrealistic, since NC binding is known to depend on the ODN sequence (44,50). To check the hypothesis of identical and non-interacting binding sites, we added increasing concentrations of 3HC-NC(11–55) to 1 µM cTAR up to a molar ratio of five peptides per cTAR molecule in a buffer with 30 mM NaCl. Due to the high affinity of NC(11–55) for cTAR and the high concentrations of cTAR and peptide used, the concentration of free 3HC-NC(11–55) is negligible in these conditions. As a consequence, if 3HC-NC(11–55) binds with identical affinity to all cTAR binding sites, the latter should be populated in parallel with increasing peptide concentration and thus, no change in the N*:T* ratio should appear. In contrast to this expectation, the N*:T* ratio regularly increases from a 0.34 value at a 1:1 molar ratio up to a 0.44 value at a 5:1 molar ratio (Figure 10). This indicates that 3HC-NC(11–55) first binds to sites giving a low N*:T* ratio while sites associated with higher N*:T* ratios are less affine, highlighting differences in the binding constants of the different sites. In contrast to the N*:T* ratio, the position of the T* band remains nearly constant during the titration, being red-shifted by about 16 nm in respect with the free 3HC-NC(11–55) protein. This absence of change of the T* band confirmed that the concentration of free protein remains negligible.
Figure 10.
Normalized fluorescence spectra of 3HC-NC(11–55) added to cTAR at different molar ratios: 1:1 (black), 2:1 (red), 4:1 (green) and 5:1 (blue). cTAR concentration was 1 µM. Experiment was performed at 30 mM NaCl concentration to ensure complete binding (73).
CONCLUSIONS
Herein, we presented a new environment-sensitive ratiometric fluorescent label for sensing peptide–ODN interactions. Being attached to the N-terminus of the NC(11–55) peptide, the label reports on the interaction with ODNs by a change in the ratio of its two emission bands. The response of the label is connected with a decrease in the exposure of the labeling site to bulk water induced by the interaction of the peptide with ODN. Using this ratiometric approach, we were able to distinguish different peptide–ODN complexes by monitoring the local properties of the peptide labeling site. This property was successfully used to evaluate quantitatively peptide–ODN interactions, to localize preferential binding sites on ODNs and to show the presence of binding sites with different affinities. This approach provided us new insights on the binding of NC to two target sequences of the (−)DNA copy of the HIV-1 genome, which are critically involved in the two obligatory strand transfers during reverse transcription. Since the N*:T* ratio of the label is sensitive to its close environment and not to the molecular weight of the peptide–ODN complex, the proposed methodology appears as a simple and complementary alternative to steady-state anisotropy for monitoring protein–ODN interactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by a grant from the Agence Nationale de la Recherche, Agence Nationale de la Recherche sur le SIDA, and ARCUS program. V.V.S. was supported by an Eiffel fellowship. Funding for open access charge: Agence Nationale de la Recherche.
Conflict of interest statement. None declared.
REFERENCES
- 1.Heyduk T. Measuring protein conformational changes by FRET/LRET. Curr. Opin. Biotechnol. 2002;13:292–296. doi: 10.1016/s0958-1669(02)00332-4. [DOI] [PubMed] [Google Scholar]
- 2.Selvin PR. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 2000;7:730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 3.Schuler B. Single-molecule fluorescence spectroscopy of protein folding. Chem. Phys. Chem. 2005;6:1206–1220. doi: 10.1002/cphc.200400609. [DOI] [PubMed] [Google Scholar]
- 4.Kajihara D, Abe R, Iijima I, Komiyama C, Sisido M, Hohsaka T. FRET analysis of protein conformational change through position-specific incorporation of fluorescent amino acids. Nat. Methods. 2006;3:923–929. doi: 10.1038/nmeth945. [DOI] [PubMed] [Google Scholar]
- 5.Cao H, Tamilarasu N, Rana TM. Orientation and affinity of HIV-1 Tat fragments in Tat-TAR complex determined by fluorescence resonance energy transfer. Bioconjug. Chem. 2006;17:352–358. doi: 10.1021/bc050277u. [DOI] [PubMed] [Google Scholar]
- 6.Jameson DM, Sawyer WH. Fluorescence anisotropy applied to biomolecular interactions. Methods Enzymol. 1995;246:283–300. doi: 10.1016/0076-6879(95)46014-4. [DOI] [PubMed] [Google Scholar]
- 7.Lakowicz JR, Gryczynski I, Piszczek G, Tolosa L, Nair R, Johnson ML, Nowaczyk K. Microsecond dynamics of biological macromolecules. Methods Enzymol. 2000;323:473–509. doi: 10.1016/s0076-6879(00)23379-x. [DOI] [PubMed] [Google Scholar]
- 8.Cohen BE, McAnaney TB, Park ES, Jan YN, Boxer SG, Jan LY. Probing protein electrostatics with a synthetic fluorescent amino acid. Science. 2002;296:1700–1703. doi: 10.1126/science.1069346. [DOI] [PubMed] [Google Scholar]
- 9.Nitz M, Mezo AR, Ali MH, Imperiali B. Enantioselective synthesis and application of the highly fluorescent and environment-sensitive amino acid 6-(2-dimethylaminonaphthoyl) alanine (DANA) Chem. Commun. 2002;8:1912–1913. doi: 10.1039/b205224e. [DOI] [PubMed] [Google Scholar]
- 10.Vazquez ME, Nitz M, Stehn J, Yaffe MB, Imperiali B. Fluorescent caged phosphoserine peptides as probes to investigate phosphorylation-dependent protein associations. J. Am. Chem. Soc. 2003;125:10150–10151. doi: 10.1021/ja0351847. [DOI] [PubMed] [Google Scholar]
- 11.Vazquez ME, Rothman DM, Imperiali B. A new environment-sensitive fluorescent amino acid for Fmoc-based solid phase peptide synthesis. Org. Biomol. Chem. 2004;2:1965–1966. doi: 10.1039/b408001g. [DOI] [PubMed] [Google Scholar]
- 12.Vazquez ME, Blanco JB, Imperiali B. Photophysics and biological applications of the environment-sensitive fluorophore 6-N,N-dimethylamino-2,3-naphthalimide. J. Am. Chem. Soc. 2005;127:1300–1306. doi: 10.1021/ja0449168. [DOI] [PubMed] [Google Scholar]
- 13.Loving G, Imperiali B. A versatile amino acid analogue of the solvatochromic fluorophore 4-N,N-dimethylamino-1,8-naphthalimide: a powerful tool for the study of dynamic protein interactions. J. Am. Chem. Soc. 2008;130:13630–13638. doi: 10.1021/ja804754y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen BE, Pralle A, Yao X, Swaminath G, Gandhi CS, Jan YN, Kobilka BK, Isacoff EY, Jan LY. A fluorescent probe designed for studying protein conformational change. Proc. Natl Acad. Sci. USA. 2005;102:965–970. doi: 10.1073/pnas.0409469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Chung NN, Lemieux C, Zelent B, Vanderkooi JM, Gryczynski I, Wilkes BC, Schiller PW. [Aladan3]TIPP: a fluorescent delta-opioid antagonist with high delta-receptor binding affinity and delta selectivity. Biopolymers. 2005;80:325–331. doi: 10.1002/bip.20200. [DOI] [PubMed] [Google Scholar]
- 16.Venkatraman P, Nguyen TT, Sainlos M, Bilsel O, Chitta S, Imperiali B, Stern LJ. Fluorogenic probes for monitoring peptide binding to class II MHC proteins in living cells. Nat. Chem. Biol. 2007;3:222–228. doi: 10.1038/nchembio868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sen S, Paraggio NA, Gearheart LA, Connor EE, Issa A, Coleman RS, Wilson D.M., 3rd, Wyatt MD, Berg MA. Effect of protein binding on ultrafast DNA dynamics: characterization of a DNA:APE1 complex. Biophys. J. 2005;89:4129–4138. doi: 10.1529/biophysj.105.062695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto A, Tainaka K, Saito I. A dielectric-sensitive fluorescent DNA probe for monitoring polarities on the interior of a DNA-binding protein. Bioconjug. Chem. 2005;16:1105–1111. doi: 10.1021/bc050077i. [DOI] [PubMed] [Google Scholar]
- 19.Yang X, Liu WH, Jin WJ, Shen GL, Yu RQ. DNA binding studies of a solvatochromic fluorescence probe 3-methoxybenzanthrone. Spectrochim. Acta A. Mol. Biomol. Spectrosc. 1999;55A:2719–2727. doi: 10.1016/s1386-1425(99)00161-4. [DOI] [PubMed] [Google Scholar]
- 20.Kumar CV, Asuncion EH. DNA binding studies and site selective fluorescence sensitization of an anthryl probe. J. Am. Chem. Soc. 1993;115:8547–8553. [Google Scholar]
- 21.Klymchenko AS, Kenfack C, Duportail G, Mely Y. Effects of polar protic solvents on dual emissions of 3-hydroxychromones. J. Chem. Sci. 2007;119:83–89. [Google Scholar]
- 22.Avilov SV, Bode C, Tolgyesi F, Klymchenko AS, Fidy J, Demchenko AP. Temperature effects on alpha-crystallin structure probed by 6-bromomethyl-2-(2-furanyl)-3-hydroxychromone, an environmentally sensitive two-wavelength fluorescent dye covalently attached to the single Cys residue. Int. J. Biol. Macromol. 2005;36:290–298. doi: 10.1016/j.ijbiomac.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Sengupta PK, Kasha M. Excited state proton-transfer spectroscopy of 3-hydroxyflavone and quercetin. Chem. Phys. Lett. 1979;68:382–385. [Google Scholar]
- 24.Chou PT, Martinez ML, Clements JH. Reversal of excitation behavior of proton-transfer vs. charge-transfer by dielectric perturbation of electronic manifolds. J. Phys. Chem. 1993;97:2618–2622. [Google Scholar]
- 25.Ormson SM, Brown RG, Vollmer F, Rettig W. Switching between charge- and proton-transfer emission in the excited state of a substituted 3-hydroxyflavone. J. Photochem. Photobiol. A: Chem. 1994;81:65–72. [Google Scholar]
- 26.Swinney TC, Kelley DF. Proton transfer dynamics in substituted 3-hydroxyflavones: Solvent polarization effects. J. Chem. Phys. 1993;99:211–221. [Google Scholar]
- 27.Klymchenko AS, Demchenko AP. Multiparametric probing of intermolecular interactions with fluorescent dye exhibiting excited state intramolecular proton transfer. Phys. Chem. Chem. Phys. 2003;5:461–468. [Google Scholar]
- 28.Strandjord AJG, Barbara PF. Proton-transfer kinetics of 3-hydroxyflavone: solvent effects. J. Chem. Phys. 1985;89:2355–2361. [Google Scholar]
- 29.McMorrow D, Kasha M. Intramolecular excited-state proton transfer in 3-hydroxyflavone. Hydrogen-bonding solvent perturbations. J. Phys. Chem. 1984;88:2235–2243. [Google Scholar]
- 30.Avilov SV, Bode C, Tolgyesi FG, Klymchenko AS, Fidy J, Demchenko AP. Heat perturbation of bovine eye lens alpha-crystallin probed by covalently attached ratiometric fluorescent dye 4′-diethylamino-3-hydroxyflavone. Biopolymers. 2005;78:340–348. doi: 10.1002/bip.20285. [DOI] [PubMed] [Google Scholar]
- 31.Klymchenko AS, Avilov SV, Demchenko AP. Resolution of Cys and Lys labeling of alpha-crystallin with site-sensitive fluorescent 3-hydroxyflavone dye. Anal. Biochem. 2004;329:43–57. doi: 10.1016/j.ab.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 32.Enander K, Choulier L, Olsson AL, Yushchenko DA, Kanmert D, Klymchenko AS, Demchenko AP, Mely Y, Altschuh D. A peptide-based, ratiometric biosensor construct for direct fluorescence detection of a protein analyte. Bioconjug. Chem. 2008;19:1864–1870. doi: 10.1021/bc800159d. [DOI] [PubMed] [Google Scholar]
- 33.Klymchenko AS, Duportail G, Mely Y, Demchenko AP. Ultrasensitive two-color fluorescence probes for dipole potential in phospholipid membranes. Proc. Natl Acad. Sci. USA. 2003;100:11219–11224. doi: 10.1073/pnas.1934603100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klymchenko AS, Duportail G, Ozturk T, Pivovarenko VG, Mely Y, Demchenko AP. Novel two-band ratiometric fluorescence probes with different location and orientation in phospholipid membranes. Chem. Biol. 2002;9:1199–1208. doi: 10.1016/s1074-5521(02)00244-2. [DOI] [PubMed] [Google Scholar]
- 35.Shynkar VV, Klymchenko AS, Duportail G, Demchenko AP, Mely Y. Two-color fluorescent probes for imaging the dipole potential of cell plasma membranes. Biochim. Biophys. Acta. 2005;1712:128–136. doi: 10.1016/j.bbamem.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Shynkar VV, Klymchenko AS, Kunzelmann C, Duportail G, Muller CD, Demchenko AP, Freyssinet JM, Mely Y. Fluorescent biomembrane probe for ratiometric detection of apoptosis. J. Am. Chem. Soc. 2007;129:2187–2193. doi: 10.1021/ja068008h. [DOI] [PubMed] [Google Scholar]
- 37.Klymchenko AS, Shvadchak VV, Yushchenko DA, Jain N, Mely Y. Excited-state intramolecular proton transfer distinguishes microenvironments in single- and double-stranded DNA. J. Phys. Chem. B. 2008;112:12050–12055. doi: 10.1021/jp8058068. [DOI] [PubMed] [Google Scholar]
- 38.Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF. Dynamical behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 39.Summers MF, Henderson LE, Chance MR, Bess J.W., Jr, South TL, Blake PR, Sagi I, Perez-Alvarado G, Sowder R.C., III, Hare DR, et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berkowitz RD, Luban J, Goff SP. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J. Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5' packaging signal of human immunodeficiency virus type 1. J. Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casas-Finet JR, Medaglia M, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darlix J.-L, Gabus C, Nugeyre M.-T, Clavel F, Barré-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Virol. 1990;216:689. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 46.Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mely Y, de Rocquigny H, Sorinas-Jimeno M, Keith G, Roques BP, Marquet R, Gerard D. Binding of the HIV-1 nucleocapsid protein to the primer tRNA(3Lys), in vitro, is essentially not specific. J. Biol. Chem. 1995;270:1650–1656. doi: 10.1074/jbc.270.4.1650. [DOI] [PubMed] [Google Scholar]
- 48.Cimarelli A, Darlix JL. Assembling the human immunodeficiency virus type 1. Cell Mol. Life Sci. 2002;59:1166–1184. doi: 10.1007/s00018-002-8495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muriaux D, Darlix JL, Cimarelli A. Targeting the assembly of the human immunodeficiency virus type I. Curr. Pharm. Des. 2004;10:3725–3739. doi: 10.2174/1381612043382701. [DOI] [PubMed] [Google Scholar]
- 50.Vuilleumier C, Bombarda E, Morellet N, Gerard D, Roques BP, Mely Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- 51.Shubsda MF, Paoletti AC, Hudson BS, Borer PN. Affinities of packaging domain loops in HIV-1 RNA for the nucleocapsid protein. Biochemistry. 2002;41:5276–5282. doi: 10.1021/bi016045+. [DOI] [PubMed] [Google Scholar]
- 52.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 53.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 54.You JC, McHenry CS. HIV nucleocapsid protein. Expression in Escherichia coli, purification, and characterization. J. Biol. Chem. 1993;268:16519–16527. [PubMed] [Google Scholar]
- 55.Urbaneja MA, Kane BP, Johnson DG, Gorelick RJ, Henderson LE, Casas-Finet JR. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J. Mol. Biol. 1999;287:59–75. doi: 10.1006/jmbi.1998.2521. [DOI] [PubMed] [Google Scholar]
- 56.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 57.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 58.You JC, McHenry CS. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]
- 59.Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 2000;74:8980–8988. doi: 10.1128/jvi.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix JL. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix JL. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernacchi S, Stoylov S, Piemont E, Ficheux D, Roques BP, Darlix JL, Mely Y. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J. Mol. Biol. 2002;317:385–399. doi: 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- 63.Azoulay J, Clamme JP, Darlix JL, Roques BP, Mely Y. Destabilization of the HIV-1 complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J. Mol. Biol. 2003;326:691–700. doi: 10.1016/s0022-2836(02)01430-4. [DOI] [PubMed] [Google Scholar]
- 64.Beltz H, Azoulay J, Bernacchi S, Clamme JP, Ficheux D, Roques B, Darlix JL, Mely Y. Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J. Mol. Biol. 2003;328:95–108. doi: 10.1016/s0022-2836(03)00244-4. [DOI] [PubMed] [Google Scholar]
- 65.Egele C, Schaub E, Ramalanjaona N, Piemont E, Ficheux D, Roques B, Darlix JL, Mely Y. HIV-1 nucleocapsid protein binds to the viral DNA initiation sequences and chaperones their kissing interactions. J. Mol. Biol. 2004;342:453–466. doi: 10.1016/j.jmb.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 66.Ramalanjaona N, de Rocquigny H, Millet A, Ficheux D, Darlix JL, Mely Y. Investigating the mechanism of the nucleocapsid protein chaperoning of the second strand transfer during HIV-1 DNA synthesis. J. Mol. Biol. 2007;374:1041–1053. doi: 10.1016/j.jmb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 67.Godet J, de Rocquigny H, Raja C, Glasser N, Ficheux D, Darlix JL, Mely Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J. Mol. Biol. 2006;356:1180–1192. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 68.Liu HW, Cosa G, Landes CF, Zeng Y, Kovaleski BJ, Mullen DG, Barany G, Musier-Forsyth K, Barbara PF. Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription. Biophys. J. 2005;89:3470–3479. doi: 10.1529/biophysj.105.065326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu HW, Zeng Y, Landes CF, Kim YJ, Zhu Y, Ma X, Vo MN, Musier-Forsyth K, Barbara PF. Insights on the role of nucleic acid/protein interactions in chaperoned nucleic acid rearrangements of HIV-1 reverse transcription. Proc. Natl Acad. Sci. USA. 2007;104:5261–5267. doi: 10.1073/pnas.0700166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cosa G, Zeng Y, Liu HW, Landes CF, Makarov DE, Musier-Forsyth K, Barbara PF. Evidence for non-two-state kinetics in the nucleocapsid protein chaperoned opening of DNA hairpins. J. Phys. Chem. B. 2006;110:2419–2426. doi: 10.1021/jp054189i. [DOI] [PubMed] [Google Scholar]
- 71.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 72.Darlix JL, Garrido JL, Morellet N, Mely Y, de Rocquigny H. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv. Pharmacol. 2007;55:299–346. doi: 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- 73.Beltz H, Clauss C, Piemont E, Ficheux D, Gorelick RJ, Roques B, Gabus C, Darlix JL, de Rocquigny H, Mely Y. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J. Mol. Biol. 2005;348:1113–1126. doi: 10.1016/j.jmb.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 74.Stoylov SP, Vuilleumier C, Stoylova E, De Rocquigny H, Roques BP, Gerard D, Mely Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–312. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 75.Amarasinghe GK, De Guzman RN, Turner RB, Summers MF. NMR structure of stem-loop SL2 of the HIV-1 psi RNA packaging signal reveals a novel A-U-A base-triple platform. J. Mol. Biol. 2000;299:145–156. doi: 10.1006/jmbi.2000.3710. [DOI] [PubMed] [Google Scholar]
- 76.Bourbigot S, Ramalanjaona N, Boudier C, Salgado GF, Roques BP, Mely Y, Bouaziz S, Morellet N. How the HIV-1 Nucleocapsid Protein Binds and Destabilises the (-)Primer Binding Site During Reverse Transcription. J. Mol. Biol. 2008;383:1112–1128. doi: 10.1016/j.jmb.2008.08.046. [DOI] [PubMed] [Google Scholar]
- 77.Morellet N, Demene H, Teilleux V, Huynh-Dinh T, de Rocquigny H, Fournie-Zaluski MC, Roques BP. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC) J. Mol. Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- 78.Baudin F, Marquet R, Isel C, Darlix JL, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 79.Johnson PE, Turner RB, Wu ZR, Hairston L, Guo J, Levin JG, Summers MF. A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–9091. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- 80.de Rocquigny H, Ficheux D, Gabus C, Fournie-Zaluski MC, Darlix JL, Roques BP. First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem. Biophys. Res. Commun. 1991;180:1010–1018. doi: 10.1016/s0006-291x(05)81166-0. [DOI] [PubMed] [Google Scholar]
- 81.Eastman JW. Quantitative spectrofluorimetry-the fluorescence quantum yield of quinine sulfate. Photochem. Photobiol. 1967;6:55–72. [Google Scholar]
- 82.Avilov SV, Piemont E, Shvadchak V, de Rocquigny H, Mely Y. Probing dynamics of HIV-1 nucleocapsid protein/target hexanucleotide complexes by 2-aminopurine. Nucleic Acids Res. 2008;36:885–896. doi: 10.1093/nar/gkm1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Livesey AK, Brochon J.-C. Analyzing the distribution of decay constants in pulse-fluorimetry using the maximum entropy method. Biophys. J. 1987;52:693–706. doi: 10.1016/S0006-3495(87)83264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klymchenko AS, Demchenko AP. 3-Hydroxychromone dyes exhibiting excited-state intramolecular proton transfer in water with efficient two-band fluorescence. New J. Chem. 2004;28:687–692. [Google Scholar]
- 85.Ramboarina S, Srividya N, Atkinson RA, Morellet N, Roques BP, Lefevre JF, Mely Y, Kieffer B. Effects of temperature on the dynamic behaviour of the HIV-1 nucleocapsid NCp7 and its DNA complex. J. Mol. Biol. 2002;316:611–627. doi: 10.1006/jmbi.2001.5379. [DOI] [PubMed] [Google Scholar]
- 86.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2nd edn. New York: Kluwer Academic/Plenum Publishers; 1999. [Google Scholar]
- 87.Bernacchi S, Piemont E, Potier N, Dorsselaer A, Mely Y. Excitonic heterodimer formation in an HIV-1 oligonucleotide labeled with a donor-acceptor pair used for fluorescence resonance energy transfer. Biophys. J. 2003;84:643–654. doi: 10.1016/S0006-3495(03)74884-X. [DOI] [PMC free article] [PubMed] [Google Scholar]